Abstract
The oxygen-sensing pathway is a crucial regulatory circuit that defines cellular conditions and is extensively exploited in cancer development. Pathogenic mutations in the von Hippel–Lindau (VHL) tumour suppressor impair its role as a master regulator of hypoxia-inducible factors (HIFs), leading to constitutive HIF activation and uncontrolled angiogenesis, increasing the risk of developing clear cell renal cell carcinoma (ccRCC). HIF hyperactivation can sequester HIF-1β, preventing the aryl hydrocarbon receptor (AHR) from correctly activating gene expression in response to endogenous and exogenous ligands such as TCDD (dioxins). In this study, we used protein–protein interaction networks and gene expression profiling to characterize the impact of VHL loss on AHR activity. Our findings reveal specific expression patterns of AHR interactors following exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and in ccRCC. We identified several AHR interactors significantly associated with poor survival rates in ccRCC patients. Notably, the upregulation of the androgen receptor (AR) and retinoblastoma-associated protein (RB1) by TCDD, coupled with their respective downregulation in ccRCC and association with poor survival rates, suggests novel therapeutic targets. The strategic activation of the AHR via selective AHR modulators (SAhRMs) could stimulate its anticancer activity, specifically targeting RB1 and AR to reduce cell cycle progression and metastasis formation in ccRCC. Our study provides comprehensive insights into the complex interplay between the AHR and HIF pathways in ccRCC pathogenesis, offering novel strategies for targeted therapeutic interventions.
Keywords: aryl hydrocarbon receptor (AHR), HIF-1A, VHL, dioxins, ccRCC
1. Introduction
The family of proteins known as basic helix–loop–helix (bHLH) PER-ARNT-SIM (PAS) transcription factors plays crucial roles in physiological adaptations to environmental signals and cancer pathogenesis [1]. Despite having distinct sets of target genes, these proteins form heterodimers by utilizing common dimerization partners within specific subfamilies, leading to intricate interactions [2]. One such protein, hypoxia-inducible factor (HIF), responds to the hypoxic microenvironment by promoting the transcription of its target genes after heterodimerization in the nucleus with the aryl hydrocarbon receptor nuclear translocator (ARNT) [3]. The dysregulation of HIF is linked to key transcriptional programmes in tumorigenesis and directly impacts patient prognosis, particularly in clear cell renal cell carcinoma (ccRCC), the most common form of kidney cancer [3,4]. In ccRCC, the loss of the von Hippel–Lindau tumour suppressor (pVHL) disrupts physiological HIF degradation, yielding constitutive HIF activation [5]. The so-called von Hippel–Lindau (VHL) disease is an inherited condition associated with increased susceptibility to various benign and malignant tumours, including retinal and cerebellar hemangioblastomas, pheochromocytomas, paragangliomas, non-functioning pancreatic neuroendocrine tumours (pNETs), and renal cell carcinoma (RCC) [6,7]. The pVHL acts as a substrate recognition component within a protein complex (VCB), consisting of elongin-B (ELOB), elongin-C (ELOC), and cullin-2 (CUL2) [8,9]. This complex exhibits E3 ubiquitin ligase activity, targeting the HIF-1α transcription factor for ubiquitination and proteasome degradation [10]. Another important protein from the bHLH-PAS family is the aryl hydrocarbon receptor (AHR), encoded by the homonymous gene localizing on chromosome 7, and acting as a 96 kDa ligand-dependent transcription factor [11]. The AHR possesses a bHLH-PAS architecture at the N-terminus and a transactivation domain at the C-terminus, allowing it to dynamically interact with multiple co-activators [11]. When inactive, the AHR forms a complex with stabilizing proteins in the cytosol, including heat shock proteins (HSP90), p23, and XAP2 [12]. Upon ligand binding, the AHR undergoes a conformational change, enabling it to translocate to the nucleus and form a heterodimer with the aryl hydrocarbon receptor nuclear translocator (ARNT). The resulting complex is known to interact with DNA via xenobiotic-responsive elements (XREs) and dioxin-responsive elements (DREs) in gene promoters [13,14]. The AHR responds to diverse ligands, including environmental chemicals, dietary components, and endogenous metabolites. Notably, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is a prototype ligand with a planar aromatic structure [15,16]. This compound typically forms as an undesired by-product in the combustion processes of organic materials as well as a secondary product in organic synthesis [17]. In the nucleus, the AHR/ARNT complex transcribes genes involved in detoxification, inflammation, immune response, and development, such as Cyp1a, Cyp1b, GSTA1, EPHX1, and PAI2 [14]. The complex’s transcriptional activity also involves other specific proteins that act as transcriptional modulators, such as p300, CREBBP, NCOA1/2/3, and NRIP1 [18]. The AHR plays diverse roles in cancer, exhibiting both tumour-promoting and tumour-suppressing activities [17]. AHR modulation affects cancer cell behaviour in a cell-specific manner; for example, in breast cancer cells, AHR inhibition enhances proliferation in ER-positive cells but has no effect in ER-negative cells [19]. AHR deletion in various cancer types influences cell proliferation, invasion, and differentiation [20]. Its interaction with signalling pathways like TGF-β, PI3K/AKT/mTOR, NF-κB, FAK/c-Src, and Wnt5a/b-β-catenin further complicates its role [20]. Ligand-activated AHR can inhibit or induce specific signalling pathways, impacting cancer cell functions [19,20,21,22,23,24,25,26]. Moreover, in the last 10 years, the role of the AHR as a therapeutic target in cancers has emerged in particular through the involvement of agonists and inhibitors in breast cancer, hepatocellular carcinoma, and melanoma [20,27,28,29,30,31,32,33]. These complex interactions indicate that the AHR’s function is context-dependent, making it a potential target for cancer-specific therapies, although more cancer-specific studies are needed for a comprehensive understanding. The identified shared transcriptional binding partner, ARNT (HIF-1β), between the AHR and HIF-1α suggests a dynamic interplay and modulation of these canonical pathways. This implies that the preferential activation of one pathway over the other may result in the impairment of specific cellular behaviour and homeostasis. VHL disease recapitulates the persistent competition between HIF1A and the AHR for heterodimerization with ARNT (HIF-1β) and subsequent DNA binding events, which influence the transcription of target genes [34]. By employing a protein–protein interaction network-based approach, we here investigated the impact of pVHL loss on the regulation of AHR activities as well as their intricate pathways interplay [35,36,37]. We identified AHR-specific interactors and used them to predict a set of biological responses that may be compromised by the constitutive activation of HIF-1α [34]. By integrating our analysis with expression data from exposures to TCDD and expression data from pVHL-defective clear cell renal cell carcinoma (ccRCC), we provide insights into the potential repercussions of an impaired AHR pathway in ccRCC and its subsequent impact on tumour progression. Furthermore, the accompanying Kaplan–Meier analysis sheds light on the association between numerous AHR interactors and unfavourable survival rates in ccRCC, collectively supporting the intricate and miscellaneous role of the AHR in the tumour microenvironment. Our data propose the AHR as a potential target for the simultaneous agonistic activation of its regulative function, such as modulating androgen receptor (AR) transcriptional activity and protecting the retinoblastoma-associated protein (RB1) against phosphorylation to reduce cell cycle progression [19,38,39,40,41,42]. These three proteins indeed present low expression levels and are linked to low survival prognoses in ccRCC. In this specific scenario, AHR-selective activation could result in an interesting therapeutic target for treatment against pVHL-null ccRCC.
2. Materials and Methods
2.1. Network Generation
The protein–protein interaction network data were retrieved from five of the most reliable databases of protein interaction, these being BIOGRID, HIPPIE, STRING, IntAct, and KEGG [43,44,45,46]. In particular, these databases were selected as they store experimentally validated data. To highlight differences in protein network composition upon exposition to TCDD only, AHR direct interactors were included, while interactions between the AHR interactors were filtered and not considered in the final network. We employed Cytoscsape 3.9.1 for network data handling, and the interaction data from databases were merged into a single network [47,48]. Each network was filtered to include only experimentally validated interactions. In the case of the KEGG network, we exploited the KEGGREST package (version 1.42.0 and Bioconductor 3.17) in R (version 4.3) to perform an integrative merge of selected KEGG pathways where the ARNT is involved, specifically the HIF1A signalling pathway (hsa04066), Cushing syndrome (hsa04934), pathways in cancer (hsa05200), chemical carcinogenesis—receptor activation (hsa05207), chemical carcinogenesis—reactive oxygen species (hsa05208), renal cell carcinoma (hsa05211), and Th17 cell differentiation (hsa04659) [49,50]. In detail, databases were searched imposing the following filters:
STRING: interactions with an “experimental score” > 0 and no text mining;
BIOGRID: interactions derived from experiments with a “Biogrid_score” > 0;
KEGG: manually curated interactions from pathways including the ARNT;
HIPPIE: this database uses a specific scoring system; we selected interactions with a score > 0.5;
IntAct: interactors defined by the terms “association” and “physical association”.
Functional annotations of AHR interactors were obtained from the manual curation of papers describing the interaction as reported in the different databases. All databases were accessed in August 2023.
2.2. NCBI-GEO Expression Profiles
The data for protein levels were retrieved from Gene Expression Omnibus (GEO), a free public database of microarray/gene profiles [51]. In this study, we employed expression profiles that describe the dioxin effect on HepaRG, MCF7, Ishikawa cells, and HepG2. The cells were subjected to 100 nM of TCDD for six hours (GSE69844, GSE69845, GSE69849, and GSE69850) [52]. The second set of expression profiles refers to datasets describing the ccRCC condition (GSE36895, GSE102101, GSE107848, and GSE186013); here, the first two datasets are from patient samples, whereas the last two datasets are from 786-O cell lines. GSE36895: the RNA of clear cell renal cell carcinoma (ccRCC) primary tumours, tumours growing in immunodeficient mice (tumorgrafts), and normal kidney cortices were labelled and hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays [53,54]. GSE102101: RNA-seq profiles of 10 patient-matched normal kidney and ccRCC pairs [55]. GSE107848: the transcriptomic profiles of 786-O under normoxia, short-term hypoxia, and long-term hypoxia were analyzed using next-generation sequencing [56]. GSE186013: transcriptomic profiles of 786-O-TR-Ctrl and 786-O-TR-VHL [57].
2.3. DEG Definition
The expression profiles were analyzed using the GEO2R online tool to compare two or more groups of samples to identify genes that are differentially expressed across experimental conditions, and default constraints were applied [51]. Genes with logFC > 0 have been defined as upregulated, and those with logFC < 0 as downregulated [58]. Moreover, to overcome the differences within the datasets, since different cell lines were treated with TCDD and the ccRCC datasets were from both patients and cell lines, proteins were defined as up- or downregulated if they had the same expression levels in at least three of the five datasets; if this was not the case, they were declared as undefined expressed and not considered.
2.4. Survival Analysis of DEGs in Renal Cell Carcinoma
Kaplan–Meier plots are commonly used for assessing the effect of a great number of genes on survival based on the EGA, TCGA database, and GEO (Affymetrix microarrays only) [59,60]. The log-rank p-values and hazard ratios (HRs) with 95% confidence intervals were computed for ccRCC and shown on the plots for each protein. The expression levels that define the effect on the patient’s survival rate were compared with the levels expressed in the GEO datasets for ccRCC. p-value evaluation and correction were performed by applying the Benjamani–Hochberg FDR method [61].
3. Results
3.1. Description of PPIN Features
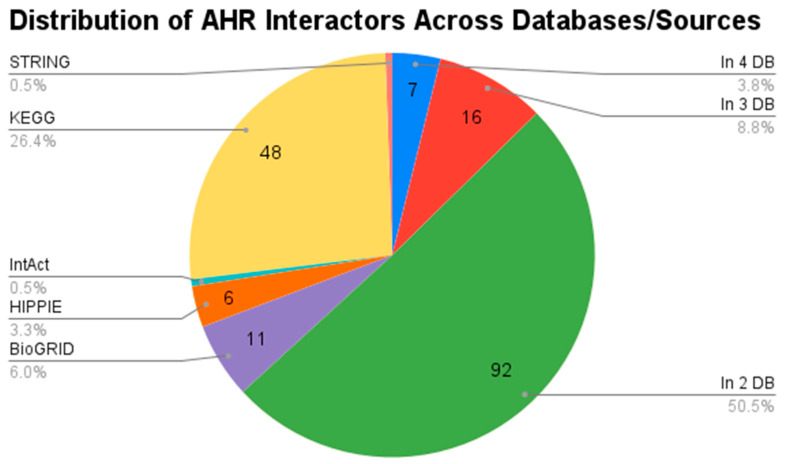
All interactions considered for constructing the AHR interactor networks underwent rigorous experimental validation and curation. The merged network, combining data from various databases, encompasses 182 nodes connected by 327 edges. This protein–protein interaction (PPI) network is centred around AHR, offering the most comprehensive view of its interactors. Figure 1 illustrates the sources of interactions in the merged network, with the majority stemming from at least two databases.
Figure 1.
Pie chart showing the distribution of AHR interactors across databases.
Notably, the database with the highest number of unique interactors was KEGG. This underscores the robustness and reliability of our AHR-centred PPI network, providing a thorough understanding of its interacting partners. The specific dimensions of each network are detailed in Table 1.
Table 1.
Derivation of nodes and edges for each network.
| BIOGRID | HIPPIE | STRING | IntAct | KEGG | Merged | |
|---|---|---|---|---|---|---|
| Nodes | 126 | 122 | 20 | 14 | 50 | 182 |
| Edges | 150 | 122 | 52 | 17 | 49 | 327 |
3.2. DEG Network Representation
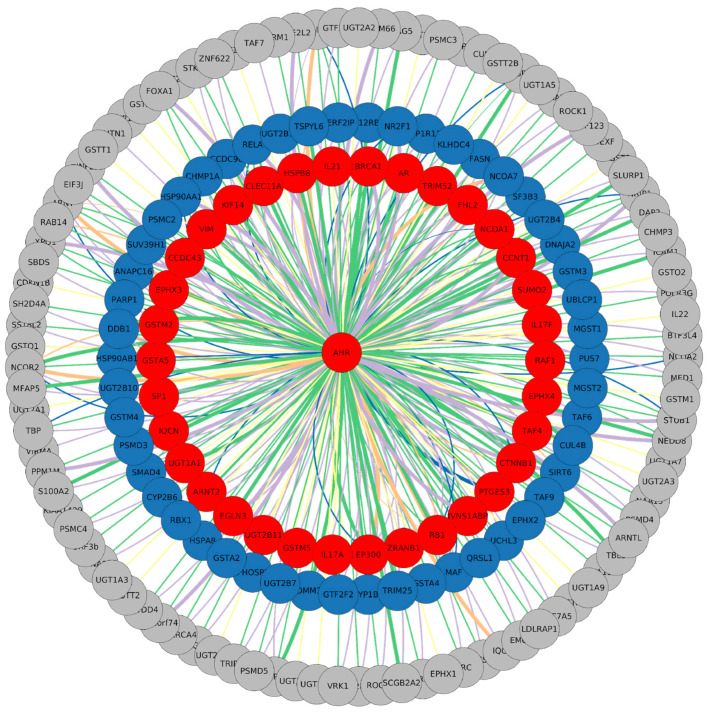
The network serves as a crucial tool for visualizing and interpreting the expression levels obtained from the GEO datasets. TCDD-related expression specifically identifies interactors that are differentially expressed following the activation of the canonical AHR pathway. The datasets under consideration involve four distinct cell lines, including HepaRG, MCF7, Ishikawa cells, and HepG2, all subjected to treatment with 100 nM of TCDD for 6 h (GSE69844, GSE69845, GSE69849, and GSE69850). The visualization of these data is shown in Figure 2, where differentially expressed genes (DEGs) are represented in various colours: 35 are upregulated, 50 are downregulated, and the majority exhibit undefined expression across the analyzed datasets.
Figure 2.
Merged AHR protein–protein interaction network. The colour code represents interactors following 6 h exposure to 100 nM of TCDD. Red is for upregulated proteins and blue is for downregulated, while nodes with an undefined expression level are represented in grey. Edges are coloured according to their node derivation: green is for interactions found in BIOGRID, violet represents data from HIPPIE, and orange for those from IntAct, while yellow and blue are for KEGG and STRING, respectively. The thickness of the lines represents the confidence score of the interactions as defined by each database, while the colours denote the different sources of the interactions, with each database assigned a distinct colour.
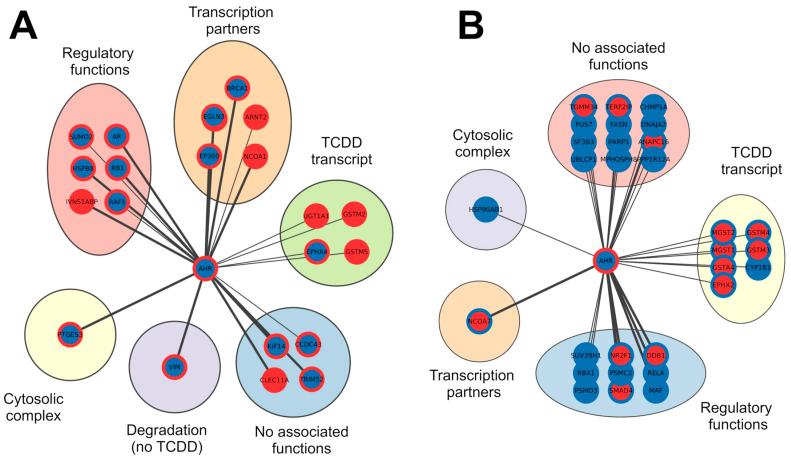
Through this representation of expression data, we can discern which AHR interactors are the most commonly associated with the activation of its canonical pathway. By mining data from different cell lines, we identified a minimal set of interactors reliably influenced by dioxin. Notably, well-known AHR target genes such as CYP1A and CYP1B are downregulated, suggesting that cell type can influence target gene expression. Conversely, other target genes implicated in xenobiotic metabolic processes, such as glutathione S-transferase A5 (GSTA5), glutathione S-transferase Mu 2 (GSTM2), and UDP-glucuronosyltransferase 2B11 (UGT2B11) [12], were found to be upregulated. This nuanced exploration sheds light on the complex relationship between the AHR and its interactors in response to TCDD across diverse cell lines. We categorized interactors into two main groups, namely upregulated and downregulated, and compared their expression levels during canonical pathway activation with their expression data derived from ccRCC, a well-known scenario characterized by the absence of functional pVHL. The previously selected upregulated and downregulated proteins were integrated with a second set of expression levels referring to the ccRCC condition. Similar to the first dataset, we applied the same colouring code and procedures defining differentially expressed genes (DEGs), resulting in proteins clustered as upregulated, downregulated, or having undefined expression. We found that within the established AHR interactor DEG pool, 35 and 30 proteins were upregulated and downregulated in ccRCC, respectively. Table 2 summarizes the DEGs that were considered for further analysis, presenting the levels of expression in the two conditions. To gain insights into the biological function of these interactions, we annotated each interactor with information derived from the literature. These interactors were grouped into six clusters, namely “Cytosolic complex”, “Regulatory functions”, “Transcription partners”, “TCDD transcript”, “Degradation (no TCDD)”, and “No function”, based on their specific interaction nature/function with the AHR (Figure 3). Moreover, in a broader context, we propose that proteins with high expression levels in both scenarios may not be induced solely by the canonical AHR pathway. Higher expression upon dioxin exposure paired with lower expression in ccRCC is likely dependent on AHR activation, while lower expression in the first dataset and higher expression in ccRCC could indicate that these genes are specifically promoted in the kidney tumour context. Finally, we were unable to discriminate low expression levels in both scenarios as they may be related either to the tumoral environment or attributed to the cell lines selected in TCDD datasets (Table 2). Nevertheless, our interactor categorization should provide a comprehensive understanding of the intricate dynamics of the AHR protein binding network in different scenarios, shedding light on potential biological functions and implications in the context of ccRCC.
Table 2.
List of proteins included in the PPIN, with expression levels found upon TCDD exposition and in ccRCC cells.
| Node Name | Expression Levels in at Least Three out of the Five Datasets | Kaplan–Meier Analysis | Expression Level Validation with the ccRCC GEO Datasets |
||
|---|---|---|---|---|---|
| Protein ID | TCDD | ccRCC | Overall Survival (p-Value FDR) |
LOW Surv. Expression Level |
|
| AHR | HIGH | LOW | LOW | X | |
| ANAPC16 | LOW | HIGH | LOW | ||
| AR | HIGH | LOW | LOW | X | |
| ARNT2 | HIGH | HIGH | LOW | ||
| BRCA1 | HIGH | LOW | LOW | ||
| CCDC43 | HIGH | LOW | LOW | X | |
| CHMP1A | LOW | LOW | HIGH | ||
| CLEC11A | HIGH | HIGH | HIGH | X | |
| CYP1B1 | LOW | LOW | HIGH | ||
| DDB1 | LOW | HIGH | LOW | ||
| DNAJA2 | LOW | LOW | LOW | X | |
| EGLN3 | HIGH | LOW | LOW | X | |
| EP300 | HIGH | LOW | LOW | X | |
| EPHX2 | LOW | HIGH | LOW | ||
| EPHX4 | HIGH | LOW | HIGH | ||
| FASN | LOW | LOW | HIGH | ||
| GSTA4 | LOW | HIGH | LOW | ||
| GSTM2 | HIGH | HIGH | HIGH | X | |
| GSTM3 | LOW | HIGH | LOW | ||
| GSTM4 | LOW | HIGH | LOW | ||
| GSTM5 | HIGH | HIGH | LOW | ||
| HSP90AB1 | LOW | LOW | LOW | X | |
| HSPB8 | HIGH | LOW | LOW | ||
| IVNS1ABP | HIGH | HIGH | LOW | ||
| KIF14 | HIGH | LOW | HIGH | ||
| MAF | LOW | LOW | LOW | X | |
| MGST1 | LOW | HIGH | HIGH | X | |
| MGST2 | LOW | HIGH | LOW | ||
| MPHOSPH8 | LOW | LOW | LOW | X | |
| NCOA1 | HIGH | HIGH | LOW | ||
| NCOA7 | LOW | HIGH | LOW | ||
| NR2F1 | LOW | HIGH | LOW | ||
| PARP1 | LOW | LOW | HIGH | ||
| PPP1R12A | LOW | LOW | LOW | X | |
| PSMC2 | LOW | LOW | HIGH | ||
| PSMD3 | LOW | LOW | HIGH | ||
| PTGES3 | HIGH | LOW | LOW | X | |
| PUS7 | LOW | LOW | HIGH | ||
| RAF1 | HIGH | LOW | HIGH | ||
| RB1 | HIGH | LOW | LOW | X | |
| RBX1 | LOW | LOW | HIGH | ||
| RELA | LOW | LOW | HIGH | ||
| SF3B3 | LOW | LOW | LOW | X | |
| SMAD4 | LOW | HIGH | LOW | ||
| SUMO2 | HIGH | LOW | HIGH | ||
| SUV39H1 | LOW | LOW | LOW | X | |
| TERF2IP | LOW | HIGH | HIGH | X | |
| TOMM34 | LOW | HIGH | HIGH | X | |
| TRIM52 | HIGH | LOW | HIGH | ||
| UBLCP1 | LOW | LOW | LOW | X | |
| UGT1A1 | HIGH | HIGH | HIGH | ||
| VIM | HIGH | LOW | HIGH | ||
Figure 3.
Networks of interactors clustered by functional relationship with the AHR. Panel (A) groups proteins upregulated after exposition to TCDD, while downregulated nodes after exposition to TCDD are reported in panel (B). Red borders mark upregulated nodes, while blue is for those that are downregulated. Fulfilled red or blue nodes are used to highlight up- or downregulated nodes in both TCDD and ccRCC samples. The thickness of the lines represents the number of sources reporting the interaction.
3.3. Analysis of the DEGs by a Kaplan–Meier Plotter
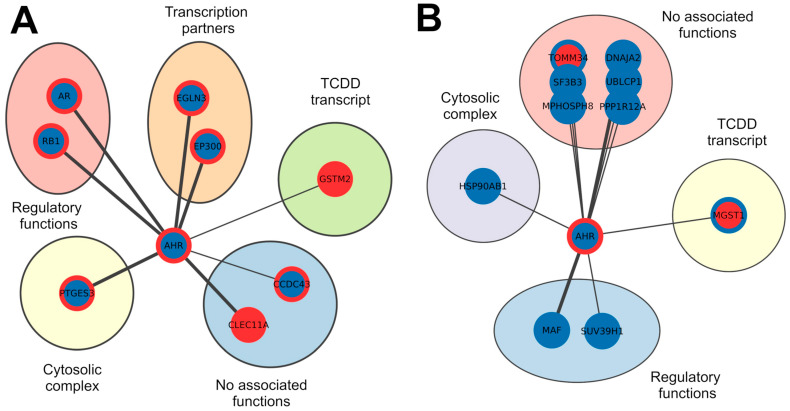
To gain a deeper understanding of the involvement of these proteins in the context of ccRCC, we employed a Kaplan–Meier plotter to assess the correlation between the expression of 52 proteins (22 upregulated and 30 downregulated in TCDD) (Table 2 and Figure 3A,B) and the survival rate of patients with this tumour type. We found that 16 upregulated and 23 downregulated proteins are significantly associated with worse survival rates, as indicated by their expression levels (p-value FDR < 0.05). To identify in our dataset which interactors are responsible for the correlation, we compared the expression levels of 39 proteins—correlated with poorer survival—with the expression levels found in the ccRCC GEO datasets. There were nine upregulated and ten downregulated proteins, whose expression levels consistently align with a worse survival rate (Table 3 and Figure 4), as determined by the Kaplan–Meier analysis.
Table 3.
Summary tables of the nodes that have significantly worse survival in ccRCC.
| Upregulated Nodes (TCDD 100 nM for 6 h) | Downregulated Nodes (TCDD 100 nM for 6 h) | ||||||
|---|---|---|---|---|---|---|---|
| Gene ID | Expression Levels in ccRCC | Overall Survival (p-Value FDR) | Expression Levels Correlated to Worse Survival Rate |
Gene ID | Expression Levels in ccRCC | Overall Survival (p-Value FDR) | Expression Levels Correlated to Worse Survival Rate |
| CCDC43 | LOW | LOW | DNAJA2 | LOW | LOW | ||
| AR | LOW | LOW | HSP90AB1 | LOW | LOW | ||
| EP300 | LOW | LOW | MAF | LOW | LOW | ||
| RB1 | LOW | LOW | MGST1 | HIGH | HIGH | ||
| EGLN3 | LOW | LOW | MPHOSPH8 | LOW | LOW | ||
| GSTM2 | HIGH | HIGH | PPP1R12A | LOW | LOW | ||
| AHR | LOW | LOW | SF3B3 | LOW | LOW | ||
| CLEC11A | HIGH | HIGH | SUV39H1 | LOW | LOW | ||
| PTGES3 | LOW | LOW | TOMM34 | HIGH | HIGH | ||
| UBLCP1 | LOW | LOW | |||||
Figure 4.
Networks of AHR interactors that have significantly worse survival from the Kaplan–Meier analysis. Panel (A) groups proteins upregulated after exposition to TCDD, while downregulated nodes after exposition to TCDD are reported in panel (B). Red borders mark upregulated nodes, while blue is for those that were downregulated. Fulfilled red or blue nodes are used to highlight up- or downregulated nodes in both TCDD and ccRCC samples. The thickness of the lines represents the number of sources reporting the interaction.
4. Discussion
The recognition of the ARNT as the transcriptional binding partner shared between the AHR and HIF1A highlights the intricate interplay and regulation within these pathways involved in environmental signal responses. The modulation of the AHR has diverse effects on cancer cell behaviour in a cell-specific manner, influencing cell proliferation, invasion, and differentiation [26]. The deletion of the AHR in different cancer types has been associated with alterations in cell proliferation, invasion, and differentiation [20]. The intricate interplay between the AHR and signalling pathways, such as TGF-β, PI3K/AKT/mTOR, NF-κB, FAK/c-Src, and Wnt5a/b-β-catenin, further complicates its role in cancer development [20]. The ligand-activated AHR is reported to either inhibit or induce specific signalling pathways, thereby influencing cancer cell functions [19,20,21,22,23,24,25,26], making it a potential target for cancer-specific therapies as it was evaluated in breast cancer, hepatocellular carcinoma, and melanoma; however, a comprehensive understanding of these contrasting functions is still to be explained. In this work, we investigated the canonical response to TCDD mediated by the aryl hydrocarbon receptor (AHR) and the expression of its interactors, comparing exposure to the receptor’s most potent ligand with the ccRCC pathology condition that precludes the main interaction with the ARNT/HIF1B. Our hypothesis is that sustained competition between the AHR and HIF1A for binding to the ARNT/HIF1B during prolonged exposure to TCDD may lead to significant deregulation of key cellular pathways. Using Cytoscape, we constructed a network centred on the AHR to visualize GEO datasets and identify differentially expressed interactors in the two conditions. Our analysis yielded 36 upregulated and 50 downregulated interactors after TCDD exposure, with 96 interactors exhibiting undefined expression. Further examination of the upregulated and downregulated sets revealed distinct expression patterns in the tumour context. We identified four clusters based on expression levels in the two conditions, classifying interactors into groups such as “Cytosolic complex”, “Regulatory functions”, “Transcription partners”, “TCDD transcript”, “Degradation (no TCDD)”, and “No function”. These data were then correlated with patient survival analysis on ccRCC using a Kaplan–Meier plotter. From the 50 investigated proteins, 9 and 10 proteins from the upregulated and downregulated sets, respectively, were associated with worse survival rates in ccRCC patients. Additionally, two proteins—the AR and RB1—were upregulated by TCDD exposure and downregulated in ccRCC, worsening patient survival rates [62]. The ambiguous role of hormone receptors, particularly the AR, has been extensively studied, revealing its involvement in metastatic migration/invasion processes and its differential regulation of VEGF-A vs. VEGF-C under different oxygen conditions in ccRCC cells [12,20,63,64,65,66,67]. RB1, a tumour suppressor, plays a crucial role in regulating the G1/S transition of the cell cycle [67]. In ccRCC, RB1 often undergoes copy number alterations, impacting cell cycle progression [42,68]. TCDD-induced G1 cell cycle arrest involves a reduction in phosphorylated RB1, facilitated by the direct interaction between the AHR and RB1, protecting RB1 from CDK2/4-mediated phosphorylation [19,69,70]. Our findings suggest that targeting the AHR could hold therapeutic potential in ccRCC; the activation of this receptor, despite the constitutive HIF1A triggering, is shown to potentiate the tumour suppressor behaviour of both the AR and RB1. In summary, this study provides insights into the condition of TCDD-activated AHR interactors within the context of ccRCC. Utilizing computational approaches and survival analysis, we identified potential therapeutic approaches, specifically AR and RB1 enhancement. Ligands of the aryl hydrocarbon receptor (AhR) are categorized into groups such as xenobiotic, endobiotic, and related compounds. It was proposed that ligands within each category may share similar functional activities, differing primarily in their relative potency. Alternatively, these ligands can be viewed as selective modulators (SAhRM), where different SAhRM groups may exhibit overlapping functions, but their genomic and biological activities can vary [71]. Previous investigations in the literature explored this concept, showing that these modulators can have specific agonist and antagonist effects on various cells and tissues. For instance, alkyl polychlorinated dibenzofurans (PCDFs) with alternative substitutions (1,3,6,8- and 2,4,6,8-) and substituted diindolylmethanes (DIMs) can bind to the AHR, leading to inhibitory interactions between the AHR and the estrogen receptor (ER). This interaction mirrors the effects observed with TCDD, including the suppression of mammary tumour growth [72]. The strategic activation of the AHR through a selective AHR modulator (SAhRM) may offer effective anti-tumour therapy in VHL-mutated ccRCC, reducing the need for surgical interventions [71,73].
Acknowledgments
We acknowledge the BiocomputingUP laboratory members for the helpful discussions.
Abbreviations
| VHL, pVHL | von Hippel–Lindau tumour suppressor |
| HIFs | Hypoxia-inducible factors |
| ccRCC | Clear cell renal cell carcinoma |
| AHR | Aryl hydrocarbon receptor |
| HIF-1β, ARNT | Aryl hydrocarbon receptor nuclear translocator |
| TCDD | Dioxin, 2,3,7,8-tetrachlorodibenzo-p-dioxin |
| AR | Androgen receptor |
| RB1 | Retinoblastoma-associated protein |
| SAhRMs | Selective AHR modulators |
| bHLH-PAS | Basic helix–loop–helix-PER-ARNT-SIM transcription factors |
| pNETs | Non-functioning pancreatic neuroendocrine tumours |
| RCC | Renal cell carcinoma |
| VCB complex | Complex formed by pVHL, elongin-B, elongin-C, and Cullin-2 |
| ELOB | Elongin-B |
| ELOC | Elongin-C |
| CUL2 | Cullin-2 |
| HIF-1α | Hypoxia-inducible factor 1-α |
| HSP90 | Heat shock protein 90 |
| p23 | Prostaglandin E synthase 3 |
| XAP2 | AH receptor-interacting protein |
| XREs | Xenobiotic-responsive elements |
| DREs | Dioxin-responsive elements |
| Cyp1a | Cytochrome P450, family 1, subfamily A, polypeptide 1 |
| Cyp1b | Cytochrome P450, family 1, subfamily B, polypeptide 1 |
| GSTA1 | Glutathione S-transferase A1 |
| EPHX1 | Epoxide hydrolase 1 |
| PAI2 | Plasminogen activator inhibitor 2 |
| p300 | Histone acetyltransferase p300 |
| CREBBP | CREB-binding protein |
| NCOA1/2/3 | Nuclear receptor coactivator 1/2/3 |
| NRIP1 | Nuclear receptor-interacting protein 1 |
| ER | Estrogen receptor |
| HepaRG | Human hepatic in vitro line |
| MCF7 | Human breast cancer cell line (Michigan Cancer Foundation-7) |
| HepG2 | Human liver cancer cell line |
| 786-O | Human renal cancer cell line |
| EGA | European Genome–Phenome Archive |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| HR | Hazard ratio |
| PPI | Protein–protein interaction |
| DEGs | Differentially expressed genes |
| GSTA5 | Glutathione S-transferase A5 |
| GSTM2 | Glutathione S-transferase Mu 2 |
| UGT2B11 | UDP-glucuronosyltransferase 2B11 |
| PPIN | Protein–protein interaction network |
| VEGF-A/C | Vascular endothelial growth factor A/C |
| CDK2/4 | Cyclin-dependent kinase 2/4 |
Author Contributions
Conceptualization, G.M. and S.C.E.T.; methodology, F.G. and G.M.; formal analysis, F.G. and G.M.; data curation, F.G.; writing—original draft preparation, F.G., G.M. and S.C.E.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by Fondazione AIRC per la Ricerca sul Cancro (AIRC), grant number IG 2019 ID 23825.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Nebert D.W. Aryl Hydrocarbon Receptor (AHR): “Pioneer Member” of the Basic-Helix/Loop/Helix per-Arnt-Sim (bHLH/PAS) Family of “Sensors” of Foreign and Endogenous Signals. Prog. Lipid Res. 2017;67:38–57. doi: 10.1016/j.plipres.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolonko-Adamska M., Uversky V.N., Greb-Markiewicz B. The Participation of the Intrinsically Disordered Regions of the bHLH-PAS Transcription Factors in Disease Development. Int. J. Mol. Sci. 2021;22:2868. doi: 10.3390/ijms22062868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziello J.E., Jovin I.S., Huang Y. Hypoxia-Inducible Factor (HIF)-1 Regulatory Pathway and Its Potential for Therapeutic Intervention in Malignancy and Ischemia. Yale J. Biol. Med. 2007;80:51–60. [PMC free article] [PubMed] [Google Scholar]
- 4.Tamukong P.K., Kuhlmann P., You S., Su S., Wang Y., Yoon S., Gong J., Figlin R.A., Janes J.L., Freedland S.J., et al. Hypoxia-Inducible Factor Pathway Genes Predict Survival in Metastatic Clear Cell Renal Cell Carcinoma. Urol. Oncol. 2022;40:e1–e495. doi: 10.1016/j.urolonc.2022.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Shenoy N., Pagliaro L. Sequential Pathogenesis of Metastatic VHL Mutant Clear Cell Renal Cell Carcinoma: Putting It Together with a Translational Perspective. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016;27:1685–1695. doi: 10.1093/annonc/mdw241. [DOI] [PubMed] [Google Scholar]
- 6.Chittiboina P., Lonser R.R. Von Hippel-Lindau Disease. Handb. Clin. Neurol. 2015;132:139–156. doi: 10.1016/B978-0-444-62702-5.00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minervini G., Pennuto M., Tosatto S.C.E. The pVHL Neglected Functions, a Tale of Hypoxia-Dependent and -Independent Regulations in Cancer. Open Biol. 2020;10:200109. doi: 10.1098/rsob.200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckley D.L., Van Molle I., Gareiss P.C., Tae H.S., Michel J., Noblin D.J., Jorgensen W.L., Ciulli A., Crews C.M. Targeting the von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the VHL/HIF-1α Interaction. J. Am. Chem. Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai W., Yang H. The Structure and Regulation of Cullin 2 Based E3 Ubiquitin Ligases and Their Biological Functions. Cell Div. 2016;11:7. doi: 10.1186/s13008-016-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chitrakar A., Budda S.A., Henderson J.G., Axtell R.C., Zenewicz L.A. E3 Ubiquitin Ligase Von Hippel-Lindau Protein Promotes Th17 Differentiation. J. Immunol. 2020;205:1009–1023. doi: 10.4049/jimmunol.2000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett P., Ramsden D.B., Williams A.C. Complete Structural Characterisation of the Human Aryl Hydrocarbon Receptor Gene. Mol. Pathol. 1996;49:M12–M16. doi: 10.1136/mp.49.1.M12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gargaro M., Scalisi G., Manni G., Mondanelli G., Grohmann U., Fallarino F. The Landscape of AhR Regulators and Coregulators to Fine-Tune AhR Functions. Int. J. Mol. Sci. 2021;22:757. doi: 10.3390/ijms22020757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao N., Whitelaw M.L., Shearwin K.E., Dodd I.B., Chapman-Smith A. Identification of Residues in the N-terminal PAS Domains Important for Dimerization of Arnt and AhR. Nucleic Acids Res. 2011;39:3695–3709. doi: 10.1093/nar/gkq1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pansoy A., Ahmed S., Valen E., Sandelin A., Matthews J. 3-Methylcholanthrene Induces Differential Recruitment of Aryl Hydrocarbon Receptor to Human Promoters. Toxicol. Sci. 2010;117:90–100. doi: 10.1093/toxsci/kfq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bock K.W. From TCDD-Mediated Toxicity to Searches of Physiologic AHR Functions. Biochem. Pharmacol. 2018;155:419–424. doi: 10.1016/j.bcp.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Tuomisto J. Dioxins and Dioxin-like Compounds: Toxicity in Humans and Animals, Sources, and Behaviour in the Environment. WikiJournal Med. 2019;6:8. doi: 10.15347/wjm/2019.008. [DOI] [Google Scholar]
- 17.Bersten D.C., Sullivan A.E., Peet D.J., Whitelaw M.L. bHLH–PAS Proteins in Cancer. Nat. Rev. Cancer. 2013;13:827–841. doi: 10.1038/nrc3621. [DOI] [PubMed] [Google Scholar]
- 18.Wang S., Hankinson O. Functional Involvement of the Brahma/SWI2-Related Gene 1 Protein in Cytochrome P4501A1 Transcription Mediated by the Aryl Hydrocarbon Receptor Complex*. J. Biol. Chem. 2002;277:11821–11827. doi: 10.1074/jbc.M110122200. [DOI] [PubMed] [Google Scholar]
- 19.Barhoover M.A., Hall J.M., Greenlee W.F., Thomas R.S. Aryl Hydrocarbon Receptor Regulates Cell Cycle Progression in Human Breast Cancer Cells via a Functional Interaction with Cyclin-Dependent Kinase 4. Mol. Pharmacol. 2010;77:195–201. doi: 10.1124/mol.109.059675. [DOI] [PubMed] [Google Scholar]
- 20.Elson D.J., Kolluri S.K. Tumor-Suppressive Functions of the Aryl Hydrocarbon Receptor (AhR) and AhR as a Therapeutic Target in Cancer. Biology. 2023;12:526. doi: 10.3390/biology12040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai M.-J., O’Malley B.W. Molecular Mechanisms of Action of Steroid/Thyroid Receptor Superfamily Members. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 22.Ohtake F., Takeyama K., Matsumoto T., Kitagawa H., Yamamoto Y., Nohara K., Tohyama C., Krust A., Mimura J., Chambon P., et al. Modulation of Oestrogen Receptor Signalling by Association with the Activated Dioxin Receptor. Nature. 2003;423:545–550. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- 23.Beischlag T.V., Perdew G.H. ERα-AHR-ARNT Protein-Protein Interactions Mediate Estradiol-Dependent Transrepression of Dioxin-Inducible Gene Transcription. J. Biol. Chem. 2005;280:21607–21611. doi: 10.1074/jbc.C500090200. [DOI] [PubMed] [Google Scholar]
- 24.Barnes-Ellerbe S., Knudsen K.E., Puga A. 2,3,7,8-Tetrachlorodibenzo- p -Dioxin Blocks Androgen-Dependent Cell Proliferation of LNCaP Cells through Modulation of pRB Phosphorylation. Mol. Pharmacol. 2004;66:502–511. doi: 10.1124/mol.104.000356. [DOI] [PubMed] [Google Scholar]
- 25.Dietrich C., Kaina B. The Aryl Hydrocarbon Receptor (AhR) in the Regulation of Cell-Cell Contact and Tumor Growth. Carcinogenesis. 2010;31:1319–1328. doi: 10.1093/carcin/bgq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanieh H., Bani Ismail M., Alfwuaires M.A., Ibrahim H.-I.M., Farhan M. Aryl Hydrocarbon Receptor as an Anticancer Target: An Overview of Ten Years Odyssey. Molecules. 2023;28:3978. doi: 10.3390/molecules28103978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell E.F., Kopparapu P.R., Koch D.C., Jang H.S., Phillips J.L., Tanguay R.L., Kerkvliet N.I., Kolluri S.K. The Aryl Hydrocarbon Receptor Mediates Leflunomide-Induced Growth Inhibition of Melanoma Cells. PLoS ONE. 2012;7:e40926. doi: 10.1371/journal.pone.0040926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell E.F., Koch D.C., Bisson W.H., Jang H.S., Kolluri S.K. The Aryl Hydrocarbon Receptor Mediates Raloxifene-Induced Apoptosis in Estrogen Receptor-Negative Hepatoma and Breast Cancer Cells. Cell Death Dis. 2014;5:e1038. doi: 10.1038/cddis.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch D.C., Jang H.S., O’Donnell E.F., Punj S., Kopparapu P.R., Bisson W.H., Kerkvliet N.I., Kolluri S.K. Anti-Androgen Flutamide Suppresses Hepatocellular Carcinoma Cell Proliferation via the Aryl Hydrocarbon Receptor Mediated Induction of Transforming Growth Factor-Β1. Oncogene. 2015;34:6092–6104. doi: 10.1038/onc.2015.55. [DOI] [PubMed] [Google Scholar]
- 30.Safe S., Cheng Y., Jin U.-H. The Aryl Hydrocarbon Receptor (AhR) as a Drug Target for Cancer Chemotherapy. Curr. Opin. Toxicol. 2017;2:24–29. doi: 10.1016/j.cotox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker J.R., Sakoff J.A., McCluskey A. The Aryl Hydrocarbon Receptor (AhR) as a Breast Cancer Drug Target. Med. Res. Rev. 2020;40:972–1001. doi: 10.1002/med.21645. [DOI] [PubMed] [Google Scholar]
- 32.Vogel C.F.A., Lazennec G., Kado S.Y., Dahlem C., He Y., Castaneda A., Ishihara Y., Vogeley C., Rossi A., Haarmann-Stemmann T., et al. Targeting the Aryl Hydrocarbon Receptor Signaling Pathway in Breast Cancer Development. Front. Immunol. 2021;12:625346. doi: 10.3389/fimmu.2021.625346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paris A., Tardif N., Galibert M.-D., Corre S. AhR and Cancer: From Gene Profiling to Targeted Therapy. Int. J. Mol. Sci. 2021;22:752. doi: 10.3390/ijms22020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lafleur V.N., Halim S., Choudhry H., Ratcliffe P.J., Mole D.R. Multi-Level Interaction between HIF and AHR Transcriptional Pathways in Kidney Carcinoma. Life Sci. Alliance. 2023;6:e202201756. doi: 10.26508/lsa.202201756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Athanasios A., Charalampos V., Vasileios T., Ashraf G.M. Protein-Protein Interaction (PPI) Network: Recent Advances in Drug Discovery. Curr. Drug Metab. 2017;18:5–10. doi: 10.2174/138920021801170119204832. [DOI] [PubMed] [Google Scholar]
- 36.Dhasmana A., Uniyal S., Anukriti, Kashyap V.K., Somvanshi P., Gupta M., Bhardwaj U., Jaggi M., Yallapu M.M., Haque S., et al. Topological and System-Level Protein Interaction Network (PIN) Analyses to Deduce Molecular Mechanism of Curcumin. Sci. Rep. 2020;10:12045. doi: 10.1038/s41598-020-69011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ba Q., Li J., Huang C., Li J., Chu R., Wu Y., Wang H. Topological, Functional, and Dynamic Properties of the Protein Interaction Networks Rewired by Benzo(a)Pyrene. Toxicol. Appl. Pharmacol. 2015;283:83–91. doi: 10.1016/j.taap.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Foersch S., Schindeldecker M., Keith M., Tagscherer K.E., Fernandez A., Stenzel P.J., Pahernik S., Hohenfellner M., Schirmacher P., Roth W., et al. Prognostic Relevance of Androgen Receptor Expression in Renal Cell Carcinomas. Oncotarget. 2017;8:78545–78555. doi: 10.18632/oncotarget.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang C., Lee S.O., Yeh S., Chang T.M. Androgen Receptor (AR) Differential Roles in Hormone-Related Tumors Including Prostate, Bladder, Kidney, Lung, Breast and Liver. Oncogene. 2014;33:3225–3234. doi: 10.1038/onc.2013.274. [DOI] [PubMed] [Google Scholar]
- 40.Zhao H., Leppert J.T., Peehl D.M. A Protective Role for Androgen Receptor in Clear Cell Renal Cell Carcinoma Based on Mining TCGA Data. PLoS ONE. 2016;11:e0146505. doi: 10.1371/journal.pone.0146505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macleod K.F. The RB Tumor Suppressor: A Gatekeeper to Hormone Independence in Prostate Cancer? J. Clin. Investig. 2010;120:4179–4182. doi: 10.1172/JCI45406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harlander S., Schönenberger D., Toussaint N.C., Prummer M., Catalano A., Brandt L., Moch H., Wild P.J., Frew I.J. Combined Vhl, Trp53 and Rb1 Mutation Causes Clear Cell Renal Cell Carcinoma in Mice. Nat. Med. 2017;23:869–877. doi: 10.1038/nm.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oughtred R., Rust J., Chang C., Breitkreutz B.-J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F., et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin H., Schaefer G.A.-L. HIPPIE v2.0: Enhancing Meaningfulness and Reliability of Protein–Protein Interaction Networks|Nucleic Acids Research|Oxford Academic. [(accessed on 18 January 2024)]. Available online: https://academic.oup.com/nar/article/45/D1/D408/2290937. [DOI] [PMC free article] [PubMed]
- 45.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otasek D., Morris J.H., Bouças J., Pico A.R., Demchak B. Cytoscape Automation: Empowering Workflow-Based Network Analysis. Genome Biol. 2019;20:185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Racine J.S. RStudio: A Platform-Independent IDE for R and Sweave. J. Appl. Econom. 2012;27:167–172. doi: 10.1002/jae.1278. [DOI] [Google Scholar]
- 50.Tenenbaum D., Volkening J., Maintainer B.P. KEGGREST: Client-Side REST Access to the Kyoto Encyclopedia of Genes and Genomes (KEGG), Bioconductor Version: Release (3.14) [(accessed on 27 August 2024)]. Available online: https://bioconductor.org/packages/release/bioc/html/KEGGREST.html.
- 51.Feng H., Gu Z.-Y., Li Q., Liu Q.-H., Yang X.-Y., Zhang J.-J. Identification of Significant Genes with Poor Prognosis in Ovarian Cancer via Bioinformatical Analysis. J. Ovarian Res. 2019;12:35. doi: 10.1186/s13048-019-0508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Abrew K.N., Kainkaryam R.M., Shan Y.K., Overmann G.J., Settivari R.S., Wang X., Xu J., Adams R.L., Tiesman J.P., Carney E.W., et al. Grouping 34 Chemicals Based on Mode of Action Using Connectivity Mapping. Toxicol. Sci. 2016;151:447–461. doi: 10.1093/toxsci/kfw058. [DOI] [PubMed] [Google Scholar]
- 53.Peña-Llopis S., Brugarolas J. Simultaneous Isolation of High-Quality DNA, RNA, miRNA and Proteins from Tissues for Genomic Applications. Nat. Protoc. 2013;8:2240–2255. doi: 10.1038/nprot.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peña-Llopis S., Vega-Rubín-de-Celis S., Liao A., Leng N., Pavía-Jiménez A., Wang S., Yamasaki T., Zhrebker L., Sivanand S., Spence P., et al. BAP1 Loss Defines a New Class of Renal Cell Carcinoma. Nat. Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao X., Tan J., Lim K.J., Koh J., Ooi W.F., Li Z., Huang D., Xing M., Chan Y.S., Qu J.Z., et al. VHL Deficiency Drives Enhancer Activation of Oncogenes in Clear Cell Renal Cell Carcinoma. Cancer Discov. 2017;7:1284–1305. doi: 10.1158/2159-8290.CD-17-0375. [DOI] [PubMed] [Google Scholar]
- 56.Chen S.-C., Chen F.-W., Hsu Y.-L., Kuo P.-L. Systematic Analysis of Transcriptomic Profile of Renal Cell Carcinoma under Long-Term Hypoxia Using Next-Generation Sequencing and Bioinformatics. Int. J. Mol. Sci. 2017;18:2657. doi: 10.3390/ijms18122657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Hu J., Fang Y., Fu Y., Liu B., Zhang C., Feng S., Lu X. Multi-Omics Profiling to Assess Signaling Changes upon VHL Restoration and Identify Putative VHL Substrates in Clear Cell Renal Cell Carcinoma Cell Lines. Cells. 2022;11:472. doi: 10.3390/cells11030472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagy Á., Munkácsy G., Győrffy B. Pancancer Survival Analysis of Cancer Hallmark Genes. Sci. Rep. 2021;11:6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Posta M., Győrffy B. Analysis of a Large Cohort of Pancreatic Cancer Transcriptomic Profiles to Reveal the Strongest Prognostic Factors. Clin. Transl. Sci. 2023;16:1479–1491. doi: 10.1111/cts.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 62.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Tissue-Based Map of the Human Proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 63.He D., Li L., Zhu G., Liang L., Guan Z., Chang L., Chen Y., Yeh S., Chang C. ASC-J9 Suppresses Renal Cell Carcinoma Progression by Targeting an Androgen Receptor-Dependent HIF2α/VEGF Signaling Pathway. Cancer Res. 2014;74:4420–4430. doi: 10.1158/0008-5472.CAN-13-2681. [DOI] [PubMed] [Google Scholar]
- 64.Huang Q., Sun Y., Zhai W., Ma X., Shen D., Du S., You B., Niu Y., Huang C.-P., Zhang X., et al. Androgen Receptor Modulates Metastatic Routes of VHL Wild-Type Clear Cell Renal Cell Carcinoma in an Oxygen-Dependent Manner. Oncogene. 2020;39:6677–6691. doi: 10.1038/s41388-020-01455-0. [DOI] [PubMed] [Google Scholar]
- 65.Ohtake F., Baba A., Fujii-Kuriyama Y., Kato S. Intrinsic AhR Function Underlies Cross-Talk of Dioxins with Sex Hormone Signalings. Biochem. Biophys. Res. Commun. 2008;370:541–546. doi: 10.1016/j.bbrc.2008.03.054. [DOI] [PubMed] [Google Scholar]
- 66.Ohtake F., Fujii-Kuriyama Y., Kato S. AhR Acts as an E3 Ubiquitin Ligase to Modulate Steroid Receptor Functions. Biochem. Pharmacol. 2009;77:474–484. doi: 10.1016/j.bcp.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 67.Luecke-Johansson S., Gralla M., Rundqvist H., Ho J.C., Johnson R.S., Gradin K., Poellinger L. A Molecular Mechanism To Switch the Aryl Hydrocarbon Receptor from a Transcription Factor to an E3 Ubiquitin Ligase. Mol. Cell Biol. 2017;37:e00630-16. doi: 10.1128/MCB.00630-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harbour J.W., Luo R.X., Dei Santi A., Postigo A.A., Dean D.C. Cdk Phosphorylation Triggers Sequential Intramolecular Interactions That Progressively Block Rb Functions as Cells Move through G1. Cell. 1999;98:859–869. doi: 10.1016/S0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 69.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H., et al. Integrated Molecular Analysis of Clear-Cell Renal Cell Carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 70.Formosa R., Borg J., Vassallo J. Aryl Hydrocarbon Receptor (AHR) Is a Potential Tumour Suppressor in Pituitary Adenomas. Endocr. Relat. Cancer. 2017;24:445–457. doi: 10.1530/ERC-17-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Safe S., Jin U., Park H., Chapkin R.S., Jayaraman A. Aryl Hydrocarbon Receptor (AHR) Ligands as Selective AHR Modulators (SAhRMs) Int. J. Mol. Sci. 2020;21:6654. doi: 10.3390/ijms21186654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Safe S., Qin C., McDougal A. Development of Selective Aryl Hydrocarbon Receptor Modulators for Treatment of Breast Cancer. Expert. Opin. Investig. Drugs. 1999;8:1385–1396. doi: 10.1517/13543784.8.9.1385. [DOI] [PubMed] [Google Scholar]
- 73.Dolciami D., Ballarotto M., Gargaro M., López-Cara L.C., Fallarino F., Macchiarulo A. Targeting Aryl Hydrocarbon Receptor for Next-Generation Immunotherapies: Selective Modulators (SAhRMs) versus Rapidly Metabolized Ligands (RMAhRLs) Eur. J. Med. Chem. 2020;185:111842. doi: 10.1016/j.ejmech.2019.111842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.