Abstract
Phylogenetic analyses showed that the virus responsible for a May 2024 Oropouche fever outbreak in Cuba was closely related to viruses from Brazil in 2023. Pools of Ceratopogonidae spp. biting midges and Culex quinquefasciatus mosquitoes were positive for Oropouche viral RNA. No cases were severe. Virus extension to new areas may increase case numbers and severity.
Keywords: Oropouche virus, orthobunyavirus, arboviruses, viruses, vector-borne infections, Cuba
On May 9, 2024, the Pan American Health Organization reported 5,193 cases of Oropouche fever (also called Oropouche virus disease) in Bolivia, Brazil, Colombia, and Peru and sent an alert with regard to autochthonous cases in areas of Brazil and Bolivia, where the disease had not been previously reported (1). During the past decade, Oropouche fever has been reported mainly in the Amazon region. However, in 2014, the first case outside that region was registered in Haiti, raising concerns about its geographic extension (2). The virus has been detected in Central and South America (e.g., Bolivia, Brazil, Colombia, Peru, and Panama); ≈500,000 cases have been diagnosed. Given the clinical picture of Oropouche fever (fever, headache, arthralgia, myalgia), the disease can be confused with dengue, Zika, or other febrile illnesses.
Oropouche virus (OROV) is a member of the family Peribunyaviridae, genus Orthobunyavirus, and was first identified in Trinidad and Tobago. OROV is maintained in nature through sylvatic and urban cycles. The urban cycle is thought to mainly involve the bite of the Culicoides paraensis midge. Another urban vector in tropical regions, where it feeds on both humans and animals, is the Culex quinquefasciatus mosquito (3). The OROV genome comprises 3 single-stranded negative-sense RNA segments (large, medium, and small). Sequencing studies of the small segment suggest the existence of 4 genotypes (I–IV) (4,5).
The source of patients for dengue surveillance in Cuba is the primary healthcare level through the search, clinical management, and notification of acute febrile illness (AFI) cases of unknown etiology or cases suspected of being dengue. A network of 30 laboratories with capacity for dengue IgM detection guarantees serologic surveillance conducted on samples collected at day 6 of fever onset. Positive samples are confirmed at the National Reference Laboratory of the Institute “Pedro Kouri” (NRL-IPK). Molecular surveillance at the NRL-IPK enables real-time PCR identification of dengue viruses in acute-phase samples from patients with suspected dengue cases. Negative samples are tested for Zika, chikungunya, and yellow fever viruses. The NRL-IPK is also in charge of virus genetic surveillance (6). Environmental and entomologic surveillance complement the national integrated arbovirus surveillance system.
The Study
On May 20, 2024, the NRL-IPK received samples from 89 patients with AFI of unknown etiology in Santiago de Cuba and Cienfuegos Provinces, Cuba, where an increase of similar cases had been reported, most negative for dengue IgM. Patients reported acute fever onset with headache and joint pain for 2–3 days and rapid recovery. Median patient age was 35 years (interquartile range [IQR] 19–50); 49.4% of patients were female and 51.6% male. Of the 89 serum samples received, 69 were from Santiago de Cuba (Boniato, n = 39; Songo la Maya, n = 30) and 20 from Cienfuegos (Abreu, n = 4; Cienfuegos, n = 11; and Rodas, n = 5). Urine (n = 6) and feces (n = 30) samples were also collected from the 89 patients.
We extracted RNA by using the commercial QIAamp Viral RNA Mini Kit (QIAGEN, https://www.qiagen.com) according to the manufacturer’s instructions. We processed extracted RNA for dengue, Zika and chikungunya viruses using by real-time PCR (7–9), and all serum samples were negative. Rapid test results for dengue nonstructural protein 1 (Bioline Dengue Duo; Abbott Laboratories, https://www.abbott.com) and chikungunya IgM (Kabla Diagnósticos, https://www.biodiagnosticos.com) were also negative. Fecal samples were negative for enterovirus (10).
We examined 89 serum and 6 urine samples for Oropouche and Mayaro viruses by using real-time PCR (11). All samples were negative for Mayaro virus, and 74 (83. 1%) serum samples (54 from Santiago de Cuba and 20 from Cienfuegos) and 5 (83.3%) urine samples were positive for OROV.
For the viral genetic characterization, we studied 9 serum samples (3 from Boniato, 3 from Songo La Maya, and 3 from Cienfuegos). We subjected extracted RNA to reverse transcription by PCR using the QIAGEN OneStep RT-PCR Kit system and primers NORO5 (5′-AAAGAGGATCCAATAATGTCAGAGTTCATTT-3′) and ORON3 (5′-AATTCGGAATTGGCATATAGTGGAATTCAC-3′) (12). We purified the 626-bp fragments obtained, which encode for the viral nucleocapsid (positions 85–718), by using the QIAquick PCR Purification Kit (QIAGEN), followed by sequencing with the Dye Terminator Cycle Sequencing Quick Start Kit (Analis, https://www.analis.com) and primers NORO5, ORON3, OROV-F (5′-TCCGGAGGCAGCATATGTG-3′), and OROV-R (5′-AAGTGCTCAATGCTGGTGTTGT-3′) (donated by the Pan American Health Organization). We purified the sequencing products and then separated the generated fragments on a CEQ 8800 Genetic Analysis System sequencer (Analis). We edited and assembled the electropherograms by using Sequencher version 4.10.1 software (Gene Codes Corporation, https://www.genecodes.com). As a reference, we used the complete sequence of the small segment of the Oropouche orthobunyavirus strain (GenBank accession no. OP244879.1, Oropouche orthobunyavirus isolate 0200178W, small segment, complete sequence). Phylogenetic analyses (MEGA version 6) (13) revealed that the isolated viruses clustered in a separate branch, closely related to those reported from Brazil in 2023 (Figure 1).
Figure 1.
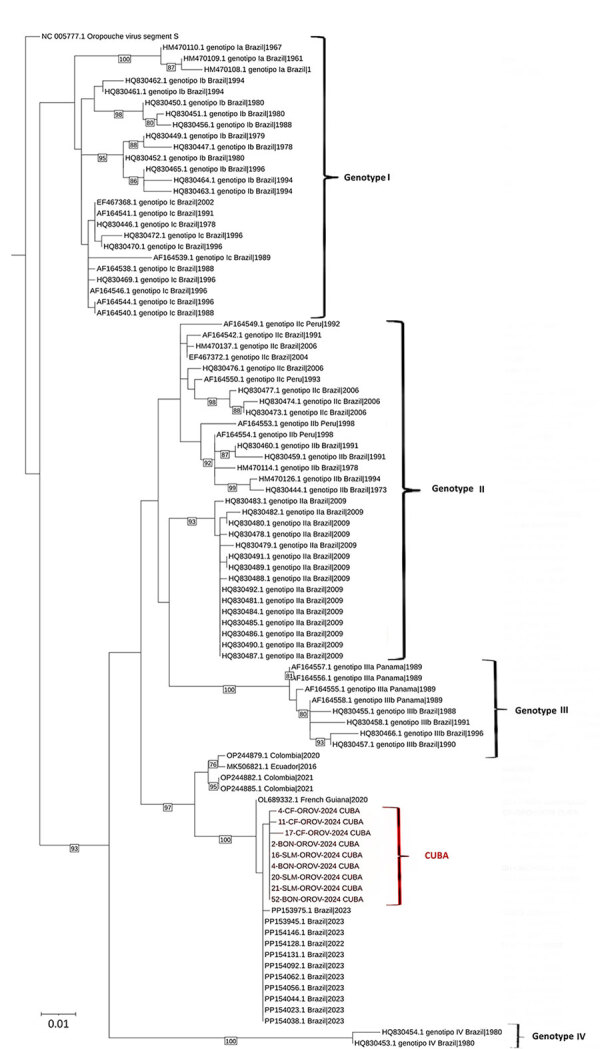
Molecular phylogenetic analysis of Oropouche viruses isolated in Cuba (red bracket) and reference sequences. The evolutionary history was inferred by using the maximum-likelihood method based on the Kimura 2-parameter model to the small segment of Oropouche orthobunyavirus from 9 patients from Boniato, Songo La Maya, and Cienfuegos (PP921382, PP921383, PP921384, PP921385, PP921386, PP921387, PP921388, PP921389, PP921390) (14). The tree with the highest log likelihood (−2,403.4997) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated by using the maximum composite likelihood approach. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.1407]). The tree is drawn to scale, with branch lengths measured in number of substitutions per site. The analysis involved 101 nt sequences deposited in GenBank (accession numbers shown) from the different outbreaks and genotypes of Oropouche virus in the Americas and the Caribbean region. All positions with <95% site coverage were eliminated (i.e., <5% alignment gaps, missing data, and ambiguous bases were allowed at any position) The final dataset contained 521 positions. Evolutionary analyses were conducted in MEGA6 (13).
After we had identified OROV transmission, national surveillance for dengue and Oropouche fever was intensified (active AFI case search). In addition, vector control intervention (adulticide treatment, source reduction, environmental management, biolarvicide application on permanent breeding sites) was applied in the areas with confirmed transmission.
During May 28–June 3, 2024, we processed 31 serum samples collected from locations with increased AFI cases (San Nicolas de Bari in Mayabeque; Perico, Cienaga de Zapata, and Jovellanos in Matanzas; and Ranchuelo in Villa Clara). OROV infection was confirmed in 25 (80.6%) samples: 7 from Matanzas, 9 from Mayabeque, and 9 from Villa Clara Provinces (Table 1). Median patient age was 34.5 years (range 4–83 years), male:female ratio was 1.1. Signs and symptoms by order of frequency were fever, headache, general malaise, myalgia, and arthralgia (Table 2; Figure 2; Appendix Table). No serious or fatal cases were reported.
Table 1. Epidemiologic characteristics of confirmed cases of Oropouche fever, Cuba, 2024*.
| Data |
Values |
| M:F ratio | 51/48 (1.1) |
| Median days of sample collection according to illness onset (range) | 2 (0–4) |
| Median age, y (range) [IQR] | 34.5 (4–83) [18–50.5] |
*IQR, interquartile range.
Table 2. Clinical characteristics of patients with confirmed Oropouche fever, Cuba, 2024.
| Clinical signs/symptoms |
No. (%) patients |
| Fever | 86 (86.9) |
| Headache | 71 (71.7) |
| General malaise | 51 (51.5) |
| Arthralgia | 22 (22.2) |
| Asthenia | 18 (18.1) |
| Anorexia | 16 (16.2) |
| Retroocular pain | 14 (14.1) |
| Abdominal pain | 8 (8) |
| Vomiting | 7 (7) |
| Diarrhea | 7 (7) |
| Chills | 4 (4) |
| Lumbar pain | 3 (3) |
Figure 2.
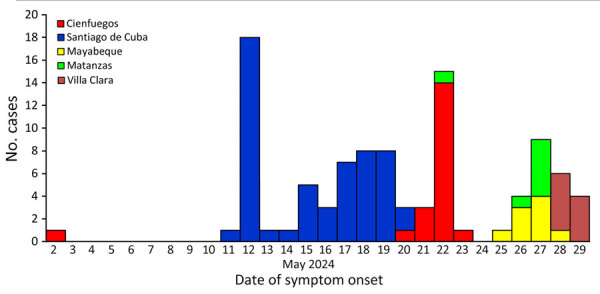
Confirmed Oropouche cases according to date of onset of signs/symptoms and provinces, Cuba, May 2024.
To investigate the vector involved with OROV transmission in Cuba, we collected 156 C. quinquefasciatus mosquitoes and 49 Ceratopogonidae spp. from suspected areas of transmission in 5 blocks of houses in Boniato. After potential vector identification, we grouped not visibly engorged female C. quinquefasciatus mosquitoes into 9 pools (10–20/pool) and Ceratopogonidae spp. specimens into 2 pools (7–19/pool). We extracted RNA and conducted real-time PCR. Positive samples were attributed to 5 (56%) pools of C. quinquefasciatus mosquitoes and 1 (50%) pool of Ceratopogonidae biting midges.
Among our study limitations was collection of signs/symptoms by using a standardized form for AFI or dengue cases; consequently, we might have missed less common presentations such as meningitis. In addition, the presence of viral RNA does not confirm the vector role in virus transmissibility. More work is needed to determine the primary vectors responsible for the current outbreak.
Conclusions
Oropouche fever is an emerging disease of concern in South and Central America. Transmission outside of the Amazon region is probably silent and not detected by public health surveillance systems. Our results confirm an outbreak of Oropouche fever in Cuba. Transmission was detected in semi-urban localities of 5 of 16 provinces located in the western, central, and eastern parts of the country. Dengue surveillance enabled us to identify cases with nondengue AFI. As of August 2024, OROV transmission had been confirmed in 7 provinces. The outbreak in Cuba alerts the Americas and the world of the need for integrated dynamic surveillance systems to detect the introduction and early transmission of OROV and consequently implement effective measures for its control.
Additional information for Figure 2.
Acknowledgments
We thank Jairo Mendez, Leticia Franco, Felipe Naveca, and Yosiel Molina for their useful comments and recommendations.
This work was funded by the Cuban Ministry of Health. Real-time PCR reagents were provided by the Pan American Health Organization.
M.G.G., S.R., and V.K. coordinated and designed the study, analyzed the results, and drafted and reviewed the manuscript; M.A. conducted the laboratory work, analyzed the results, and cleaned the data; A.J.B., S.S., and M.P. performed all real-time PCRs; L.P., R.G., and M.P. performed the genetic characterization; G.G., Y.M., and A.C. performed the vector studies; and C.P., J.R.A., and M.R. coordinated the epidemiologic and field studies; and D.C. and I.B. collected the clinical and epidemiologic data. All authors reviewed the draft and approved the final version.
Biography
Ms. Benitez is a microbiologist at the Institute of Tropical Medicine “Pedro Kouri” in Havana, Cuba, pursuing a master’s degree in virology. Her primary research interests are the arboviral infections, including viral molecular diagnosis.
Footnotes
Suggested citation for this article: Benitez AJ, Alvarez M, Perez L, Gravier R, Serrano S, Hernandez DM, et al. Oropouche fever, Cuba, May 2024. Emerg Infect Dis. 2024 Oct [date cited]. https://doi.org/10.3201/eid3010.240900
These authors contributed equally to this article.
References
- 1.Pan American Health Organization/World Health Organization. Alerta Epidemiológica: oropouche en la región de las Américas, 9 de mayo del 2024. [cited 2024 May 12]. https://www.paho.org/es/documentos/alerta-epidemiologica-oropouche-region-americas-9-mayo-2024
- 2.Elbadry MA, Durães-Carvalho R, Blohm GM, Stephenson CJ, Loeb JC, White SK, et al. Orthobunyaviruses in the Caribbean: Melao and Oropouche virus infections in school children in Haiti in 2014. PLoS Negl Trop Dis. 2021;15:e0009494. 10.1371/journal.pntd.0009494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso BF, Serra OP, Heinen LB, Zuchi N, Souza VC, Naveca FG, et al. Detection of Oropouche virus segment S in patients and inCulex quinquefasciatus in the state of Mato Grosso, Brazil. Mem Inst Oswaldo Cruz. 2015;110:745–54. 10.1590/0074-02760150123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travassos da Rosa JF, de Souza WM, Pinheiro FP, Figueiredo ML, Cardoso JF, Acrani GO, et al. Oropouche virus: clinical, epidemiological, and molecular aspects of a neglected orthobunyavirus. Am J Trop Med Hyg. 2017;96:1019–30. 10.4269/ajtmh.16-0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Files MA, Hansen CA, Herrera VC, Schindewolf C, Barrett ADT, Beasley DWC, et al. Baseline mapping of Oropouche virology, epidemiology, therapeutics, and vaccine research and development. NPJ Vaccines. 2022;7:38. 10.1038/s41541-022-00456-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guzmán MG, Vázquez S, Álvarez M, Pelegrino JL, Ruiz Amores D, Martínez PA, et al. Vigilancia de laboratorio de dengue y otros arbovirus en Cuba, 1970–2017. Rev Cubana Med Trop. 2019;71. [Google Scholar]
- 7.Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, et al. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis. 2013;7:e2311. 10.1371/journal.pntd.0002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–9. 10.3201/eid1408.080287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson BW, Russell BJ, Goodman CH. Laboratory diagnosis of chikungunya virus infections and commercial sources for diagnostic assays. J Infect Dis. 2016;214(suppl 5):S471–4. 10.1093/infdis/jiw274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nijhuis M, van Maarseveen N, Schuurman R, Verkuijlen S, de Vos M, Hendriksen K, et al. Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by real-time PCR. J Clin Microbiol. 2002;40:3666–70. 10.1128/JCM.40.10.3666-3670.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naveca FG, Nascimento VAD, Souza VC, Nunes BTD, Rodrigues DSG, Vasconcelos PFDC. Multiplexed reverse transcription real-time polymerase chain reaction for simultaneous detection of Mayaro, Oropouche, and Oropouche-like viruses. Mem Inst Oswaldo Cruz. 2017;112:510–3. 10.1590/0074-02760160062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasconcelos HB, Nunes MRT, Casseb LMN, Carvalho VL, Pinto da Silva EV, Silva M, et al. Molecular epidemiology of Oropouche virus, Brazil. Emerg Infect Dis. 2011;17:800–6. 10.3201/eid1705.101333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information for Figure 2.


