Abstract
Trypsin enhances rotavirus infectivity by an unknown mechanism. To examine the structural basis of trypsin-enhanced infectivity in rotaviruses, SA11 4F triple-layered particles (TLPs) grown in the absence (nontrypsinized rotavirus [NTR]) or presence (trypsinized rotavirus [TR]) of trypsin were characterized to determine the structure, the protein composition, and the infectivity of the particles before and after trypsin treatment. As expected, VP4 was not cleaved in NTR particles and was cleaved into VP5∗ and VP8∗ in TR particles. However, surprisingly, while the VP4 spikes were clearly visible and well ordered in the electron cryomicroscopy reconstructions of TR TLPs, they were totally absent in the reconstructions of NTR TLPs. Biochemical analysis with radiolabeled particles indicated that the stoichiometry of the VP4 in NTR particles was the same as that in TR particles and that the VP8∗ portion of NTR, but not TR, particles is susceptible to further proteolysis by trypsin. Taken together, these structural and biochemical data show that the VP4 spikes in the NTR TLPs are icosahedrally disordered and that they are conformationally different. Structural studies on the NTR TLPs after trypsin treatment showed that spike structure could be partially recovered. Following additional trypsin treatment, infectivity was enhanced for both NTR and TR particles, but the infectivity of NTR remained 2 logs lower than that of TR particles. Increased infectivity in these particles corresponded to additional cleavages in VP5∗, at amino acids 259, 583, and putatively 467, which are conserved in all P serotypes of human and animal group A rotaviruses and also corresponded with a structural change in VP7. These biochemical and structural results show that trypsin cleavage imparts order to VP4 spikes on de novo synthesized virus particles, and these ordered spikes make virus entry into cells more efficient.
Rotaviruses are the leading cause of severe gastroenteritis in young children worldwide (16). Structural and biochemical analyses show that rotaviruses are large, icosahedral particles having a complex architecture consisting of three concentric capsid layers surrounding a genome of 11 segments of double-stranded RNA (41, 42). The innermost capsid layer, composed of VP2, encloses the genomic double-stranded RNA. A significant portion of the genomic RNA, particularly that in close contact with the inner surface of the VP2 layer, is icosahedrally ordered (43). Anchored to the inner surface of VP2 at the icosahedral vertices are two proteins, VP1 and VP3, involved in transcription of the genome within the intact particle. The intermediate capsid layer is composed of trimers of VP6 organized on a T=13 (levo) icosahedral lattice (44). The outermost layer in the infectious virus is composed of the major capsid glycoprotein VP7 and the hemagglutinin spike protein VP4 (16). VP7 is present as 780 molecules grouped as 260 trimers at the local and strict threefold axes of a T=13 left-handed icosahedral lattice (44, 51). The VP7 layer is perforated with 132 aqueous channels located at all the 5- and 6-coordinated positions of the T=13 icosahedral lattice. These channels are ∼140 Å deep and span the outer two capsid layers. VP4 is present as 60 homodimeric spikes that are located at one edge of the peripentonal channels (40). These spikes extend ∼100 Å from the VP7 surface. Further structural studies revealed a large globular domain of VP4 below the VP7 layer that interacts extensively with VP6 (47, 50). The expression and purification of 2/4/6- and 2/4/6/7-virus-like particles (VLPs) provide further evidence for the interactions between VP6 and VP4 (11). Antibody binding studies with these VLPs and studies of reassortant rotaviruses suggest that VP7 has a stabilizing effect on the structure of VP4 (6, 11).
It is not yet clear how rotaviruses gain entry into host cells. Rotaviruses may enter either through an endocytotic pathway or through direct penetration. Although earlier studies implicated VP7 in cell attachment (19, 46), recent studies have increasingly indicated that VP4 is the major player in the entry process (11, 26, 30) while VP7 may modulate the functions of VP4 (3, 10, 35, 49).
VP4 is susceptible to proteolysis and is cleaved by trypsin into VP8∗ (26 kDa) and VP5∗ (60 kDa). The two trypsin cleavage products remain associated with the virion, although the precise topographical locations of VP5∗ and VP8∗ in the VP4 spike are still uncertain. Structural studies with escape mutants using a monoclonal antibody which maps within VP5∗ indicate that the distal end of the spike contains a domain within the VP5∗ region (40). VP5∗ contains a putative fusion domain similar to that seen in enveloped viruses such as alphaviruses and influenza viruses. Although direct evidence that this domain is critical for infectivity is lacking, this domain may facilitate rotavirus penetration into cells. VP8∗ contains the hemagglutination or sialic acid binding domain that appears necessary for the subset of rotaviruses that require sialic acid for infectivity (8a, 18, 22, 23).
Trypsin cleavage of VP4 alters a number of the biological functions of rotaviruses. Proteolytic cleavage of VP4 enhances viral infectivity severalfold (1, 17). Trypsinized viruses enter host cells more rapidly, possibly without using the endosomal pathway, than nontrypsinized virions (24, 25). In vitro experiments show that particles containing cleaved VP4 possess lipophilic activity (37, 45). The ability of rotaviruses to induce syncytia in cholesterol-supplemented MA104 cells is dependent on trypsin cleavage of VP4 at amino acid 247 (21). The molecular mechanisms of increased infectivity by proteolysis and membrane penetration are not well understood, but the importance of proteolysis in rotavirus replication is particularly relevant since these viruses grow in the enterocytes of the small intestine, an environment rich in proteases. To determine the structural basis for trypsin-enhanced infectivity of rotaviruses, we examined the SA11 4F rotavirus strain grown in the absence and presence of trypsin to characterize the structural, biochemical, and biologic differences of triple-layered particles (TLPs) before and after additional trypsin treatment.
MATERIALS AND METHODS
Cells and virus.
TLPs of simian rotavirus strain SA11 4F (4, 34, 39) were prepared by infecting fetal African green monkey kidney (MA104) cells at a multiplicity of infection of 20 PFU/cell with SA11 4F viruses previously treated with 10 μg of trypsin (2× crystallized bovine trypsin; Worthington Biochemical Corp., Freehold, N.J.)/ml for 30 min at 37°C. The virus was adsorbed for 90 min at 37°C, and then M199 medium containing 1 μg of trypsin/ml was added to obtain SA11 4F particles grown in the presence of trypsin (trypsinized rotavirus [TR]). To obtain SA11 4F TLPs containing an intact, uncleaved VP4 (nontrypsinized rotavirus [NTR]), the MA104 cells were washed three times with phosphate-buffered saline after virus adsorption and incubated in M199 medium containing 0.5 μg of aprotinin (Sigma Chemical Co., St, Louis, Mo.)/ml. NTR TLPs were also prepared without the addition of aprotinin. 35S-labeled particles were prepared by growing the virus in methionine-free medium containing 15 μCi of Redivue Pro-Mix (Amersham Pharmacia Biotech, Piscataway, N.J.)/ml. The cells and medium for TR or NTR TLPs were harvested at 24 h postinfection, pooled, frozen and thawed three times, and then sonicated for 3 min using a Cup Horn sonicator (Heat Systems-Ultrasonics, Plainview, N.Y.). The medium was clarified by centrifugation for 30 min at 12,000 rpm in a Beckman JA-14 rotor. The virus was pelleted for 1 h at 40,000 rpm in a 50.2 Ti rotor. The resulting pellet was suspended in TNC buffer (10 mM Tris-HCl [pH 7.4], 140 mM NaCl, 10 mM CaCl2), and then CsCl was added to obtain a refractive index of 1.3690 and the mixture was centrifuged for 18 h at 35,000 rpm in an SW50.1 rotor. The gradients were fractionated, and fractions containing TLPs were pooled, diluted in TNC buffer, and then pelleted by centrifugation for 2 h at 35,000 rpm in an SW41 rotor and suspended in TNC buffer.
Trypsin treatment of the SA11 4F TLPs.
Purified SA11 4F TLPs grown in the presence or absence of trypsin were digested with 25, 50 or 75 μg of 2× crystallized trypsin (180 p-toluene-sulfonyl-l-arginine methyl ester [TAME] units per mg) or l-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin (180 TAME units/mg)/ml for 30 min at 37°C. Following digestion, soybean trypsin inhibitor (Worthington; 216 TAME units inhibited/mg) was added to the TLPs treated with TPCK-treated trypsin. Immediately after trypsin digestion, aliquots of mock- and trypsin-digested TLPs were analyzed concurrently by electron cryomicroscopy (cryo-EM), fluorescent focus assay, and Western blot analysis.
Characterization of SA11 4F TLPs.
SA11 4F TLPs were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a modification of the method of Laemmli with 8 or 10% separating and 4% stacking gels as previously described (32). Samples were dissociated by boiling for 3 min in sample buffer containing 1% SDS, 10% 2-mercaptoethanol, 0.05 M Tris-HCl (pH 6.8), 10% glycerol, and 0.0025% phenol red. Western blot analysis using a mouse hyperimmune serum prepared against triple-layered SA11 4F virus grown in the presence of trypsin, anti-VP5∗ monoclonal antibodies 3D8, 1D8, and 5B10, and a rabbit hyperimmune serum prepared against a peptide corresponding to amino acids 160 to 186 of SA11 4F VP8∗ was performed as previously described (5). The peptide and peptide antiserum were prepared as described previously (2).
N-terminal amino acid protein sequence analysis.
For N-terminal amino acid sequence analysis, proteins separated by SDS-PAGE were transferred onto a polyvinylidene fluoride membrane and stained with Coomassie blue, and the bands corresponding to the trypsin cleavage products, detected by Western blot analysis, were excised (33, 36). N-terminal microsequence analysis was performed on an Applied Biosystems 477A protein sequencer in the Protein Sequencing Core Facility of Baylor College of Medicine.
Fluorescent focus assay.
Virus infectivity was determined by fluorescent focus assay as described previously (9). Briefly, preparations of NTR and TR SA11 4F particles were treated with 0 (mock), 25, or 50 μg of trypsin/ml (see above). Two or three aliquots of each of the mock- and trypsin-treated virus samples were assayed for virus infectivity in triplicate. Following virus absorption onto a confluent monolayer of MA104 cells, the virus inoculum was removed and the monolayer was washed three times with M199 medium prior to the addition of M199 medium without trypsin. The titer of each sample was determined and converted to log10 values, and Student's t test (two-tailed test) was performed to determine if trypsin treatment significantly increased virus infectivity. The concentration of each virus preparation was calculated using the formula 1 optical density unit at 260 nm = 185 μg/ml. The specific infectivity was expressed as focus-forming units per microgram of virus.
Cryo-EM.
TLPs for microscopy were embedded in a thin layer of vitreous ice on holey carbon films using standard procedures (15, 28). Frozen hydrated specimens were imaged in a JEOL 1200 electron cryomicroscope, using a 100-kV electron beam with a dose of ∼5e−/Å2 at a magnification of ×30,000. For each specimen area, focal pairs were recorded, the first one at a defocus of ∼1 μm and the subsequent one at a defocus of ∼2 μm. Images were recorded on Kodak SO-163 electron films with a 1-s exposure time. Micrographs were developed for 12 min in a Kodak D-19 developer at 21°C and fixed for 10 min in Kodak fixer.
Three-dimensional structural analysis.
Micrographs were chosen for structural analysis based on the criteria of ice quality, particle concentration, and optimum defocus. Images were digitized with a Perkin-Elmer microdensitometer at a raster-scanning interval of 5.33 Å in the object. Particles from both images of digitized focal pairs were boxed into individual particle images with a pixel area of 256 by 256 and masked with a suitable radius. While the closer-to-focus images were used for the three-dimensional reconstruction, the further-from-focus images were used to confirm the orientations of the particles. Orientations of the particles were determined using the common lines procedure (12) and refined using the cross-common lines procedure (20) as described previously (27). The three-dimensional reconstructions were computed using cylindrical expansion methods (12). The distribution of the particle orientations was assessed by plotting them on the icosahedral asymmetric unit represented in terms of φ and θ and by estimating mean inverse eigenvalues (13). The defocus value of each micrograph was estimated from the positions of the contrast transfer function rings in the sum of individual particle image Fourier transforms. The final reconstructions were computed to a resolution that contained information within the first zero of the contrast transfer function. Corrections for contrast transfer function were carried out using a Wiener filter assuming an amplitude contrast factor of 0.14 and a signal/noise ratio of 0.2 as described by Zhou et al. (52). Effective resolution of each reconstruction was estimated using Fourier cross correlation coefficients and using equation 3 from a report by van Heel (48), and phase residuals were estimated using equation 6 between independent reconstructions of the same specimen or between two independent reconstructions obtained by randomly dividing the data from a single micrograph into two sets. The reconstructions were visualized using IRIS Explorer (NAG, Inc.) with several customized modules (Lawton and Prasad, unpublished data). The contour level used in all the structural representations was chosen to yield approximately 780 molecules of VP6 between the radii of 260 and 350 Å.
RESULTS
Cryo-EM reconstructions of SA11 4F TR and NTR TLPs.
To determine the effect of trypsin on the structure of rotaviruses, the SA11 4F rotavirus was grown in the presence of trypsin or without exogenously added trypsin in the presence of the serine protease inhibitor aprotinin to inhibit residual trypsin and then was purified. The cryo-images of the TR TLPs at appropriate defocus values (2 to 3 μm) showed that the spikes could be visualized on the particles as described earlier (40). We carried out several independent reconstructions of TR TLPs to a resolution of 23 Å, including as many as 300 particles and as few as 50 particles. In these reconstructions, the spikes were always well defined, and all the features of the spikes, including the lower domain described for SA11 4F and other rotavirus strains, were observed (40, 47, 50). As an example, the three-dimensional structure of the TR TLPs reconstructed from 76 particle images to a resolution of 23 Å is shown in Fig. 1A.
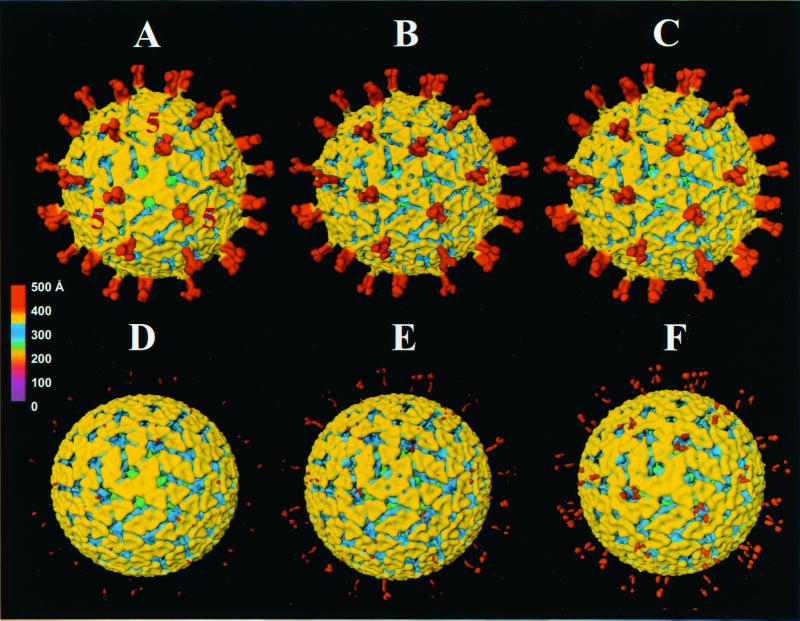
FIG. 1.
Three-dimensional reconstructions, viewed along the icosahedral threefold axis, of SA11 4F TLPs grown in the presence (top) and absence (bottom) of trypsin and treated with 0 (A and D), 25 (B and E), 50 (C), and 75 (F) μg of trypsin/ml. Fivefold axial positions defining one of the icosahedral facets of the T=13 lattice are indicated in panel A. Reconstructions are radially colored according to the chart. Reconstructions were carried out from electron cryo-images of individual specimens embedded in a thin layer of vitreous ice as described in Materials and Methods. Figures 3 to 5 show biochemical analyses of these particles.
The cryo-images of the NTR particles showed regular rotavirus morphology with a smooth outer periphery as previously observed (44). Surprisingly, the spikes on the particles were not obvious in these images even at lower defocus values (2 to 3 μm). Three-dimensional reconstruction using 123 particle images to a resolution of 23 Å, as in the case of TR TLPs, showed no VP4 spikes (Fig. 1D). The distribution of the particle orientations was adequate, as 98% of the mean inverse eigenvalues were <0.01. Even at lower threshold levels, amidst increasing background, mass density due to the spikes was not evident. The rest of the virus structure, however, had all the features that are normally observed. Similar results were obtained with different micrographs from these TLPs and also with two other preparations of NTR TLPs (data not shown).
Cryo-EM reconstructions of TR and NTR TLPs after exogenous trypsin treatment.
The TR TLPs were further treated with additional trypsin to examine if structural changes could be observed following exogenous trypsin treatment. The concentration of the particles used for cryo-EM ranged from 1 to 6 mg/ml; therefore, trypsin concentrations of 25 and 50 μg/ml were used to ensure that every VP4 spike would be cleaved, including cleavage of arginine 247, which requires high concentrations of trypsin (1). The particles were treated with 2× crystallized trypsin for 30 min at 37°C, and then aliquots were immediately taken for cryo-EM or used for biochemical or infectivity analysis.
No obvious structural changes were observed for the TR spike structure in the 25-μg/ml trypsin-treated specimen (Fig. 1B). A difference map between the 0- and 25-μg/ml trypsin-treated TR TLPs showed an alteration in the VP7 capsid layer, particularly near the channels surrounding the icosahedral threefold axes. The mass density is apparently translocated from beneath VP7, at the interface of VP7 and VP6, into the channels (type III) surrounding the icosahedral threefold axes (Fig. 2). The same structural changes were seen when 50 μg of trypsin/ml was added to the TR viruses. Although other small changes were observed between the mock- and trypsin-treated particles, these changes were not reproducibly seen in three independent reconstructions.
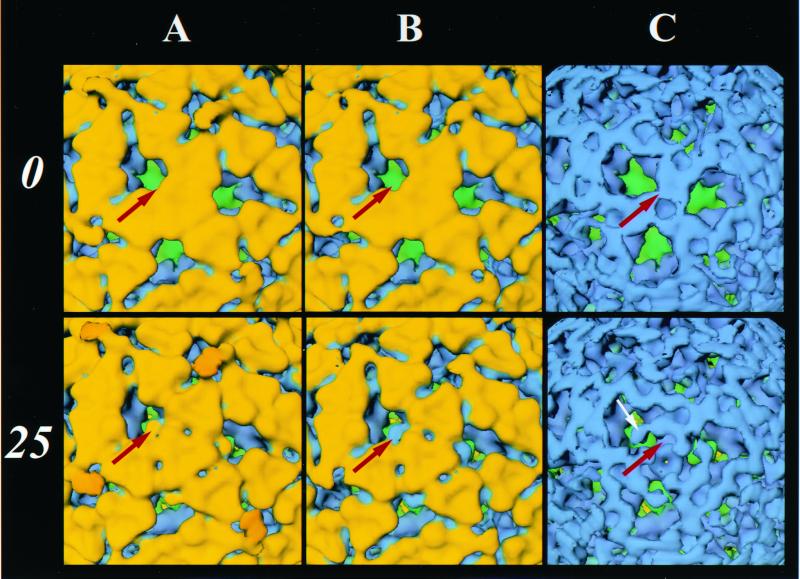
FIG. 2.
Structural changes in the TLPs induced by exogenous trypsin. Radial cutaways at ∼405 Å (A), ∼385 Å (B), and ∼360 Å (C) of TLP structures grown in the presence of trypsin treated with 0 (top) and 25 (bottom) μg of trypsin/ml. Red arrows point to the location where significant and reproducible changes from three independent reconstructions are observed in the trypsin-treated structure in relation to the particles without added trypsin. An interior portion of VP7, which is tucked inside between the VP7 trimers, swivels out (shown by a white arrow in bottom panel C) when virions are treated with exogenous trypsin. Identical structural changes are also seen in NTR particles upon exogenous trypsin treatment.
Structural analysis of the NTR TLPs after additional trypsin treatment (25 or 50 μg of trypsin/ml) showed that some mass density due to the spikes was clearly visible in these reconstructions (Fig. 1E [25 μg/ml]; 50-μg/ml data not shown). The dimeric shape and asymmetry of the VP4 spikes were evident. However, the spikes were clearly not as well defined as those on the TR TLPs, and the lower internal domain was not evident. The ability to see spikes on the reconstruction of NTR TLPs after exogenous trypsin treatment suggested that trypsin cleavage stabilized or ordered the spikes. Interestingly, the addition of exogenous trypsin produced the same VP7-associated structural changes observed between the 0- and 25-μg/ml trypsin-treated TR TLPs.
To determine if higher concentrations of trypsin were necessary to obtain a spike structure similar to that on TR TLPs, the particles were treated with 75 μg of trypsin/ml. An improvement in the definition of the spikes (Fig. 1F) was observed, but they were still not as well defined as the spikes on the TR TLPs. Although the spike density above the VP7 layer became more apparent after trypsin treatment of NTR TLPs (Fig. 1D or E), the mass density corresponding to the internal domain of VP4 below the VP7 layer, as seen in the TR TLPs and other rotavirus strains (40, 47, 50), was not observed in the NTR TLP reconstructions.
To determine if the antiproteolytic agent aprotinin used during virus propagation adversely affected the assembly of VP4 resulting in the disordered VP4, NTR TLPs were grown in the absence of aprotinin. Reconstructions of these TLPs using 100 particles to a resolution of 24 Å showed weak and scattered density only in the distal end of the spike structure (data not shown). After trypsin treatment (50 μg/ml) of these NTR TLPs grown in the absence of aprotinin, the VP4 spikes looked similar to the spikes on trypsinized NTR particles; they were still not as well defined as spikes on the TR TLPs. This indicates that the VP4 spikes on these virions, similar to NTR TLPs grown in the presence of aprotinin, have a different conformation than the VP4 spikes on the TR TLPs.
Determination of the infectivity of TR and NTR TLPs.
VP4 has been shown to be the rotavirus attachment protein, and trypsin cleavage of VP4 increases the infectivity of rotaviruses (1, 11, 17, 24). Our inability to reconstruct VP4 on NTR TLPs suggested that VP4 is icosahedrally disordered but regains some order after trypsin treatment; however, VP4 on TR TLPs is well ordered. To correlate the structure of each particle to infectivity, infectivity assays were performed on aliquots of the same TR and NTR TLPs analyzed biochemically and used for cryo-reconstructions. Immediately after trypsin treatment, the virus was diluted and infectivity was assayed. A fluorescent focus assay was used in our analysis, instead of plaque assays, to determine virus titers. Previous studies showed that virus binding to cells is independent of the cleavage of the VP4 spike but that enhanced infectivity of viruses bound to cells is due to cleavage of VP4 with proteolytic enzymes. The fluorescent focus method assays directly the amount of infectious virus during a single cycle of replication, whereas the plaque assay, with proteolytic enzymes used in the overlay, detects the infectious virus and the activated virus bound to cells and produced over multiple rounds of replication.
The specific infectivity (focus-forming units per microgram) of the NTR particles was approximately 3 logs lower than that of the TR particles before trypsin treatment (Fig. 3). Treatment of TR and NTR particles with 25 or 50 μg of trypsin/ml significantly increased the infectivity of each of the viruses over mock-treated virus (P < 0.05; Fig. 3). Although the infectivity of the NTR particles increased upon trypsin treatment for each concentration of trypsin tested, the specific infectivity of NTR particles was approximately 2 logs lower than for TR particles.
FIG. 3.
Determination of infectivity of SA11 4F TLPs by fluorescent focus assay. Preparations of NTR and TR SA11 4F TLPs were treated with 0, 25, or 50 μg of trypsin/ml for 30 min at 37°C, and the titer was determined by fluorescent focus assay. Significant differences in titer between mock- and trypsin-treated preparations are indicated (∗; Student's t test [P < 0.05]). Error bars represent the standard error of the mean. FFU, focus-forming units.
Biochemical characterization of TR and NTR TLPs.
Aliquots of the same NTR and TR particles used for cryo-reconstructions and analyzed for infectivity were examined by Western blot using a polyclonal antibody raised against SA11 4F TLPs to determine the protein composition of the TR and NTR TLPs before and after exogenous trypsin treatment. No discernible change was observed in the amount of VP5∗ or VP8∗ present between the pre- and post-trypsin-treated TR samples (Fig. 4A). However, two new bands with apparent relative molecular weights of 24,000 and 22,000 (24K and 22K, respectively) appeared in the trypsin-digested samples compared with the starting mock-treated TR particles (Fig. 4A). Particles were also treated with TPCK-treated trypsin for 30 min at 37°C, followed by the immediate addition of soybean trypsin inhibitor to inhibit further trypsin activity and to evaluate if trace amounts of chymotrypsin in the 2× crystallized trypsin or further digestion with trypsin was responsible for the cleavage products. The same cleavage products were detected with either treatment (data not shown).
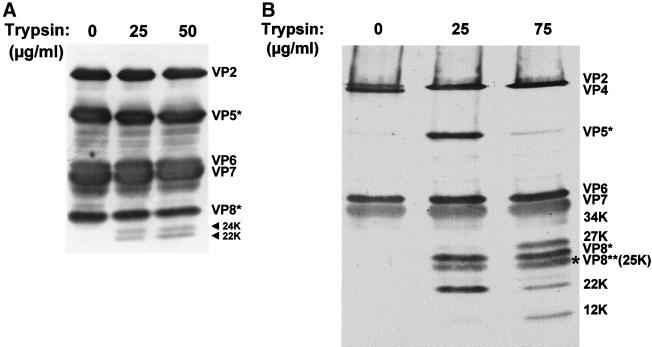
FIG. 4.
SDS-PAGE and Western blot analysis of SA11 4F TR and NTR particles. SA11 4F TLPs grown in the presence (A) and absence (B) of trypsin were purified and treated with increasing concentrations of trypsin as indicated. The proteins were separated by SDS-PAGE, transferred to nitrocellulose, and detected with a hyperimmune anti-SA11 4F TLP mouse serum. The location of the individual proteins is indicated on the right of each panel. The approximate molecular weights for the new bands representing previously uncharacterized cleavage products from viruses exposed to trypsin after purification are indicated. The arrows highlight two new bands representing previously uncharacterized VP5∗ cleavage products of 24K and 22K, and the asterisk highlights a new VP8∗-specific cleavage product of 25K from viruses exposed to trypsin.
To determine if these new bands were VP7 cleavage products, since structural changes were observed in the VP7 capsid layer after exogenous trypsin treatment, monoclonal (60 from H. B. Greenberg) and polyclonal antibodies against VP7 were used for Western blot analysis (31). None of the VP7 antibodies reacted with these new bands (data not shown), indicating these were either not VP7 cleavage products or could not be detected with the antiserum used. Therefore, these bands were subjected to N-terminal microsequencing analysis.
The two new bands were identified as trypsin cleavage products of VP5∗. The sequence of the 24K band began at amino acid 259 and of the 22K band began at amino acid 583. The migration of the cleavage products, amino acids 583 to 776 and 259 to 582, and the predicted molecular weight of each of these cleavage products, 21,963 and 36,302, respectively, suggested that another cleavage must have occurred to produce the 24K band. Cleavage at the arginine at position 467 to yield a product consisting of amino acids 259 to 467 would result in a peptide with the correct predicted molecular weight for the 24K band, 23,855, and another peptide with a molecular weight of 12,620 (amino acids 467 to 582). The antisera used to detect the rotavirus proteins in the Western blot did not detect this 12,620-molecular-weight cleavage product. Attempts to N-terminally microsequence a potential cleavage product migrating in this range were unsuccessful. These cleavage sites, lysine at amino acid 258, arginine (lysine for rotavirus strain 1076) at amino acid 582, and arginine at the predicted cleavage site at amino acid 467, are conserved in human and animal rotavirus strains belonging to all described P genotypes to date. Table 1 shows the conserved cleavage sites for a subset of human and animal rotavirus strains with different P and G types.
TABLE 1.
New trypsin cleavage sites are conserved in human and animal rotaviruses
| Rotavirus straina | P typeb | G type | Sequence at confirmed trypsin cleavage sitec
|
Sequence at putative trypsin cleavage site at amino acid 467 | |
|---|---|---|---|---|---|
| Amino acid 258 | Amino acid 582 | ||||
| Si/SA11 4F | 6[1] | 3 | DIVVSKTSLWK | RSASVRSVSST | YEIAGRFSLIS |
| Si/SA11 cl3 | 5B[2] | 3 | DIVISKTSLWK | RGSSIRSIGSS | YEIAGRFSLIS |
| Si/RRV | 5B[3] | 3 | DIVVSKTSLWK | RGASIRSVGSS | YEVAGRLSLIS |
| Hu/RV-5 | 1B[4] | 2 | DITISKTSLWK | RSASIRSNLST | YEMSGRFSLIS |
| Bo/UK | 7[5] | 6 | DIVVSKTSLWK | RSTSVRSIGST | YEVAGRFSFIS |
| Hu/1076 | 2A[6] | 2 | DIIISKTSLWK | RSSSIKSNISS | YEIAGRFSFIL |
| Po/OSU | 9[7] | 5 | DIVVSKTSLWK | RSSSIRSIGSS | YEISGRFSLIS |
| Po/YM | 9[7] | 11 | DIVVSKTSLWK | RSSSIRSIGSS | YEISGRLSLIS |
| Hu/Wa | 1A[8] | 1 | DIIVSKTSLWK | RNVSIRSNLSA | YEISGRFSLIY |
| Hu/K8 | 3[9] | 1 | DIVISKTSLWK | RLTSVRSVGTI | YEIAGRFSLIS |
| Hu/69M | 4[10] | 8 | DIIVSKTSLWK | RGSSIRSIGSS | YEVAGRFSLIS |
| Bo/B223 | 8[11] | 10 | DIIISKASLWK | RSSTLRSVNSA | YEAAGRFSLIL |
| Eq/H-2 | 4[12] | 3 | DIVISKTSLWK | RNSSIRSVSSS | YEIAGRFSLIS |
| Po/MDR-13 | [13] | 3/5 | DIVISKASLWK | RGSTIRSIGSS | YEINGRFSLIS |
| La/ALA | 11[14] | 3 | DIVVSKTSLWK | RSVSVRSIGGN | YEIAGRFSLIS |
| Ov/Lp14 | [15] | 10 | DIVISKASLWK | RNSSIRSIGST | YEIAGRFSLIS |
| Mu/EB | 10[16] | 3 | DIVVSKTSLWK | RSNTIRSISST | YELAGRFSLIS |
| Av/PO-13 | [17] | 7 | DIVISKTSLWK | RASSIRSNASV | YETAGRFSLIS |
| Eq/L338 | 12[18] | 13 | DILISKASLWK | RSSSIRSIGST | YEIAGRFSLIS |
| Po/PRV 4F | [19] | 3 | DIVISKASPWK | RNSSIRSNVSS | YEIAGRFSLIS |
| Mu/EHP | [20] | 3 | DVVISKASLWK | RSGSIRSISSN | YEVAGRFSLIS |
Species of origin for rotavirus strains are indicated by the following abbreviations: Hu, human; Si, simian; Mu, murine; Ca, canine; La, lapine; Eq, equine; Po, porcine; Bo, bovine; Av, avian; Ov, ovine.
Each P genotype is designated by the number in square brackets. Not all viruses in each P genotype have been characterized into a P serotype by neutralization assays.
Sequences were obtained with the following Genbank accession numbers: SA11 4F, X57319; SA11cl3, M23188; RRV, J03567; RV-5, M32559; UK, M22306; 1076, M88480; OSU, X13190; YM, M63231; Wa, M96825; K8, D90260; 69M, M60600; B223, D13394; H-2, L04638; MDR-13, L07886; ALA, U62149; Lp14, L11599; EB, U08419; PO-13, AB009632; L338, D13399; PRV 4F, L10359; EHP, U08424. The sites of cleavage are indicated in bold.
The three preparations of SA11 4F NTR TLPs used for cryo-EM reconstructions were also analyzed by Western blotting. For each NTR TLP preparation, Western blot analysis showed that VP4 was intact and uncleaved before trypsin treatment (Fig. 4B, 0 μg of trypsin/ml). After trypsin treatment, VP4 was cleaved into VP5∗, VP8∗, and additional cleavage products with apparent molecular weights of 25K and 22K (Fig. 4B and 5A, 25 μg of trypsin/ml).
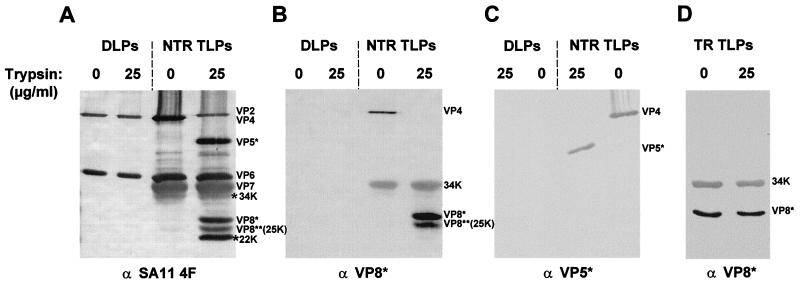
FIG. 5.
SDS-PAGE and Western blot analysis of NTR and TR SA11 4F virus. NTR or TR SA11 4F TLPs or DLPs were purified and treated with increasing concentrations of trypsin as indicated. The proteins were separated by SDS-PAGE, transferred to nitrocellulose, and detected with a hyperimmune anti-SA11 4F TLP mouse serum (A), a VP8∗ (amino acids 160 to 186) peptide antiserum (B and D), or monoclonal antibodies against VP5∗ (C). The location of the individual proteins is indicated on the right of each panel. The approximate molecular weights for the new bands representing previously uncharacterized cleavage products from viruses exposed to trypsin after purification are indicated.
Monoclonal antibodies to VP5∗ and a VP8∗ peptide antiserum were used in a Western blot of the 0- and 25-μg/ml trypsin-treated NTR TLPs to identify the additional cleavage products (Fig. 5). Interestingly, the previously uncharacterized cleavage product with a molecular weight of approximately 25,000 (25K) was detected with the VP8∗ antiserum in trypsin-treated NTR TLPs but was not detected in the trypsin-treated TR TLP preparations (Fig. 5B and D). The VP8∗ peptide antiserum also detected another protein which migrated at a molecular weight of approximately 34,000 (34K) below the VP7 band in NTR and TR TLPs but not in double-layered particles (DLPs) (Fig. 5B and D). The lack of detection of the 34K band in the DLPs confirmed the specificity of the VP8∗ antiserum (Fig. 5B). The pool of VP5∗ monoclonal antibodies detected only VP4 and VP5∗ (Fig. 5C); however, the antiserum against SA11 4F detected 22K- and 24K-apparent-molecular-weight bands, possibly VP5∗ cleavage products (Fig. 4B, 25 and 75 μg of trypsin/ml). The precise locations in the VP4 sequence at which the 34K, VP8∗, and 25K cleavages occurred could not be determined because the N termini of these proteolytic fragments were blocked (data not shown). The VP8∗ and 25K VP8∗-specific cleavage products may be the same proteins observed by Arias et al. in experiments with rotaviruses grown in the absence of trypsin to study trypsin cleavage sites that enhanced infectivity (1).
Trypsin treatment of NTR TLPs with 75 μg of trypsin/ml resulted in almost complete digestion of VP5∗ and the appearance of 27K- and 12K-molecular-weight cleavage products as well as the 26K and 25K VP8∗-specific cleavage products (Fig. 4B, 75 μg of trypsin/ml). For each of the three trypsin-treated NTR TLP preparations we characterized and reconstructed, the appearance of the 25K and 26K VP8∗ cleavage products never varied; however, the amount of each of the 24K and 22K VP5∗ cleavage products did vary among preparations.
Protease sensitivity of VP4 in nonpurified NTR TLPs.
The enhanced protease sensitivity in VP8∗ observed only in the NTR particles suggested that the conformation of VP4 in these particles is not the same as that in the TR particles. One major difference during virus cultivation of the NTR and TR particles is that the latter are released into the medium containing 1 μg of trypsin/ml. It is possible that this immediate exposure to trypsin may induce a critical proteolytic event that renders the VP4 in TR particles more stable and resistant to further trypsinization. In contrast, in the NTR particles without such critical trypsin cleavage, VP4 may be less stable and become further destabilized by subsequent CsCl purification and become more sensitive to trypsin.
To determine if exposure to low concentrations of trypsin prior to CsCl gradient purification could account for the contrasting properties of VP4 in terms of protease resistance and structural stability, we conducted biochemical experiments treating NTR particles with low concentrations of trypsin prior to CsCl purification. In these experiments, the NTR virus released into the medium was collected, the medium was clarified, and the virus was concentrated by ultracentrifugation and then suspended in TNC buffer as described previously. This viral suspension was treated with 0, 1, and 10 μg of trypsin/ml. Samples treated with 0 and 10 μg of trypsin/ml were incubated for 30 min at 37°C, whereas samples treated with 1 μg of trypsin/ml were incubated for 30 min or 1 h. Western blot analysis revealed the additional VP8∗-specific, 25K trypsin cleavage product for NTR particles treated with both 1 and 10 μg of trypsin/ml (Fig. 6). These results clearly indicated that the exposure to low concentrations of trypsin prior to CsCl purification does not account for the contrasting properties of VP4 in the NTR and TR particles.
FIG. 6.
SDS-PAGE and Western blot analysis of NTR SA11 4F virus. The medium from SA11 4F-infected MA104 cells grown in the absence of trypsin with aprotinin was clarified, and the virus was concentrated and then treated with increasing concentrations of trypsin for the time indicated. The proteins were separated by SDS-PAGE, transferred to nitrocellulose, and detected with a hyperimmune anti-SA11 4F TLP mouse serum (A) or a VP8∗ (amino acids 160 to 186) peptide antiserum (B). The location of the individual proteins is indicated on the right. The new VP8∗-specific band [VP8∗∗(25K)] representing a previously uncharacterized cleavage product from NTR exposed to trypsin is indicated.
Stoichiometric determination of the ratio of spike protein on TR and NTR TLPs.
Our inability to visualize the spikes in the reconstructions of NTR particles may indicate either that VP4 is present in low copy numbers in these particles or that the spikes are icosahedrally disordered. To determine which of these possibilities is contributing to the absence of spikes in the reconstructions, we determined the stoichiometric ratio of VP4 or VP5∗ to VP6 for two preparations each of purified radiolabeled TR and NTR particles using PhosphorImager analysis. The Storm system captures images from both strong and weak signals in a single exposure, and the linear dynamic range is 1,000 times greater than that of film. Equal amounts of each preparation were treated with 1, 10, or 25 μg of TPCK-treated trypsin/ml for 30 min at 37°C, and then soybean trypsin inhibitor was added to inhibit further trypsin activity. Five micrograms of each NTR and TR preparation, before and after trypsin treatment, was separated by SDS-PAGE, and the gel was dried and exposed to the phosphorimager screen (Fig. 7A). The quantitative representation of the proteins was determined using ImageQuantNT software. The ratio of VP4 or VP5∗ to VP6 was similar for each of the TR and NTR preparations before and after trypsin treatment, as was found by Chen and Ramig (8). Additionally, the new VP8** 25K cleavage product was clearly seen in the 1-, 10-, and 25-μg/ml trypsin-treated NTR but not TR TLPs (Fig. 7A; 0- and 25-μg/ml trypsin-treated samples are shown). These results indicate that the absence of spikes in the NTR reconstructions is not because of a lack of VP4 on these particles but because the VP4 spikes are icosahedrally disordered.
FIG. 7.
SDS-PAGE and fluorography of 35S-labeled NTR and TR SA11 4F virus. Radiolabeled NTR and TR SA11 4F TLPs were purified and treated with increasing concentrations of trypsin as indicated. The proteins were separated by SDS–10% PAGE (A) or SDS–8% PAGE (B), and the gel was dried and exposed to X-ray film. The location of the individual proteins is indicated on the right of each panel. The approximate molecular weight for the new VP8∗-specific band [VP8∗∗(25K)] representing a previously uncharacterized cleavage product from viruses exposed to trypsin after purification is indicated.
To determine whether the stoichiometry of VP5∗ changed or remained the same with increasing concentrations of trypsin, both NTR and TR radiolabeled particles were treated with various concentrations of trypsin and the ratio of VP5∗ to VP6 was quantitated (Fig. 7). These experiments indicated that the ratio of VP5∗ to VP6 for both TR and NTR remained the same for 0 and 1 μg of trypsin/ml but decreased by approximately 10 and 30% for the 10- and 25-μg/ml trypsin concentrations, respectively. The observed decrease in the amount of VP5∗ correlates with the appearance of two proteolytic products of VP5∗ as detected by Western blots and with the observed increase in the infectivity measured using the same samples. These results in terms of VP5∗ cleavage and the increase in infectivity are consistent with observations made by Arias et al. (1).
DISCUSSION
We present here a comparison of the biologic, biochemical, and structural properties of the SA11 4F strain of rotavirus TLPs grown in the complete absence or in the presence of trypsin. In addition, we examined the effects of adding trypsin exogenously to these TLPs. The rationale for these studies stems from the importance of protease treatment on the infectivity of rotaviruses. The infectivity of rotaviruses increases severalfold upon protease treatment (1, 17). Our aim is to understand the structural basis for trypsin-enhanced infectivity in rotaviruses. Our simple and straightforward strategy was to purify nontrypsinized and trypsinized rotaviruses, to determine the structure, infectivity, and protein composition of the particles before and after additional trypsin treatment, and to examine if changes observed in the structure and protein composition correlated with increased infectivity. We chose the SA11 4F rotavirus strain for these studies because this strain grows to moderate yield in the complete absence of trypsin (4).
Our present study reveals that spikes on NTR SA11 4F, well characterized for the lack of VP4 proteolytic degradation products, cannot be visualized even after including as many as 123 particles in the reconstruction. In contrast, the spikes on TR viruses are readily visualized in reconstructions by combining data from as few as 30 particles. Although the spikes on the NTR particles could be visualized after trypsin treatment of NTR and increased definition of the spikes was obtained with increasing concentrations of trypsin, under no condition were the spikes as well defined as seen in the virus grown with trypsin. Because of these unanticipated results, these experiments were repeated with three different NTR preparations.
The absence of spikes in the reconstruction of the NTR particles indicates either that these particles have a lower copy number of spikes or that the spikes are icosahedrally disordered. The icosahedral averaging imposed by the computational procedures for three-dimensional reconstruction strengthens icosahedrally well-ordered features and weakens features that are icosahedrally inconsistent. Our biochemical analyses clearly indicate that VP4 is not lost in the NTR particles and also that the stoichiometry of VP4 is essentially the same as that found in the TR particles. Therefore, the absence of spike density in the NTR reconstruction strongly indicates that the VP4 proteins are icosahedrally disordered. It is possible that the orientation of the VP4 spike differs considerably within the particle and also between particles and thereby the signal due to the spikes is not coherently reinforced in the reconstruction. Upon the addition of increasing concentrations of trypsin, the spikes do however become visible in the reconstructions, indicating that a small number of spikes become icosahedrally ordered.
Icosahedral disorder in the VP4 spikes of the NTR particles suggests that VP4 is conformationally flexible. In addition, the VP4 proteins in these particles may have a different conformation from that in the TR particles. The enhanced protease sensitivity of VP4 in the NTR particles indeed indicates that the conformations of VP4 in the NTR and TR particles are different. This conformational difference is not dependent on treatment either with low or high trypsin concentrations, before or after CsCl purification. Our results thus indicate that the contrasting conformational properties of VP4 in the NTR and TR particles cannot be accounted for by an extracellular trypsin treatment, thus suggesting that trypsin may have an intracellular role in ensuring the correct conformation of VP4 and its proper assembly onto the particles. Such an effect of trypsin on the de novo particles would require that trypsin added exogenously during virus propagation enters into cells. It is possible that trypsin may enter the cells during virus infection due to altered cell permeability (38) or may coenter cells with virus, as was shown for the toxin α-sarcin (14, 29), and subsequently affect particle assembly.
Although trypsin treatment of NTR results in a better definition of the VP4 spikes, such a treatment did not result in any discernible changes with respect to VP4 spikes in the TR particles. Interestingly, in both NTR and TR particles, a significant and reproducible structural difference was detected in the VP7 trimer at the icosahedral threefold axis upon trypsin treatment. An internal portion of VP7 near the VP7-VP6 interface which is tucked inside the VP7 trimer swings out into the type III channels. Since no VP7 cleavage products were detected, it is likely that this structural change corresponds to a mass translocation or a domain movement in VP7 due to the additional trypsin. The origin of the conformational change in VP7 is not clear; perhaps it is a direct effect of trypsin or an indirect effect of subtle conformational changes in VP4 that are not detected at the present resolution of structural analysis.
Biochemically, additional trypsin treatment of NTR and TR particles also resulted in the identification of previously unrecognized cleavage products of VP5∗ that coincided with a loss of VP5∗ density and a significant rise in viral infectivity. These cleavage sites in VP5∗, at lysine 258, arginine 582, and putatively at arginine 467, are conserved in all group A rotavirus strains, are also observed in the NTR particles, and take place only after the initial cleavage of VP4 into VP5∗ and VP8∗. It is likely that the initial cleavage of VP4 at positions 231, 241, or 247 allows these cleavage sites to become accessible to further protease treatment. Previous studies have shown that specific trypsin cleavage after arginine 247 correlates with virus activation for cell entry and is required for induction of fusion-from-without by VLPs (1, 21). From our present work, it is unclear whether the structural change in VP7 or the generation of these newly described VP5∗-specific cleavage products may also be responsible for the concurrent rise in infectivity since detection of these minor cleavage products is dependent on the use of a hyperimmune serum raised against purified TR TLPs. A systematic examination will be necessary to determine what early step of the infectious cycle, i.e., attachment, penetration, or uncoating, is enhanced by additional trypsin cleavage of TLPs. This can be tested by mutation of these sites in VP4 and transcapsidating native core particles with expressed VP4-VP6 complexes (7, 21).
In summary, we have shown here that the conformation and assembly of VP4 spikes are significantly altered in TLPs grown in the absence of trypsin. In these viruses the spikes are icosahedrally disordered and more protease sensitive, indicating that they are conformationally different from the VP4 spikes, which are well ordered in TLPs grown in the presence of trypsin. The contrasting properties of VP4 in the TLPs grown in the absence of trypsin cannot be simply accounted for by the exogenous treatment of TLPs with trypsin, thereby suggesting that trypsin cleavage may have an intracellular role to play in the proper assembly of VP4. The initial trypsin cleavage event may be critical for conferring proper structural characteristics to VP4, for subsequent proteolysis, as also shown by others, and perhaps for structural alterations in VP7, as shown here, for enhancement of infectivity. Based on these studies, we hypothesize that the presence of trypsin during virus cultivation by a mechanism that is not yet clear imparts order to the spikes and that trypsin treatment of these ordered spikes makes virus entry into cells more efficient by facilitating direct penetration of the plasma membrane.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI 36040 (B.V.V.P.), DK 30144 (M.K.E.), and AI 16687 (R.F.R.).
REFERENCES
- 1.Arias C F, Romero P, Alvarez V, López S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J M, Tian P, Zeng C Q Y, Morris A P, Estes M K. Age dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 3.Beisner B, Kool D A, Marich A, Holmes I H. Characterization of G serotype dependent non-antibody inhibitors of rotavirus in normal mouse serum. Arch Virol. 1998;143:1277–1294. doi: 10.1007/s007050050375. [DOI] [PubMed] [Google Scholar]
- 4.Burns J W, Chen D, Estes M K, Ramig R F. Biological and immunological characterization of a simian rotavirus SA11 variant with an altered genome segment 4. Virology. 1989;169:427–435. doi: 10.1016/0042-6822(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 5.Burns J W, Greenberg H B, Shaw R D, Estes M K. Functional and topographical analyses of epitopes on the hemagglutinin (VP4) of the simian rotavirus SA11. J Virol. 1988;62:2164–2172. doi: 10.1128/jvi.62.6.2164-2172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Burns J W, Estes M K, Ramig R F. Phenotypes of rotavirus reassortants depend upon the recipient genetic background. Proc Natl Acad Sci USA. 1989;86:3743–3747. doi: 10.1073/pnas.86.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D, Ramig R F. Rescue of infectivity by sequential in vitro transcapsidation of rotavirus core particles with inner capsid and outer capsid proteins. Virology. 1993;194:743–751. doi: 10.1006/viro.1993.1315. [DOI] [PubMed] [Google Scholar]
- 8.Chen D Y, Ramig R F. Determinants of rotavirus stability and density during CsCl purification. Virology. 1992;186:228–237. doi: 10.1016/0042-6822(92)90077-3. [DOI] [PubMed] [Google Scholar]
- 8a.Ciarlet M, Estes M K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol. 1999;80:943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 9.Ciarlet M, Hidalgo M, Gorziglia M, Liprandi F. Characterization of neutralization epitopes on the VP7 surface protein of serotype G11 porcine rotaviruses. J Gen Virol. 1994;75:1867–1873. doi: 10.1099/0022-1317-75-8-1867. [DOI] [PubMed] [Google Scholar]
- 10.Coulson B S, Londrigan S L, Lee D J. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc Natl Acad Sci USA. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford S E, Labbe M, Cohen J, Burroughs M H, Zhou Y-J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowther R A. Three-dimensional reconstruction and the architecture of spherical viruses. Endeavour. 1971;30:124–129. doi: 10.1016/0160-9327(71)90038-x. [DOI] [PubMed] [Google Scholar]
- 13.Crowther R A, DeRosier D J, Klug A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc R Soc Lond. 1970;317:319–340. doi: 10.1098/rspb.1972.0068. [DOI] [PubMed] [Google Scholar]
- 14.Cuadras M A, Arias C F, López S. Rotaviruses induce an early membrane permeabilization of MA104 cells and do not require a low intracellular Ca2+ concentration to initiate their replication cycle. J Virol. 1997;71:9065–9074. doi: 10.1128/jvi.71.12.9065-9074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubochet J, Adrian M, Chang J J, Homo J C, Lepault J, McDowall A W, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 16.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1625–1655. [Google Scholar]
- 17.Estes M K, Graham D Y, Mason B B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuentes-Pananá E M, López S, Gorziglia M, Arias C F. Mapping the hemagglutination domain of rotaviruses. J Virol. 1995;69:2629–2632. doi: 10.1128/jvi.69.4.2629-2632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuhara N, Yoshie O, Kitaoka S, Konno T. Role of VP3 in human rotavirus internalization after target cell attachment via VP7. J Virol. 1988;62:2209–2218. doi: 10.1128/jvi.62.7.2209-2218.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller S D. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert J M, Greenberg H B. Cleavage of Rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J Virol. 1998;72:5323–5327. doi: 10.1128/jvi.72.6.5323-5327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewish M J, Takada Y, Coulson B S. Integrins alpha2beta1 and alpha4beta1 can mediate SA11 rotavirus attachment and entry into cells. J Virol. 2000;74:228–236. doi: 10.1128/jvi.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isa P, López S, Segovia L, Arias C F. Functional and structural analysis of the sialic acid-binding domain of rotaviruses. J Virol. 1997;71:6749–6756. doi: 10.1128/jvi.71.9.6749-6756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaljot K, Shaw R D, Rubin D H, Greenberg H B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988;62:1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keljo D J, Kuhn M, Smith A. Acidification of endosomes is not important for the entry of rotavirus into the cell. J Pediatr Gastroenterol Nutr. 1988;7:257–263. doi: 10.1097/00005176-198803000-00016. [DOI] [PubMed] [Google Scholar]
- 26.Kirkwood C D, Bishop R F, Coulson B S. Attachment and growth of human rotaviruses RV-3 and S12/85 in Caco-2 cells depend on VP4. J Virol. 1998;72:9348–9352. doi: 10.1128/jvi.72.11.9348-9352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawton J A, Prasad B V V. Automated software package for icosahedral virus reconstruction. J Struct Biol. 1996;116:209–215. doi: 10.1006/jsbi.1996.0032. [DOI] [PubMed] [Google Scholar]
- 28.Lawton J A, Zeng C Q Y, Mukherjee S K, Cohen J, Estes M K, Prasad B V V. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liprandi F, Moros Z, Gerder M, Ludert J E, Pujol F H, Ruiz M-C, Michelangeli F, Charpilienne A, Cohen J. Productive penetration of rotavirus in cultured cells induces coentry of the translation inhibitor alfa-sarcin. Virology. 1997;237:430–438. doi: 10.1006/viro.1997.8803. [DOI] [PubMed] [Google Scholar]
- 30.Ludert J E, Feng N, Yu J H, Broome R L, Hoshino Y, Greenberg H B. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vivo and in vitro. J Virol. 1996;70:487–493. doi: 10.1128/jvi.70.1.487-493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maass D R, Atkinson P H. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J Virol. 1990;64:2632–2641. doi: 10.1128/jvi.64.6.2632-2641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason B B, Graham D Y, Estes M K. Biochemical mapping of the simian rotavirus SA11 genome. J Virol. 1983;46:413–423. doi: 10.1128/jvi.46.2.413-423.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 34.Mattion N M, Estes M K. Sequence of a rotavirus gene 4 associated with unique biological properties. Arch Virol. 1991;120:109–113. doi: 10.1007/BF01310953. [DOI] [PubMed] [Google Scholar]
- 35.Méndez E, Arias C F, López S. Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J Virol. 1996;70:1218–1222. doi: 10.1128/jvi.70.2.1218-1222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moos M J, Nguyen N Y, Liu T Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988;263:6005–6008. [PubMed] [Google Scholar]
- 37.Nandi P, Charpilienne A, Cohen J. Interaction of rotavirus particles with liposomes. J Virol. 1992;66:3363–3367. doi: 10.1128/jvi.66.6.3363-3367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obert G, Peiffer I, Servin A L. Rotavirus-induced structural and functional alterations in tight junctions of polarized intestinal Caco-2 cell monolayers. J Virol. 2000;74:4645–4651. doi: 10.1128/jvi.74.10.4645-4651.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira H G, Azeredo R S, Fialho A M, Vidal M N. Genomic heterogeneity of simian rotavirus SA11. J Gen Virol. 1984;65:815–818. doi: 10.1099/0022-1317-65-4-815. [DOI] [PubMed] [Google Scholar]
- 40.Prasad B V V, Burns J W, Marietta E, Estes M K, Chiu W. Localization of VP4 neutralization sites in rotavirus by 3D cryo-electron microscopy. Nature. 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 41.Prasad B V V, Estes M K. Molecular basis of rotavirus replication: structure-function correlations. In: Chiu W, Burnett M, Garcea R, editors. Structural biology of viruses. Oxford, United Kingdom: Oxford University Press; 1997. pp. 239–278. [Google Scholar]
- 42.Prasad B V V, Estes M K. Electron cryomicroscopy and computer image processing techniques: use in structure-function studies of rotavirus. In: Gray J, Desselberger U, editors. Rotaviruses: methods and protocols. Vol. 34. Totowa, N.J: Humana Press Inc.; 2000. pp. 9–31. [DOI] [PubMed] [Google Scholar]
- 43.Prasad B V V, Rothnagel R, Zeng C Q Y, Jakana J, Lawton J A, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 44.Prasad B V V, Wang G J, Clerx J P M, Chiu W. Three-dimensional structure of rotavirus. Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz M-C, Abad M J, Charpilienne A, Cohen J, Michelangeli F. Cell lines susceptible to infection are permeabilized by cleaved and solubilized outer layer proteins of rotavirus. J Gen Virol. 1997;78:2883–2893. doi: 10.1099/0022-1317-78-11-2883. [DOI] [PubMed] [Google Scholar]
- 46.Sabara M, Gilchrist J E, Hudson G R, Babiuk L A. Preliminary characterization of an epitope involved in neutralization and cell attachment protein that is located on the major bovine rotavirus glycoprotein. J Virol. 1985;53:58–66. doi: 10.1128/jvi.53.1.58-66.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw A L, Rothangel R, Chen D, Ramig R F, Chiu W, Prasad B V V. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993;74:693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Heel M. Similarity measures between images. Ultramicroscopy. 1987;21:95–99. [Google Scholar]
- 49.Xu Z, Woode G N. Studies on the influence of the VP7 gene on rotavirus replication. Virology. 1994;198:394–398. doi: 10.1006/viro.1994.1049. [DOI] [PubMed] [Google Scholar]
- 50.Yeager M, Berriman J A, Baker T S, Bellamy A R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeager M, Dryden K A, Olson N H, Greenberg H B, Baker T S. 3D structure of rhesus rotavirus by cryoelectron microscopy and image reconstruction. J Cell Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z H, Prasad B V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]