Abstract
Hepatitis C virus (HCV), a major cause of liver disease worldwide, is frequently resistant to the antiviral alpha interferon (IFN). The HCV nonstructural 5A (NS5A) protein has been implicated in HCV antiviral resistance in many studies. NS5A antagonizes the IFN antiviral response in vitro, and one mechanism is via inhibition of a key IFN-induced enzyme, the double-stranded-RNA-activated protein kinase (PKR). In the present study we determined if NS5A uses other strategies to subvert the IFN system. Expression of full-length NS5A proteins from patients who exhibited a complete response (FL-NS5A-CR) or were nonresponsive (FL-NS5A-NR) to IFN therapy in HeLa cells had no effect on IFN induction of IFN-stimulated gene factor 3 (ISGF-3). Expression of mutant NS5A proteins lacking 110 (NS5A-ΔN110), 222 (NS5A-ΔN222), and 334 amino-terminal amino acids and mutants lacking 117 and 230 carboxy-terminal amino acids also had no effect on ISGF-3 induction by IFN. Expression of FL-NS5A-CR and FL-NS5A-NR did not affect IFN-induced STAT-1 tyrosine phosphorylation or upregulation of PKR and major histocompatibility complex class I antigens. However, NS5A expression in human cells induced interleukin 8 (IL-8) mRNA and protein, and this effect correlated with inhibition of the antiviral effects of IFN in an in vitro bioassay. NS5A induced transcription of a reporter gene driven by the IL-8 promoter, and the first 133 bp of the IL-8 promoter made up the minimal domain required for NS5A transactivation. NS5A-ΔN110 and NS5A-ΔN222 stimulated the IL-8 promoter to higher levels than did the full-length NS5A protein, and this correlated with increased nuclear localization of the proteins. Additional mutagenesis of the IL-8 promoter suggested that NF-κB and AP-1 were important in NS5A-ΔN222 transactivation in the presence of tumor necrosis factor alpha and that NF–IL-6 was inhibitory to this process. This study suggests that NS5A inhibits the antiviral actions of IFN by at least two mechanisms and provides the first evidence for a biological effect of the transcriptional activity of the NS5A protein. During HCV infection, viral proteins may induce chemokines that contribute to HCV antiviral resistance and pathogenesis.
Chronic hepatitis C virus (HCV) infection is a significant clinical problem affecting an estimated 150 million individuals worldwide and 3.9 million individuals in the United States. About 85% of people infected with HCV develop chronic infection, and approximately 70% of patients develop histological evidence of chronic liver disease (41).
Alpha interferon (IFN) is a Food and Drug Administration-approved treatment for chronic HCV infection. Only 8 to 12% of patients with HCV genotype 1 have a sustained clinical virological response to IFN therapy (4, 43, 61). Recently, combination therapy with interferon and the guanosine analogue ribavirin was shown to be superior to IFN monotherapy in producing sustained biochemical and virological responses (9, 45, 62). However, despite the significant improvement in rates of sustained response, as many as 60% of patients with high-titer HCV genotype 1 infection are nonresponsive to combination therapy.
When IFN binds to its receptor, two receptor-associated tyrosine kinases of the STAT/JAK family, Tyk2 and Jak1, become activated. These activated kinases phosphorylate STAT-1 and STAT-2 on a single conserved tyrosine residue (8). STAT-1 and STAT-2 form heterodimers and combine with the p48 protein to form an active transcription factor known as IFN-stimulated gene factor 3 (ISGF-3). ISGF-3 binds to a common element termed the interferon-stimulated response element (ISRE), found in the promoter regions of all IFN-stimulated genes, whereupon transcription occurs. Expression of the entire HCV polyprotein has been shown to inhibit IFN-induced STAT/JAK signaling in human U2-OS osteosarcoma cells (25). It was not reported which HCV protein was responsible for this effect.
Recent studies have led to exciting discoveries in the emerging research area of the roles of HCV proteins in antiviral resistance. Two examples are the interaction of the HCV nonstructural 5A protein (NS5A) and the second envelope (E2) glycoprotein with the IFN-induced, double-stranded-RNA-activated protein kinase (PKR). PKR is one of the major intracellular enzymes that mediate the antiviral action of IFN (32). Both the NS5A and E2 proteins inhibit PKR activity, which is postulated to allow HCV replication to continue in the presence of an IFN-induced antiviral response (19, 77). For E2, the interaction with PKR requires a 12-amino-acid domain (77), which is a highly stable element that does not mutate during antiviral therapy (1, 57). For NS5A, the interaction with PKR requires the IFN-sensitivity-determining region (ISDR) on NS5A, a region that is associated with clinical IFN resistance in Japanese and Spanish patients (6, 12, 13, 36, 67). Since accumulation of mutations in the ISDR also prevented the NS5A-PKR interaction (17), ISDR-dependent inhibition of PKR seemed to provide a molecular explanation of HCV resistance to IFN. However, subsequent studies have shown that there is no correlation between ISDR mutations and IFN response in France, Germany, Italy, and the United States (7, 11, 15, 22, 27, 56, 60, 64, 65, 72, 73, 79), which has caused considerable debate (26). In a prospective study that sequenced entire NS5A genes, we demonstrated that the response to antiviral therapy correlates with mutations in two regions in the C terminus of the NS5A protein (52), the previously described V3 region (11, 30), and a region that that overlaps the proline-rich region of NS5A.
NS5A has also been intensely studied in vitro, and we (58) and others (18, 55, 71) have established in vitro bioassays showing that expression of NS5A in vitro inhibits the antiviral actions of IFN against IFN-sensitive viruses. Intriguingly, these in vitro bioassays have shown that the regions on NS5A required for inhibiting PKR activity may not be required for inhibiting antiviral actions of IFN (55, 58). Using a similar bioassay, a recent study found that expression of the entire HCV polyprotein also inhibited the antiviral actions of IFN independently of PKR (14). Cumulatively, these studies suggest that NS5A-mediated inhibition of the IFN system may involve mechanisms other than targeting of PKR.
The CXC chemokine interleukin-8 (IL-8) is a 71-amino-acid chemotactic cytokine. IL-8 is induced primarily by the cytokines IL-1 and tumor necrosis factor alpha (TNF-α) and is produced by many cells, including fibroblasts and hepatocytes. IL-8 is a proinflammatory cytokine that induces neutrophil, T-lymphocyte, and basophil chemotaxis and degranulation (49). IL-8 has been shown to be a principal mediator of the inflammatory response to many viruses and bacteria and has recently been shown to inhibit the antiviral actions of IFN-α in vitro. It was demonstrated that addition of recombinant human IL-8 could rescue the replication of encephalomyocarditis virus (EMCV) in the presence of IFN (33).
In this study, we studied the interaction of NS5A with the STAT/JAK pathway and the IL-8 system as potential mechanisms for inhibition of the antiviral actions of IFN.
MATERIALS AND METHODS
Cells, cytokines, and viruses.
Human HeLa Tet-Off cells were obtained from Clontech (Palo Alto, Calif.). HeLa Tet-Off cells were grown in Hela Tet-Off media: Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS; Hyclone, Logan, Utah), 2 mM l-glutamine (Gibco-BRL, Grand Island, N.Y.), 1× PSF (100 U of penicillin G per ml, 100 μg of streptomycin per ml, 0.25 μg of fungizone [Gibco-BRL] per ml), 200 μg of G418 (Calbiochem, San Diego, Calif.) per ml, 1 μg of tetracycline or doxycycline (Sigma, St. Louis, Mo.) per ml. In this study, two stable human HeLa cell lines expressing full-length NS5A from genotype 1b-infected IFN-nonresponsive and completely responsive patients (termed FL-NS5A-NR and FL-NS5A-CR, respectively) were used. NS5A expression in both cell lines was under the control of the tetracycline-regulated promoter (24) and was generated as described previously (58). Cells were grown in HeLa Tet-Off media supplemented with 100 μg of hygromycin B (Calbiochem) per ml. Murine L929 cells (obtained from the American Type Culture Collection, Manassas, Va.) were grown in Dulbecco's modified Eagle medium containing 10% FBS, 2 mM l-glutamine, 1× PSF, and 1 mM nonessential amino acids (Gibco-BRL). All cells were grown in a humidified 37°C 5% CO2 incubator (Forma, Marietta, Ohio). Recombinant IFN (Intron A; kindly provided by Schering-Plough, Kenilworth, N.J.) or human leukocyte IFN (Sigma) was used in some experiments. The stocks had activities of 6 × 106 U per ml. Recombinant human IL-8 and TNF-α (R&D Systems, Minneapolis, Minn.) at stock concentrations of 100 μg/ml were also used in some experiments. For virus rescue assays, EMCV (American Type Culture Collection) was used. The virus stock had a titer of 109 PFU per ml.
Plasmids.
A full-length NS5A construct obtained from a genotype 1b IFN-nonresponsive patient was cloned into pTRE as described previously (58) and designated pTRE-FL-NS5A-NR. A series of amino- and carboxy-terminal deletion mutants were generated by PCR with Pfu polymerase, using pTRE-FL-NS5A-NR as a template. The primers NS5A-1b-335 (5′ TTG ACC ATG GCG CTG TGG CGA GTG GCT G), NS5A-1b-671 (5′ TTG ACC ATG GGG TCC CCC CCC TCC TTG), and NS5A-1b-1007 (5′ TTG ACC ATG GAC TAC GTC CCT CCG GG) were used in separate PCR amplifications with NS5A-1b-1363 (5′ TGT CTA GAT TAG GAC ATT GAG CAG CAG ACG) to generate NS5A proteins lacking 110, 222, and 334 amino-terminal amino acids. The primers NS5A-1b-1009 (5′ TCT CTA GAT TAG TCC GGG TCT TTC CAG GG) and NS5A-1b-670 (5′ TGT CTA GAT TAC CTG GCC AGC TTA CGC TTC G) were used in separate PCR amplifications with NS5A-1b-1 (5′ TGA GGA TCC ACC ATG GGC TCC GGC TCG TGG CTA) to generate NS5A proteins lacking 117 and 230 carboxy-terminal amino acids. The PCR products were digested with NcoI and XbaI and cloned into the NcoI and XbaI sites of pTRE-pUC (see below). The resulting constructs were designated pTRE-NS5A-ΔN110, pTRE-NS5A-ΔN222, pTRE-NS5A-ΔN334, pTRE-NS5A-ΔC117, and pTRE-NS5A-ΔC230, respectively. A full-length NS5A construct obtained from a patient with genotype 1b who showed a complete response to IFN was cloned into pMOS Blue (Amersham-Pharmacia, Piscataway, N.J.) as described previously (11). The NS5A insert was subcloned into pTRE as follows. First, the SpeI-SpeI multiple-cloning site from pUC21 was ligated into the XbaI site of pTRE to generate pTRE-pUC. NS5A was released from pMOS Blue first by SphI digestion, which was subsequently blunted, and then by NcoI digestion. NS5A was directly ligated into pTRE-pUC at the NcoI and EcoRV sites. This plasmid was designated pTRE-FL-NS5A-CR. Plasmid constructs containing the full-length (542-luc) and truncated (133-luc and 98-luc) IL-8 promoters controlling the expression of the luciferase gene were generated as described previously (50). The 133-luc plasmid was also mutated at the NF-κB, AP-1, and NF–IL-6 binding sites to generate plasmids NF-κB–luc, AP-1–luc, and NF–IL-6–luc, respectively, as described previously (50). All plasmids were isolated with an Endofree Mega plasmid kit (Qiagen, Santa Clarita, Calif.). pTRE-E3L expresses the vaccinia virus E3L protein, a known inhibitor of the IFN antiviral response (5), and was cloned as described previously (58). pSVE1A expresses the adenovirus type 5 E1A protein. pcDNA/NEO/PKR expresses wild-type PKR and was cloned as previously described (46). Cells were transfected by electroporation as described previously (58).
Trans rescue assay.
Cells (5 × 105 per well) (stable cell line or transiently transfected cells) were plated in a six-well tissue culture plate and grown in induction media in the presence or absence of 2 μg of tetracycline per ml for 24 to 48 h. Recombinant IFN (Intron A) was then added at various concentrations for 24 h to induce the IFN antiviral state. Infection with EMCV at a multiplicity of infection (MOI) of 0.01 was then performed, and supernatants were harvested at various times postinfection. Changes in infectious virus yields were measured by a standard virus plaque assay with L929 cells as described previously (58).
Western blot analysis.
HeLa Tet-Off cell lines expressing FL-NS5A-NR and FL-NS5A-CR were induced and then treated with human leukocyte IFN at various time points. Protein extracts were prepared in lysis buffer I (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 400 mM NaCl, 1% [vol/vol] NP-40, 1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 U of aprotinin per ml, 10 U of leupeptin per ml, and 25 mM NaF). Protein concentration was determined with Coomassie Protein Assay reagent (Pierce Chemicals, Rockford, Ill.). Cell extracts were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitocellulose. Proteins were detected with primary antibodies to human PKR (a gift from Ara Hovanessian, Pasteur Institute), β-tubulin (Sigma), phosphorylated STAT-1 (Biosource International, Camarillo, Calif.), and human antiserum from HCV-positive patients for the detection of NS5A (58). Secondary antibodies to mouse, rabbit, and human were conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, Pa.) and visualized with a chemiluminescence substrate (Pierce Chemicals).
Flow cytometry and immunofluoresence analyses.
HeLa Tet-Off cells expressing FL-NS5A-NR and FL-NS5A-CR were induced to express NS5A and then treated with human leukocyte IFN for 24 h. Cells were detached with 50 mM EDTA in phosphate-buffered saline (PBS). They were washed twice in PBS and then incubated on ice for 30 min with anti-HLA-A, -B, and -C fluorescein isothiocyanate-conjugated antibody in fluorescence-activated cell sorter (FACS) buffer (1× PBS, 1% FBS, and 0.02% [wt/vol] NaN3). Untreated cells were incubated with anti-mouse myeloma protein immunoglobulin G1-FITC-conjugated antibody as an isotype control (PharMingen, San Diego, Calif.). Cells were then washed three times with FACS buffer and then analyzed by flow cytometry. HeLa cells expressing FL-NS5A-NR, NS5A-ΔN222, and NS5A-ΔC117 were fixed and stained with a monoclonal NS5A antibody (Austral Biologicals, San Ramon, Calif.), followed by Cy3-coupled goat anti-mouse secondary antibodies (Amersham, Pharmacia) as described previously (58). Cells were visualized with a Delta Vision immunofluorescence microscope (Applied Precision, Issaquah, Wash.).
Determination of IL-8 mRNA.
IL-8 mRNA was detected and quantitated by reverse transcription-PCR (RT-PCR) and an RNase protection assay. For RT-PCR, cytoplasmic RNA was isolated from cells expressing or not expressing NS5A in the absence and presence of Intron-A using the RNeasy system (Qiagen). RNA samples were subsequently treated with DNase I to remove contaminating DNA. cDNA was synthesized using random hexanucleotides (pdN6) as described previously (60). To quantitate the amount of IL-8 mRNA, a semiquantitative PCR for IL-8 mRNA was performed using serial dilutions of input cDNA. The sequence of the upstream primer was 5′ ATG ACT TCC AAG CTG GCC GTG GCT-3′, and that of the downstream primer was 5′ TCT CAG CCC TCT TCA AAA ACT TCT C-3′. The primers span introns on the IL-8 gene and produce a PCR fragment of 289 bp if mRNA is amplified. If genomic DNA is amplified, then a fragment of approximately 1,500 bp is generated. For RNase protection assays, the assays were performed according to protocols of the manufacturer (Riboquant System; PharMingen). Cellular RNAs were hybridized to radiolabeled single-stranded riboprobe cocktails and digested with RNase. Protected RNAs were electrophoretically separated on denaturing urea-acrylamide gels, and autoradiography was performed.
ELISA.
To measure IL-8 protein levels in cell culture supernatants, a commercially available double-sandwich enzyme-linked immunosorbent assay (ELISA) was used according to the specifications of the manufacturer (Endogen, Woburn, Mass.).
Luciferase reporter gene experiments.
To determine the ability of NS5A to transactivate the IL-8 promoter, luciferase reporter gene experiments were performed. Luciferase reporter plasmids under the control of the IL-8 promoter were either transfected alone into NS5A-expressing cell lines or cotransfected with pTRE-FL-NS5A-NR, or pTRE plasmids were transfected into HeLa Tet-Off cells. Transfected cells were split into triplicate wells and grown in the absence or presence of tetracycline for various times to induce or repress NS5A expression, respectively. Cell lysates were prepared with the Luciferase Assay System (Promega, Madison, Wis.). Luciferase activity was determined by mixing 20 μl of cell lysate with 100 μl of luciferase assay buffer on an Autolumat LB 953 automated luminometer (Berthold Instruments, Bad Wildbad, Germany).
ISGF-3 electromobility shift assays.
NS5A proteins were transfected into HeLa Tet-Off cells and grown in the absence and presence of tetracycline to induce and repress NS5A expression, respectively, for 24 h. During the course of these experiments, we noticed that HeLa cells expressed low levels of p48, an essential component of ISGF-3 treated with IFN. Thus, after the 24-h induction of NS5A expression, all cells were treated with 5 ng of recombinant IFN-γ (R&D Systems) for 15 h. This has been demonstrated to increase the level of p48 (40). Cells were then treated or not treated with 500 U of IFN (Intron-A; Schering-Plough) per ml for 15 min. Crude cytoplasmic and nuclear extracts were prepared using a procedure modified from Dignam et al. (10). Briefly, cells were washed twice with cold PBS and scraped into 1.0 ml of RSB (10 mM Tris-Cl [pH 7.4], 10 mM NaCl, 3 mM MgCl2). Cells were pelleted by centrifugation at 4,000 rpm for 3 min in an Eppendorf model 5415C centrifuge (Brinkmann Instruments, Westbury, N.Y.), and the swelled cell volume was estimated. Cells were resuspended and lysed in a 2× concentration of the swelled cell volume of RSBG40 (RSB plus 10% glycerol, 0.25% NP-40, 0.5 mM DTT, 0.5 mM PMSF). Nuclei were pelleted by centrifugation at 10,000 rpm for 5 min, and the supernatant was saved as a crude cytosolic extract. The packed nuclear volume was estimated, and nuclei were resuspended in a 1× concentration of the packed nuclear volume of extraction buffer (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, 0.5 mM PMSF, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 1 mM sodium vanadate) and incubated on ice for 30 min. Debris were pelleted by centrifugation at 13,000 rpm for 5 min, and the supernatant was saved as crude nuclear extract. Equal amounts of cell extracts were then incubated with 2 × 105 to 5 × 105 cpm of the double-stranded probe.
The probe corresponded to the ISRE of the ISG-15 gene and had the sequence 5′ GAT CCT CGG GAA AGG GAA ACC GAA ACT GAA GCC-3′. The ISRE probe was radiolabeled with [α-32P]dGTP and [α-32P]dATP using Klenow enzyme. Following incubation at room temperature for 20 min in a hybridization buffer consisting of 40 mM KCl, 20 mM K+ HEPES [pH 7.6], 1 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 40 mg of Ficoll per ml, 0.32 mg of poly(dI-dC) per ml, 20 μg of pGEM plasmid DNA per ml, and 4 mM AMP, the DNA-protein complexes were separated on nondenaturing 6% acrylamide–20 mM Tris-borate-EDTA gels at 4°C and detected by autoradiography. To ensure the specificity of the complex, we performed competition experiments with unlabeled double-stranded oligonucleotides in the binding reaction mixture at a 50× molar excess. To further ensure that the DNA-protein complex was indeed ISGF-3, binding reactions were also performed in the presence of a p48 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), which resulted in the formation of a supershift of the ISGF-3 complex.
Statistics.
IL-8 levels in culture supernatants and in the reporter gene assay results were determined in triplicate, and data are expressed as means ± standard deviations. Student's t tests were used to compare differences in IL-8 levels and luciferase activities in cell cultures.
Nucleotide sequence accession numbers.
The accession numbers for the sequences of FL-NS5A-NR and FL-NS5A-CR are AF034151 and AY008261, respectively.
RESULTS
Effects of NS5A expression on IFN signal transduction.
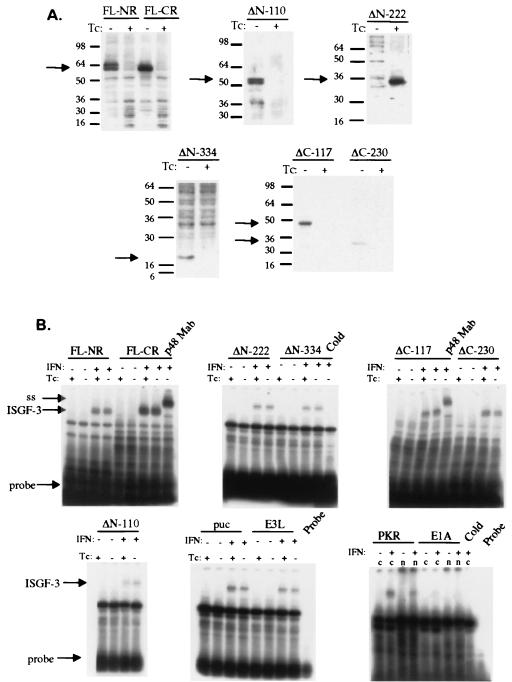
Since the NS5A protein has been controversially implicated in inhibition of the IFN system in clinical and in vitro studies, the following experiments were performed to investigate other potential mechanisms of IFN resistance in chronic hepatitis C. In theory, a viral protein can inhibit the IFN system at the receptor, signal transduction, transcriptional, and posttranscriptional levels. We first examined the effect of NS5A expression on IFN signal transduction, since a previous report had shown that expression of all HCV proteins inhibits signal transduction (25). Figure 1 presents the results of expression of full-length and mutant NS5A proteins on IFN induction of ISGF-3. We have previously established a tissue culture system to express NS5A under the control of the tetracycline-regulated promoter (24). In this system, NS5A can be selectively induced or repressed by the removal or addition of tetracycline to the tissue culture medium, and intermediate levels of NS5A protein expression can be achieved by titrating the dose of tetracycline (58). We cloned full-length NS5A proteins from two patients, one who was a complete, long-term responder (FL-NS5A-CR) and one who was a nonresponder (FL-NS5A-NR) to IFN therapy. Using FL-NS5A-NR as a template, we generated a series of amino- and carboxy-terminal deletions of the proteins, as described in Materials and Methods. Panel A depicts expression of full-length NS5A proteins from the complete responder and nonresponder patients and of mutant NS5A proteins lacking 110 (NS5A-ΔN110), 222 (NS5A-ΔN222), and 334 (NS5A-ΔN334) amino-terminal amino acids and 117 (NS5A-ΔC117) and 230 (NS5A-ΔC230) carboxy-terminal amino acids. All proteins of the expected sizes were expressed, and the expression was tightly regulated by the addition of tetracycline to the tissue culture media. Figure 1B depicts the effects of NS5A on IFN induction of ISGF-3, the principal transcription factor induced by IFN. As can be seen, expression of FL-NS5A-NR, FL-NS5A-CR, and all amino- and carboxy-terminal NS5A mutant proteins had no effect on IFN induction of ISGF-3. For controls, we expressed the vaccinia virus E3L protein, a known inhibitor of the IFN system (5, 58), and PKR and also found no effect on ISGF-3 induction. Transfection of the parental pTRE-pUC plasmid also had no effect on ISGF-3 induction. Expression of the adenovirus E1A protein, previously shown to inhibit STAT/JAK signaling (39), resulted in a near complete inhibition of ISGF-3. Expression of E3L, PKR, and E1A was verified by Western blot analysis (data not shown). The specificity of the electromobility shift was verified in two ways. First, the inclusion of a monoclonal antibody to the p48 protein produced a supershift. Second, the inclusion of excess unlabeled oligonucleotide completely inhibited complex formation. Thus, these data indicate that expression of the HCV NS5A protein does not affect STAT/JAK signaling via induction of ISGF-3.
FIG. 1.
Effect of NS5A expression on IFN signal transduction. (A) Tetracycline (Tc)-regulated expression of full-length and mutant NS5A proteins. FL-NR denotes a full-length NS5A construct derived from an IFN-nonresponsive patient, while FL-CR denotes a full-length NS5A construct derived from an IFN-responsive patient. ΔN-110, ΔN-222, ΔN-334, ΔC-117, and ΔC-230 represent NS5A deletion mutants lacking 110, 222, 334, amino acids from the amino terminus and 117 and 230 amino acids from the carboxy terminus, respectively. Protein positions are shown by arrows. Western blots were probed with HCV-infected patient serum as described in Materials and Methods. (B) Effect of NS5A expression on ISGF-3 induction. Plasmids were transiently transfected into HeLa Tet-Off cells, grown in the absence and presence of tetracycline to induce and repress NS5A, respectively, and treated with IFN to induce ISGF-3. Protein extracts were hybridized to a 32P-labeled oligonucleotide corresponding to the consensus ISRE. “puc” represents a control transfection with the pTRE-puc parental plasmid with no insert. “E3L,” “PKR,” and “E1A” represent expression of the E3L, PKR, and E1A proteins, respectively. The p48 monoclonal antibody used to form supershifts (denoted by “ss” and an arrow) is denoted by “p48 Mab”. “n” and “c” represent nuclear and cytoplasmic extracts, respectively. ISGF-3 is induced only with IFN treatment and is indicated with arrows.
To further verify this result, we determined the effect of NS5A expression on IFN-induced changes in protein phosphorylation and gene expression. We performed Western blot analysis on protein extracts from cells expressing or not expressing FL-NS5A-NR or FL-NS5A-CR and treated with IFN for various times. Figure 2A depicts the results. The top panels of Fig. 2A demonstrate that expression of FL-NS5A-NR and FL-NS5A-CR is again tightly regulated by doxycycline in this experiment. The membranes were then probed with antibodies for phosphorylated STAT-1 (second panels), PKR (third panels), and, as a control, β-tubulin (lower panels). As can be seen, expression of the FL-NS5A-NR and FL-NS5A-CR proteins had no effect on IFN-induced tyrosine phosphorylation of STAT-1 (second panels) or on IFN-induced expression of PKR (third panels). Similar results were obtained with a genotype 1a full-length protein (FL-NS5A-1a) and an ISDR deletion mutant protein (NS5A-1a-ΔISDR) (data not shown).
FIG. 2.
Effect of NS5A expression on IFN-induced posttranslational modifications and upregulation of gene expression. (A) Effect of NS5A expression on IFN induction of STAT-1 phosphorylation and PKR expression. FL-NS5A-NR and FL-NS5A-CR proteins were repressed or induced by growing cells in the presence or absence of doxycycline (Dox) for 24 h and treated with IFN, and protein extracts were harvested at various times thereafter. Protein extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, respectively, followed by Western blotting with antibodies to NS5A (first panels), phosphorylated STAT-1 (second panels), PKR (third panels), and β-tubulin (fourth panels) as described in Materials and Methods. (B) Effect of NS5A expression on IFN-induced upregulation of MHC antigen. HeLa cells expressing FL-NS5A-NR (left panels) and FL-NS5A-CR (right panels) proteins for 24 h were treated with IFN, and cells were prepared for flow cytometry as described in Materials and Methods. Top panels are from cells grown in the presence of doxcycline (NS5A expression repressed), while bottom panels are from cells grown in the absence of doxcycline (NS5A expression induced). The gray histogram on the left represents a control staining with an isotype-matched control. To the right of the control histogram in each panel are the specific staining patterns. IFN-treated cells are depicted by the gray curve, while cells not treated with IFN are depicted by the black curve.
Major histocompatibility complex (MHC) upregulation is a hallmark immunostimulatory consequence of IFN stimulation of cells. Figure 2B depicts the effects of NS5A expression on IFN-induced upregulation of MHC antigen expression on HeLa cells. Cells were grown in the absence or presence of doxycycline to induce or repress NS5A, respectively, and treated with IFN, and MHC antigen was measured by flow cytometry. FL-NS5A-NR and FL-NS5A-CR proteins did not affect IFN-induced upregulation of MHC antigen expression on the surfaces of human cells. Expression of FL-NS5A-1a or NS5A-1a-ΔISDR also had no effect on IFN upregulation of MHC (data not shown). Collectively, the data in Fig. 1 and 2 indicate that NS5A expression does not affect the activation of the IFN system, at least at the level of IFN signal transduction through the STAT-JAK pathway involving ISGF-3 and tyrosine phosphorylation of STAT-1. NS5A does not appear to affect the expression of genes induced by IFN. These data suggested that NS5A might modulate the IFN-induced antiviral response by a postinduction mechanism. Since it had been previously demonstrated that IL-8 could inhibit the antiviral actions of IFN against certain viruses in vitro, we examined the role of IL-8 in NS5A-mediated antagonism of the IFN system.
NS5A induces IL-8 mRNA.
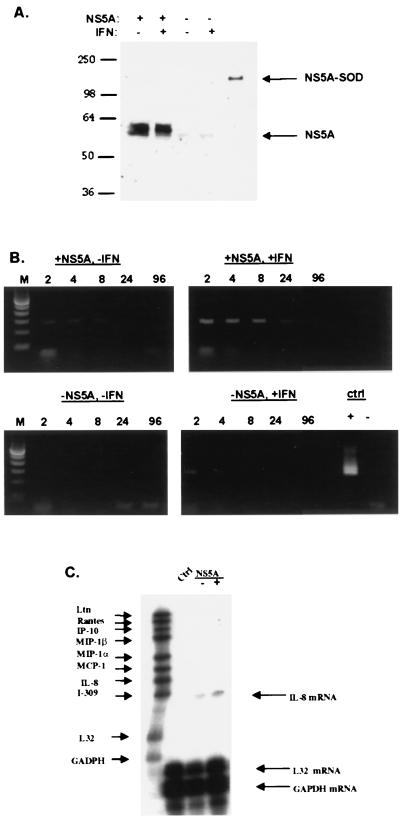
In the absence of tetracycline, NS5A expression was detected by Western blot analysis (Fig. 3A). Following addition of tetracycline to the tissue culture medium, NS5A expression was no longer detected. The addition of IFN did not affect the level of expression of NS5A. A semiquantitative PCR for IL-8 mRNA was then performed using serial dilutions of cDNA samples. Figure 3B (top panels) indicates that NS5A expression was associated with an approximately eightfold induction of IL-8 mRNA, as detected by semiquantitative RT-PCR. IL-8 cDNA signals were not due to genomic DNA contamination, as verified by the absence of larger IL-8 PCR products and by the absence of PCR products when reverse transcriptase was omitted from the cDNA synthesis reaction mixture (Fig. 3B). The results of an RNase protection assay (Fig. 4C) indicate that NS5A selectively induced IL-8 approximately eightfold among several chemokines. HeLa cells did not appear to express any of the other chemokines such as RANTES, IP-10, and MIP-1β. In summary, NS5A expression was associated with increases in IL-8 mRNA expression in vitro.
FIG. 3.
NS5A expression increases the expression of IL-8 mRNA. (A) Tetracycline-regulated expression of NS5A in human cells. HeLa cells expressing FL-NS5A-NR were incubated in medium with or without tetracycline for 48 h to repress or induce NS5A expression, respectively. Cells were then treated or not treated with IFN (20 U/ml) for 24 h. Cell extracts were prepared, and equal amounts of total cellular proteins were analyzed by Western blotting. The positions of the 58-kDa NS5A and positive control, NS5-(superoxide dismutase [SOD]), proteins are shown with arrows. (B) Semiquantitative detection of IL-8 mRNA by RT-PCR. cDNA was prepared from HeLa cells expressing or not expressing FL-NS5A-NR and treated or not treated with IFN. Serial dilutions of 1:2, 1:4, 1:8, 1:24, and 1:96 of cDNA (depicted above each lane with 2, 4, 8, 24, and 96, respectively) were subjected to PCR using IL-8-specific primers. Positive and negative controls (ctrl) are depicted with + and − signs, respectively. Amplification of cDNA generated without reverse transcriptase produced no PCR signal, indicating that the observed IL-8 PCR products were derived from IL-8 mRNA. Lanes M, molecular size markers. (C) Detection of IL-8 mRNA by RNase protection assay. Equal amounts of total cellular RNA from HeLa cells expressing or not expressing FL-NS5A-NR were hybridized to a 32P-labeled riboprobe cocktail containing probes for various human chemokines, including IL-8. Hybrids resistant to RNase digestion were separated on denaturing acrylamide gels, and autoradiography was performed. The positions of IL-8, control L32, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs are indicated with arrows. Note that the size of the protected fragment is smaller than the probe size because the probe contains vector-derived sequences that do not hybridize with target mRNA.
FIG. 4.
NS5A expression is associated with increased IL-8 protein production and inhibition of the antiviral effects of IFN. (A) Correlation among levels of IL-8 in culture supernatants, amounts of NS5A protein expression, and levels of EMCV rescue in the trans rescue assay. HeLa cells expressing FL-NS5A-NR were grown for 48 h in medium containing 0, 0.001, 0.01, and 1.0 μg of tetracycline per ml for 48 h, treated with 20 U of IFN per ml for 24 h, and infected with EMCV at an MOI of 0.01 for 24 h. The amount of IL-8 protein in culture supernatants, determined by ELISA, is indicated in the bar graph. Changes in EMCV titers were determined 24 h postinfection by viral plaque assay. The levels of NS5A protein expression were determined by Western blot analysis of equal amounts of total cellular protein extracts and are presented as +++, ++, +, and −, which correspond to high, intermediate, low, and undetectable levels of NS5A protein, respectively, as determined by computerized scanning of the chemiluminescent signal. Fold EMCV rescue represents the difference in EMCV titers in the presence of IFN among cells treated with 0, 0.001, and 0.01 μg of tetracycline per ml versus the 1.0-μg/ml concentration. (B) IL-8 inhibits the antiviral actions of IFN in vitro. HeLa Tet-Off cells were pretreated with or without 33 ng of recombinant human IL-8 per ml for 6 h and then with 20 U of IFN per ml for 18 h. Cells were then infected with EMCV at an MOI of 0.1 for 24 h. Supernatants were harvested, and the amount of EMCV was determined by titration on L929 cells. Error bars represent standard deviations, and P values derived from Student's t tests are indicated.
NS5A, the trans rescue assay, and IL-8.
Since IL-8 has been shown to inhibit the antiviral action of IFN (33) we tested the hypothesis that IL-8 is induced by NS5A in the trans rescue assay (58). The principle of the trans rescue assay is as follows. In the absence of HCV gene expression, IFN treatment of cells inhibits the replication of the IFN-sensitive virus EMCV (a positive-stranded RNA virus like HCV), measured as a reduction in viral titers. If HCV proteins inhibit the antiviral actions of IFN, higher titers of EMCV are produced in the presence of IFN. Figure 4A demonstrates titration of the amount of tetracycline, which resulted in a range of NS5A protein levels. In the absence of tetracycline, NS5A expression (determined by Western blotting) was highest, and this was associated with the highest levels of IL-8 protein (determined by ELISA) in culture supernatants. Increasing the tetracycline concentration in the medium reduced the amounts of both NS5A and IL-8 protein. Moreover, EMCV replication in the presence of IFN decreased along with NS5A and IL-8 levels. From the opposite perspective, EMCV replication during IFN treatment increased approximately eightfold during NS5A and IL-8 induction. The data indicate a direct correlation between the level of EMCV replication in the presence of IFN and levels of both NS5A and IL-8 protein. We attempted to neutralize endogenous IL-8 using neutralizing anti-IL-8 monoclonal antibodies. This was not successful because prolonged incubation of significant amounts of anti-IL-8 antibodies was toxic to the cells (data not shown). Nonetheless, it was previously shown that antibodies to IL-8 reversed the IL-8-mediated inhibition of IFN antiviral action (33).
To determine if IL-8 could directly inhibit the antiviral actions of IFN in our system, EMCV replication was examined in HeLa cells pretreated with or not treated with recombinant human IL-8. Cells were then treated or not treated with IFN and then infected with EMCV. As shown in Fig. 4B, in the absence of IL-8 pretreatment, IFN inhibited EMCV replication by >3 log units. However, when cells were pretreated with IL-8, an 11.6-fold increase in EMCV yields during IFN challenge was observed. Thus, the IFN-induced antiviral activity was reduced by greater than 10-fold. This indicates that recombinant human IL-8 can partially inhibit the antiviral actions of IFN in our system.
Transactivation of the IL-8 promoter by NS5A.
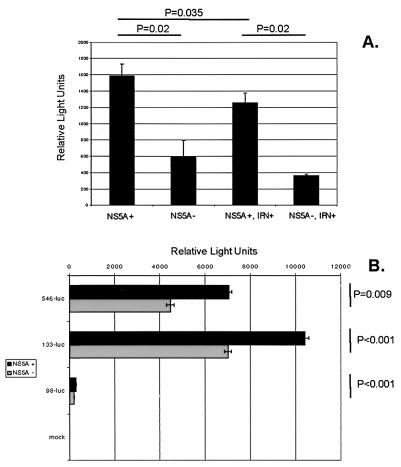
Since NS5A has been shown to be a transcriptional activator (16, 31, 75), we next determined if NS5A expression could transactivate the IL-8 promoter. For these experiments, the luciferase reporter protein under the control of the IL-8 promoter was used (50). Figure 5A indicates that NS5A expression resulted in a significant 2.5-fold increase of luciferase activity in the absence of IFN, when the reporter gene was under the control of the 546-bp IL-8 promoter. In the presence of IFN, NS5A expression was associated with a significant threefold increase in luciferase activity. Because there are a number of response elements in the IL-8 promoter with binding sites for hepatocyte nuclear factor-1, AP-1, NF–IL-6, and NF-κB (49), we next determined the minimal promoter domains for NS5A-mediated transcriptional activation of the IL-8 gene using a set of truncated IL-8 promoters. Figure 5B indicates that NS5A transactivated both the 546- and 133-bp IL-8 promoters. NS5A transactivation resulted in higher levels of luciferase activity with the 133-bp promoter than with the 546-bp promoter. Further truncation of the IL-8 promoter to include only 98 bp abrogated the ability of NS5A to transactivate the reporter gene. These results suggest that NS5A increases IL-8 mRNA and protein expression via transcriptional activation and further suggest that the minimal IL-8 promoter required for NS5A transactivation consists of 133 bp immediately upstream of the IL-8 gene. This region of the IL-8 promoter contains binding sites for the transcription factors NF-κB, AP-1, and NF–IL-6.
FIG. 5.
NS5A transactivates the IL-8 promoter. (A) Effect of NS5A expression on luciferase expression under the control of the 546-luc IL-8 promoter. The 546-luc plasmid and pTRE-FL-NS5A-NR (10 μg each) were transfected into HeLa Tet-Off cells, and cells were split in triplicate and grown in media with or without tetracycline for 48 h. Cells were then treated or not treated with IFN for 24 h. The amount of luciferase activity in cell lysates was determined using a luminometer. (B) Effect of NS5A expression on luciferase activity under the control of truncated IL-8 promoters. Ten micrograms of the relevant reporter plasmid was transfected into HeLa FL-NS5A-NR cells, and cells were grown in media with or without tetracycline for 48 h. The amount of luciferase activity in cell lysates was determined using a luminometer. 546-luc, 133-luc, and 98-luc contain 546, 133, and 98 bases of the IL-8 promoter, respectively. Error bars represent standard deviations, and P values derived from Student's t tests are indicated.
Analysis of NS5A mutants lacking the amino terminus.
The transcriptional activity reported for NS5A has been observed only when 100 or more amino acids are deleted from the amino terminus of the protein (16, 31, 68, 75). We therefore compared the effects of expressing the FL-NS5A-NR, NS5A-ΔN110, NS5A-ΔN222, and NS5A-ΔC117 proteins on transactivation of the IL-8 promoter. Figure 6A indicates that expression of NS5A-ΔN110 and NS5A-ΔN222 resulted in higher levels of luciferase activity than did expression of the full-length protein. The NS5A-ΔC117 protein did not transactivate the IL-8 promoter, suggesting a requirement for C-terminal amino acids in NS5A transactivation. We hypothesized that these differences in transcriptional activity may be related to the subcellular localization of the full-length and truncated NS5A proteins. We therefore performed immunofluoresence experiments to localize the FL-NS5A-NR, NS5A-ΔN222, and NS5A-ΔC117 proteins in HeLa cells. Figure 6B depicts the results. Expression of FL-NS5A-NR in HeLa cells was localized to the perinuclear region (top panels), consistent with our previous report (58). Expression of NS5A-ΔN222 in HeLa cells produced a diffuse staining throughout both the cytoplasm and the nucleus (middle panels), whereas expression of the NS5A-ΔC117 protein resulted in a punctate cytoplasmic staining pattern (lower panels). Positive staining was seen only when cells were grown in the absence and not in the presence of tetracycline, indicating that the expression of each NS5A protein was tightly regulated. These results suggest that the increase in transcriptional activity by NS5A proteins lacking amino termini may be due to increased migration of the protein to the nucleus, while the lack of IL-8 transactivation by the carboxy-terminally deleted NS5A protein may be due to increased retention of the protein in the cytoplasm. Based on this result, we used NS5A-ΔN222 to determine the minimal regions of the IL-8 promoter to gain further insight into the transcription factors required for NS5A transactivation. NS5A-ΔN222 was cotransfected with wild-type and various mutant IL-8 promoters. After 48 h, cells were treated with TNF-α for 6 h and luciferase activity was measured. Similar to our previous results, the 546-luc and 133-luc constructs had increased luciferase activities when NS5A was expressed in the presence of TNF-α. Deletion of the IL-8 promoter to 98 bp inhibited reporter gene activity upon TNF-α stimulation. The expression of NS5A could not overcome the lack of a response from the 98-bp promoter, although a twofold increase in luciferase activity was noted when NS5A was expressed. Mutation of the NF-κB and AP-1 binding sites also inhibited luciferase activity in the presence of TNF-α and NS5A expression, although the effect of mutating NF-κB was more pronounced than that of mutating AP-1. In contrast, mutation of the NF–IL-6 binding site resulted in higher levels of luciferase activity in the presence of TNF-α. In this case, NS5A expression resulted in even higher levels of luciferase activity. These data suggest that, after stimulation of HeLa cells with TNF-α or expression of NS5A, NF-κB and, to a lesser extent, AP-1 are required for transactivation of the IL-8 promoter and that the binding of NF–IL-6 is inhibitory to this process.
FIG. 6.
Characterization of domains on NS5A and the IL-8 promoter required for transactivation. (A) Effect of NS5A mutant proteins on luciferase expression under the control of the full-length (546-luc) IL-8 promoter. Ten-microgram quantities of the 546-luc plasmid and pTRE-FL-NS5A-NR, NS5A-ΔN110, NS5A-ΔN222, or NS5A-ΔC117 were transfected into HeLa Tet-Off cells, and cells were split in triplicate and grown in media with or without tetracycline for 48 h. The amount of luciferase activity in cell lysates was determined using a luminometer. The data shown are the fold changes in luciferase activity with the gene in the induced versus the uninduced state. (B) Im- munofluoresence analysis of HeLa cells expressing FL-NS5A-NR, NS5A-ΔN222, or NS5A-ΔC117. HeLa Tet-Off cells stably transfected with FL-NS5A-NR (top panels), NS5A-ΔN222 (middle panels), or NS5A-ΔC117 (lower panels) were grown in the absence or presence of tetracycline (Tc) to induce or repress NS5A expression for 48 h. Cells were fixed and stained with a monoclonal NS5A antibody and then with Cy3-linked anti-mouse secondary antibodies, as described in Materials and Methods. Images are at a ×100 amplification. (C) Effect of NS5A-ΔN222 expression on luciferase activity under the control of mutated IL-8 promoters. Ten-microgram quantities of NS5A-ΔN222 and the relevant reporter plasmid were cotransfected into HeLa Tet-Off cells; cells were split in triplicate and grown in media with or without tetracycline for 48 h. Cells were then treated with 4 ng of TNF-α for 6 h. The amount of luciferase activity in cell lysates was determined using a luminometer. 546-luc, 133-luc, and 98-luc contain 546, 133, and 98 bases of the IL-8 promoter, respectively. NF-κB–luc, AP-1–luc, and NF–IL-6–luc represent the 133-luc plasmid with point mutations in the NF-κB, AP-1, and NF–IL-6 binding sites.
DISCUSSION
Antiviral therapy for HCV infection is still rather primitive, with only two Food and Drug Administration-approved drugs available. While newer-generation therapies for HCV are clearly more effective than conventional IFN monotherapy, over 60% of patients with HCV genotype 1 infection are not cured of their infection. In order to improve the success of antiviral intervention in HCV, it is important to understand the molecular mechanisms of antiviral resistance. In recent years, insight into the molecular mechanisms involved in the intrinsic resistance of HCV to IFN therapy has been gained. The emerging trend in the literature is that HCV proteins, when overexpressed in vitro, disrupt normal biological processes by interaction with cellular proteins. Two examples of this is the interaction of HCV NS5A and E2 proteins with the IFN-induced enzyme PKR (19, 77). However, since the roles of mutations in the NS5A protein and in the clinical response to IFN therapy and the effects of NS5A on the IFN system in vitro are controversial, it has been suggested that NS5A may use other mechanisms to perturb the IFN-induced antiviral response (59).
The present study provides detailed mechanistic evaluation of an HCV protein in the context of the broad IFN response pathway. The present report found no evidence for a role of NS5A in inhibiting signal transduction via the STAT/JAK pathway, at least in terms of tyrosine phosphorylation of STAT-1 and induction of ISGF-3, the principal factor induced by IFN. This finding was supported by the observations that NS5A did not affect the induction of expression of two IFN-induced genes, those for PKR and MHC antigen. It is possible that levels of serine phosphorylation on STAT proteins are affected by NS5A expression (78). Evidence was provided that NS5A can induce the expression of IL-8 mRNA and protein. IL-8 expression was associated with NS5A-induced inhibition of the IFN-α antiviral response in vitro, and the addition of recombinant human IL-8 also partially rescued EMCV replication during IFN challenge. Using a luciferase reporter gene under the control of the IL-8 promoter, it was also shown that NS5A expression could transactivate the IL-8 promoter.
It is currently unknown if NS5A-mediated transcriptional activation of the IL-8 promoter occurs directly or indirectly. NS5A may in theory bind to the IL-8 promoter if it can enter the nucleus. Evidence discounting a direct effect of NS5A are derived from observations that full-length NS5A proteins have a cytoplasmic perinuclear distribution when expressed in human cells (29, 35, 48, 58, 76). It is possible, however, that a small amount of the NS5A protein, beyond the level of detection in these systems, may migrate to the nucleus. In this regard, it has recently been shown that processing of NS5A occurs naturally in transfected cells in vitro and that N-terminally deleted forms of the protein migrate to the nucleus (68). The C terminus of NS5A also contains a nuclear localization signal that does not by itself direct NS5A to the nucleus but is nonetheless functional in directing nuclear translocation when it is placed at the amino terminus of a reporter gene (29). In agreement with previous reports, we found that deletion of the amino terminus from NS5A resulted in increased transactivation of the IL-8 promoter, which was associated with increased nuclear staining of the protein. Deletion of C-terminal amino acids from NS5A created a protein that failed to transactivate the IL-8 promoter, and the protein appeared to be retained in the cytoplasm. These results suggest that the NS5A protein may be able to enter the nucleus under certain physiological instances.
Promoter mutagenesis experiments suggested that the minimal region for NS5A transactivation involved the 133 bp of the IL-8 promoter. Previous reports indicate that the transcription factors NF-κB, AP-1, and NF–IL-6 are involved in TNF-α-induced stimulation of the IL-8 promoter and that the involvement of each transcription factor can be cell line dependent (49). In this report, we found that TNF-α induction of the IL-8 promoter in HeLa cells required NF-κB and AP-1 but that NF–IL-6 appeared inhibitory to this process. NS5A expression could not overcome deletion of the NF-κB and AP-1 binding sites to activate luciferase activity. Thus, it is possible that NS5A induces IL-8 transcription by interacting with transcription factors. Indeed, NS5A has been shown to interact with a novel transcription factor (23). We are investigating further the domains of NS5A responsible for IL-8 transactivation and the subcellular localizations of the proteins and characterizing the transcription factors required for this process.
The results presented in this and other reports (17, 28, 63, 74) suggest that NS5A is a pleiotropic molecule, possessing multiple activities in vivo through its interaction with various cellular proteins and signaling pathways. Specifically, it appears that NS5A employs multiple mechanisms to achieve antiviral resistance, implying that inhibition of PKR by NS5A is but one way by which NS5A subverts the IFN system. This may be a plausible hypothesis since NS5A-ISDR mutations are not universally associated with IFN sensitivity (7, 11, 15, 27, 52, 56, 60, 64, 65, 72, 73, 79). More recent data suggest that a region in the C terminus of NS5A, termed the variable 3 (V3) region, may be associated with IFN resistance, at least in France and North America (11), and with a distinct region within the proline-rich domain of NS5A (52). A recent study also demonstrated that expression of the entire HCV polyprotein inhibits the activity of IFN against EMCV independently of PKR (14). Moreover, HCV replicons selected for increased replicative ability in vitro harbor mutations in NS5A, including a deletion of the entire ISDR (3). It should be emphasized that NS5A gene-encoded mechanisms for inhibition of the IFN system need not be mutually exclusive. It is possible that the different activities of NS5A are related to the location of the protein in the cell. In this scenario, cytoplasmic NS5A may inhibit PKR while nuclear NS5A may interact with transcription factors and influence gene expression.
Why NS5A would upregulate a chemokine that induces an inflammatory response is currently not known. One might speculate that NS5A-induced increases in IL-8 affects the activity of the intrahepatic immune response to HCV, through T-cell chemotaxis to the HCV-infected liver. Since T-cell infiltration is a hallmark histological finding of chronic hepatitis C, NS5A induction of IL-8 may ultimately be involved in HCV persistence and pathogenesis. Since IL-8 is elevated in alcoholic hepatitis (44), it is tempting to speculate that NS5A induction of IL-8 may exacerbate the deleterious effects of ethanol on the liver, contributing to the increase in liver disease activity and poor antiviral responses in HCV-infected patients who abuse alcohol (69). Moreover, since lymphocytes have been shown to harbor replicating HCV (37, 38, 47), NS5A induction of IL-8, followed by lymphocyte recruitment to the infected liver, may also facilitate the spread of HCV to extrahepatic reservoirs. It is also possible that NS5A-mediated induction of IL-8 may contribute to HCV antiviral resistance, through the interaction between the chemokine- and IFN-signaling pathways. Indeed, there are several reports of IFN-induced downregulation of IL-8 (2, 53, 70). Thus, NS5A may subvert the IFN signaling pathway to promote the expression of cellular genes that may counteract the IFN-induced antiviral response. These concepts will remain speculative until they can be formally addressed in a small animal model or in in vitro expression and replication systems.
Currently, the mechanism of IL-8-mediated inhibition of the IFN system is not known. Khabar and colleagues (33) demonstrated that the anti-IFN effect of IL-8 was detectable as late as 20 h after the addition of IFN. This suggests that IL-8 inhibits the IFN-induced antiviral response at a posttranscriptional level and is consistent with this report showing that NS5A does not affect IFN signal transduction. Preliminary data suggest that IL-8 inhibited 2′-5′-oligoadenylate synthetase (OAS), a major pathway for IFN action against RNA viruses (34). We are currently exploring the hypothesis that NS5A induction of IL-8 perturbs the OAS and other IFN-induced effector molecule pathways. It is also possible that the anti-IFN effect of IL-8 involves the high-effinity IL-8 receptor CXCR1. Indeed, HeLa cells which are susceptible to IL-8 proviral action express CXCR1 but not CXCR2 (K. S. A. Khabar, unpublished data; 34).
Viral modulation of chemokine expression represents the continuous battle between viral invaders and antiviral, inflammatory, and immune responses. Perhaps the best examples are the recent discoveries that human immunodeficiency virus (HIV) entry into macrophages and T lymphocytes requires interaction with distinct chemokine receptors and that soluble chemokines can inhibit the entry of HIV (20). In HIV infection, IL-8 is frequently elevated and the HIV Tat and Vpr proteins can induce IL-8 expression (54, 66). Other examples include human cytomegalovirus, which encodes a chemokine receptor that may facilitate viral replication by regulating cell cycle progression or inhibiting apoptosis (51). Recent studies have demonstrated that Sendai virus-infected cells induce the CC chemokine RANTES and that this effect is mediated by interferon regulatory factor 3 and NF-κB (21, 42). Thus, it will be important to determine the mechanisms involved in the induction of IL-8 by the HCV NS5A protein and how IL-8 inhibits the antiviral actions of IFN. If this interaction between HCV and the chemokine system can be proven to occur in vivo, then its selective disruption may increase therapeutic response rates and reduce the pathogenicity associated with chronic hepatitis C.
ACKNOWLEDGMENTS
We thank Robert L. Carithers, Jr., Anne M. Larson, and Jean-Michel Pawlotsky for providing serum specimens for cloning; Ara Hovanessian for PKR antiserum; and Jeffery Vieira, Christine Posavad, and Lawrence Corey for helpful discussions.
This research was partially supported by an NIH Hepatitis C Cooperative Center pilot feasibility grant and Schering-Plough (S.J.P.); the University of Washington Royalty Research Fund and NIH grants AI41320-02, AI39049-02 (D.R.G.), and AI28900 (D.E.L.); the King Faisal Specialist Hospital and Research Centre (K.S.A.K.); and grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (N.M.). S.J.P. is a Liver Scholar of the American Liver Foundation.
REFERENCES
- 1.Abid K, Quadri R, Negro F. Hepatitis C virus, the E2 envelope protein, and the alpha-interferon response. Science. 2000;287:1555. doi: 10.1126/science.287.5458.1555a. [DOI] [PubMed] [Google Scholar]
- 2.Aman M J, Rudolf G, Goldschmitt J, Aulitzky W E, Lam C, Huber C, Peschel C. Type-I interferons are potent inhibitors of interleukin-8 production in hematopoietic and bone marrow stromal cells. Blood. 1993;15:2371–2378. [PubMed] [Google Scholar]
- 3.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 4.Carithers R L J, Emerson S S. Therapy of hepatitis C: meta-analysis of interferon alpha-2b trials. Hepatology. 1997;26:83S–88S. doi: 10.1002/hep.510260715. [DOI] [PubMed] [Google Scholar]
- 5.Chang H W, Watson J C, Jacobs B L. The E3L gene of vaccinia virus encodes and inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase H, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 7.Chung R T, Monto A, Dienstag J L, Kaplan L M. Mutations in the NS5A region do not predict interferon-responsiveness in American patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58:353–358. [PubMed] [Google Scholar]
- 8.Darnell J E, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 9.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alpha-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373–1381. doi: 10.1099/0022-1317-79-6-1373. [DOI] [PubMed] [Google Scholar]
- 12.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A gene. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 14.Francois C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum H E, Gatignol A, Wychowski C, Moradpour D, Meurs E F. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J Virol. 2000;74:5587–5596. doi: 10.1128/jvi.74.12.5587-5596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frangeul L, Cresta P, Perrin M, Lunel F, Opolon P, Agut H, Huraux J M. Mutations in NS5A region of hepatitis C virus genome correlate with presence of NS5A antibodies and response to interferon therapy for most common European hepatitis C virus genotypes. Hepatology. 1998;28:1674–1679. doi: 10.1002/hep.510280630. [DOI] [PubMed] [Google Scholar]
- 16.Fukuma T, Enomoto N, Marumo F, Sato C. Mutations in the interferon-sensitivity determining region of hepatitis C virus and transcriptional activity of the nonstructural region 5A protein. Hepatology. 1998;28:1147–1153. doi: 10.1002/hep.510280433. [DOI] [PubMed] [Google Scholar]
- 17.Gale M, Blakely C M, Kwieciszewski B, Tan S L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale M, Kwieciszewski B, Dossett M, Nakao H, Katze M G. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 20.Garzino-Demo A, Devico A L, Gallo R C. Chemokine receptors and chemokines in HIV infection. J Clin Immunol. 1998;18:243–255. doi: 10.1023/A:1027329721892. [DOI] [PubMed] [Google Scholar]
- 21.Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-κB and IFN-regulatory factor transcription factors. J Immunol. 2000;164:5352–5361. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- 22.Gerotto M, Sullivan D C, Polyak S J, Chemello L, Cavalletto L, Pontisso P, Alberti A, Gretch D R. Effect of retreatment with interferon alone or interferon plus ribavirin on hepatitis C virus quasispecies diversification in nonresponder patients with chronic hepatitis C. J Virol. 1999;73:7241–7247. doi: 10.1128/jvi.73.9.7241-7247.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh A K, Majumder M, Steele R, Yaciuk P, Chrivia J, Ray R, Ray R B. Hepatitis C virus NS5A protein modulates transcription through a novel cellular transcription factor, SRCAP. J Biol Chem. 2000;275:7184–7188. doi: 10.1074/jbc.275.10.7184. [DOI] [PubMed] [Google Scholar]
- 24.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heim M H, Moradpour D, Blum H E. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herion D, Hoofnagle J H. The interferon sensitivity determining region: all hepatitis C virus isolates are not the same. Hepatology. 1997;25:769–771. doi: 10.1002/hep.510250346. [DOI] [PubMed] [Google Scholar]
- 27.Hofgärtner W T, Polyak S J, Sullivan D G, Carithers R L, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 28.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-alpha catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 29.Ide Y, Zhang L W, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene. 1996;182:203–211. doi: 10.1016/s0378-1119(96)00555-0. [DOI] [PubMed] [Google Scholar]
- 30.Inchauspe G, Zebedee S, Lee D H, Sugitani M, Nasoff M, Prince A M. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci USA. 1991;88:10292–10296. doi: 10.1073/pnas.88.22.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato N, Lan K H, OnoNita S K, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katze M G. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 33.Khabar K S A, Al-Zoghaibi F, Al-Ahdal M N, Murayama T, Dhalla M, Mukaida N, Taha M, Al-Sedairy S T, Siddiqui Y, Kessie G, Matsushima K. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J Exp Med. 1997;186:1077–1085. doi: 10.1084/jem.186.7.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khabar K S A, Murayama T, al-Ahdal M N, Mukaida N, Matsushima K. Interleukin-8 is induced by viruses, enhances cytomegalovirus and picornavirus replication, and inhibits the antiviral action of inteferon-alpha. Enfield, N.H.: Science Publishers, Inc.; 2000. [Google Scholar]
- 35.Kim J E, Song W K, Chung K M, Back S H, Jang S K. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch Virol. 1999;144:329–343. doi: 10.1007/s007050050507. [DOI] [PubMed] [Google Scholar]
- 36.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209–2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 37.Laskus T, Radkowski M, Wang L F, Vargas H, Rakela J. The presence of active hepatitis C virus replication in lymphoid tissue in patients coinfected with human immunodeficiency virus type 1. J Infect Dis. 1998;178:1189–1192. doi: 10.1086/515682. [DOI] [PubMed] [Google Scholar]
- 38.Laskus T, Radkowski M, Wang L F, Vargas H, Rakela J. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: specific detection of negative-strand viral RNA in various tissues. Hepatology. 1998;28:1398–1401. doi: 10.1002/hep.510280531. [DOI] [PubMed] [Google Scholar]
- 39.Leonard G T, Sen G C. Effects of adenovirus E1A protein on interferon signaling. Virology. 1996;224:25–33. doi: 10.1006/viro.1996.0503. [DOI] [PubMed] [Google Scholar]
- 40.Levy D E, Lew D J, Decker T, Kessler D S, Darnell J E., Jr Synergistic interaction between interferon-alpha and interferon-gamma through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang T J, Rehermann B, Seeff L B, Hoofnagle J H. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 42.Lin R, Heylbroeck C, Genin P, Pitha P M, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindsay K L. Therapy of hepatitis C: overview. Hepatology. 1997;26:71S–77S. doi: 10.1002/hep.510260713. [DOI] [PubMed] [Google Scholar]
- 44.McClain C J, Barve S, Deaciuc I, Kugelmas M, Hill D. Cytokines in alcoholic liver disease. Semin Liver Dis. 1999;19:205–219. doi: 10.1055/s-2007-1007110. [DOI] [PubMed] [Google Scholar]
- 45.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alpha-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 46.Meurs E, Chong K, Galabru J, Thomas N S, Kerr I M, Williams B R G, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 47.Moldvay J, Deny P, Pol S, Brechot C, Lamas E. Detection of hepatitis C virus RNA in peripheral blood mononuclear cells of infected patients by in situ hybridization. Blood. 1994;83:269–273. [PubMed] [Google Scholar]
- 48.Moradpour D, Kary P, Rice C M, Blum H E. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology. 1998;28:192–201. doi: 10.1002/hep.510280125. [DOI] [PubMed] [Google Scholar]
- 49.Mukaida N, Harada A, Yasumoto K, Matsushima K. Properties of pro-inflammatory cell type-specific leukocyte chemotactic cytokines, interleukin 8 (IL-8) and monocyte chemotactic and activating factor (MCAF) Microbiol Immunol. 1992;36:773–789. doi: 10.1111/j.1348-0421.1992.tb02080.x. [DOI] [PubMed] [Google Scholar]
- 50.Murayama T, Ohara Y, Obuchi M, Khabar K S A, Higashi H, Mukaida N, Matsushima K. Human cytomegalovirus induces interleukin-8 production by a human monocytic cell line, THP-1, through acting concurrently on AP-1- and NF-κB-binding sites of the interleukin-8 gene. J Virol. 1997;71:5692–5695. doi: 10.1128/jvi.71.7.5692-5695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy P M. Chemokine receptors: structure, function and role in microbial pathogenesis. Cytokine Growth Factor Rev. 1996;7:67–64. doi: 10.1016/1359-6101(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 52.Nousbaum J-B, Polyak S J, Sullivan D G, Larson A M, Carithers R L, Jr, Gretch D R. Prospective characterization of full-length HCV NS5A quasispecies during induction and combination antiviral therapy. J Virol. 2000;74:9028–9038. doi: 10.1128/jvi.74.19.9028-9038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveira I C, Sciavolino P J, Lee T H, Vilcek J. Downregulation of interleukin 8 gene expression in human fibroblasts: unique mechanism of transcriptional inhibition by interferon. Proc Natl Acad Sci USA. 1992;89:9049–9053. doi: 10.1073/pnas.89.19.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ott M, Lovett J L, Mueller L, Verdin E. Superinduction of IL-8 in T cells by HIV-1 Tat protein is mediated through NF-κB factors. J Immunol. 1998;160:2872–2880. [PubMed] [Google Scholar]
- 55.Paterson M, Laxton C D, Thomas H C, Ackrill A M, Foster G R. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology. 1999;117:1187–1197. doi: 10.1016/s0016-5085(99)70405-1. [DOI] [PubMed] [Google Scholar]
- 56.Pawlotsky J M, Germanidis G, Neumann A U, Pellerin M, Frainais P O, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polyak S J, Nousbaum J B, Larson A M, Cotler S, Carithers R L, Jr, Gretch D R. The protein kinase-interacting domain in the hepatitis C virus envelope glycoprotein-2 gene is highly conserved in genotype 1-infected patients treated with interferon. J Infect Dis. 2000;182:397–404. doi: 10.1086/315720. [DOI] [PubMed] [Google Scholar]
- 58.Polyak S J, Paschal D, McArdle S, Gale M, Moradpour D, Gretch D R. Characterization of the effects of hepatitis C virus non-structural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262–1271. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- 59.Polyak S J, Gerotto M. Molecular basis for responsiveness to anti-viral therapy in hepatitis C. Forum (Genova) 2000;10:46–58. [PubMed] [Google Scholar]
- 60.Polyak S J, McArdle S, Liu S L, Sullivan D G, Chung M J, Hofgartner W T, Carithers R L, McMahon B J, Mullins J I, Corey L, Gretch D R. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poynard T, Leroy V, Cohard M, Thevenot T, Mathurin P, Opolon P, Zarski J P. Meta-analysis of interferon randomized trials in the treatment of viral hepatitis C: effects of dose and duration. Hepatology. 1996;24:778–789. doi: 10.1002/hep.510240405. [DOI] [PubMed] [Google Scholar]
- 62.Poynard T, Marcellin P, Lee S S, Niederau C, Minuk G S, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, Albrecht J. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group. Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 63.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rispeter K, Lu M J, Zibert A, Wiese M, deOliveira J M, Roggendorf M. A suggested extension of the HCVISDR does not alter our former conclusions on its predictive value for IFN response. J Hepatol. 1999;30:1163–1164. doi: 10.1016/s0168-8278(99)80276-1. [DOI] [PubMed] [Google Scholar]
- 65.Rispeter K, Lu M J, Zibert A, Wiese M, deOliveira J M, Roggendorf M. The “interferon sensitivity determining region” of hepatitis C virus is a stable sequence element. J Hepatol. 1998;29:352–361. doi: 10.1016/s0168-8278(98)80051-2. [DOI] [PubMed] [Google Scholar]
- 66.Roux P, Alfieri C, Hrimech M, Cohen E A, Tanner J E. Activation of transcription factors NF-κB and NF–IL-6 by human immunodeficiency virus type 1 protein R (Vpr) induces interleukin-8 expression. J Virol. 2000;74:4658–4665. doi: 10.1128/jvi.74.10.4658-4665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saiz J C, LopezLabrador F X, Ampurdanes S, Dopazo J, Forns X, SanchezTapias J M, Rodes J. The prognostic relevance of the nonstructural 5A gene interferon sensitivity determining region is different in infections with genotype 1b and 3a isolates of hepatitis C virus. J Infect Dis. 1998;177:839–847. doi: 10.1086/515243. [DOI] [PubMed] [Google Scholar]
- 68.Satoh S, Hirota M, Noguchi T, Hijikata M, Handa H, Shimotohno K. Cleavage of hepatitis C virus nonstructural protein 5A by a caspase-like protease(s) in mammalian cells. Virology. 2000;270:476–487. doi: 10.1006/viro.2000.0287. [DOI] [PubMed] [Google Scholar]
- 69.Schiff E R. Hepatitis C and alcohol. Hepatology. 1997;26:39S–42S. doi: 10.1002/hep.510260707. [DOI] [PubMed] [Google Scholar]
- 70.Schnyder-Candrian S, Strieter R M, Kunkel S L, Walz A. Interferon-alpha and interferon-gamma down-regulate the production of interleukin-8 and ENA-78 in human monocytes. J Leukoc Biol. 1995;57:929–935. doi: 10.1002/jlb.57.6.929. [DOI] [PubMed] [Google Scholar]
- 71.Song J, Fujii M, Wang F, Itoh M, Hotta H. The NS5A protein of hepatitis C virus partially inhibits the antiviral activity of interferon. J Gen Virol. 1999;80:879–886. doi: 10.1099/0022-1317-80-4-879. [DOI] [PubMed] [Google Scholar]
- 72.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alpha. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 73.Squadrito G, Orlando M E, Cacciola I, Rumi M G, Artini M, Picciotto A, Loiacono O, Siciliano R, Levrero M, Raimondo G. Long-term response to interferon alpha is unrelated to “interferon sensitivity determining region” variability in patients with chronic hepatitis C virus-1b infection. J Hepatol. 1999;30:1023–1027. doi: 10.1016/s0168-8278(99)80255-4. [DOI] [PubMed] [Google Scholar]
- 74.Tan S L, Nakao H, He Y P, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997;236:360–364. doi: 10.1006/bbrc.1997.6967. [DOI] [PubMed] [Google Scholar]
- 76.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M C. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 78.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 79.Zeuzem S, Lee J H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alpha. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]