Abstract
A 157-amino-acid fragment of Moloney murine leukemia virus reverse transcriptase encoding RNase H is shown to rescue the growth-defective phenotype of an Escherichia coli mutant. In vitro assays of the recombinant wild-type protein purified from the conditionally defective mutant confirm that it is catalytically active. Mutagenesis of one of the presumptive RNase H-catalytic residues results in production of a protein variant incapable of rescue and which lacks activity in vitro. Analyses of additional active site mutants demonstrate that their encoded variant proteins lack robust activity yet are able to rescue the bacterial mutant. These results suggest that genetic complementation may be useful for in vivo screening of mutant viral RNase H gene fragments and in evaluating their function under conditions that more closely mimic physiological conditions. The rescue system may also be useful in verifying the functional outcomes of mutations based on protein structural predictions and modeling.
RNases H degrade the RNA strand of RNA-DNA heteroduplexes, and functional isoforms of the enzymes such as reverse transcriptase (RT) have been identified in many systems (2, 4, 6, 9). RT activity encoded by retroviral pol genes converts genomic RNA into double-stranded DNA and requires the concerted actions of its N-terminal polymerase and C-terminal RNase H domains (5, 19). Viral first strand DNA synthesis is initiated by a tRNA primer annealed to the 5′ end of template viral genomic RNA which forms the substrate for RT (5, 19). First-strand DNA synthesis is completed following transfer to and elongation of the strong-stop nascent DNA from the 3′ end of the genomic RNA. During first-strand DNA synthesis, template RNA is degraded to yield the polypurine tract RNA primer necessary for second-strand DNA synthesis (5, 19). These reactions all require the actions of RNase H, and therefore this activity is essential in the replicative life cycle of retroviruses (19). Despite sequence differences among RTs, the functional interrelationship and coordination of their polymerase and RNase H domains during reverse transcription appear to be very similar (1, 2, 7, 8, 10–12, 16, 19, 22, 24–26). Differences relating to the function of their RNase H domains do, however, exist. In contrast to human immunodeficiency virus (HIV) RNase H, which requires physical contact with its polymerase domain, the isolated Moloney murine leukemia virus (Mo-MuLV) RT RNase H domain retains enzymatic activity (10, 16, 21, 24, 25). This observation has made studies of the isolated Mo-MuLV domain possible. In spite of these functional differences in polymerase and RNase H interactions, the remarkable overall similarity in RT biological function among retroviral RNases H may permit studies of Mo-MuLV RNase H to model studies of related viral RNases H. Mo-MuLV RNase H belongs to the type I family of RNases H (RNases HI), whose members include Escherichia coli, Trypanosoma brucei, Crithidia fasciculata, and Saccharomyces cerevisiae RNase H, among others, as well as all retroviral RTs (4, 10, 13, 14, 24).
The major RNase H of E. coli, encoded by the rnhA gene, is required for specifying replication initiation from oriC and for directing replication of ColE1 plasmids (6, 14). In the mutant MIC3001 (rnh-339::cat recB270), rnhA is nonfunctional as a result of chloramphenicol acetyltransferase gene insertion (rnh-339:: cat) (14). Additionally, these cells carry a temperature-sensitive recB gene, recB270(Ts), which functions as a suppressor of the rnh-339::cat insertion allele at 30°C. At 42°C the combined failure to synthesize the rnhA product and while producing a recB(Ts) protein, which yields a nonfunctional recBCD enzyme, blocks MIC3001 growth. This temperature-sensitive phenotype, however, is alleviated by supplying cellular RNase H activity in trans at 42°C (4, 13, 14, 17). The ability of viral RNases H to alleviate this no-growth phenotype has thus far not been demonstrated.
The goal of this study is to develop an in vivo assay for retroviral RNase H activity by determining whether Mo-MuLV RNase H can rescue the growth deficiency of the E. coli mutant, MIC3001. By developing such a system it may be possible to more reliably characterize the structural requirements of viral-RNases H for function under conditions that more closely mimic physiological conditions. Additionally, such a system may facilitate qualitative and quantitative screens of Mo-MuLV RNase H gene variants encoding enzymes possessing attenuated function.
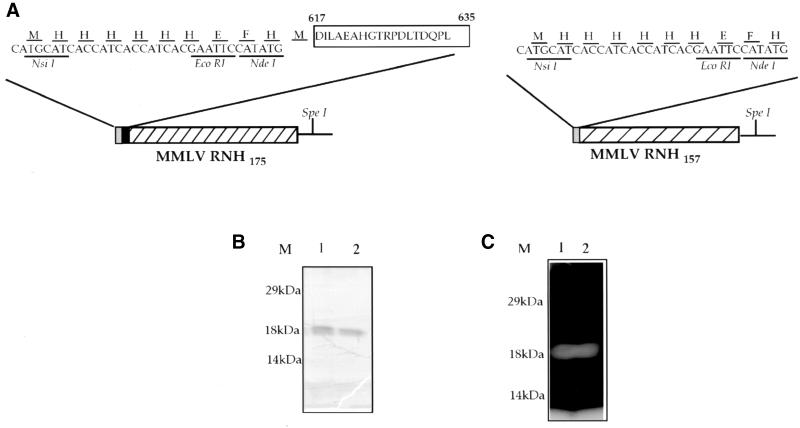
For complementation assays, MIC3001 is transformed with plasmid pAC9-157 and pAC9-175 (Fig. 1). pAC9-157 was constructed by ligating the EcoRI-SpeI insert fragment of pJKH0103-1, an unpublished derivative of pJKH0103 (11) into the EcoRI-SpeI vector fragment of pTRI (pAC-2) (13). pAC9-157 encodes the C-terminal 157-amino-acid RNase H fragment of Mo-MuLV RT. pAC9-175, which encodes the C-terminal 175-amino-acid fragment of RT, was similarly constructed. Controls for complementation assays included pTR1, which encodes the T. brucei RNase HI (TbbRNHI) (13); p6His-3; pAC2Δ120; and pHTRΔP. p6His-3 represents the insertless pAC-2 vector, which expresses a histidine tag alone and was constructed by EcoRl digestion of pTR1 and religation of the resulting vector fragment. pAC2Δ120 and pHTRΔP, which express inactive C- and N-terminal fragments, respectively (amino acids 121 to 301 and amino acids 1 to 123) of TbbRNHI were constructed as described previously (17–18).
FIG. 1.
Expression, purification, and RNase H assay of recombinant Mo-MuLV RNase H. (A) Schematic arrangement of predicted proteins expressed by pAC9-157 and pAC9-175. pAC9-157 and pAC9-175 encode the respective 471- and 525-bp RNase HI fragments of Mo-MuLV RT that encode the protein fragments Pro635 to Leu791 and Asp617 to Leu791. The EcoRI-SpeI insert fragments of pJKH0103-1 and pEG200-1 were cloned into EcoRI-SpeI-cut pAC2 vector, yielding an ORF encoding the recombinant proteins Mo-MuLVrnh157 and Mo-MuLVrnh175, schematically shown above. The sequence of the N-terminal histidine tag is shown above its nucleotide sequence using standard one-letter amino acid designations. This sequence is encoded by an XbaI-NsiI-EcoRI-NdeI synthetic linker. Restriction endonuclease sites used in cloning the various fragments are shown. (B and C) Protein purified from extracts of pAC9-157- and pAC9-175-transformed MIC3001 cells on nickel chelate agarose columns as described (13). Lanes: M, prestained molecular weight marker (Gibco-BRL); 1, wild-type Mo-MuLVrnh157 protein produced prior to complementation analyses; 2, Mo-MuLVrnh157 protein produced following four rounds of complementation analysis in MIC3001 by plasmid pAC9-157. (B) Coomassie blue-stained gel of purified recombinant proteins. (C) In situ RNase H gel renaturation assay of the proteins shown in panel B. Gel assays were performed in the presence of 1 mM MnCl2. The assay for Mo-MuLVrnh175 is not shown.
MIC3001 transformants, grown at 30°C in Luria-Bertani (LB) broth–50 μg of carbenicillin ml−1, were plated onto modified LB-agar plates which contained 10 g of Bacto Peptone (Difco Laboratories, Detroit, Mich.), 5 g of Bacto Yeast Extract (Difco) liter, 0.1 to 1.0 mM MnSO4 and 50 μg of carbenicillin ml−1 and were incubated 48 h at 42°C. Table 1 shows that pAC9-157 and pAC9-175 rescue the growth deficiency of these cells. Plasmid DNAs, isolated from transformants growing at 42°C, were used in second-round transformation of freshly competent MIC3001 cells followed by a second round of complementation testing. Plasmid DNAs isolated from the resulting cells growing at 42°C were used in subsequent third- and fourth-round transformation and complementation testing. The sequence of the insert DNAs of plasmids isolated from cells following these assays was confirmed by sequencing. Complementation analyses were performed by plating sufficient transformants, initially grown at 30°C, to yield ∼100 cells per plate. The numbers of colonies formed by pAC9-157 and pAC9-175 transformants at 42°C were similar to the numbers formed under growth conditions that did not require exogenous RNase H activity for cell growth (30°C), and this result indicates the high frequency of genetic rescue (Table 1). Complementation of MIC3001 by pAC9-157 and pAC9-175 required reduction in the NaCl ionic strength and was optimal at 0 to 10 mM NaCl. At low NaCl ionic strength, however, genetic rescue also required the addition of Mn2+ to 0.1 to 1.0 mM. In contrast to pAC9-157 and pAC9-175, pAC2Δ120 and pHTRΔP failed to alleviate the temperature-sensitive phenotype of MIC3001, indicating that simple protein overexpression was insufficient to rescue these cells. These control constructs failed to yield viable colonies at 42°C even after extended incubation. Purification and gel analysis of recombinant enzyme showed that pAC9-157 and pAC9-175 encoded the proteins Mo-MuLVmh157 and Mo-MuLVmh175, respectively, whose migrations through gels were consistent with their predicted sizes (Fig. 1B). RNase H gel assays also verified that these proteins were the sole source of RNase H activity in the recombinant protein preparation (Fig. 1C). For subsequent genetic rescue analyses, only pAC9-157 was used since this construct defined a minimal RNase H gene fragment.
TABLE 1.
Complementation analysis of wild-type and mutant Mo-MuLV RNase Ha
| Expression plasmid | Growth of colonies at:
|
Complementation frequency | |
|---|---|---|---|
| 30°C | 42°C | ||
| p6His-3 | + (103) | − (0) | <0.01 |
| pHTRΔP | + (97) | − (0) | <0.01 |
| pBluescript | + (110) | − (0) | <0.01 |
| pAC9-157 | + (98) | + (87) | 0.86 |
| pAC9-175 | + (89) | + (93) | 1.04 |
| pE48A | + (95) | − (0) | <0.01 |
| pE48Q | + (91) | +b (ND)c | ND |
| pD69A | + (74) | +d (ND) | ND |
| pAC2 | + (101) | + (96) | 0.95 |
Mo-MuLV RNase HI dependent growth of E. coli MIC3001. Transformed cells grown in LB-carbenicillin or LB-carbenicillin-choramphenicol were poured onto MLB-agar plates supplemented with 0.1 to 1.0 mM Mn2+. p6His-3 represents a control plasmid lacking an RNase H gene insert. pHTRΔP represents a control plasmid expressing the inactive N-terminal fragment (amino acids 1 to 123) of the T. brucei RNase HI enzyme. pAC-2 (positive control) encodes the full-length active T. brucei type I RNase H. Wild-type Mo-MuLV RNase H is encoded by pAC9-157 and pAC9-175. pE48A, pE48Q, and pD69A encode active site variants of pAC9-157. Growth (+) and failure to grow at the restrictive temperature (−) are indicated. The actual numbers of colonies yielded upon culture dilution and platings are indicated in parentheses. Complementation frequency is calculated by dividing the number of colonies formed at 42°C by the number formed at 30°C.
Microcolony formation after 48 h.
ND, Not determined (colonies too small to count reliably).
Small colony formation after 48 h.
To determine whether an active Mo-MuLV RNase H was required to rescue MIC3001, independent mutations at catalytic residues of Mo-MuLV RNase H were constructed and tested. Plasmid pE48A was constructed by digesting pAC9-157 with restriction endonuclease KpnI and SacI and ligating the resulting vector fragment to the linker with the sequence pair 5′ CTCGGCCCCAGCGTGCTGACCT3′-3′ CATGGAGCCGGGGTCGCACGAC5′. These deoxyoligonucleotides reconstituted the entire Mo-MuLV RNase H open reading frame (ORF) introducing a single amino acid substitution, changing Glu682 (Glu48 in the isolated RNase H domain) to Ala. Plasmid pE48Q encodes a mutant Mo-MuLV RNase H in which Glu48 was changed to Gln (isolated RNase H domain numbering). pE48Q was assembled by ligating the linker pair 5′ CTCGGCCCCAGCGTGCTCAGCT3′-3′ CATGGAGCCGGGTCGCACGAG5′ into the KpnI/SacI-double-digested vector fragment of pAC9-157. The underlined sequence corresponds to the nucleotide changes in the resulting ORF. Plasmid pD69A was similarly constructed by digesting pAC9-157 with restriction endonucleases SacI and MluI and reconstituting the viral RNase H sequence with synthetic deoxyoligonucleotides containing a single base pair substitution. The resulting pD69A construct yielded the single amino acid substitution Asp69 to Ala in the Mo-MuLV RNase H ORF. All oligonucleotides were synthesized by Life Technologies (Gaithersburg, Md.). Results presented in Table 1 show that the mutation substituting alanine for glutamate (E48A) abolishes the ability of Mo-MuLV RNase H to rescue the RNase deficiency of MIC3001. Mutation of the corresponding catalytic Glu residue of E. coli RNase HI also abolishes activity in the E. coli enzyme (15). The inability of MIC3001, producing catalytically inactive Mo-MuLV RNase H, to grow at 42°C suggests that no genetic reversion of this host cell occurred. This result also indicated that no change occurred in the sequence of the input DNA that affected its function during the course of these assays. Other active site mutants were studied, and interestingly, expression of the E48Q mutation rescued MIC3001, yielding small microcolonies after extended incubation of transformants at 42°C (Table 1). Similarly, mutation of Asp69 to Ala was also able to rescue MIC3001, yielding colonies slightly larger than those formed by the E48Q mutant. It is unlikely that colonies formed by the E48Q mutant at 42°C represent revertants of the original mutation since these colonies never approach the size of colonies formed by the wild-type ORF even after extended incubation. The size of the E48Q mutant microcolonies formed at 42°C did not permit individual colony isolation for subsequent plasmid DNA sequence analyses.
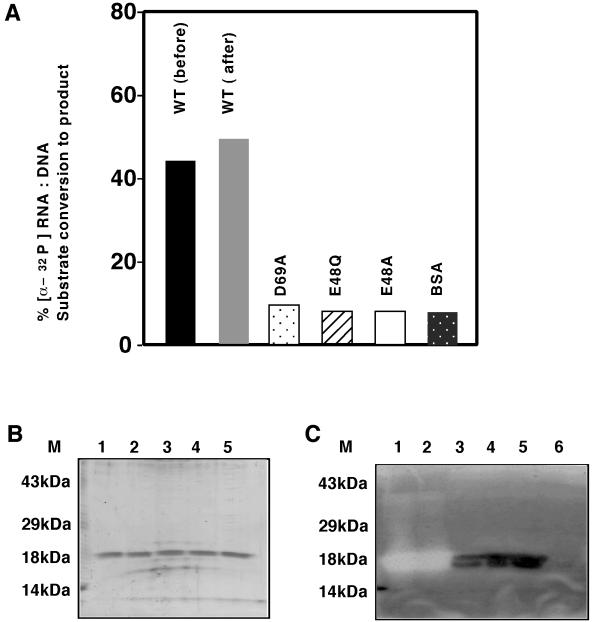
To evaluate activity associated with E48A, E48Q, and D69A mutants in vitro, the recombinant proteins were purified from MIC3001 cells as hexahistidine-tagged fusion proteins as described (13) and tested for function using [α-32p]poly(rA):poly (dT)5500 substrate prepared as described (13, 17). The results of these assays are shown in Fig. 2. Sufficient wild-type Mo-MuLVrnh157 was used in RNase H solution assays to convert 50% of the substrate to product, and equivalent amounts of each mutant protein were tested similarly. These assays show that the levels of activity of the wild-type protein expressed by pAC9-157 prior to and following four rounds of complementation were essentially unchanged (Fig. 2A). Although functional, the activity of the D69A mutant in these assays was significantly lower. Despite its ability to minimally alleviate the temperature-sensitive phenotype of MIC3001 at 42°C, the mutant protein E48Q failed to show activity in these assays, suggesting that it may be inactive or possess activity below the limits of experimental detection. In situ gel RNase H renaturation assays were also performed to determine whether RNase H activity detected in solution was indeed derived from the predicted recombinant proteins (Fig. 2B and C). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining confirmed that each protein produced was consistent with the size of its predicted ORF. In renaturation assays, wild-type Mo-MuLVrnh157 displayed robust renaturable RNase H activity under the conditions of the assay, and this protein appeared to be the species responsible for activity in the solution assays shown in Fig. 2A. The mutant protein D69A displayed extremely low RNase H activity, degrading small amounts of RNA-DNA heteroduplexes in situ (Fig. 2C). Mutant proteins E48A and E48Q failed to show RNase H activity in these gel assays (Fig. 2C). Interestingly, mutants D69A, E48Q, and E48A showed strong RNA-DNA duplex binding based on the level of immobilized duplex remaining associated with each protein in gels. The results of these assays verified that RNase H activity is indeed associated with the recombinant enzyme purified from the RNase HI-deficient cells and that alteration in amino acid residues which affected genetic rescue also affected activity in vitro.
FIG. 2.
RNase H assays of purified recombinant proteins. Mo-MuLVrnh157 and mutant E48A, E48Q, and D69A proteins were expressed and purified as histidine fusions from MIC3001 transformants. All fractions analyzed corresponded to the 100 mM imidazole eluate of nickel chelate agarose columns. (A) RNase H solution assay. Aliquots (2.8 μg) of proteins were assayed at 37°C for activity against [α-32P]poly(rA):(dT)5500 heteroduplex in the presence of 1.0 mM MnCl2. Conversion of substrate to product is indicated as percent conversion. Abbreviations: WT (before), wild type Mo-MuLVrnh157 protein expressed and purified prior to four rounds of complementation assay; WT (after), wild type MoMuLVrnh157 protein expressed and purified after to four rounds of complementation assay in MIC3001; BSA, bovine serum albumin. (B) Coomassie blue-stained gel of purified recombinant proteins used in panel A (350 ng of protein per lane). Lane 6, 100 mM nickel chelate agarose eluate of extracts prepared from p6His-3-transformed MIC3001. (C) In situ RNase H gel renaturation assays of protein shown in panel B. (B and C) Lanes: M, prestained molecular weight markers (Gibco-BRL) 1, Mo-MuLVrnh157 before complementation; 2, Mo-MuLVrnh157 after complementation; 3, D69A mutant protein; 4, E48Q mutant protein; 5, E48A mutant protein. Renaturation assays were performed in 1 mM MnCl2.
The relationship of growth media constituents to viral RNase H function is also noteworthy and may reflect physiological conditions necessary for Mo-MuLV RNase H activity. Growth of Mo-MuLV RNase H transformants under conditions of low NaCl and in the presence of Mn2+ as described here does not affect the RNase H-independent cell growth at 30°C. These changes in medium constituents, however, are consistent with what is known of the requirements of Mo-MuLV RNase H for proper folding, structural stabilization, and function (11).
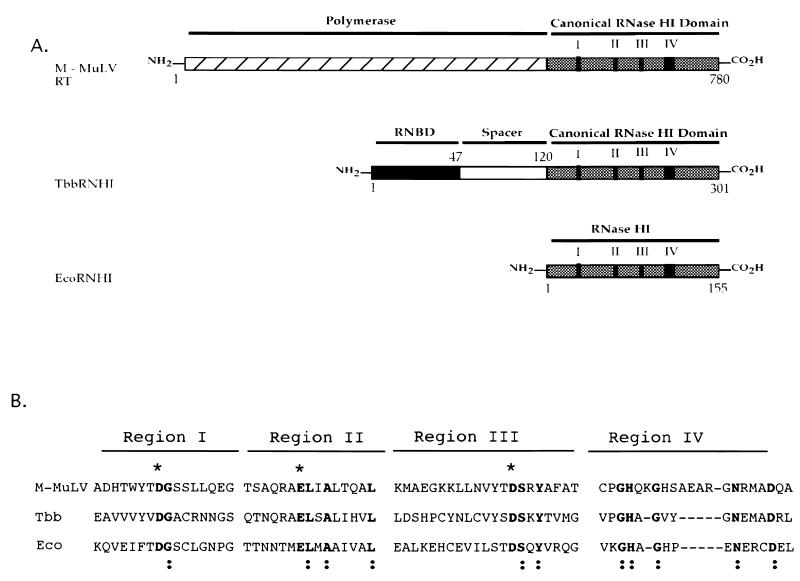
Sequence analyses of retroviral and cellular proteins possessing RNase H activity indicate that all have in common a conserved C-terminal canonical RNase HI domain (Fig. 3). These canonical domains are similar in size, and that in the case of E. coli RNase HI represents a natural stand-alone functional domain. Within these domains reside a limited number of amino acid residues which define the RNase HI motifs. Sequence homology in these motifs is centered around the catalytic triad of residues, aspartate, glutamate, and aspartate (Fig. 3B). In the isolated Mo-MuLV RNase H domain of RT, these correspond to Asp10, Glu48, and Asp69. These residues have been reported to be critical for activity in E. coli RNase HI (15) and without exception, they occur in all cellular RNases HI and all retroviral RNases H (20). More importantly, based on the ability of a number of divergent RNase HI genes to rescue MIC3001, these residues appear to be more important to function than global sequence homology.
FIG. 3.
Schematic arrangement and alignment of representative proteins encoding RNase HI activities. (A) Alignment of Mo-MuLV RT with cellular RNases HI. TbbRNHI, T. brucei RNase HI with its conserved eukaryote-specific RNA binding domain (RNBD), divergent spacer domain (spacer) and canonical RNase HI domain. Collinear and conserved protein sequence blocks in the RNase HI domains are indicated as I to IV, and their sequences are also shown. EcoRNHI, E. coli RNase HI. (B) Alignment of select collinear and noncontiguous amino acid sequences of Mo-MuLV, HIV eukaryotic, and E. coli RNases HI. Gene sequences are available from GenBank. The noncontiguous sequences are shown as regions I, II, III, and IV, and their overall collinear relationship is shown in Fig. 1. Sequence numbering is as follows. Regions I, II, III, and IV of Mo-MuLV RT correspond to residues 637 to 652, 676 to 690, 691 to 711, and 754 to 774. For Tbb (TbbRNHI) these regions correspond to residues 120 to 135, 168 to 182, 185 to 205, and 275 to 290. For Eco these regions correspond to residues (EcoRNHI) 3 to 18, 42 to 77, and 121 to 141. Residues shown in boldface lettering and marked with asterisks represent predicted catalytic residues conserved among all RNases HI. Residues shown in boldface and marked with a colon represent invariant residues shared by all the enzymes shown.
In studies completed here substitution of Mo-MuLV RNase H Glu48 with Gln, however, surprisingly results in an enzyme that has sufficient activity to weakly rescue the RNase H deficiency of MIC3001 yet that fails to display detectable activity in vitro. Genetic rescue by the E48Q mutant occurs despite the change in residue charge from −1 for Glu to 0 for the uncharged polar residue, Gln. The unexpected growth of E48Q-expressing cells may be related to an unusual modification occurring under physiological conditions. It is well established that glutamine and arginine deamidation occurs in many biological systems (23, 27). The E48Q mutant protein produced during complementation assays may be subject to deamidation in vivo over the extended 48-h incubation period at 42° C. Because deamidation results in the formation of a negative charge, such a change in a small fraction of E48Q mutant protein produced in vivo would restore the correct charge to the mutant proteins' active site and may restore enzyme function. The failure of purified recombinant E48Q protein used in in vitro assays to show activity suggests that it may not undergo deamidation due to its short duration in cells during the induction and purification process. Attempts are under way to induce in vitro deamidation in E48Q mutant protein to confirm that this event is responsible for restoring enzymatic activity. An alternate possibility is that physiologically low levels of enzyme activity detected by cell growth may not be readily detectable in vitro under conditions of the assays performed here.
Ongoing work on the in vitro activity of viral RNases H has helped to create a highly refined biochemical profile of these proteins (2, 10, 11). These studies continue to be critical in establishing the conditions and sequence basis of enzyme function. The ability of the isolated Mo-MuLV RNase H domain to functionally substitute for cellular deficiencies in RNase typically alleviated by cellular RNases HI is an important observation. It permits extensive genetic analysis of individual amino acid residues and their contribution to enzymatic function in vivo. Additionally, it helps to confirm what has been learned of the enzyme in vitro.
Finally, the ability of Mo-MuLV RNase H to rescue an E. coli RNase-deficient mutant indicates that the barriers to retroviral RNase H gene function in E. coli are small. Other RNases H, particularly hepatitis B virus and HIV RNases H, represent promising viral drug targets that remain underexploited. The studies completed here may be important in developing hepatitis B virus and HIV RNase H-based rescue systems for more-comprehensive genetic studies of these important enzymes.
Acknowledgments
I thank S. Marqusee (University of California, Berkeley) for providing the constructs pJKH0103 and pEG200.
This work was supported by National Science Foundation Career Development Award MCB-9600920 and by a LifeSpan/Brown University/Tufts University Center for AIDS Research Center Grant (P30-AI 42853) Developmental Award to A.G.C.
REFERENCES
- 1.Blain S W, Goff S P. Differential effects of Moloney murine leukemia virus reverse transcriptase mutations on RNase H activity in Mg2+ and Mn2+ J Biol Chem. 1996;271:1448–1454. doi: 10.1074/jbc.271.3.1448. [DOI] [PubMed] [Google Scholar]
- 2.Blain S W, Goff S P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 3.Borkow G, Fletcher R S, Barnard J, Arion D, Motakis D, Dmitrienko I, Parniak M P. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry. 1997;36:3179–3185. doi: 10.1021/bi9624696. [DOI] [PubMed] [Google Scholar]
- 4.Campbell A G, Ray D S. Functional complementation of an Escherichia coli ribonuclease H mutation by a cloned genomic fragment from the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci USA. 1993;90:9350–9354. doi: 10.1073/pnas.90.20.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin J M. Retroviridae: the viruses and their replication, p., 763–843. In: Fields B, Knipe D, Howley P, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 6.Crouch R J. Ribonuclease H: from discovery to 3D structure. New Biol. 1990;2:771–777. [PubMed] [Google Scholar]
- 7.Davies J F, 2d, Hostomska Z, Hostomsky Z, Jordan S R, Matthews D A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 8.Fassati A, Goff S P. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippov V, Filippova M, Gill S S. Functional characterization of RNase H1 from Drosophila melanogaster. Biochem Biophys Res Commun. 1997;240:844–849. doi: 10.1006/bbrc.1997.7756. [DOI] [PubMed] [Google Scholar]
- 10.Gao H Q, Boyer P L, Arnold E, Hughes S H. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 11.Goedken E R, Marqusee S. Folding the ribonuclease H domain of Moloney murine leukemia virus reverse transcriptase requires metal binding or a short N-terminal extension. Proteins. 1998;33:135–143. [PubMed] [Google Scholar]
- 12.Goedken E R, Marqusee S. Metal binding and activation of the ribonuclease H domain from moloney murine leukemia virus. Protein Eng. 1999;12:975–980. doi: 10.1093/protein/12.11.975. [DOI] [PubMed] [Google Scholar]
- 13.Hesslein D G T, Campbell A G. Molecular cloning and expression of a ribonuclease H from the kinetoplastid, Trypanosoma brucei. Mol Biochem Parasitol. 1997;86:121–126. doi: 10.1016/s0166-6851(97)90014-1. [DOI] [PubMed] [Google Scholar]
- 14.Itaya M, McKelvin D, Chatterjie S K, Crouch R J. Selective cloning of genes encoding RNase H from Salmonella typhimurium, Saccharomyces cerevisiae and Escherichia coli rnh mutant. Mol Gen Genet. 1991;227:438–445. doi: 10.1007/BF00273935. [DOI] [PubMed] [Google Scholar]
- 15.Kanaya S, Kohara A, Miura Y, Sekiguchi A, Iwai S, Inoue H, Ohtsuka E, Ikehara M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990;265:4615–4621. [PubMed] [Google Scholar]
- 16.Keck J L, Marqusee S. Substitution of a highly basic helix/loop sequence into the RNase H domain of human immunodeficiency virus reverse transcriptase restores its Mn(2+)-dependent RNase H activity. Proc Natl Acad Sci USA. 1995;92:2740–2744. doi: 10.1073/pnas.92.7.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobil J H, Campbell A G. Trypanosoma brucei RNase HI requires its divergent spacer subdomain for enzymatic function and its conserved RNA binding motif for nuclear localization. Mol Biochem Parasitol. 2000;107:135–142. doi: 10.1016/s0166-6851(00)00182-1. [DOI] [PubMed] [Google Scholar]
- 18.Kobil J H, Campbell A G. Functional analysis of the domain organization of Trypanosoma brucei RNase HI. Biochem Biophys Res Commun. 2000;270:336–342. doi: 10.1006/bbrc.2000.2397. [DOI] [PubMed] [Google Scholar]
- 19.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B, Knipe D, Howley P, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 845–916. [Google Scholar]
- 20.Ohtani N, Haruki M, Morikawa M, Kanaya S. Molecular diversities of RNases H. J Biosci Bioeng. 1999;88:12–19. doi: 10.1016/s1389-1723(99)80168-6. [DOI] [PubMed] [Google Scholar]
- 21.Palaniappan C, Wisniewski M, Jacques P S, Le Grice S F, Fay P, J, Bambara R A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 22.Rausch J W, Le Grice S F. Substituting a conserved residue of the ribonuclease H domain alters substrate hydrolysis by retroviral reverse transcriptase. J Biol Chem. 1997;272:8602–8610. doi: 10.1074/jbc.272.13.8602. [DOI] [PubMed] [Google Scholar]
- 23.Robinson A B, McKerrow J H, Cary P. Controlled deamidation of peptides and proteins: an experimental hazard and a possible biological timer. Proc Natl Acad Sci USA. 1970;66:753–757. doi: 10.1073/pnas.66.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanese N, Goff S P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci USA. 1988;85:1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telesnitsky A, Goff S P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tisdale M, Schulze T, Larder B A, Moelling K. Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J Gen Virol. 1991;72:59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- 27.Wright H T. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]