Abstract
The envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) is extensively glycosylated, containing approximately 23 asparagine (N)-linked glycosylation sites on its gp120 subunit. In this study, specific glycosylation sites on gp120 of a dualtropic primary HIV-1 isolate, DH12, were eliminated by site-directed mutagenesis and the properties of the resulting mutant envelopes were evaluated using a recombinant vaccinia virus-based cell-to-cell fusion assay alone or in the context of viral infections. Of the glycosylation sites that were evaluated, those proximal to the V1/V2 loops (N135, N141, N156, N160) and the V3 loops (N301) of gp120 were functionally critical. The glycosylation site mutations near the V1/V2 loop compromised the use of CCR5 and CXCR4 equally. In contrast, a mutation within the V3 loop preferentially inhibited the usage of CCR5; although this mutant protein completely lost its CCR5-dependent fusion activity, it retained 50% of the wild-type fusion activity with CXCR4. The replication of a virus containing this mutation was severely compromised in peripheral blood mononuclear cells, MT-4 cells, and primary monocyte-derived macrophages. A revertant virus, which acquired second site changes in the V3 loop that resulted in an increase in net positive charge, was isolated. The revertant virus fully recovered the usage of CXCR4 but not of CCR5, thereby altering the tropism of the parental virus from dualtropic to T-tropic. These results suggest that carbohydrate moieties near the V1/V2 and the V3 loops play critical roles in maintaining proper conformation of the variable loops for optimal interaction with receptors. Our results, combined with those of previously reported studies, further demonstrate that the function of individual glycans may be virus isolate dependent.
Human immunodeficiency virus type 1 (HIV-1) primarily infects CD4+ T lymphocytes and cells of monocyte-macrophage lineage. The cellular tropism of HIV-1 is determined largely at the level of virus entry, which depends on a series of interactions between viral envelope glycoprotein and cellular receptors. The gp120 surface glycoprotein subunit is thought to interact first with CD4 (the primary receptor) and then with one (or more) of the recently identified coreceptors, which include chemokine receptors CXCR4 and CCR5 (for reviews, see references 15, 16, 26, and 32). These interactions are thought to trigger conformational changes in the complex multimeric viral envelope glycoprotein structure, allowing the hydrophobic domain of transmembrane glycoprotein gp41 subunit to interact with the cellular membrane and induce virus-cell fusion.
While the exact mechanism of membrane fusion is still unclear, the interactions between gp120 and cellular receptors are slowly beginning to be understood. The detailed molecular nature of the interactions between gp120 and CD4 has been elucidated from analysis of a crystal structure of gp120 core complexed with CD4 (two N-terminal domains) and the 17b neutralizing monoclonal antibody (Fab fragment), which interacts with the putative coreceptor binding domain of gp120 (22). The interactions between gp120 and its coreceptors have been investigated using a multitude of indirect experimental approaches, including site-directed mutational analyses, functional and infectivity studies with chimeric proteins and viruses, and biochemical competition experiments with site-specific antibodies and chemokines (5–7, 10, 11, 14, 21, 23, 35, 40–42, 45–47, 49, 52). The accumulated data from these studies suggest that the variable loops V1/V2 and V3, which likely form a pocket surrounding the four-stranded antiparallel β-sheet (bridging sheet), play important roles in coreceptor interactions. Depending on the conformation of these loops, gp120 binds to CCR5, CXCR4, or both, thus determining the cellular tropism of the virus isolate (macrophage [M]-tropic, T-cell line [T]-tropic, or dualtropic, respectively).
HIV-1 gp120 is one of the most extensively glycosylated proteins (33). It contains 23 or 24 N-linked glycosylation sites, and the glycans attached to these sites account for approximately one-half of the protein's total mass (based on polyacrylamide gel mobility). Numerous studies using glycosylation and glycosidase inhibitors have revealed the importance of the carbohydrate moieties in determining the conformation of the HIV-1 envelope glycoprotein, a property that undoubtedly affects its processing, intracellular transport, and ability to interact with CD4 (13, 17, 19, 29, 38, 50). The gross modifications resulting from the use of these inhibitors, however, are not as informative as site-directed mutagenesis, which permits evaluation of the effects of individual glycans on protein structure and function. For example, site-directed mutagenesis of all 24 individual N-linked glycosylation sites of HIV-1HXB2 indicated that most of the glycosylation sites were individually dispensable (25). Of the 24 sites, only 5 (amino acids 88, 141, 197, 262, and 276), all of which are located in the amino-terminal half of gp120, affected virus infectivity.
Most site-directed mutagenesis studies have been conducted with gp120s of T-tropic laboratory-adapted HIV-1 strains (e.g., HXB2 or NL4-3). Several studies have demonstrated that these envelope glycoproteins have biochemical and immunological properties which differ from those of primary HIV-1 isolates (e.g., greater gp120 shedding and increased susceptibility to neutralizing antibodies or CD4 [1, 12, 30, 31, 48, 55; for a review, see reference 39]). We have previously characterized cellular tropism and coreceptor usage of a primary isolate, HIV-1DH12 (7, 23, 44), a dualtropic virus that can utilize CXCR4 and CCR5 almost equally and can infect both T-cell lines and primary monocyte-derived macrophages (MDM). In the present study, we examined the role of carbohydrate moieties in cellular tropism and coreceptor usage of HIV-1DH12 by mutagenizing specific N-linked glycosylation sites throughout gp120. The mutant envelope proteins were examined for their capacity to induce cell-to-cell fusion. Some of the mutants were further analyzed in the context of virus infectivity in both T-cell line and primary cells. Our results indicate that the N-linked glycosylation sites near the V1/V2 and V3 variable loops are critical for the induction of membrane fusion and virus entry.
MATERIALS AND METHODS
Envelope glycoprotein mutagenesis.
Mutations affecting the N-linked glycosylation sites were introduced into plasmid pTM-DHgp120H (24), which encodes HIV-1DH12 gp120. Asparagine (N) codons AAT or AAC were changed to glutamine (Q) codon CAG by using the mutagenic oligonucleotides listed in Table 1. Codon changes were made using the Quik Change site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene). N-linked glycosylation site mutations (μ) were subsequently transferred to pNVV-DHenv (7), which encodes the entire gp160 of HIV-1DH12. The mutations in pTM-DHgp120H μ1-μ3 were transferred to pNVV-DHenv by replacing the 488-bp KpnI-StuI fragment (nucleotides [nt] 120 to 608) to generate pNVV-DHenv μ1 to μ3. For pNVV-DHenv μ4 and pNVV-DHenv μ5 to μ10, the 787-bp EcoNI fragments (nt 600 to 1387) and the 559-bp BglII fragments (nt 817 to 1376), respectively, were transferred from the corresponding pTM-DHgp120H clones. The plasmids pNVV-DHenv μ2-1, μ2-2, μ3-1, μ3-2, μ8-1, and μ8-2 were generated directly from pNVV-DHenv with the same site-directed mutagenesis protocol using the oligonucleotides shown in Table 1.
TABLE 1.
N-linked glycosylation mutants of HIV-1DH12 envelope glycoprotein
| Mutation no. | Amino acid site | Amino acid change | Mutagenic oligonucleotide (5′ to 3′)a |
|---|---|---|---|
| 1 | 48 | Asn to Gln | GTGTGGAAAGAAGCACAGACCACTCTATTTTGTGC |
| 2 | 135 | Asn to Gln | GCACTGATTTGAAGCAGGGTACTAATTTGAAGCAGGGTACTAAAATCATTGGG |
| 141 | Asn to Gln | ||
| 2-1 | 135 | Asn to Gln | GCACTGATTTGAAGCAGGGTACTAATTTG |
| 2-2 | 141 | Asn to Gln | CTAATTTGAAGCAGGGTACTAAAATC |
| 3 | 156 | Asn to Gln | GGAGAAATAAAACAGTGCTCTTTCCAGGTCACCAAAAACATAATAG |
| 160 | Asn to Gln | ||
| 3-1 | 156 | Asn to Gln | GGAGAAATAAAACAGTGCTCTTTCAATG |
| 3-2 | 160 | Asn to Gln | CTGCTCTTTCCAGGTCACCAAAAAC |
| 4 | 241 | Asn to Gln | GGACCATGTACACAGGTCAGTACAGTACAATGTAC |
| 5 | 276 | Asn to Gln | GTAATTAGATCTAGCCAGTTCACGGACAATGCTAAAATC |
| 6 | 289 | Asn to Gln | CATAATAGTACAGCTGCAGGAAACTGTAGAAATTAATTG |
| 7 | 289 | Asn to Gln | CATAATAGTACAGCTGCAGGAAACTGTAGAAATTCAGTCTACAAGACCCAAC |
| 295 | Asn to Gln | ||
| 8 | 295 | Asn to Gln | CTGTAGAAATTCAGTGTACAAGACCCAACCAGAATACAAGAAAAGGG |
| 301 | Asn to Gln | ||
| 8-1 | 295 | Asn to Gln | GAAACTGTAGAAATTCAGTGTACAAGACCCAAC |
| 8-2 | 301 | Asn to Gln | GTACAAGACCCAACCAGAATACAAGAAAAGGG |
| 9 | 354 | Asn to Gln | GAAAAATTTGAACAGAAAACAATAGTCTTTCAGAAATCCTCAGGGGGGG |
| 360 | Asn to Gln | ||
| 10 | 390 | Asn to Gln | CAAAAAAACTGTTTCAGAGTACTTGGCAGGGTACTGAAGGGTC |
| 394 | Asn to Gln |
Mutations introduced in the oligonucleotides are underlined.
The Asn-to-Gln mutations in pNVV-DHenv μ2, μ7, μ8, μ8-1, and μ8-2 were transferred to an infectious molecular clone of chimeric virus AD8-DHenv (pAD8-DHenv [8]), which encodes the envelope glycoprotein of HIV-1DH12 in the background of HIV-1AD8. Mutations in μ2, μ8-1, and μ8-2 were transferred using the 1,883-bp DraIII-SalI fragment (nt 368 to 2251), while those in μ7 and μ8 were transferred using the 1,643-bp StuI-SalI fragment (nt 608 to 2251). The revertant envelope clone pNVV-DHenv μ8-R was constructed by transferring the 559-bp BglII fragment (nt 817 to 1376) from pCR-μ8-R (see below). All of the mutations were verified by DNA sequence analysis. The nucleotide numbering was based on the sequence of HIV-1DH12 gp160.
Protein expression and Western blotting.
The recombinant vaccinia viruses vvDHenv μ1 to μ10 were generated from corresponding pNVV-DHenv μ1 to μ10, following protocols previously described (9). Confluent monolayers of HeLa cells in 6-well plates were infected with recombinant vaccinia viruses at a multiplicity of infection of 10. Cell lysates were prepared by adding lysis buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 1.5 mM MgCl2, 1% NP-40) 48 h postinfection. Culture supernatants and cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by electrotransfer to nitrocellulose (Hybond; Amersham Life Science) for a Western immunoblot assay. Envelope glycoproteins were detected by blotting with rabbit anti-gp160 antiserum (53) followed by goat anti-rabbit immunoglobulin G-peroxidase conjugate and visualizing with an ECL Western blot detection kit according to the manufacturer's protocol (Amersham Life Science).
Fusion assay.
A highly sensitive secreted alkaline phosphatase (SEAP) reporter gene-based assay was used to quantitate cell-cell fusion events as previously described (23). To prepare target cells, recombinant vaccinia viruses encoding CCR5 (vvCCR5) or CXCR4 (vBD4 [3]), T7 RNA polymerase (vTF7-3 [18]), and human CD4 (vCB-3 [4]) were used to coinfect Mus dunni cells. To generate vvCCR5, the CCR5 gene was PCR amplified from a DNA preparation from human peripheral blood mononuclear cells PBMC using the following primers: (+), 5′-CTGAGGATCCCATATGGATTATCAAGTGTCA-AGT-3′, and (−), 5′-GATCTTAAGCTTCTAGATCAGTGATGGTGATGGTGATGCGATCC-TCTCAAGCCCACAGATATTTC-3′. The conditions for PCR were as follows: 94°C for 7 min; 3 cycles of 94°C for 1 min, 40°C for 1 min, and 72°C for 3 min; then 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min; and finally 72°C for 7 min. The PCR product was digested with BamHI and HindIII (incorporated in the primers) and cloned into the same restriction sites in pGEM-3Zf(−) (Promega, Madison, Wis.) to generate pGEM-CCR5. Subsequently, the 1,100-bp SmaI-XbaI fragment from pGEM-CCR5, which contains the CCR5 gene, was cloned into SmaI-AvrII sites of pNVV-3 (provided by M. Oldstone). The resulting plasmid, pNVV-CCR5, was used to generate recombinant vaccinia virus vvCCR5 following protocols previously described (9). To prepare the effector cells, M. dunni cells were coinfected with recombinant vaccinia viruses encoding SEAP under the control of T7 promoter (vTM-SEAP [23]) and either wild-type (vvDHenv [7]) or mutant (vvDHenv μ1 to μ10) envelope glycoproteins. Although some envelope mutants exhibited reduced levels of membrane-associated gp120, we used the same multiplicity of infection of the recombinant vaccinia viruses to express the same amount of the total envelope glycoprotein. Infected cells were incubated at 37°C for 5 h and then trypsinized. After the cells were washed twice with the culture medium (Dulbecco's minimal essential medium containing 10% fetal bovine serum), duplicate samples containing 5 × 104 (each) target and effector cells were mixed in a 96-well plate. Cells were cultured in the medium containing 80 μg of cytosine arabinoside per ml for 8 to 10 h at 37°C. SEAP activity in the culture supernatant was measured as previously described (23).
Virus stocks and infections.
The HIV-1 viruses used in this study were generated from an infectious molecular clone, pAD8-DHenv (8), which contains the env gene of HIV-1DH12 in the background of HIV-1AD8. Wild-type and mutant viruses were generated by transfecting respective plasmids into HeLa cells using the calcium phosphate-based Profection transfection system (Promega). Virus stocks were concentrated by ultracentrifugation (Beckman SW 55Ti rotor; 35,000 rpm for 30 min). The relative amounts of viruses were determined by measuring reverse transcriptase (RT) activity in the stocks as previously described (54). Viral infections were performed in 96-well plates using either phytohemagglutinin-blasted human PBMC, MT-4 cells, or MDM essentially as previously described (7).
Cloning the revertant env gene.
Human PBMC were infected with AD8-DHenv μ8-R virus, which was initially isolated from PBMC culture supernatants at peak RT activity (day 22). Infected cells were harvested and Hirt DNA was prepared as described previously (20). A 3-kb fragment spanning the env gene of HIV-1DH12 was amplified by PCR from the Hirt DNA using the following primers: (+), 5′-CAGTAGATCCTAGACTAGAGCCCTGG-3′ (387 nt upstream from the env initiation codon), and (−), 5′-GCTGCTCCCACCCCATCTG-CTGCTG-3′ (98 nt downstream from the env stop codon). The conditions for PCR were as follows: 94°C for 2 min followed by 10 cycles 94°C for 20 s, 63°C for 30 s, and 68°C for 4 min; 20 cycles of 94°C for 20 s, 63°C for 30 s, and 68°C for 4 min, plus 20 additional s for each incremental cycle; and finally 68°C for 7 min. PCR was performed using the Expand long template PCR system (Boehringer Mannheim), and the PCR product was cloned using a Topo TA cloning kit (Invitrogen). The env gene of the resulting plasmid (pCR-μ8-R) was subsequently sequenced.
RESULTS
Mutagenesis and expression of envelope glycoproteins.
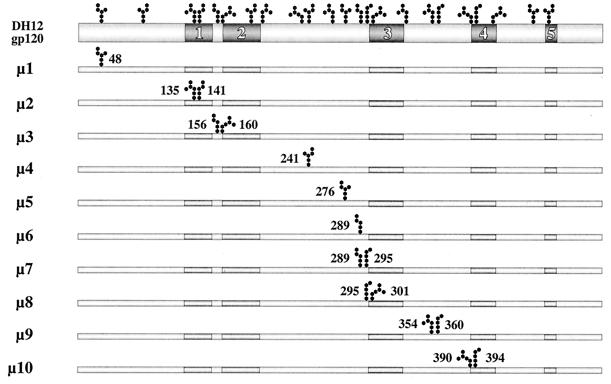
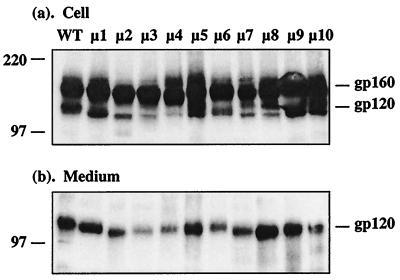
To examine whether carbohydrate moieties of HIV-1 gp120 played any role in its function or structural integrity, mutations (asparagine to glutamine) were introduced into the conserved as well as variable regions of HIV-1DH12 gp120, as depicted in Fig. 1. Either one (μ1, μ4, μ5, and μ6) or two (μ2, μ3, μ7, μ8, μ9, and μ10) potential N-linked glycosylation sites were changed throughout gp120. To determine whether these mutations had any effect on the expression and/or processing of gp160, recombinant vaccinia viruses encoding either wild-type or mutant envelope glycoproteins were used to infect HeLa cells. Cell lysates and culture supernatants were subjected to SDS-PAGE and Western immunoblotting with rabbit anti-gp160 antiserum as shown in Fig. 2a and b, respectively. Although the level of gp120 was somewhat lower for a few of the mutants (e.g., μ3 and μ4) compared to the wild-type protein, all of the mutant gp160s were expressed and processed to gp120. An increase in electrophoretic mobility was observed for each of the deglycosylated mutant glycoproteins compared to the wild type. This was more apparent for the mutants containing two mutations (μ2, μ3, μ7, μ8, μ9, and μ10). These results suggest that each of the asparagine residues we had mutated was indeed being utilized for N-linked glycosylation.
FIG. 1.
A schematic diagram of N-linked glycosylation site mutant constructs. The positions of 23 potential N-linked glycosylation sites and the variable regions of HIV-1DH12 gp120 are identified at the top. Ten initial mutant constructs (μ1 through μ10) are shown with the locations of the mutated glycosylation site(s). Mutants μ1 and μ4 through 6 have only a single site removed while the others have two sites removed.
FIG. 2.
Western immunoblot of envelope glycoproteins expressed by recombinant vaccinia viruses. Cell lysates (a) and culture medium (b) of HeLa cells infected with either wild-type or mutant envelope-expressing vaccinia viruses were subjected to SDS-PAGE followed by Western immunoblotting. The positions of molecular weight markers, gp160, and gp120 are indicated.
Coreceptor usage of N-linked glycosylation site mutants.
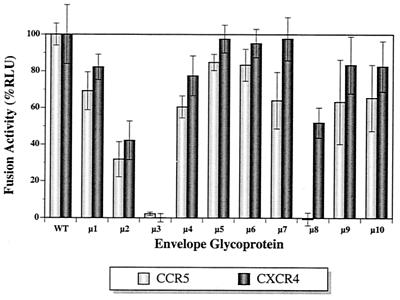
HIV-1DH12 is a dualtropic virus, utilizing both CCR5 and CXCR4 as coreceptors for virus entry. To examine whether deglycosylation had affected its ability to induce membrane fusion and/or coreceptor usage, we performed a highly sensitive cell-to-cell fusion assay using SEAP as an indicator (23). As shown in Fig. 3, the mutations did not appreciably disrupt coreceptor usage, except for μ2, μ3, and μ8. The fusion activity decreased to 30 to 40% of that of the wild-type protein for μ2, whereas the activity was completely abolished for μ3 with either CCR5 or CXCR4. This loss of fusion activity by μ3 was not necessarily due to reduced gp160-processing efficiency of this mutant since μ4, which exhibited a similar level of processing, induced only slightly less cell-to-cell fusion activity compared to the wild type. A different pattern of coreceptor fusion dysfunction was observed for μ8; while retaining approximately 50% of the fusion activity with CXCR4 compared to the wild type, it completely lost the activity with the CCR5 receptor. These results indicate that the carbohydrate moieties near the V1/V2 (μ2 and μ3) and V3 (μ8) loops of HIV-1 gp120 influence its capacity to induce membrane fusion, possibly by altering the structure of envelope domain(s) interacting with either CD4 and/or the coreceptors. A soluble CD4 (sCD4) binding assay indicated that the interaction of μ3 gp120 with CD4 was reduced approximately 50% compared to wild-type gp120. In contrast, μ2 and μ8 gp120s exhibited binding properties to sCD4 similar to those of the wild-type protein (data not shown).
FIG. 3.
Cell-to-cell fusion activity. Fusion activity of mutant envelope proteins with CCR5 or CXCR4 is shown as percentage of relative light units (RLU) in comparison to the wild-type protein.
Replication profile of viruses containing μ2 and μ8.
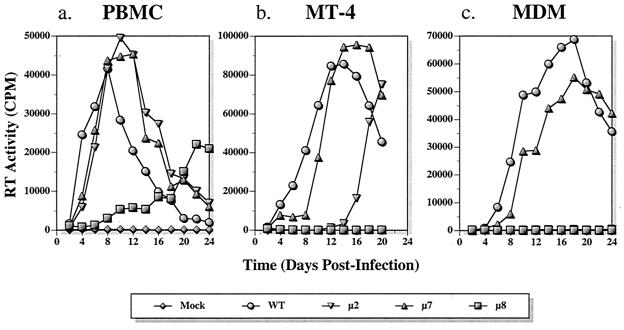
To characterize the effects of glycosylation site mutations μ2 and μ8 in the context of an infectious virus, the mutations were transferred to a full-length molecular clone, pAD8-DHenv (8). This chimeric virus, which contains the HIV-1DH12 env gene in the background of HIV-1AD8, was used because its capacity to produce progeny virus was superior to that of the parental HIV-1DH12 (8). The replication profiles of μ2 and μ8 viruses in human PBMC, MT-4 cells, and human primary MDM were compared to those of the wild-type virus and of another mutant virus (μ7), which did not exhibit defective cell-to-cell fusion (Fig. 4).
FIG. 4.
Replication of viruses carrying mutant envelopes. The replication of three mutant viruses (μ2, μ7, and μ8) was compared to that of the wild type in PBMC (a), MT-4 T cells (b), and MDM (c). Virion-associated RT activity in the culture medium of virus-infected cells was determined as described in Materials and Methods.
The replication kinetics of mutants μ2 and μ7 in PBMC were slightly delayed compared to that of wild-type virus (Fig. 4a). Thus, although μ2 exhibited only 30 and 40% of the fusion activity with CCR5 and CXCR4, respectively, the mutation had minimal effects on virus replication in PBMC. In contrast, the replication of μ8, which had retained 50% of the fusion activity with CXCR4 but completely lost its ability to utilize CCR5, was significantly delayed in PBMC. In some experiments, replication of μ8 was not observed (data not shown). In MT-4 cells, μ7 replicated with only a slight delay compared to the wild type, similar to what was observed in PBMC (Fig. 4b). In contrast, the replication of μ2 was markedly delayed (first detected 16 days postinfection). Surprisingly, no replication of μ8 was ever detected in MT-4 cells even though μ8 exhibited fusion activity similar to or greater than that of μ2 with CXCR4. Only wild-type and μ7 viruses were infectious in MDM, with μ7 lagging slightly behind the wild type (Fig. 4c). No replication of μ2 virus, which exhibited approximately 30% of the wild-type fusion activity with CCR5, was detected in MDM.
Fine mutagenesis of the V1/V2 and V3 regions of gp120.
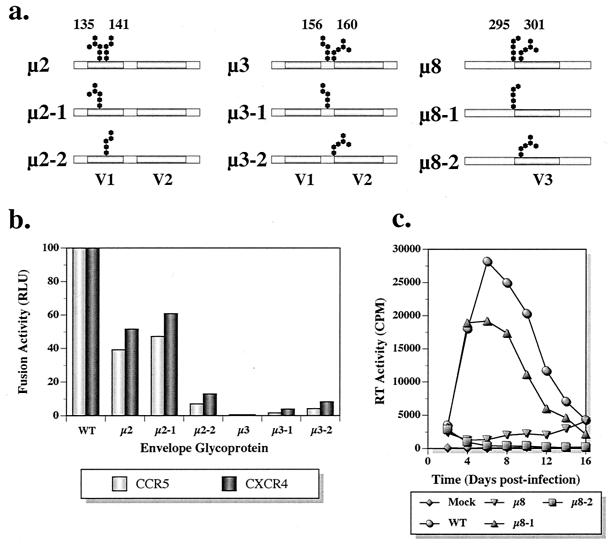
Membrane fusion and virus infectivity assays indicated that the sites mutated in the mutants μ2, μ3, and μ8 very likely play an important role(s) in envelope glycoprotein function. All three mutants carry mutations at two adjacent glycosylation sites. To ascertain the relative importance of each glycosylation site within the pair, six additional single-site mutants were constructed by individually mutating N135 (μ2-1), N141 (μ2-2), N156 (μ3-1), N160 (μ3-2), N295 (μ8-1), and N301 (μ8-2) (Fig. 5a). For the first four mutants (μ2-1, μ2-2, μ3-1, and μ3-2), recombinant vaccinia viruses were generated and the fusogenic properties of the mutant envelope glycoproteins were examined. As shown in Fig. 5b, μ2-1 exhibited fusion activity indistinguishable from that of μ2 with both CCR5 and CXCR4. In contrast, the defect was more severe for μ2-2, which retained only 10% of the wild-type fusion activity. Fusion activity was severely affected for both μ3-1 and μ3-2, suggesting that the carbohydrate moieties at both glycosylation sites are critical for this protein function.
FIG. 5.
Further analyses of mutants μ2, μ3, and μ8. The two altered glycosylation sites in mutants μ2, μ3, and μ8 were individually mutagenized to evaluate the relative importance of each site. (a) Mutant constructs with the corresponding glycosylation site that was mutated. (b) Cell-to-cell fusion activity levels of mutants μ2-1, μ2-2, μ3-1, and μ3-2 were compared with those of the wild type, μ2, and μ3. (c) Replication kinetics of the mutants μ8-1 and μ8-2 in PBMC are shown compared to those of the wild type and μ8.
For μ8-1 and μ8-2, the mutations were characterized in the context of virus replication in human PBMC. As shown previously, the replication of μ8 virus was markedly delayed by the mutation (Fig. 5c). However, virus bearing the μ8-1 mutation exhibited replication kinetics similar to that of its wild-type parent, although a somewhat lower progeny virus yield was observed. This result was somewhat anticipated since the μ7 virus, which also carries the mutation at N295, replicated quite efficiently (Fig. 4a) and demonstrated nearly wild-type coreceptor usage (Fig. 3). In contrast, the virus containing the μ8-2 mutation failed to replicate. These results strongly indicate that carbohydrate moiety on N301 is critical for several envelope glycoprotein functions and is primarily responsible for the replication defects observed in μ8.
Characterization of the μ8 revertant.
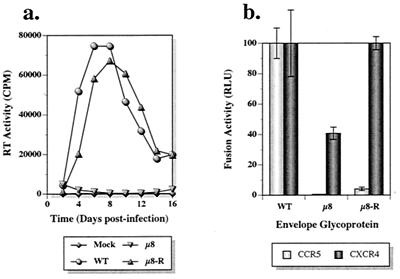
A characteristic property of HIV-1 mutants undergoing second-site revertant changes is the delayed appearance of progeny virus, which usually regains partial or even wild-type infection kinetics. In this study, μ2 virus harvested at the markedly delayed peak of its infection of MT-4 cells (day 20) (Fig. 4b) replicated with wild-type kinetics in subsequent infection of MT-4 cells (data not shown), indicating the very likely emergence of a μ2 revertant virus. Because of the unusual coreceptor usage phenotype of μ8 (viz., CCR5−/CXCR4+) (Fig. 3), we were interested in ascertaining whether second-site revertants might have arisen during the extended μ8 infection of PBMC (Fig. 4a). Progeny virus present in day 22 culture supernatants from this infection was used to reinfect fresh PBMC. As shown in Fig. 6a, the progeny virus (designated μ8-R) replicated almost as efficiently as the wild-type virus, indicating that a reversion had, in fact, occurred. The revertant virus, μ8-R, also exhibited infection kinetics similar to that of the wild-type virus in MT-4 cells (data not shown), suggesting that the reversion permitted efficient utilization of CXCR4 as a coreceptor. In contrast, μ8-R failed to replicate in MDM (data not shown).
FIG. 6.
Characterization of second-site revertant. Replication kinetics (a) and cell-to-cell fusion activity levels (b) of the revertant μ8-R and its parental μ8 virus are compared to those of the wild type. (a) Virus replication in PBMC. CPM, counts per minute; WT, wild type.
To elucidate the nature of reversion, the env gene of the revertant virus was PCR amplified, cloned, and sequenced. As shown in Table 2, μ8-R had acquired two mutations within the V3 loop (N300Y and G306R) located near the original mutations (N295Q and N301Q), which were still present. To confirm that the two amino acid substitutions observed were indeed responsible for the phenotypic change, both mutations were transferred into the pNVV-DHenv plasmid and recombinant vaccinia virus was generated. As shown in Fig. 6b, the second-site mutations completely restored CXCR4 coreceptor usage to wild-type levels, as measured in a fusion assay. However, the revertant envelope glycoprotein still failed to utilize CCR5, consistent with the inability of the revertant virus to replicate in MDM.
TABLE 2.
HIV-1DH12 gp120 V3 loop amino acid sequence
| Virus | V3 amino acid sequencea |
|---|---|
| 295 301 311 321 | |
| DH12 | NCTRPN NNTRKGITLG PGRVFYTTGE IVGDIRKAHC |
| DH12μ8 | Q----- Q--------- ---------- ---------- |
| DH12μ8-R | Q----Y Q----R---- ---------- ---------- |
Asparagines (N) that were mutated to glutamine (Q) are in bold type; −, identical amino acids.
DISCUSSION
Carbohydrate moieties on HIV-1 gp120 are known to play several important roles in the virus life cycle. Characterization of the biochemical properties of the envelope glycoproteins synthesized in the presence of various inhibitors of glycosylation or glycosidases have shown that proper attachment and trimming of glycans on gp120 are important for its folding and, therefore, for its biological function (13, 17, 19, 29, 38, 50). However, our study clearly demonstrates that gp120-associated N-linked glycosylation sites do not contribute equally during virus entry, a result consistent with previously reported site-directed mutagenesis studies for HIV-1 (25) or simian immunodeficiency virus (36). Among the glycosylation sites we have evaluated, only those that are near or within the V1/V2 (amino acids N135, N141, N156, N160) and V3 (amino acid N301) loops were critical for membrane fusion. These results are also consistent with numerous reports showing that the V1/V2 and the V3 loops are critical determinants of coreceptor usage and cellular tropism.
Although the amounts of total envelope glycoprotein expressed were quite comparable, the levels of membrane-associated gp120 were lower for some of the mutant envelopes (i.e., μ2, μ3, μ4, μ6, μ7, and μ8) compared to the wild-type protein (Fig. 2a). This could be due to decreased gp160 processing, increased shedding of gp120 from gp41, or both. Thus, the defect in fusion activity for some of the mutants could be due to the reduced levels of functional gp120-gp41 complexes on the membrane in addition to (or separate from) the inherent biochemical properties of the mutant protein. We performed our fusion assays based on the same amount of total envelope protein expressed (i.e., by using the same amount of vaccinia virus) rather than normalizing the level of membrane-associated gp120 for two reasons. First, because the altered level of membrane-associated gp120 in itself is a phenotype of the mutation, comparing different envelopes based on the same amount of total protein expressed, rather than the amount of membrane-associated gp120, would be more appropriate. Second, it is our observation that cell-to-cell membrane fusion is quite efficient and that very little envelope protein is required for this process. So, even the low amount of membrane-associated gp120 observed for some of the mutant envelopes should be more than sufficient to induce cell-to-cell fusion. For example, μ4, which has the lowest amount of membrane-associated gp120 (Fig. 2a), exhibited quite efficient fusion activity (Fig. 3). Thus, the contribution of the low level of membrane-associated gp120 on the reduced fusion activity exhibited by μ2, μ3, and μ8 is not likely to be significant.
We have identified five gp120-associated N-linked glycosylation sites that play important roles for dualtropic HIV-1DH12. The corresponding glycosylation sites in other virus isolates may or may not serve equally important functions. For example, mutation of N301 in DH12 gp120 (sixth residue of the V3 loop) completely eliminated the usage of CCR5 and severely impaired (reducing by 50%) CXCR4 usage. Similarly, mutation of the corresponding residue on NL4-3 (T-tropic) and SF13 (dualtropic) compromised CXCR4-dependent fusion activity (34). However, no reduction of CCR5 usage was observed when the same residue was mutated on SF13 and SF162 (M-tropic) (34) or ConB (M-tropic) (51). Additionally, the corresponding mutation had no effect on infectivity of other T-tropic stains (LAI, BRU, and HXB2 [2, 27, 43]). These apparent discrepancies are likely due to specific differences in amino acid sequences in the V3 loop for different isolates, which together with carbohydrate residues create a V3 loop structure that interacts with receptors. This notion is supported by the emergence of the second-site revertant of μ8 virus (μ8-R), which acquired compensating mutations in the V3 loop without glycosylation site replacement. This revertant regained full usage of CXCR4 and replicated efficiently in PBMC and MT-4 cells, although CCR5 usage remained severely impaired. Of the two amino acid changes observed in μ8-R, a glycine-to-arginine substitution at the 11th position of the V3 loop increased the net positive charge of the V3 loop, thought to be important for CXCR4 usage (37). Coincidentally, a similar change at the 11th position of the V3 loop has been reported to rescue the defect in CXCR4 usage by a mutation affecting the glycosylation site at the 6th position of the HIV-1NL4-3 V3 loop (34). These results indicate that depending on the amino acid residues of the neighboring region, the carbohydrate moieties may or may not play critical roles in gp120 function.
The properties exhibited by different glycosylation site mutants were not identical with respect to the extent or the type of defect. While mutating N135 (μ2-1) reduced fusion activity with both CXCR4 and CCR5 to 50% of the wild-type level, mutating N141, N156, or N160 (μ2-2, μ3-1, and μ3-2, respectively) almost completely disrupted the fusion activity with both coreceptors. Mutant μ2, which carries mutations both at N135 and N141, exhibited a phenotype similar to μ2-1. In contrast, μ8 (N295 and N301), and presumably μ8-2 (N301), exhibited preferential defect in CCR5 usage. Furthermore, unlike μ3, which exhibited reduced CD4 binding activity, mutants μ2 and μ8 interacted with CD4 as efficiently as the wild-type gp120. At present, we can only speculate about which step of the envelope glycoprotein-induced membrane fusion process is impaired. For example, the primary defect of mutant μ3, and presumably of μ3-1 and μ3-2, appears to be CD4 binding. On the other hand, μ2 seems to be impaired at one of the post-CD4 binding steps, since the mutant gp120 bound CD4 as efficiently as the wild-type protein. This defect could include the inability to undergo conformational change that occurs when gp120 binds CD4 (i.e., from closed to open conformation where the bridging sheet becomes exposed) or the inability of gp120 to bind the coreceptors. In the case of μ8, the interaction of gp120 with CCR5 is affected more than that with CXCR4. Future analyses of direct binding between different mutant gp120s and CCR5 or CXCR4 may provide additional information on the nature of the defects observed in this study.
At first glance, some of the cell-to-cell fusion and virus infectivity assay results may seem inconsistent. For example, the replication kinetics of the virus carrying the μ2 mutation was very similar to that of the wild-type virus in PBMC. In contrast, the replication of μ2 virus in MT-4 T cells was severely impaired and no evidence of its replication was detected in MDM. These results could simply reflect possible differences in the levels of CD4 and/or chemokine receptors expressed in different cell types as previously demonstrated by Ly and Stamatatos (28), who reported that mutation of glycosylation sites at the base of the V2 loop of gp120 affects viral replication kinetics in a cell-dependent manner; mutants replicated efficiently in cells expressing high levels of receptors but not in cells expressing lower levels.
Considering that recombinant vaccinia viruses express high levels of envelope glycoprotein, CD4, and coreceptors, it was somewhat surprising to observe a marked reduction in cell-to-cell fusion activity for μ2 despite its efficient replication in PBMC. A possible explanation is that both CCR5 and CXCR4 are present on primary CD4+ T cells while only a single coreceptor is expressed on MT-4 cells, MDM, and the M. dunni cells used for the fusion assay. The dynamics of the interactions between a dualtropic gp120 with CCR5, CXCR4, or both on the surface of primary CD4+ T lymphocytes remain largely unknown. Given the multiple receptor binding sites on the virion surface, the interactions between virus and cell are likely to be highly complex.
ACKNOWLEDGMENTS
We thank Bernard Moss, Ed Berger, Bob Doms, and Michael Oldstone for providing valuable reagents. Recombinant soluble CD4 was obtained from R. Sweet, SmithKline Beecham Pharmaceuticals, through the AIDS Research and Reference Reagent Program, NIAID, NIH.
REFERENCES
- 1.Ashkenazi A, Smith D H, Marsters S A, Riddle L, Gregory T J, Ho D D, Capon D J. Resistance of primary isolates of human immunodeficiency virus type 1 to soluble CD4 is independent of CD4-rgp120 binding affinity. Proc Natl Acad Sci USA. 1991;88:7056–7060. doi: 10.1073/pnas.88.16.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back N K T, Smit L, de Jong J-J, Keulen W, Schutten M, Goudsmit J, Tersmette M. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199:431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- 3.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broder C C, Dimitrov D S, Blumenthal R, Berger E A. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 5.Cann A J, Churcher M J, Boyd M, O'Brien W, Zhao J Q, Zack J, Chen I S. The region of the envelope gene of human immunodeficiency virus type 1 responsible for determination of cell tropism. J Virol. 1992;66:305–309. doi: 10.1128/jvi.66.1.305-309.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesebro B, Nishio J, Perryman S, Cann A, O'Brien W, Chen I S, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho M W, Shibata R, Martin M A. Infection of chimpanzee peripheral blood mononuclear cells by human immunodeficiency virus type 1 requires cooperative interaction between multiple variable regions of gp120. J Virol. 1996;70:7318–7321. doi: 10.1128/jvi.70.10.7318-7321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho M W, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- 10.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 12.Daar E S, Li X L, Moudgil T, Ho D D. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dedera D, Vander Heyden N, Ratner L. Attenuation of HIV-1 infectivity by an inhibitor of oligosaccharide processing. AIDS Res Hum Retrovir. 1990;6:785–794. doi: 10.1089/aid.1990.6.785. [DOI] [PubMed] [Google Scholar]
- 14.de Jong J-J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doranz B J, Berson J F, Rucker J, Doms R W. Chemokine receptors as fusion cofactors for human immunodeficiency virus type 1 (HIV-1) Immunol Res. 1997;16:15–28. doi: 10.1007/BF02786321. [DOI] [PubMed] [Google Scholar]
- 16.D'Souza M P, Harden V A. Chemokines and HIV-1 second receptors. Confluence of two fields generates optimism in AIDS research. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 17.Fennie C, Lasky L A. Model for intracellular folding of the human immunodeficiency virus type 1 gp120. J Virol. 1989;63:639–646. doi: 10.1128/jvi.63.2.639-646.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruters R A, Neefjes J J, Tersmette M, de Goede R E, Tulp A, Huisman H G, Miedema F, Ploegh H L. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature. 1987;330:74–77. doi: 10.1038/330074a0. [DOI] [PubMed] [Google Scholar]
- 20.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 22.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M K, Heaton J, Cho M W. Identification of determinants of interaction between CXCR4 and gp120 of a dual-tropic HIV-1DH12 isolate. Virology. 1999;257:290–296. doi: 10.1006/viro.1999.9686. [DOI] [PubMed] [Google Scholar]
- 24.Lee M K, Martin M A, Cho M W. Higher western blot immunoreactivity of glycoprotein 120 from R5 HIV type 1 isolates compared with X4 and X4R5 isolates. AIDS Res Hum Retrovir. 2000;16:765–775. doi: 10.1089/088922200308765. [DOI] [PubMed] [Google Scholar]
- 25.Lee W R, Syu W J, Du B, Matsuda M, Tan S, Wolf A, Essex M, Lee T H. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:2213–2217. doi: 10.1073/pnas.89.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Littman D R. Chemokine receptors: keys to AIDS pathogenesis? Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 27.Losman B, Biller M, Olofsson S, Schonning K, Lund O S, Svennerholm B, Hansen J E, Bolmstedt A. The N-linked glycan of the V3 region of HIV-1 gp120 and CXCR4-dependent multiplication of a human immunodeficiency virus type 1 lymphocyte-tropic variant. FEBS Lett. 1999;454:47–52. doi: 10.1016/s0014-5793(99)00740-1. [DOI] [PubMed] [Google Scholar]
- 28.Ly A, Stamatatos L. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J Virol. 2000;74:6769–6776. doi: 10.1128/jvi.74.15.6769-6776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montefiori D C, Robinson W E, Jr, Mitchell W M. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1988;85:9248–9252. doi: 10.1073/pnas.85.23.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore J P, Burkly L C, Connor R I, Cao Y, Tizard R, Ho D D, Fisher R A. Adaptation of two primary human immunodeficiency virus type 1 isolates to growth in transformed T cell lines correlates with alterations in the responses of their envelope glycoproteins to soluble CD4. AIDS Res Hum Retrovir. 1993;9:529–539. doi: 10.1089/aid.1993.9.529. [DOI] [PubMed] [Google Scholar]
- 31.Moore J P, McKeating J A, Huang Y X, Ashkenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 33.Myers G, Lenroot R. HIV glycosylation: what does it portend? AIDS Res Hum Retrovir. 1992;8:1459–1460. doi: 10.1089/aid.1992.8.1459. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama E E, Shioda T, Tatsumi M, Xin X, Yu D, Ohgimoto S, Kato A, Sakai Y, Ohnishi Y, Nagai Y. Importance of the N-glycan in the V3 loop of HIV-1 envelope protein for CXCR-4- but not CCR-5-dependent fusion. FEBS Lett. 1998;426:367–372. doi: 10.1016/s0014-5793(98)00375-5. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien W A, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 36.Ohgimoto S, Shioda T, Mori K, Nakayama E E, Hu H, Nagai Y. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J Virol. 1998;72:8365–8370. doi: 10.1128/jvi.72.10.8365-8370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada T, Patterson B K, Otto P A, Gurney M E. HIV type 1 infection of CD4+ T cells depends critically on basic amino acid residues in the V3 domain of envelope glycoprotein 120. AIDS Res Hum Retrovir. 1994;10:803–811. doi: 10.1089/aid.1994.10.803. [DOI] [PubMed] [Google Scholar]
- 38.Pal R, Hoke G M, Sarngadharan M G. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:3384–3388. doi: 10.1073/pnas.86.9.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parren P W, Moore J P, Burton D R, Sattentau Q J. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13:S137–S162. [PubMed] [Google Scholar]
- 40.Pleskoff O, Sol N, Labrosse B, Alizon M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR-4 (fusin) J Virol. 1997;71:3259–3262. doi: 10.1128/jvi.71.4.3259-3262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 42.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schonning K, Jansson B, Olofsson S, Hansen J E. Rapid selection for an N-linked oligosaccharide by monoclonal antibodies directed against the V3 loop of human immunodeficiency virus type 1. J Gen Virol. 1996;77:753–758. doi: 10.1099/0022-1317-77-4-753. [DOI] [PubMed] [Google Scholar]
- 44.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth R J, Yi Y, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 50.Walker B D, Kowalski M, Goh W C, Kozarsky K, Krieger M, Rosen C, Rohrschneider L, Haseltine W A, Sodroski J. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc Natl Acad Sci USA. 1987;84:8120–8124. doi: 10.1073/pnas.84.22.8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W K, Dudek T, Zhao Y J, Brumblay H G, Essex M, Lee T H. CCR5 coreceptor utilization involves a highly conserved arginine residue of HIV type 1 gp120. Proc Natl Acad Sci USA. 1998;95:5740–5745. doi: 10.1073/pnas.95.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willey R L, Klimkait T, Frucht D M, Bonifacino J S, Martin M A. Mutations within the human immunodeficiency virus type 1 gp160 envelope glycoprotein alter its intracellular transport and processing. Virology. 1991;184:319–329. doi: 10.1016/0042-6822(91)90848-6. [DOI] [PubMed] [Google Scholar]
- 54.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wrin T, Loh T P, Vennari J C, Schuitemaker H, Nunberg J H. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]