Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is related to the development of Kaposi's sarcoma. Open reading frame K9 of KSHV encodes viral interferon regulatory factor 1 (vIRF1), which functions as a repressor of interferon- and IRF1-mediated signal transduction. In addition, vIRF1 acts as an oncogene to induce cellular transformation. Here we show that vIRF1 directly associates with the tumor suppressor p53 and represses its functions. The vIRF1 interaction domains of p53 are the DNA binding domain (amino acids [aa] 100 to 300) and the tetramerization domain (aa 300 to 393). p53 interacts with the central region (aa 152 to 360) of vIRF1. vIRF1 suppresses p53-dependent transcription and deregulates its apoptotic activity. These results suggest that vIRF1 may regulate cellular function by inhibiting p53.
The Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also called human herpesvirus 8, is a novel human gammaherpesvirus that plays a role in the development of KS lesions, primary effusion lymphoma, and a subset of multicentric Castleman's disease (2, 25). Analysis of the genomic sequence of KSHV shows significant homologies with herpesvirus saimiri and Epstein-Barr virus. The KSHV genome contains a unique set of nonstructural genes that have homologies with genes encoding cellular proteins (22). For example, the K9 open reading frame of KSHV, which encodes viral interferon regulatory factor 1 (vIRF1), shows significant homology with the cellular IRF. vIRF1 is a 449-amino-acid (aa) protein whose synthesis is induced by tetradecanoyl phorbol acetate (TPA) (20, 23). The expression of antisense RNA to vIRF1 down-regulates the expression of specific KSHV genes in BCBL-1 cells, suggesting that vIRF1 may regulate KSHV gene expression (13).
In transient-transfection assays, vIRF1 represses cellular interferon- and IRF1-mediated transcriptional activation (13, 26). vIRF1 transforms NIH 3T3 cells, and NIH 3T3 cells that stably express vIRF1 display features of a malignant fibrosarcoma in nude mice (4, 13). In addition, vIRF1 inhibits tumor necrosis factor alpha-mediated apoptosis (1). These facts strongly suggest that vIRF1 augments the tumorigenicity of KSHV. vIRF1 also associates with p300/CBP (CREB binding protein), thus inhibiting the transactivation of CBP and the histone acetyltransferase activity of p300 (1, 12, 24).
The tumor suppressor p53 is absent from numerous types of human cancer cells (7). p53 is a key regulator of a wide range of cellular activities, including cell cycle regulation, apoptosis, response to DNA damage, differentiation, and angiogenesis (11, 15). Several viral oncoproteins (simian virus 40 large T antigen, papillomavirus E6, adenovirus E1B, and hepatitis B virus X protein) bind to p53 and modulate its function. p53 contains four distinct functional domains: the transcriptional activation domain at the amino terminus (aa 1 to 42), the DNA binding domain (DBD) in the central region (aa 102 to 292), the tetramerization domain (TD; aa 323 to 356), and the regulatory domain (aa 363 to 393) (15).
vIRF1 associates with p53 in vivo.
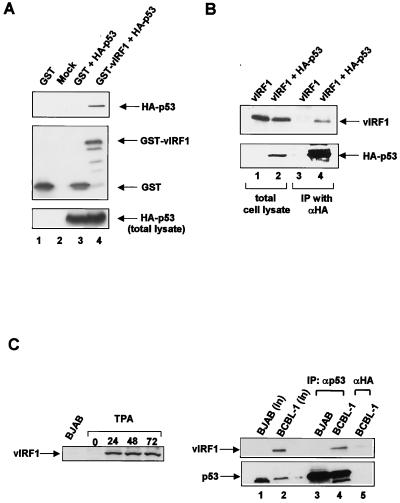
Because vIRF1 causes transformation and inhibits tumor necrosis factor-mediated apoptosis, we investigated whether vIRF1 interacts with p53. By using the calcium phosphate method (6), 293T cells were transfected with glutathione S-transferase (GST), GST-vIRF, and hemagglutinin (HA)-p53 expression plasmids (pEBG, pEBG/vIRF, and pcDNA3/HA-53, respectively). Forty-eight hours after transfection, the cells were lysed with EBC buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 200 μM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride). The cell extracts were then incubated with glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech, Uppsala, Sweden) to precipitate the GST and GST-vIRF1 proteins, and the precipitated proteins were immunoblotted with an anti-HA antibody. HA-p53 coprecipitated with GST-vIRF1 but did not coprecipitate with GST (Fig. 1A, top). To show that the GST, GST-vIRF1, and HA-p53 proteins were expressed properly in the transfected cells, we performed separate immunoblot experiments with anti-GST and anti-HA monoclonal antibodies (Fig. 1A, middle and bottom, respectively). 293T cells were also transfected with vIRF1 (pcDNA3/vIRF1) and HA-p53 (pcDNA3/HA-p53) expression plasmids. The cell extracts were immunoprecipitated with an anti-HA monoclonal antibody, and again the vIRF1 protein coimmunoprecipitated with HA-p53 (Fig. 1B).
FIG. 1.
In vivo interaction of vIRF1 with p53. (A) A GST expression plasmid (pEBG) or a GST-vIRF1 expression plasmid (pEBG/vIRF1) was cotransfected with an HA-p53 expression plasmid (pcDNA3/HA-p53) into 293T cells. Whole-cell extracts were prepared 48 h after transfection and precipitated with glutathione-Sepharose 4B beads. The precipitated proteins were washed and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). GST fusion protein and HA-p53 were detected by Western blotting with anti-HA (top and bottom) and anti-GST (middle). Lanes: 1, GST alone; 2, no expression plasmid; 3, GST with HA-p53; 4, GST-vIRF1 with HA-p53. (B) A vIRF1 expression plasmid (pcDNA3/vIRF1) was cotransfected with or without the HA-p53 expression plasmid. Whole-cell extracts were incubated with protein G resin after preincubation with anti-HA. The resulting immunoprecipitates were washed and resolved by SDS-PAGE. vIRF1 and HA-p53 were detected by Western blotting with anti-vIRF1 rabbit polyclonal antibody (top) or with anti-HA (αHA) antibody (bottom). Lanes: 1 and 3, vIRF1 alone; 2 and 4, vIRF1 with HA-p53. IP, immunoprecipitation. Anti-vIRF1 polyclonal antibody was obtained from postimmune sera collected from rabbits immunized with the GST-vIRF1 fusion protein. (C) In vivo interaction of vIRF1 with p53 in KSHV-infected BCBL-1 cells. BCBL-1 cells were treated with TPA as previously described (20), and whole-cell extracts were prepared after the indicated number of hours. vIRF1 expression was detected by Western blotting with anti-vIRF1 antibody (left). Whole-cell extracts from BJAB and BCBL-1 cells were immunoblotted with anti-vIRF1 and anti-p53 (lanes 1 and 2). Direct coimmunoprecipitation (IP) was performed by using BCBL-1 cells after TPA stimulation. BCBL-1 and BJAB cells (1 × 107 cells) were harvested 48 h after TPA stimulation, and whole-cell extracts were incubated with protein G resin after being preincubated with either anti-p53 (αp53) (lanes 3 and 4) or anti-HA (αHA) (lane 5) antibody. vIRF1 was detected by Western blotting with anti-vIRF1 (right top) or anti-p53 (right bottom) antibody.
To determine whether vIRF1 and p53 interact in a KSHV-infected cell line, we performed coimmunoprecipitation assays with KSHV-infected BCBL-1 cells. First, expression of vIRF1 was stimulated by treatment of the cells with TPA (Fig. 1C, left). Total lysates of BJAB and BCBL-1 cells were immunoblotted with an anti-vIRF1 polyclonal antibody and an anti-p53 monoclonal antibody, DO-I (Santa Cruz Biotechnology, Santa Cruz, Calif.) (Fig. 1C, lanes 1 and 2). The cells were then immunoprecipitated with either anti-p53 antibody or anti-HA antibody. In addition, vIRF1 coimmunoprecipitated with p53 in the KSHV-infected BCBL-1 cells (Fig. 1C, lane 4). In contrast, vIRF1 was not detected in KSHV-negative BJAB cell extracts or by immunoprecipitation with anti-HA (Fig. 1C, lanes 3 and 5). These results demonstrate that vIRF1 interacts with p53.
vIRF1 interacts with the DBD and TD of p53, whereas p53 interacts with the central region of vIRF1.
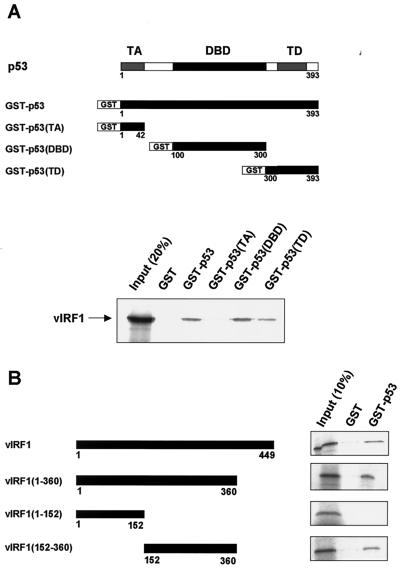
To map the vIRF1 binding domain in p53, we performed GST pulldown assays (10) by using 35S-labeled, in vitro-translated vIRF1 and a series of GST-p53 deletion mutants (Fig. 2A). The transcriptional activation (TA) domain (aa 1 to 42) of p53 did not interact with vIRF1, whereas the p53 DBD (aa 100 to 300) and TD (aa 300 to 393) interacted with vIRF1. The vIRF1 binding affinity of the p53 DBD was slightly stronger than that of the TD. These results show that vIRF1 directly interacts with p53 through its DBD and TD.
FIG. 2.
Identification of domains involved in vIRF1-p53 interaction. (A) Top, schematic representation of p53 and its functional domains. The TA domain (aa 1 to 42), DBD (aa 1 to 42), and TD (aa 300 to 393) are shown. Bottom, GST pulldown assays (10) performed with wild-type and mutant GST-p53 fusion proteins by using 35S-labeled, in vitro-translated vIRF1. The input (20%) and GST pulldown mixtures were resolved by SDS-PAGE, and vIRF1 was visualized by autoradiography. (B) Left, schematic representation of vIRF1 and its deletion mutants. Right, an experiment similar to that described for panel A that was performed by using the GST-p53 and 35S-labeled, in vitro-translated vIRF1 and vIRF1 mutant proteins.
To determine the p53 binding domain within vIRF1, we constructed the vIRF1 deletion mutants vIRF1(1-360), vIRF1(1-152), and vIRF1(152-360) (Fig. 2B). GST pulldown assays were performed by using 35S-labeled, in vitro-translated vIRF1 (and vIRF1 mutants) and GST-p53 (Fig. 2B). Only vIRF1(1-360) and vIRF1(152-360) interacted with GST-p53 (Fig. 2C). These results indicate that the p53 interaction domain within vIRF1 is located in the central region (aa 152 to 360) of vIRF1.
vIRF1 represses p53-dependent transcription.
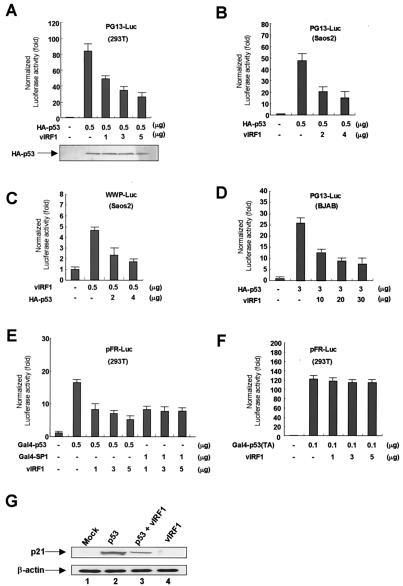
To test whether the vIRF1 protein modulates p53-mediated transcription, we transiently cotransfected 293T cells with a reporter plasmid that contained synthetic p53 response elements fused to the gene for luciferase (PG13-Luc) and a vIRF1 expression plasmid (pcDNA3-vIRF1) with or without an HA-p53 expression plasmid (pcDNA3/HA-p53). The cells were transfected by the calcium phosphate method (6), harvested 24 h after transfection, and resuspended in cell lysis buffer. The insoluble fraction was removed by centrifugation, and the luciferase activity in the supernatant was measured with a luminometer (EG&G Berthold, Pforzheim, Germany). In each transfection assay, a Rous sarcoma virus β-galactosidase expression plasmid was cotransfected and β-galactosidase activity was measured as an internal control for transfection efficiency. In the presence of vIRF1, p53-driven transcription of the luciferase gene was inhibited in a dose-dependent manner (Fig. 3A). Expression of HA-p53 was monitored by a Western blot assay, which showed that the level of HA-p53 was not changed by the presence of vIRF1. Similarly, we carried out cotransfection assays in p53-null Saos2 cells by using either the PG13-Luc reporter or the WWP-Luc reporter, which contains an authentic p21 promoter. For both reporters, cotransfection with the vIRF1 expression plasmid repressed luciferase activity in a dose-dependent manner (Fig. 3B and C). These results indicate that HA-p53 can substitute for endogenous p53. Transfection experiments were also performed with the BJAB B-cell lymphoma cell line. BJAB cells were transfected by electroporation as described previously (14). As in 293T and Saos2 cells, vIRF1 repressed p53-mediated transcription in BJAB cells (Fig. 3D).
FIG. 3.
vIRF1 represses p53-dependent transcription. Luciferase activity was measured with a luminometer. The total amount of transfected DNA in each experiment was kept constant by the addition of a blank vector (pcDNA3). The activity of the reporter alone was normalized to a value of 1, and each luciferase measurement was normalized to the internal control, β-galactosidase activity. Each experiment was carried out at least three times. (A) A synthetic p53 response element fused to a luciferase gene (PG13-Luc) was inhibited by vIRF1 in the presence of p53 in 293T cells. 293T cells were cotransfected with PG13-Luc (1 μg), a β-galactosidase expression plasmid (0.5 μg), a p53 expression plasmid (pcDNA3/HA-p53) (0.5 μg), and increasing amounts of an expression plasmid encoding vIRF1 (pcDNA3/vIRF1). Equal amounts of total cellular extracts were resolved by SDS-PAGE and subjected to HA-specific immunoblotting. (B) PG13-Luc was inhibited by vIRF1 in the presence of p53. Saos2 cells were cotransfected with PG13-Luc (1 μg), a β-galactosidase expression plasmid (RSV/β-gal) (0.5 μg), a p53 expression plasmid (pcDNA3/HA-p53) (0.5 μg), and increasing amounts of an expression plasmid encoding vIRF1 (pcDNA3/vIRF1). (C) The WWP-Luc plasmid was inhibited by vIRF1 in the presence of p53. Saos2 cells were cotransfected with WWP-Luc (1 μg) and the other plasmids listed in panel B. (D) vIRF1 represses p53-dependent transcription in BJAB cells. BJAB cells were cotransfected by electroporation (14) with PG13-Luc (5 μg), an HA-p53 expression plasmid, and a vIRF1 expression plasmid. (E) vIRF1 represses transcriptional activation by p53 and does not influence an unrelated transcriptional activator. 293T cells were cotransfected with a Gal4-Luc reporter plasmid (pFR-Luc) (1 μg), a vIRF1 expression plasmid, and either a Gal4-p53 expression plasmid (0.5 μg) or a Gal4-SP1 expression plasmid (0.5 μg). (F) vIRF1 does not repress transcriptional activation by p53(TA) (aa 1 to 42). 293T cells were cotransfected with pFR-Luc (1 μg), a Gal4-p53(TA) expression plasmid, and a vIRF1 expression plasmid. (G) vIRF1 represses p53-induced p21 expression in 293T cells. A vIRF1 expression plasmid (pcDNA3/vIRF1) and an HA-p53 expression plasmid (pcDNA3/HA-p53) were cotransfected into 293T cells by the calcium phosphate method. Cells were harvested 48 h after transfection. Lanes: 1, no expression plasmid; 2, pcDNA3/HA-p53 (7 μg) plus pcDNA3 (15 μg); 3, pcDNA3/HA-p53 (7 μg) plus pcDNA3/vIRF1 (15 μg); 4, pcDNA3 (7 μg) plus pcDNA3/vIRF1 (15 μg). p21 was detected with a monoclonal antibody to p21 (top), and β-actin was detected with a monoclonal antibody to β-actin (bottom).
To test whether vIRF1 directly represses the transcriptional activation activity of p53, we carried out cotransfection assays with 293T cells by using a Gal4-p53 expression plasmid and pFR-Luc, which contains five Gal4 binding sites, as the reporter. Cotransfection with a vIRF1 expression plasmid repressed Gal4-p53-driven luciferase expression in a dose-dependent manner, indicating that vIRF1 can repress the transcriptional activation activity of p53 (Fig. 3E). To test whether the effect of vIRF1 on the transcriptional activity of p53 is specific, we replaced Gal4-p53 with a Gal4-SP1 fusion protein in the transfection assay. Cotransfection with the vIRF1 expression plasmid did not alter Gal4-SP1-driven luciferase activity expression.
To test whether vIRF1-p53 interaction is important in the repression of p53 transactivation, we carried out cotransfection assays by using Gal4-p53(TA) (aa 1 to 42) in 293T cells. The TA domain of p53 did not interact with vIRF1 in GST pulldown assays (Fig. 2A). Unlike Gal4-p53, Gal4-p53(TA) was not inhibited by a vIRF1 expression plasmid (Fig. 3E and F). These data show that protein-protein interaction between p53 and vIRF1 is necessary for inhibition of the transactivation of p53.
To test whether vIRF1 down-regulates p21 expression in vivo, we measured the p21 expression level in 293T cells by immunoblotting. BJAB cells in 100-mm-diameter dishes were cotransfected by the calcium phosphate method (6), harvested 48 h after transfection, and immunoblotted with an anti-p21 monoclonal antibody (Santa Cruz Biotechnology). In the presence of p53, p21 expression was dramatically increased whereas p21 expression was decreased upon cotransfection with the vIRF1 expression plasmid (Fig. 3G, top). To show that the total protein concentration in each lane was the same, we performed Western blot assays with an antibody to β-actin (Fig. 3G, bottom). These results demonstrate that vIRF1 suppresses p53-mediated p21 expression in 293T cells and are consistent with the above results showing that vIRF1 inhibits p53-mediated transcription.
vIRF1 deregulates p53-induced apoptosis in Saos2 cells.
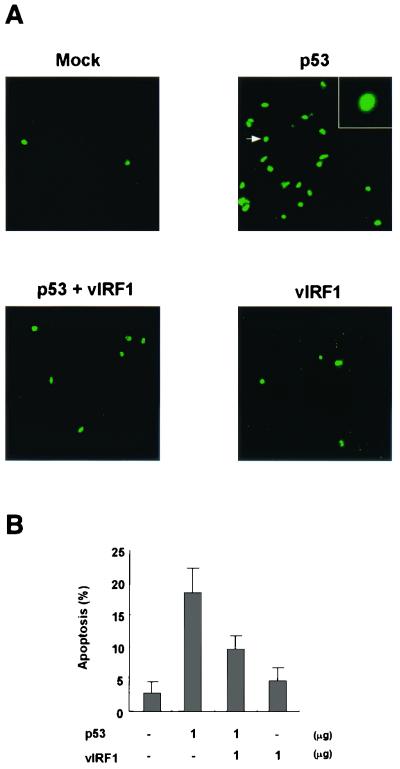
To examine the effect of vIRF1 on p53-mediated apoptosis, we performed terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays (5) with p53-null Saos2 cells. The TUNEL reaction was performed by using an in situ cell detection kit as specified by the manufacturer (Roche, Mannheim, Germany). Forty-eight hours after transfection, successfully transfected Saos2 cells were subjected to the TUNEL reaction. The TUNEL reaction used fluorescein-5-isothiocyanate as the labeling reagent, and TUNEL-positive (apoptotic) cells showed green nuclei. Transfection of cells with a p53 expression plasmid increased the number of TUNEL-positive cells, while cotransfection of vIRF1 with a p53 expression plasmid decreased the number of TUNEL-positive cells (Fig. 4A). TUNEL-positive cells represented 3.1, 18.7, 9.2, and 4.6% of the total cell population when expressing no protein, p53, p53 in combination with vIRF1, and vIRF1 alone, respectively (Fig. 4B). These data show that vIRF1 inhibits the apoptotic activity of p53.
FIG. 4.
vIRF1 deregulates p53-induced apoptosis in Saos2 cells. Saos2 cells grown on coverslips in 35-mm-diameter dishes were transfected with Superfect transfection reagent (Qiagen, Hilden, Germany). Forty-eight hours after transfection, cells were fixed and the TUNEL reaction was performed as recommended by the manufacturer (Roche). The analyses were performed with a Zeiss confocal microscope with fluorescein isothiocyanate filter sets. (A) Left top, mock transfection (no expression plasmid); right top, pcDNA3/HA-p53 (1 μg) plus pcDNA3 (1 μg); bottom left, pcDNA3/HA-p53 (1 μg) plus pcDNA3/vIRF1 (1 μg); bottom right, pcDNA3/vIRF1 (1 μg) plus pcDNA3 (1 μg). Magnification, ×100. In the insert at the top right, the magnification of the cell indicated by the arrow is ×400. (B) Schematic representation of TUNEL-positive cell percentages. The bars represent the percentages of transfected cells that showed apoptosis; apoptosis was determined by counting the green dead cells. The values shown are means calculated from two duplicate experiments. A total of 1,000 cells were counted in each experiment.
Our results suggest that vIRF1 interacts with tumor suppressor p53 and is capable of repressing p53-mediated transcription and apoptosis. The observation that vIRF1 interacts with the DBD and TD of p53 suggests that vIRF1 inhibits the transcriptional activation activity of p53 by interfering with the DNA binding or tetramerization of p53. Therefore, we tested the ability of p53 to bind DNA in the presence and absence of vIRF1. vIRF1 did not diminish the DNA binding affinity of p53 in vitro (data not shown). In addition, we showed that vIRF1 also inhibited the transactivation of Gal4-p53 (Fig. 3D), indicating that the suppression of p53 transcriptional activation by vIRF1 does not require the direct DNA binding ability of p53. Another potential mechanism for vIRF1 inhibition of p53 activity might be blocking of the tetramerization of p53. It is well known that p53 forms tetramers and that tetramerization is required for efficient cell growth suppression by p53 (9, 19). Through binding to the TD of p53, vIRF1 may block tetramerization, resulting in the inhibition of p53 function. A third possible mechanism for the repression of p53-regulated promoters by vIRF1 might be sequestration of a common coactivator, such as p300/CBP. Because vIRF1 is known to interact with p300/CBP, vIRF1 may inhibit transcriptional activation by p53 by sequestering this coactivator. Like papillomavirus E6, vIRF1 interacts with p53 and p300/CBP. In the case of E6, repression of p53-regulated promoters is dependent on both the E6-p53 and E6-p300/CBP interactions (18). In the case of vIRF1, vIRF1-p53 interaction is important for the repression of p53-regulated promoters since vIRF1 does not repress the transactivation of p53(TA), which does not interact with vIRF1 (Fig. 3F). Further study is needed to decipher the precise mechanism of vIRF1 inhibition of p53 function.
p53 is known to be a key regulator of growth arrest through the induction of p21 and growth arrest- and DNA damage-inducible protein 45. p21 was identified as a potent inhibitor of several cyclin-dependent kinases (CDKs), including cyclin D-CDK4/6, cyclin E-CDK2, and cyclin A-CDK2 (15). p21 leads to inhibition of the cyclin D-CDK4/6 complex and subsequent accumulation of the unphosphorylated form of the retinoblastoma protein, which arrests cells in the G1 phase of the cell cycle. Because vIRF1 repressed p53-induced p21 expression (Fig. 3G), vIRF1 might induce cell proliferation by diminishing p21 expression. In addition, vIRF1 deregulates p53-induced apoptosis (Fig. 4). It is well known that stable expression of vIRF1 leads to the transformation of NIH 3T3 cells, resulting in morphologic changes, and the induction of malignant fibrosarcoma in nude mice (4, 13); however, the molecular mechanism of this phenomenon has not been fully elucidated. To transform cells and induce tumors, tumor suppressor genes must be inactivated and this inactivation can occur in a number of ways. Because p53 induces not only growth arrest but also apoptosis and DNA repair, p53 is the major target for tumorigenic viral proteins such as E6 and the simian virus 40 large T antigen (15). Our results begin to explain the transforming activity of vIRF1.
Previous reports showed that latency-associated nuclear protein 1 (LANA1), K-bZIP, and vIRF3/LANA2 of KSHV interact with p53 and that these interactions result in the repression of p53-mediated transcription and apoptosis (3, 16, 21). Recently, Rivas et al. (21) reported that KSHV LANA2 is a B-cell-specific latent viral protein and that it inhibits p53. Both vIRF1 and vIRF3/LANA2 bind to the TD of p53, but vIRF1 also binds to the DBD of p53. The p53-inhibiting activity of vIRF1 is similar to that of vIRF3/LANA2, suggesting that IRF homologues of KSHV may regulate the cell cycle by targeting p53. Although vIRF1 is not generally expressed in KS, it seems to be important in multicentric Castleman's disease (8, 17). vIRF1 may contribute to the development of B-cell hyperplasia in Castleman's disease by repressing p53-induced apoptosis.
Acknowledgments
This work was supported in part by grants from the National Research Laboratory Program of the Korea Institute of Science and Technology Evaluation and Planning (KISTEP), the Korea Science and Engineering Foundation (KOSEF) through the Protein Network Research Center at Yonsei University, and the BK21 Program of the Ministry of Education, Korea.
REFERENCES
- 1.Burýšek L, Yeow W-S, Lubyová B, Kellum M, Schafer S L, Huang Y Q, Pitha P M. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol. 1999;73:7334–7342. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Friborg J, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 4.Gao S, Boshoff J C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 5.Gorczyca W, Bigman K, Mittelman A, Ahmed T, Gong J, Melamed M R, Darzynkiewicz Z. Induction of DNA strand breaks associated with apoptosis during treatment of leukemias. Leukemia. 1993;7:659–670. [PubMed] [Google Scholar]
- 6.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 7.Hollstein M, Sidransky D, Vogelstein B, Harris C C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 8.Katano H, Sato Y, Kurata T, Mori S, Sata T. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 2000;269:335–344. doi: 10.1006/viro.2000.0196. [DOI] [PubMed] [Google Scholar]
- 9.Kraiss S, Quaiser A, Oren M, Montenarh M. Oligomerization of oncoprotein p53. J Virol. 1988;62:4737–4744. doi: 10.1128/jvi.62.12.4737-4744.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D, Lee B, Kim J, Kim D W, Choe J. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J Biol Chem. 2000;275:7045–7051. doi: 10.1074/jbc.275.10.7045. [DOI] [PubMed] [Google Scholar]
- 11.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 12.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung J U. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol. 2000;20:8254–8263. doi: 10.1128/mcb.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung J U. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol. 1998;72:5433–5440. doi: 10.1128/jvi.72.7.5433-5440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May P, May E. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 16.Park J, Seo T, Hwang S, Lee D, Gwack Y, Choe J. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J Virol. 2000;74:11977–11982. doi: 10.1128/jvi.74.24.11977-11982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected disease: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel D, Huang S M, Baglia L A, McCance D J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietenpol J A, Tokino T, Thiagalingam S, el-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 21.Rivas C, Thlick A, Parravicini C, Moore P S, Chang Y. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol. 2001;75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo T, Lee D, Lee B, Chung J H, Choe J. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) binds to and inhibits transactivation of CREB-binding protein. Biochem Biophys Res Commun. 2000;270:23–27. doi: 10.1006/bbrc.2000.2393. [DOI] [PubMed] [Google Scholar]
- 25.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 26.Zimring J C, Goodbourn S, Offermann M K. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol. 1998;72:701–707. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]