Abstract
The human cytomegalovirus (HCMV) US28 gene product, pUS28, is a G protein-coupled receptor that interacts with both CC and CX3C chemokines. To date, the role of pUS28 in immune evasion and cell migration has been studied only in cell types that can establish productive HCMV infection. We show that HCMV can latently infect THP-1 monocytes and that during latency US28 is transcribed. We also show that the transcription is sustained during differentiation of the THP-1 monocytes. Since cells expressing pUS28 were previously shown to adhere to immobilized CX3C chemokines (C. A. Haskell, M. D. Cleary, and I. F. Charo, J. Biol. Chem. 275:34183–34189, 2000), we hypothesize that latently infected circulating monocytes express pUS28, thereby enabling adhesion of these cells to CX3C-exposing endothelium. Consequently, the US28-encoded chemokine receptor may play an important role in dissemination of latent HCMV.
The human cytomegalovirus (HCMV) genome contains four G-protein-coupled receptor-like genes, UL33, UL78, US27, and US28 (8, 11). The US28-encoded G-protein-coupled receptor pUS28 appears to be functional in many different respects. (i) It was identified as a chemokine receptor capable of binding the CC chemokines RANTES, MCP-1, MCP-3, MIP1-α, and MIP1-β (2, 10, 21, 26), as well as soluble forms of the CX3C chemokine fractalkine (18). Upon interaction with the CC chemokines, pUS28 induces Ca2+ mobilization and extracellular signal-related kinase 2 activation (2, 10, 21, 26). In addition, transient expression of pUS28 leads to constitutive activation of both phospholipase C and NF-κB signaling (6). In this system, fractalkine acts as an inverse agonist for pUS28 (6). (ii) Transient, high-level expression of pUS28 in smooth muscle cells induces chemokinesis in the presence of MCP-1 and chemotaxis within a RANTES gradient (38). (iii) Expression of pUS28 leads to internalization of RANTES and MCP-1 in both HCMV-infected fibroblasts and endothelial cells (3, 4). Thus, this receptor acts as a CC-chemokine sink, possibly enabling HCMV-infected cells to evade immune surveillance. (iv) The chemokine receptor pUS28 is a coreceptor for several human immunodeficiency virus strains (27, 32) and can elicit cell-to-cell fusion upon interaction with several types of viral envelope proteins (33). (v) The expression of pUS28 on the cell surface of murine pre-B-cell line 300-19 can establish cell rolling and adhesion to a fixed fractalkine surface (15). If pUS28 is expressed on the surface of HCMV-infected leukocytes, then it could play an important role in virus trafficking from the circulation to inflammatory sites. Particularly, the receptor could mediate adhesion of circulating HCMV-infected monocytes to endothelial cells and subsequent transmission of HCMV from infected monocytes to endothelial cells or, alternatively, transendothelial migration of infected monocytes toward subendothelial tissues. To date, only a few reports on US28 expression exist. US28-specific transcripts were detected in peripheral blood mononuclear cells of some naturally infected individuals (31). In addition, US28-specific transcripts were detected from 2 h to at least 24 h post-HCMV infection (p.i.) of human foreskin fibroblasts (HFF) in vitro (4, 42, 44). Finally, US28-specific transcripts were detected in HCMV-infected myeloid cell cultures—at 4 h p.i. in infected U373 MG astrocytoma cells and at 4 and 24 h p.i. in infected THP-1 monocytic cells (44). Interestingly, in many cell types of myeloid origin, HCMV establishes latent infection, i.e., persistence of nonreplicating viral genomic DNA in infected cells. Latent infection occurs in progenitors of granulocytes, macrophages, and dendritic cells (13, 20), as well as in peripheral blood monocytes (5) and immature macrophages (37). Recently, HCMV major immediate early (MIE) gene-derived transcripts were identified in latently infected monocyte or granulocyte progenitor cells (20), as well as in bone marrow (BM)-derived CD33+ CD14+ and CD33+ CD15+ cells (13). These cytomegalovirus latency-associated transcripts (CLTs) have either a sense or an antisense orientation relative to the conventional MIE coding region. The sense CLTs have two specific transcription start sites (LSS1 and LSS2) within the MIE promoter-enhancer region, upstream of the productive-infection transcription start site (PSS) (Fig. 1). The function of neither CLTs nor their corresponding gene products is known. To date, CLTs are the only transcripts identified in latently infected cells. Here we report a system for studying HCMV gene expression in latently infected monocytic THP-1 cells. Using this system, we demonstrate that US28, like the MIE gene, is transcribed during both latent and productive HCMV infection.
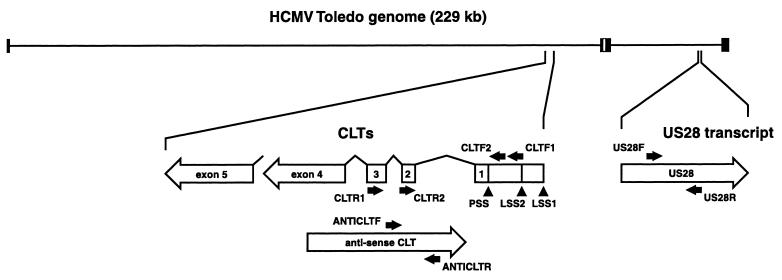
FIG. 1.
RT-PCR primers and their target transcripts. Schematic representation of the HCMV Toledo genome and the relative positions of the MIE gene and US28. Black boxes represent repeat regions of the Toledo genome. The US28-specific transcripts and CLTs are indicated below the genome at a smaller scale by open arrows. The positions and polarity of the US28-, as well as the CLT-specific RT-PCR primers (Table 1) are indicated by black arrows. Transcription starts of the sense CLTs (LSS1 and LSS2) and the PSS are indicated by arrowheads.
MATERIALS AND METHODS
Cells and virus.
Primary HFF, the human fibroblast cell line MRC5, and the myeloid cell lines K562, KG1a, HL-60, U937, and THP-1 cells were cultured as described previously (4, 14, 44). Stocks of wild-type (Toledo) and a US27-US28 double deletion mutant virus (RV101) (4) were generated by propagation in HFF and MRC5 (4). For reverse transcription (RT)-PCR analysis, virions from HFF culture medium samples, each containing 3 × 106 PFU of either Toledo or RV101 HCMV per ml, were separated from cellular debris by low-speed centrifugation (10 min at 10,000 × g, 4°C) and pelleted by ultracentrifugation (30 min at 100,000 × g, 4°C). A sample of Toledo virus was inactivated by irradiation with 4.7 J of UV using a Stratalinker UV Crosslinker 1800 (Stratagene Cloning Systems, Amsterdam, The Netherlands).
RT-PCR.
Primers specific for either the HCMV US28 gene (US28F and US28R), both sense (CLTF1, CLTF2, CLTR1, and CLTR2) and anti-sense CLTs (ANTICLTF and ANTICLTR); the HCMV DNA polymerase gene UL54 (DNAPOLF* and DNAPOLR*, and a supplemental set, DNAPOLF and DNAPOLR); or the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (GAPDHF and GAPDHR) were obtained from Eurogentec (Eurogentec, Seraing, Belgium). The nucleotide sequences of these primers are shown in Table 1. The positions of the CLT-specific and US28-specific primers relative to their corresponding transcripts are shown in Fig. 1. Primers CLTR1 and CLTR2 were derived from nucleotide sequences within CLT exon 3 and exon 2, respectively (Fig. 1). Primers CLTF1 and CLTF2 were derived from the sequence between the latency-specific transcription start site 2 (LSS2) and the PSS (Fig. 1). The sequences of primer ANTICLTF and ANTICLTR are collinear with MIE intron sequences and can therefore not detect mature sense CLTs (Fig. 1). However, it may be possible that these primers amplify either sense-polarized MIE-specific transcripts that have not been identified before, or immature CLTs containing intron sequences. Both possibilities should be taken into consideration for every reference to antisense CLTs in this report. Each target sample for RT-PCR was obtained by isolation of poly(A)+ RNA from 5 × 106 cells using a QuickPrep Micro mRNA purification kit (Amersham Pharmacia Biotech, Saclay, France). Subsequently, poly(A)+ RNA samples were treated with DNase I (Amersham Pharmacia Biotech) according to the manufacturer's protocol. They were then reverse transcribed using an Advantage RT-for-PCR Kit (Clontech-Ozyme, Montigny-Le-Bretonneux, France). RT was primed with oligo(dT) primers included in the kit. Aliquots of the resulting cDNA that corresponded to 2 × 105 cells were added to PCR mixtures. PCR mixtures were prepared using AdvanTaq Plus DNA Polymerase kit (Clontech-Ozyme) according to the manufacturer's protocol. Thermal cycling conditions were similar for PCRs with all primer sets mentioned (2 min of denaturation at 94°C, followed by 50 cycles of 5 s at 94°C and 30 s at 70°C), except for those for the GAPDH- and HCMV DNA polymerase-specific (DNAPOLF and DNAPOLR) primer sets, which were 2 min of denaturation at 94°C, followed by 50 cycles of 5 s at 94°C and 30 s at 68°C. Thermal cycling was done with a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer, Courtaboeuf, France). PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining.
TABLE 1.
PCR primers used in this study
| Gene | Primer sequence |
|---|---|
| GAPDHF | GGGGAGCCAAAAGGGTCATCATCT |
| GAPDHR | GAGGGGCCATCCACAGTCTTCT |
| US28F | CGTCGGATTCAATGCTCCGGCGATGTTTAC |
| US28R | GAATGGCGATGATCACGGCAAAGATCCACC |
| CLTF1 | TAGTCTGCAGGAACGTCGTGGCCTTGGT |
| CLTF2 | TCGTCAGGATTATCAGGGTCCATCTTTCTCTT |
| CLTR1 | TACATCAATGGGCGTGGATAGCGGTTTGAC |
| CLTR2 | AAATGGGCGGTAGGCGTGTACGGTGG |
| ANTICLTF | ACTCATGGTCGCTCGGCAGCTCCTTGCTC |
| ANTICLTR | GCACAAACCCCGACACGTACCGTGGCA |
| DNAPOLF* | TGGCTAAAATTCCGTTGCGGCGTGTCAT |
| DNAPOLR* | TGCAAGGGCGGCGACATCTGAAACATA |
| DNAPOLF | GGGCACAGCGGCGGTAGAGATGAT |
| DNAPOLR | TCCCGCGTTGTTTCGTGGCTAATG |
Indirect immunocytometry.
The following monoclonal antibodies (MAb) were used for HCMV antigen detection in both MRC5 and THP-1 cells: (i) MAb E13 (a kind gift from M. C. Mazeron, Hôpital Lariboisière, Paris, France), which detects both HCMV immediate early 1 and 2 antigens; (ii) MAb F6a, which detects the HCMV early antigen ppUL83 (pp65) (Michelson et al., unpublished data); and (iii) an anti-pp150 MAb (a kind gift from H. P. Hartus, Universität Erlangen-Nürnberg, Erlangen, Germany), which detects the HCMV late antigen ppUL32 (pp150) (17). The secondary antibodies used were fluorescein-conjugated sheep anti-mouse immunoglobulin G (Amersham Pharmacia Biotech) for the detection of MAb F6a and fluorescein-conjugated goat anti-mouse immunoglobulin G (CALTAG Laboratories, Burlingame, Calif.) for the detection of MAb E13 and MAb anti-pp150. Samples containing 106 uninfected or HCMV-infected cells of either MRC5 or THP-1 origin were permeabilized in permeabilization buffer (phosphate-buffered saline with 0.2% bovine serum albumin [Sigma-Aldrich Chemie, Steinheim, Germany] and 0.05% saponin [Sigma-Aldrich Chemie]) for 20 min at room temperature. All subsequent incubations were done in permeabilization buffer at 4°C. Cells were incubated for 30 min with 5 μg of primary MAb per ml, washed two times, and stained with secondary MAb according to the manufacturer's protocol. Finally, the cells were fixed in phosphate-buffered saline with 4% formaldehyde and subjected to flow cytometry using a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, La Pond-de Claix, France). Data were analyzed using the CELL Quest flow cytometry analysis program (version 3.3; Becton Dickinson Immunocytometry Systems).
RESULTS
Detection of HCMV-specific transcripts in various infected myeloid cell lines.
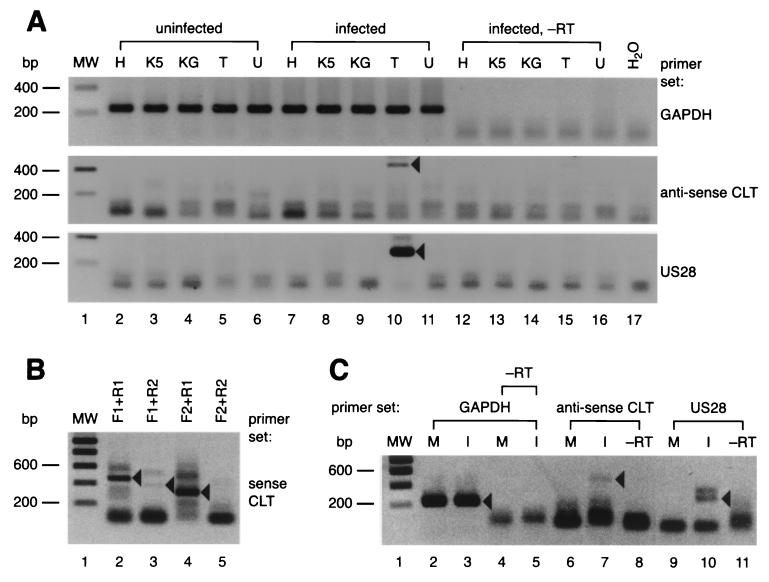
Previously, both HCMV-specific DNA and RNA in latently infected individuals were detected using PCR techniques. Genomic HCMV DNA can be detected in approximately 0.01% of mononuclear cells in blood and BM samples from naturally infected individuals (19, 36, 39), whereas between 0.01 and 0.001% of BM-derived CD33+ CD14+ and CD33+ CD15+ cells contain CLTs (13). Similar to the techniques used for the detection of genomic HCMV DNA in these cells, we developed an RT-PCR system to enable detection of HCMV-specific transcripts during latent infection in vitro. In order to determine the type of cell that would be suitable for establishing latent HCMV infection in vitro, we screened several infected myeloid cells lines for the presence of US28-specific transcripts and antisense CLTs. Each of these lines represents a specific differentiation stage in hematopoiesis toward monocytes: K562 cells (very early pluripotent hematopoietic stem cells), KG1a (pluripotent CD34+ hematopoietic stem cell-like), HL-60 (monocyte or granulocyte progenitor-like), U937 (promonocytic), and THP-1 (monocytic) (reviewed by Harris 14). Cells (5 × 107 per sample) were infected with HCMV strain Toledo at a multiplicity of infection (MOI) of 4. On day 4 p.i., culture medium was refreshed. Finally, on day 8 p.i., cells were harvested and subjected to RT-PCR. Whereas all cDNA samples were positive for GAPDH transcripts with the expected size of 235 bp (Fig. 2A, lanes 2 to 11, upper panel), only the sample containing cDNA from the infected THP-1 monocytic cell line was PCR positive for both US28 transcripts (Fig. 2A, lane 10, middle panel; expected size = 298 bp) and antisense CLT (Fig. 2A, lane 10, lower panel; expected size = 469 bp). Fresh samples were subjected to a new round of PCR in which various combinations of sense CLT-specific primer pairs were included (Fig. 1). As shown in Fig. 2B, PCR mixtures containing either primer pairs CLTF1 and CLTR1, CLTF2 and CLTR1, or CLTF1 and CLTR2 each produced products that matched the expected sizes of 436, 368, and 297 bp, respectively (Fig. 2B, lanes 2 to 4). In contrast, PCR samples containing the primer combination of CLTF2 and CLTR2 (expected PCR product size = 229 bp) remained negative, possibly due to incompatibility of the PCR primer pair (Fig. 2B, lane 5). Nonetheless, these results indicate that HCMV-infected THP-1 cells harbor US28-specific transcripts, as well as sense and anti-sense CLTs at day 8 p.i. We were also able to detect these transcripts at a later time point. Both US28- and antisense CLT-specific RT-PCR signals were obtained from samples of infected THP-1 cells at day 15 p.i. (Fig. 2C, lanes 7 and 10). We therefore focused on HCMV gene expression in THP-1 cells in subsequent experiments.
FIG. 2.
The US28 and MIE genes are transcribed in HCMV-infected THP-1 cells. The figure shows ethidium bromide-stained 2% agarose gels in which RT-PCR samples were separated. All agarose gel images shown in this report were digitized and contrast-inverted for clarity using a video scanner (Virbert Lourmat, Marne la Vallée, France). Molecular weight (MW) marker sizes are indicated on the left of each panel, and the primer sets corresponding to each panel are indicated on the right. Black arrowheads denote the relevant PCR products. (A) Detection of GAPDH-specific transcripts, antisense CLT-specific, and US28-specific transcripts in a panel of HCMV-infected myeloid cell types at day 8 p.i. The cell types used are indicated above the panel using the following abbreviations: H, HL-60; K5, K562; KG, KG1a; T, THP-1; U, U937. −RT, sample not treated with reverse transcriptase. (B) Detection of sense CLTs from HCMV-infected THP-1 cells at day 8 p.i. The primer combinations that were used in this experiment (see also Table 1 and Fig. 1) are indicated above each lane. (C) Detection of GAPDH-specific transcripts, anti-sense CLT-specific, and US28-specific transcripts in HCMV-infected THP-1 cells at day 15 p.i. M, mock infected; I, HCMV Toledo infected.
US28 is transcribed de novo in THP-1 cells at late time points p.i.
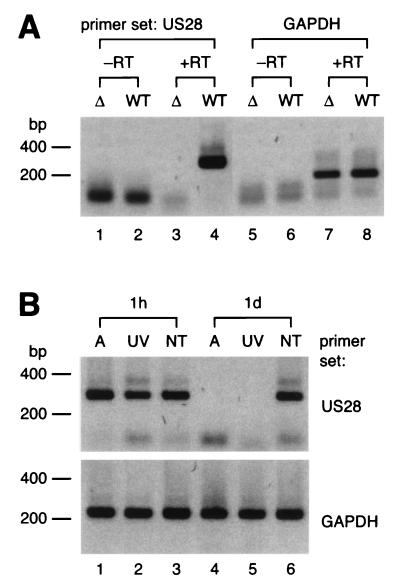
Although we were able to detect viral transcripts in THP-1 cells by RT-PCR, we did not exclude the possibility that these transcripts could be deposited by virions into the target cells upon inoculation. To examine whether US28-specific mRNA can be deposited in THP-1 cells, we prepared virions from both HCMV Toledo and a (negative control) US27-US28 deletion mutant strain, RV101 (4), and subjected these to RT-PCR analysis. Poly(A)+ RNA was isolated from the virion preparations and subsequently reverse transcribed and included in PCR mixtures containing either US28- or GAPDH-specific primers. Surprisingly, both US28- and GAPDH-specific transcripts were detected in mixtures from the Toledo samples (Fig. 3A), indicating that the viral inoculum contains both viral and host cell-specific poly(A)+ RNA. Consequently, it is possible that both types of RNA can be deposited either at the cell surface of inside THP-1 cells upon inoculation. To examine transfer of mRNA from the inoculum to the target cells, we infected THP-1 cells that were treated with actinomycin D (10 μg/ml, 1 h before, during, and after infection) to block de novo RNA synthesis. In addition, untreated THP-1 cells were inoculated with either UV-inactivated or intact HCMV. Samples of these THP-1 cultures were analyzed by RT-PCR at 1 h and 1 day p.i. Both US28- and GAPDH-specific transcripts could be detected in actinomycin-D-treated cells at 1 h p.i. (Fig. 3B, lane 1), whereas the actinomycin-D-treated cells at day 1 p.i. were PCR negative (Fig. 3B, lane 4). This indicates that although both US28- and GAPDH-specific mRNA are deposited either at the cell surface or inside THP-1 cells upon infection, the transcripts cannot persist for 1 day in these cells. Similar results were obtained by infection with UV-inactivated virus. US28-specific transcripts could be detected in THP-1 cells treated with inactivated virus at 1 h p.i. but not at day 1 p.i. (Fig. 3B, lanes 2 and 5, respectively). In contrast, transcripts were detected at each of these time points in cells infected with untreated virus (Fig. 3B, lanes 3 and 6). Taken together, we conclude that US28 is transcribed de novo in THP-1 cells at very late time points (8 to at least 15 days) p.i.
FIG. 3.
US28-specific transcripts are deposited either inside or at the surface of THP-1 cells immediately after infection but transcribed de novo at later times p.i. RT-PCR samples are visualized on agarose gels as described in Fig. 2. (A) Detection of viral and cellular poly(A)+ RNA in virus inoculum from HCMV-infected fibroblasts. The primer sets that were included in the RT-PCR samples are indicated on top of the panel. (B) Detection of US28-specific transcripts either in actinomycin-D-treated HCMV-infected THP-1 cells or in THP-1 cells infected with UV-inactivated HCMV. The primer sets that were included in the RT-PCR samples are indicated at the right of each panel. Samples from which reverse transcriptase enzyme was omitted remained PCR negative (data not shown). Abbreviations: −RT, sample not treated with reverse transcriptase; +RT, reverse transcriptase treated; Δ, HCMV RV101 (US27-US28 deletion mutant) virions; WT, wild-type HCMV; h, hours p.i.; d, days p.i.; A, actinomycin D treated; UV, UV-treated; NT, not treated.
Detection of lytic HCMV antigens in infected THP-1 cells.
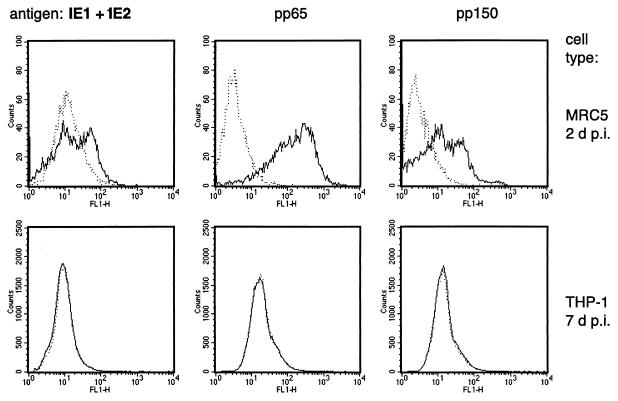
HCMV infection in unstimulated THP-1 cells was previously defined as abortive. This was based on experiments indicating the absence of HCMV antigen expression (16, 22, 41, 44), probably due to repression of the MIE promoter-enhancer (35). However, subtle differences in culture conditions may result in the presence of small numbers of differentiated cells harboring replicating HCMV in an infected THP-1 population. Consequently, US28-specific transcripts and CLTs could be synthesized in a small fraction of productively rather than latently infected cells. To determine whether in our system a fraction of the HCMV-treated THP-1 culture contains reproductively infected cells, we subjected a large quantity (5 × 105 per sample) of HCMV-treated THP-1 cells to immunocytometric analysis. An assay was set up to enable detection of antigens representing all phases of the lytic HCMV infection program, including pUL122 and pUL123 (the MIE 1 and 2 transactivator antigens), ppUL83 (a dominant early-phase tegument phospoprotein pp65), and ppUL32 (a dominant late-phase tegument phosphoprotein pp150). By using this assay, each of these antigens was detected at day 2 p.i. in MRC5 fibroblasts that were infected at an MOI of 0.1 but not at day 7 p.i. in THP-1 monocytes that were infected at an MOI of 4 (Fig. 4). These results indicate that the absence of lytic HCMV antigens in THP-1 cells is inadequate to define abortive infection, considering that both US28-specific transcripts and CLTs were detected in these cells. Instead, the detection of gene transcription in the absence of lytic HCMV antigens may indicate latent infection. Nevertheless, productively infected THP-1 cells may be present at levels below the level of antigen detection by immunocytometry. Therefore, as described in the section below, an alternative standard is needed to determine whether the US28-specific transcripts occur in productively or latently infected cells.
FIG. 4.
Expression of lytic-phase antigens in HCMV-infected THP-1 cells at day 7 p.i. The top panels show immunocytometric histograms of HCMV-infected MRC5 fibroblasts (total cell count = 10,000) at day 2 p.i. The lower panels show histograms of infected THP-1 cells (total cell count = 500,000) at day 7 p.i. FL1-H, relative intensity of fluorescence. Dotted lines represent uninfected cells.
Quantitative assessment of HCMV transcripts in infected THP-1 cells.
Latent HCMV infection, as opposed to productive infection, implicates the absence of factors enabling viral replication, possibly including those required for viral genomic DNA replication. Here, we set out to determine whether US28-transcribing HCMV-infected THP-1 cells contain such factors. For this purpose, we designed an RT-PCR assay to compare the transcription levels of US28 with those of the UL54 HCMV DNA polymerase gene in infected THP-1 cells. Initially, the sensitivity of both US28- and UL54-specific RT-PCR assays (each utilizing either US28F-US28R or DNAPOLF*-DNAPOLR*, respectively [Table 1]) were determined. This was done by applying RT-PCR to target cDNA serially diluted in a cDNA background corresponding to 2 × 105 of uninfected THP-1 cells. Positive RT-PCR signals were obtained in samples initially containing a minimum of 10 copies of either US28specific (Fig. 5A, lanes 1 to 5, upper panel) or UL54-specific (Fig. 5A, lanes 1 to 5, lower panel; expected size = 245 bp) cDNA. These results indicate that the US28- and UL54-specific RT-PCR assays have similar sensitivities. Subsequently, we determined the amount of HCMV-infected THP-1 cells containing either US28- or UL54-specific transcripts at day 7 p.i.: cDNA samples were obtained from a panel of HCMV-treated THP-1 cells serially diluted with untreated cells corresponding to a total of 2 × 105 cells. These samples were subjected to RT-PCR analysis. Consequently, we found that the HCMV-treated THP-1 cell culture could be diluted at least 10-fold before the corresponding US28-specific RT-PCR signal was lost (Fig. 5, lanes 1 to 6, upper panel). This indicates that US28 is transcribed in at least 1 out of 2 × 104 HCMV-treated THP-1 cells. Surprisingly, we were also able to detect UL54-specific transcripts in HCMV-infected THP-1 cells. However, these transcripts could only be detected in the undiluted culture sample (Fig. 5, lanes 1 to 6, middle panel). Since both assays were shown to have similar sensitivities, we conclude that US28-transcribing cells are in 10-fold excess of UL54-transcribing cells within an HCMV-infected THP-1 culture at day 7 p.i. Next, we set out to compare the levels of US28 and UL54 transcription in the total cDNA fraction corresponding to 2 × 105 THP-1 cells. For this purpose, a series of diluted cDNA samples from THP-1 cells at day 7 p.i. was subjected to RT-PCR analysis. In effect, US28-specific transcripts could be detected in cDNA samples that were diluted up to 1,000-fold (Fig. 5C, lanes 1 to 6, upper panel). In contrast, UL54-specific transcripts could only be detected in undiluted samples (Fig. 5C, lanes 1 to 6, middle panel), indicating that the level of US28 transcription is 1,000-fold that of UL54.
FIG. 5.
Quantitative comparison of US28 and HCMV DNA polymerase gene transcription in HCMV-infected THP-1 cells at day 7 p.i. RT-PCR samples are visualized on agarose gels as described in Fig. 2. The primer sets that correspond to each of the panels are indicated on the right of each panel. Black arrowheads denote the relevant PCR products. (A) Sensitivity of RT-PCR for the detection of US28 and HCMV DNA polymerase cDNA. The amount of target cDNA molecules that was included in each RT-PCR sample is indicated at the top. (B) Quantification of HCMV-infected THP-1 cells transcribing either US28 or the HCMV DNA polymerase gene at day 7 p.i. The quantities on top correspond to the amount of cells that were taken from the original HCMV-infected THP-1 culture and mixed with uninfected THP-1 cells to a total of 2 × 105 prior to cDNA preparation and RT-PCR. (C) Quantification of US28- and HCMV DNA polymerase-specific transcripts in untreated and PAA-treated HCMV-infected THP-1 cells at day 7 p.i. The dilution factor of each cDNA sample relative to an initial sample representing 2 × 105 cells is indicated on the top. Abbreviations: M, mock-infected; I, HCMV-infected; I/PAA, infected and PAA-treated; −RT, sample not treated with reverse transcriptase.
Since the amount of cells transcribing US28 was estimated to be 10-fold that of UL54-transcribing cells, it is possible that the HCMV-treated THP-1 culture contains both a latently and a productively infected fraction. Alternatively, the US28-positive–UL54-negative fraction could represent THP-1 cells that absorbed HCMV particles that were passed on by neighboring UL54-positive cells at a late time point in culture. The US28-positive–UL54-negative fraction might therefore be productively infected, albeit at an early stage of infection in which UL54 transcription is not yet manifest. To determine whether US28 transcription is the result of such a productive infection phenomenon, we assessed US28 transcription in HCMV-treated THP-1 cells treated with a viral DNA replication inhibitor. THP-1 cells were infected as described above. At 4 h p.i., phosphonoacetic acid (PAA) was added to the culture medium at a final concentration of 200 μg/ml. PAA treatment was maintained until the cells were harvested at day 7 p.i. The inhibitory potential of PAA was confirmed by assessing inhibition of pp150 synthesis in HCMV-infected MRC5 fibroblasts using immunocytometry (data not shown). RT-PCR was performed on serially diluted cDNA samples derived from HCMV-infected PAA-treated THP-1 cells. Interestingly, US28-specific signals were obtained in samples diluted as much as 1,000-fold (Fig. 5C, lanes 7 to 12, upper panel), similar to what was found for infected cells that were not treated with PAA (Fig. 5C, lanes 1 to 6, upper panel). To check whether PAA treatment had no effect on the natural cellular transcription level, the dilution series from both untreated and PAA-treated cells were subjected to RT-PCR using GAPDH-specific primers. Levels of GAPDH transcription were found to be similar for both samples (Fig. 5C, lanes 1 to 12, lower panel), indicating that US28 transcript levels are also similar. Surprisingly, RT-PCR signals for UL54 could not be observed in cDNA samples from PAA-treated cells, whereas the undiluted sample of untreated HCMV-infected cells was RT-PCR positive (Fig. 5, lanes 1 to 12, middle panel). Although PAA treatment affects HCMV DNA polymerase activity, we did not expect UL54 transcript levels to be lower in PAA-treated cells than in untreated cells. This might indicate that UL54 transcription is propelled by a late-phase positive feedback mechanism. We did not investigate this possibility further. Nevertheless, we have shown that (i) US28 is transcribed in HCMV-infected THP-1 cells at least until day 15 p.i., (ii) the amount of US28-transcribing THP-1 cells is approximately 10-fold that of UL54-transcribing cells at day 7 p.i., and (iii) detection of US28 transcription is irrespective of viral replication in HCMV-infected THP-1 cells. These results indicate that US28 is transcribed in latently infected THP-1 monocytes.
US28 transcription in differentiated THP-1 cells.
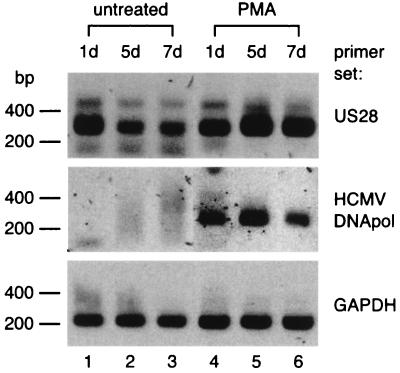
In addition to demonstrating that the HCMV US28 gene is transcribed during latent infection in undifferentiated THP-1 monocytes, we set out to determine whether US28 transcription also occurs in differentiated cells. Previously, HCMV-infected monocyte-derived macrophages were shown to support viral replication (22, 37, 40). Therefore, we also assessed the replication status of the HCMV-infected cells by determining whether the HCMV DNA polymerase gene UL54 would be upregulated upon differentiation. First, THP-1 cells were infected with HCMV as described above, and then a culture sample was treated with phorbol-12-myristate-13-acetate (PMA) (50 ng/ml) from 0 to 72 h p.i. In contrast to untreated THP-1 cells, PMA-treated THP-1 cells became adherent after several hours of stimulation (data not shown). Such an adhesion phenotype is typical for differentiation of monocytic cells (14). At days 1, 5, and 7 p.i., both US28 and UL54 transcription were assessed in cDNA samples derived from untreated and PMA-treated THP-1 cultures. Alternative primers for UL54 transcript detection, DNAPOLF and DNAPOLR (Table 1), were used for RT-PCR, resulting in an assay less sensitive than RT-PCR with the aforementioned DNAPOLF* and DNAPOLR* primers (Table 1). As a result, UL54-transcripts could not be detected in undifferentiated HCMV-infected THP-1 cells (Fig. 6, lanes 1 to 3, middle panel). In contrast, by using these primers, UL54 transcripts were detected in differentiated THP-1 cells at all time points (Fig. 6, lanes 4 to 6, middle panel). These results confirm upregulation of UL54 transcription and possibly HCMV replication upon differentiation of THP-1 cells. Finally, we were able to detect US28 transcript in both untreated and PMA-treated HCMV-infected cells at all time points mentioned (Fig. 6, lanes 1 to 6, upper panel). We therefore conclude that, in addition to transcription in latently infected THP-1 monocytes, US28 transcription is sustained in infected THP-1 cells throughout differentiation into adherent, macrophage-like cells.
FIG. 6.
Transcription of US28 and the HCMV DNA polymerase gene in undifferentiated (untreated) and differentiated (PMA-treated) THP-1 cells. RT-PCR samples are visualized on agarose gels as described in Fig. 2. The primer sets that correspond to each of the panels are indicated on the right. The time points (p.i.) at which samples were taken for RT-PCR are shown at the top. d, days p.i.
DISCUSSION
To date, both US28 transcripts and CLTs are the only transcripts identified in myeloid cells that are latently infected with HCMV. Yet, they are also present in productively infected cells (4, 24, 42, 43, 44) and are therefore not explicit markers of latency. To determine whether cells are latently infected with HCMV, the presence of either viral DNA or RNA from candidate latency-associated genes should be demonstrated in parallel to the absence of virus production. In this report, we considered the presence of HCMV DNA polymerase UL54 transcripts to be a determinant of productive infection. However, this determinant could prove to be too stringent; in addition to being a marker for productive infection, it could indicate viral DNA replication during latency, such as in a self-renewing HCMV-infected myeloid cell population. Nevertheless, we showed that the majority of US28-transcribing THP-1 cells does not contain UL54-specific transcripts. Moreover, US28 transcription levels were not affected in the presence of an HCMV DNA replication inhibitor. These findings indicate that this majority of US28-transcribing THP-1 cells are latently infected. Similar conditions could be used as a standard for identifying other latency-associated transcripts. Thus, a strict definition of latency could comprise (i) the absence of UL54-specific transcripts and (ii) candidate HCMV gene transcription irrespective of viral DNA replication. Nonetheless, UL54 transcription does not necessarily exclude latency. Its role in viral genome replication during latent infection will have to be addressed in future studies.
We demonstrated that the lytic phase antigens IE1-IE2, pp65, and pp150 were present at levels below the detection level in undifferentiated THP-1 cells and that HCMV DNA polymerase UL54 transcription levels were higher in differentiated cells than in undifferentiated THP-1 cells. Similarly, it was previously shown that MIE protein levels are higher in differentiated THP-1 cells that were infected with HCMV than in less differentiated cells (23). In addition, the generation of HCMV particles could not be demonstrated in monocytes, whereas replication was demonstrated in monocyte-derived macrophages and dendritic cells (34, 37). Thus, the relative degree of monocyte differentiation may be an important factor for regulation of HCMV gene expression. This explains why in our study US28 transcripts were not detected in K562, KG1a, HL-60, and U937 cells, which are less differentiated than the monocyte-type THP-1 cells. In addition to containing US28 and the MIE gene, the HCMV genome may contain many other genes that could concert latency and replication in myeloid cells. However, the low amount of infected (relative to unifected) myeloid precursors and the low-level transcription of HCMV genes during latent infection currently limit the study of HCMV gene expression in vivo to RT-PCR analysis. In the future, a sensitive immunological assay will have to be developed to confirm expression of viral proteins during latent HCMV infection in monocytes.
In this report we demonstrated that HCMV inoculum contains both viral and host cell RNA. We did not determine further whether the poly(A)+ RNA originated from virions or cellular debris associated with the virions. However, Greijer et al. (12) recently reported that both viral and host cell RNA molecules can be detected in sucrose density step-gradient-purified HCMV virions. Furthermore, they showed that virion-associated RNA can be deposited either at the surface of or inside inoculated fibroblasts upon inoculation. RNA could be detected after 1 h, but not at 4 h or at later time points p.i.—similar to our observation that virus-associated transcripts can be deposited in or on THP-1 cells upon infection but that these transcripts do not persist in or on these cells for more than 1 day p.i.
Kledal et al. (18) reported that pUS28 is capable of binding soluble forms of the CX3C chemokine fractalkine (or neurotactin) with subnanomolar affinity. This chemokine exists both in a soluble and a cell membrane-bound form. The membrane-bound form consists of a chemokine domain, a mucin stalk, a transmembrane and an intracellular domain (1, 28). Interestingly, it appears to combine the properties of both chemoattractants and adhesion molecules. Fractalkine is expressed at the surface of activated endothelium, thereby enabling leukocyte capture, firm integrin-independent adhesion, and attachment of circulating monocytes under flow conditions (7, 9). Additionally, it was shown that pUS28-expressing 300-19 murine pre-B cells could be captured by a fractalkine- or stalk-coated surface under physiological flow conditions (15). Although many CC chemokines can bind pUS28 (4, 10, 21, 26), fractalkine has the highest affinity, as well as a slow off-rate for this receptor (15, 18). In this report it was shown that US28 is transcribed in monocytic cells that harbor latent HCMV. Thus, expression of the US28 gene could play a role in the tethering of latently infected circulating monocytes to endothelial tissue expressing membrane-bound fractalkine. Alternatively, pUS28 could guide latently infected monocytes to other tissues expressing fractalkine or CC chemokines by chemotaxis. Fractalkine is shed from intestinal epithelial cells upon stimulation with interleukin 1β (25). Moreover, membrane-bound fractalkine is expressed by skin-derived dendritic and Langerhans cells (30), as well as by skin endothelial cells and dermal dendrocytes (29). These cell types are permissive for productive HCMV infection and therefore potential targets for monocyte-mediated HCMV dissemination. Finally, monocytes mature into either macrophages or dendritic cells upon migration into target tissues (14). We showed that transcription of US28 persists during differentiation of THP-1 cells by stimulation with PMA. It is therefore possible that pUS28 continues to guide infected monocytes during their differentiation in the target tissues. Further studies require the development of suitable antibodies against the US28 gene product. By using these antibodies, pUS28 surface expression on latently infected monocytes from healthy individuals could be confirmed, as well as pUS28-dependent interaction of these cells with the endothelium and other potential target tissues. Hence, an intriguing function of pUS28 in the dissemination of latent HCMV may be established.
ACKNOWLEDGMENTS
We thank Maria-Paola Landini for the kind gift of Toledo virus and Tom Jones for providing the deletion mutant RV101. We also thank Françoise Bachelerie and Agustin Valenzuela-Fernandez for critically reading the manuscript.
P.S.B. is a beneficiary of a research fellowship from the European Molecular Biology Organization, Heidelberg, Germany. This work was supported by the Agence National de la Recherche contre le SIDA.
REFERENCES
- 1.Bazan J F, Bacon K B, Hardiman G, Wang W, Soo K, Rossi D, Greaves D R, Zlotnik A, Schall T J. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 2.Billstrom M A, Johnson G L, Avdi N J, Worthen G S. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J Virol. 1998;72:5535–5544. doi: 10.1128/jvi.72.7.5535-5544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billstrom M A, Lehman L A, Worthen G S. Depletion of extracellular RANTES during human cytomegalovirus infection of endothelial cells. Am J Respir Cell Mol Biol. 1999;21:163–167. doi: 10.1165/ajrcmb.21.2.3673. [DOI] [PubMed] [Google Scholar]
- 4.Bodaghi B, Jones T R, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier J L, Michelson S. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J Exp Med. 1998;188:855–866. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolovan-Fritts C A, Mocarski E S, Wiedeman J A. Peripheral blood CD14+ cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood. 1999;93:394–398. [PubMed] [Google Scholar]
- 6.Casarosa P, Bakker R A, Verzijl D, Navis M, Timmerman H, Leurs R, Smit M J. Constitutive signalling of the human cytomegalovirus-encoded chemokine receptor US28. J Biol Chem. 2001;276:1133–1137. doi: 10.1074/jbc.M008965200. [DOI] [PubMed] [Google Scholar]
- 7.Chapman G A, Moores K E, Gohil J, Berkhout T A, Patel L, Green P, Macphee C H, Stewart B R. The role of fractalkine in the recruitment of monocytes to the endothelium. Eur J Pharmacol. 2000;392:189–195. doi: 10.1016/s0014-2999(00)00117-5. [DOI] [PubMed] [Google Scholar]
- 8.Chee M S, Satchwell S C, Preddie E, Weston K M, Barrell B G. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 9.Fong A M, Robinson L A, Steeber D A, Tedder T F, Yoshie O, Imai T, Patel D D. Fractalkine and CX3CR1 mediate a novel mechanism of leukocyte capture, firm adhesion, and activation under physiologic flow. J Exp Med. 1998;188:1413–1419. doi: 10.1084/jem.188.8.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J L, Murphy P M. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 11.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 12.Greijer A E, Dekkers C A J, Middeldorp J M. Human cytomegalovirus virions differentially incorporate viral and host cell RNA during the assembly process. J Virol. 2000;74:9078–9082. doi: 10.1128/jvi.74.19.9078-9082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris P. Human myeloid cell lines. In: Herzenberg L, Blackwell C, editors. The integrated immune system. 5th ed. IV. Cambridge, Mass: Blackwell Science; 1996. pp. 173.1–173.16. [Google Scholar]
- 15.Haskell C A, Cleary M D, Charo I F. Unique role of the chemokine domain of fractalkine in cell capture: kinetics of receptor dissociation correlate with cell adhesion. J Biol Chem. 2000;275:34183–34189. doi: 10.1074/jbc.M005731200. [DOI] [PubMed] [Google Scholar]
- 16.Huang T H, Oka T, Asai T, Okada T, Merrills B W, Gertson P N, Whitson R H, Itakura K. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahn G, Harthus H P, Broker M, Borisch B, Platzer B, Plachter B. Generation and application of a monoclonal antibody raised against a recombinant cytomegalovirus-specific polypeptide. Klin Wochenschr. 1990;68:1003–1007. doi: 10.1007/BF01646545. [DOI] [PubMed] [Google Scholar]
- 18.Kledal T N, Rosenkilde M M, Schwartz T W. Selective recognition of the membrane-bound CX3C chemokine, fractalkine, by the human cytomegalovirus-encoded broad-spectrum receptor US28. FEBS Lett. 1998;441:209–214. doi: 10.1016/s0014-5793(98)01551-8. [DOI] [PubMed] [Google Scholar]
- 19.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo K, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn D E, Beall C J, Kolattukudy P E. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem Biophys Res Commun. 1995;211:325–330. doi: 10.1006/bbrc.1995.1814. [DOI] [PubMed] [Google Scholar]
- 22.Lashmit P E, Stinski M F, Murphy E A, Bullock G C. A cis repression sequence adjacent to the transcription start site of the human cytomegalovirus US3 gene is required to down regulate gene expression at early and late times after infection. J Virol. 1998;72:9575–9584. doi: 10.1128/jvi.72.12.9575-9584.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C H, Lee G C, Chan Y J, Chiou C J, Ahn J H, Hayward G S. Factors affecting human cytomegalovirus gene expression in human monocyte cell lines. Mol Cells. 1999;9:37–44. [PubMed] [Google Scholar]
- 24.Lunetta J M, Wiedeman J A. Latency-associated sense transcripts are expressed during in vitro human cytomegalovirus productive infection. Virology. 2000;278:467–476. doi: 10.1006/viro.2000.0666. [DOI] [PubMed] [Google Scholar]
- 25.Muehlhoefer A, Saubermann L J, Gu X, Luedtke-Heckenkamp K, Xavier R, Blumberg R S, Podolsky D K, MacDermott R P, Reinecker H C. Fractalkine is an epithelial and endothelial cell-derived chemoattractant for intraepithelial lymphocytes in the small intestinal mucosa. J Immunol. 2000;164:3368–3376. doi: 10.4049/jimmunol.164.6.3368. [DOI] [PubMed] [Google Scholar]
- 26.Neote K, DiGregorio D, Mak J Y, Horuk R, Schall T J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 27.Ohagen A, Li L, Rosenzweig A, Gabuzda D. Cell-dependent mechanisms restrict the HIV type 1 coreceptor activity of US28, a chemokine receptor homolog encoded by human cytomegalovirus. AIDS Res Hum Retrovirus. 2000;16:27–35. doi: 10.1089/088922200309575. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Lloyd C, Zhou H, Dolich S, Deeds J, Gonzalo J A, Vath J, Gosselin M, Ma J, Dussault B, Woolf E, Alperin G, Culpepper J, Gutierrez-Ramos J C, Gearing D. Neurotactin, a membrane-anchored chemokine upregulated in brain inflammation. Nature. 1997;387:611–617. doi: 10.1038/42491. [DOI] [PubMed] [Google Scholar]
- 29.Papadopoulos E J, Fitzhugh D J, Tkaczyk C, Gilfillan A M, Sassetti C, Metcalfe D D, Hwang S T. Mast cells migrate, but do not degranulate, in response to fractalkine, a membrane-bound chemokine expressed constitutively in diverse cells of the skin. Eur J Immunol. 2000;30:2355–2361. doi: 10.1002/1521-4141(2000)30:8<2355::AID-IMMU2355>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 30.Papadopoulos E J, Sassetti C, Saeki H, Yamada N, Kawamura T, Fitzhugh D J, Saraf M A, Schall T, Blauvelt A, Rosen S D, Hwang S T. Fractalkine, a CX3C chemokine, is expressed by dendritic cells and is up-regulated upon dendritic cell maturation. Eur J Immunol. 1999;29:2551–2559. doi: 10.1002/(SICI)1521-4141(199908)29:08<2551::AID-IMMU2551>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 31.Patterson B K, Landay A, Andersson J, Brown C, Behbahani H, Jiyamapa D, Burki Z, Stanislawski D, Czerniewski M A, Garcia P. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol. 1998;153:481–490. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 33.Pleskoff O, Treboute C, Alizon M. The cytomegalovirus-encoded chemokine receptor US28 can enhance cell-cell fusion mediated by different viral proteins. J Virol. 1998;72:6389–6397. doi: 10.1128/jvi.72.8.6389-6397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riegler S, Hebart H, Einsele H, Brossart P, Jahn G, Sinzger C. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J Gen Virol. 2000;81:393–399. doi: 10.1099/0022-1317-81-2-393. [DOI] [PubMed] [Google Scholar]
- 35.Sinclair J H, Baillie J, Bryant L A, Taylor-Wiedeman J A, Sissons J G. Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J Gen Virol. 1992;73:433–435. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 36.Slobedman B, Mocarski E S. Quantitative analysis of latent human cytomegalovirus. J Virol. 1999;73:4806–4812. doi: 10.1128/jvi.73.6.4806-4812.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Söderberg-Naucler C, Fish K N, Nelson J A. Growth of human cytomegalovirus in primary macrophages. Methods. 1998;16:126–138. doi: 10.1006/meth.1998.0650. [DOI] [PubMed] [Google Scholar]
- 38.Streblow D N, Söderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson J A. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99:511–520. doi: 10.1016/s0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka N, Kimura H, Iida K, Saito Y, Tsuge I, Yoshimi A, Matsuyama T, Morishima T. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J Med Virol. 2000;60:455–462. doi: 10.1002/(sici)1096-9071(200004)60:4<455::aid-jmv14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 40.Turtinen L W, Seufzer B J. Selective permissiveness of TPA differentiated THP-1 myelomonocytic cells for human cytomegalovirus strains AD169 and Towne. Microb Pathog. 1994;16:373–378. doi: 10.1006/mpat.1994.1037. [DOI] [PubMed] [Google Scholar]
- 41.Weinshenker B G, Wilton S, Rice G P. Phorbol ester-induced differentiation permits productive human cytomegalovirus infection in a monocytic cell line. J Immunol. 1988;140:1625–1631. [PubMed] [Google Scholar]
- 42.Welch A R, McGregor L M, Gibson W. Cytomegalovirus homologs of cellular G protein-coupled receptor genes are transcribed. J Virol. 1991;65:3915–3918. doi: 10.1128/jvi.65.7.3915-3918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White K L, Slobedman B, Mocarski E S. Human cytomegalovirus latency-associated protein pORF94 is dispensable for productive and latent infection. J Virol. 2000;74:9333–9337. doi: 10.1128/jvi.74.19.9333-9337.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zipeto D, Bodaghi B, Laurent L, Virelizier J L, Michelson S. Kinetics of transcription of human cytomegalovirus chemokine receptor US28 in different cell types. J Gen Virol. 1999;80:543–547. doi: 10.1099/0022-1317-80-3-543. [DOI] [PubMed] [Google Scholar]