Abstract
Most of the Epstein-Barr virus genome in latently infected cells is in a standard nucleosomal structure, but the region encompassing oriP and the Epstein-Barr virus-encoded small RNA (EBER) genes shows a distinctive pattern when digested with micrococcal nuclease. This pattern corresponds to a previously mapped nuclear matrix attachment region. Although the EBER genes are adjacent to oriP, there is only a two- to fourfold effect of oriP on EBER expression. However, sequences containing a consensus ATF site upstream of EBER1 are important for EBER1 expression.
The relationship between gene expression, local chromatin structure, and proximity to origins of DNA replication are all of current interest in the study of gene regulation. The Epstein-Barr virus (EBV) chromosome may be a useful system to study these topics, since it is a relatively large circular double-stranded DNA genome (about 172 kb in length) which is maintained in cell nuclei as a multicopy episome (7, 18). The EBV DNA contains about 85 genes, most of which are transcribed by cell RNA polymerase II, but the two EBV-encoded small RNA (EBER) genes are transcribed by RNA polymerase III. Sensitivity of RNA polymerase III transcription to control by proteins conventionally considered to regulate transcription factors for genes transcribed by RNA polymerase II has been studied recently (20). The EBER genes are located immediately adjacent to the oriP plasmid origin of replication, which is required for the maintenance of the EBV episome in human cells. oriP contains two segments of reiterated binding sites for the EBV EBNA1 protein, the family of repeats (FR) and the dyad symmetry element. In addition to its role in plasmid maintenance, the FR portion of oriP also acts as a transcription enhancer and has been shown to increase transcription from the EBV Cp promoter (23) and the promoter for the LMP1 gene (9), both of which are transcribed by RNA polymerase II.
The EBERs are the most abundant EBV transcripts in latently infected cells. EBER1 is 167 nucleotides and EBER2 is 172 nucleotides in length. Neither is thought to be translated into protein, and their functions are uncertain (for a review, see reference 4). Although the EBER genes are not essential for EBV infection of human B lymphocytes and establishment of lymphoblastoid cell lines (24), the EBERs are expressed in several types of EBV-associated cancer. Analysis of the cell growth properties of the Akata Burkitt's lymphoma cell line indicated that EBER genes conferred a more-transformed phenotype on clones of Akata cells that lack EBV, converting their growth properties to be more similar to EBV-positive clones of Akata cells (19). EBERs are not expressed to a significant degree in the lytic EBV infection observed in oral hairy leukoplakia (10), and the transcription of the EBER genes has been shown to be reduced when infected cells are induced to the lytic cycle (11). EBER genes are transcribed by RNA polymerase III, and the EBER genes contain internal promoter sequences similar to those in other polymerase III genes, such as tRNA and 5S RNA. However, unlike transcription in tRNA and 5S RNA, EBER transcription is also regulated by sequences 5′ of each of the EBER genes (14). These sequences appear to bind transcription factors normally associated with the regulation of genes transcribed by RNA polymerase II. The promoter for EBER2 has been analyzed by testing mutated promoters in transfection assays (13, 14). The results showed that deleting 5′ sites that would be expected to bind SP1 (box I) and ATF (box II) together reduced transcription by more than 90%. Additionally, deleting a sequence resembling a TATA box (box III) reduced expression even further. Point mutations in either box I or box III reduced expression by 80%, whereas mutating the ATF-like box II site reduced expression by 50%.
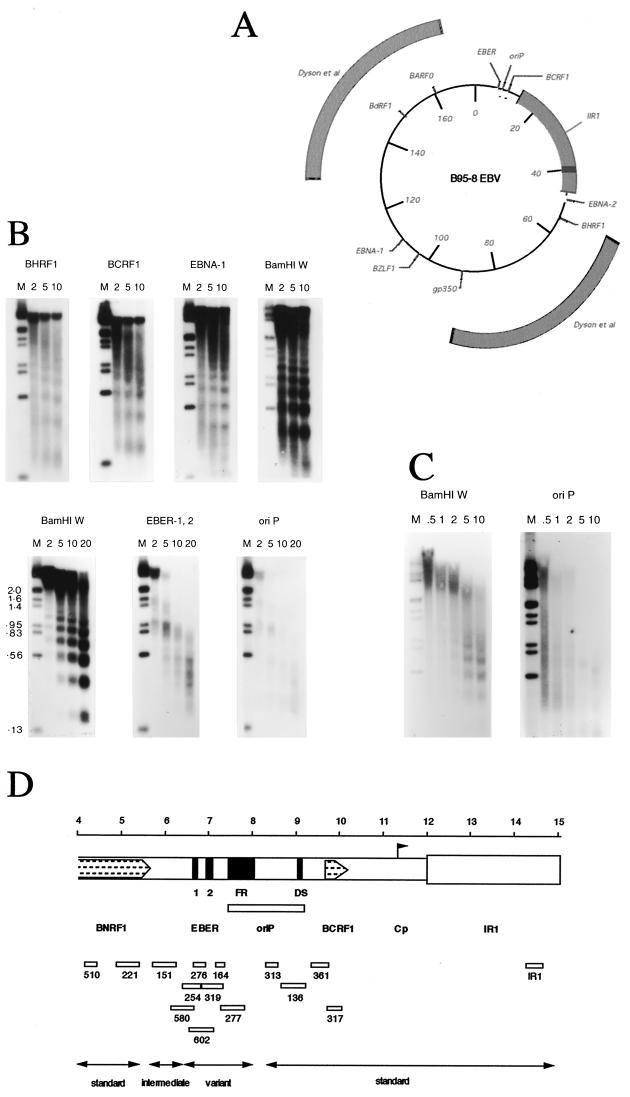
EBV DNA has been shown previously to be organized in nucleosomes in latently infected cells (6, 22), and the relationship between histone acetylation at the Zp promoter and reactivation of the viral genome from latency have recently been demonstrated (17). The earlier studies of the nucleosomal structure of EBV DNA in latently infected cells used very large probes (6), so the nucleosome patterns were averaged over large regions of the genome. To investigate chromatin structure locally in more-defined regions of the viral genome, we probed Southern blots of DNA obtained from partial micrococcal nuclease digests of nuclei from EBV-positive Akata cells with shorter DNA probes (Fig. 1A). Probes at most locations in the viral genome gave the same regular nucleosomal pattern superimposed on a background smear from the lytically replicated viral DNA (Fig. 1B, top). The lytically replicated viral DNA, which appears not to be in a regular nucleosomal structure, was present because of the low level of spontaneous lytic viral replication that occurs in Akata cells; a similar pattern was observed previously in B95-8 cells (6). As with B95-8 cells, it was possible to reduce the background by growing the cells in a low concentration of phosphonoacetic acid (PAA) to prevent lytic replication of EBV. After this treatment, a much clearer pattern could be observed (Fig. 1B, bottom, BamHI W).
FIG. 1.
(A) Summary of probes used to determine nucleosome patterns in Akata EBV, shown on a circular map representing the B95-8 strain (172,281 bp). Map coordinates are shown in kilobases at 20-kb intervals. The regions of the genome probed previously (6) (positions 54853 to 92708 and 128848 to 166619) are compared with the probes used in this study. Coordinates of probes shown were as follows: EBER, positions 6629 to 7096; oriP, 7271 to 7818; BCRF1, 9705 to 10138; IR1, 14265 to 14585; EBNA2, 48810 to 49059; BHRF1, 54406 to 54891; gp350, 89621 to 89958; BZLF1, 102725 to 102980; EBNA1, 109299 to 109877; BdRF1, 148814 to 149389; and BARF0, 160301 to 160760. (B) Southern blots of micrococcal nuclease digestion patterns (6) of Akata 6 (17) and AM58 chromatin. Nuclei from Akata 6 cells were digested with micrococcal nuclease for 2, 5, 10, or 20 min, and DNA was extracted and analyzed by Southern blotting with the indicated probes (details of probes are given in the legend to panel A). For the bottom three panels, the Akata 6 cells were grown in 0.2 to 0.4 mM PAA for 10 days prior to analysis to reduce the level of spontaneous lytic EBV replication in the culture. DNA size markers were coelectrophoresed (lanes M), and sizes are shown in kilobases on the bottom panels. (C) Micrococcal nuclease digestion patterns as shown in panel B but from AM58 cells, which contain an EBV strain lacking the EBER genes (24). (D) Analysis of boundaries of variant micrococcal nuclease digestion pattern around oriP and EBER gene regions. Part of the genome of B95-8 EBV is represented under a scale in kilobases, and the positions of the EBER genes, oriP, the major internal repeat IR1, and open reading frames BCRF1 and BNRF1 (part) are shown. Probes marked below were derived from the BC series of M13 clones used in the EBV sequencing program (3) and are drawn to scale; boundaries are as follows: 510.BC, positions 4176 to 4425; 221.BC, 4920 to 5430; 151.BC, 5712 to 6259; 580.BC, 6121 to 6684; 254.BC, 6463 to 6835; 602.BC, 6587 to 7150; 276.BC, 6656 to 6933; 319.BC, 6865 to 7343; 164.BC, 7184 to 7380; 277.BC, 7271 to 7818; 313.BC, 8339 to 8602; 136.BC, approximately 8699 to 9277; 361.BC, 9368 to 9746; 317.BC, 9708 to 10065; IR1, 14265 to 14585.
The exceptions to the regular nucleosomal pattern were those probes hybridizing to EBV sequences near the oriP and EBER gene regions (Fig. 1B). Those probes gave a weak and smeared pattern on the Southern blots. By using various short probes around this region (Fig. 1D and data not shown), it was possible to identify approximate boundaries of the part of the EBV genome that do not give a regular nucleosomal pattern, which corresponded approximately to nucleotides 6400 to 8300 on the viral genetic map. This region of EBV approximates the matrix attachment region (MAR) (16), which has previously been found by other techniques to be attached to the nuclear matrix during latency. It is unclear whether the unusual micrococcal nuclease profile of this part of the viral DNA is due to extreme sensitivity to micrococcal nuclease or to a chromatin structure that is different from the rest of the viral episome (15). The EBER genes are adjacent to oriP, within the region of unusual chromatin structure. The different chromatin structure was, however, not simply a consequence of the high level of transcription of the EBER genes because a recombinant EBV in which the EBER genes had been deleted (AM58) (24) also showed the same poorly defined micrococcal nuclease digestion pattern using an oriP probe (Fig. 1C).
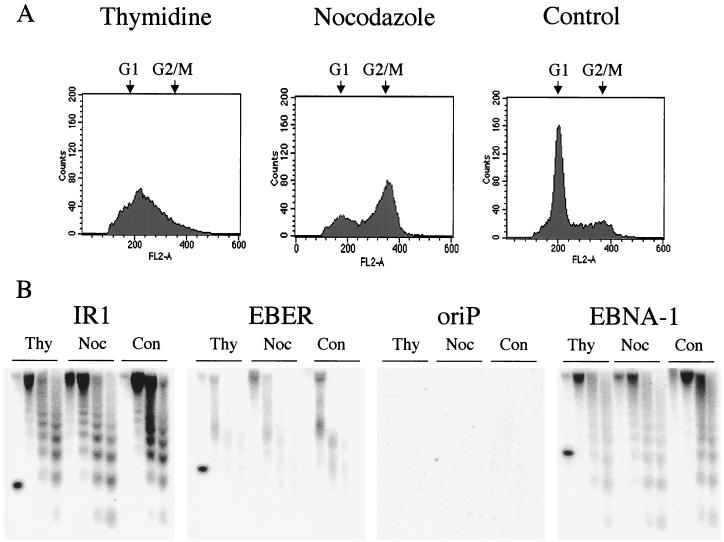
We considered the possibility that the variant EBV chromatin pattern might be affected by the cell cycle stage of the cells. EBV DNA synthesis occurs during the cell S phase, and the adherence of EBV episomes to host cell chromosomes has been visualized in mitotic chromosomes so a cell cycle relationship could be envisaged. Burkitt's lymphoma cells are generally difficult to synchronize in the cell cycle (2), but populations enriched for S or M phase were obtained by treating the cells with thymidine or nocodazole, respectively. Propidium iodide staining of the nuclei and flow cytometry demonstrated the effectiveness of these procedures (Fig. 2A). However, Southern blotting of comparable micrococcal nuclease partial digests of the chromatin from these cells showed no effect of the thymidine or nocodazole in the regions studied (Fig. 2B). The IR1 and EBNA1 probes have the standard 200-bp nucleosomal ladder, and the EBER gene region and oriP display the weak hybridization patterns shown in Fig. 1. We thus found no evidence for a relationship of the variant structure to a particular phase of the cell cycle.
FIG. 2.
Micrococcal nuclease digestion patterns are not affected by enriching for cells in the S or M phase of the cell cycle. Akata cells grown in 0.2 mM PAA were treated for 48 h with 2 mM thymidine (S-phase block) or 24 h with 0.5 μg of nocodazole (5)/ml for an M-phase block or left as a control. (A) Samples of cells were stained with propidium iodide and analyzed by flow cytometry for DNA content. Positions of G1 and G2/M DNA content are indicated. (B) Cell nuclei were prepared and digested with micrococcal nuclease as shown in Fig. 1. The extracted DNA was analyzed by Southern blotting with replica filters as shown in Fig. 1 with probes IR1, EBV (positions 14265 to 14585), EBER (6629 to 7096), and EBNA1 (109299 to 109877). Ten picograms of the PCR product used to make the probe was spiked into the left-hand lane of each of these filters to act as a positive hybridization control. The EBER filter was then stripped in boiling water for 10 min and reprobed with the oriP probe (277.BC, 7271 to 7818).
Although the EBERs are transcribed by RNA polymerase III with the normal intragenic control elements, the unusual additional control of the promoter by upstream elements which may bind transcription factors normally associated with RNA polymerase II transcription is of considerable interest. The previously reported deletion analysis of the promoter for the EBER2 gene (14), identifying elements box I, II, and III (Fig. 3A) was performed in the absence of oriP and EBNA1, and no detailed mapping has been reported for EBER1. We therefore sought to test the effect of 5′ deletions of the EBER1 promoter and the contribution of oriP and/or EBNA1 to EBER expression. Although we have recently advocated the use of established cell lines containing episomal vectors in the analysis of EBV gene regulation (17), problems were encountered with the EBER genes because of progressive loss of EBER expression. We were able to prepare transfected cell lines which initially showed expression of the EBER1 gene comparable with that found in normal Akata cells (Fig. 3B), but expression of the EBER genes and EBNA1 was invariably lost as these EBER lines were cultured. In contrast to our studies with other oriP plasmids, there was presumably some selection against the p294:EcoJ plasmid during extended culture, although the cells were maintained in hygromycin. This variation rendered the stable transfection system unsuitable for quantitative analysis of EBER promoter elements, so we instead used a transient assay system similar to that employed for EBER2 analysis (14). Assessment of the effect of oriP on EBER expression can evidently only be done with a transient assay system, since the plasmids lacking oriP would not be maintained as episomes.
FIG. 3.
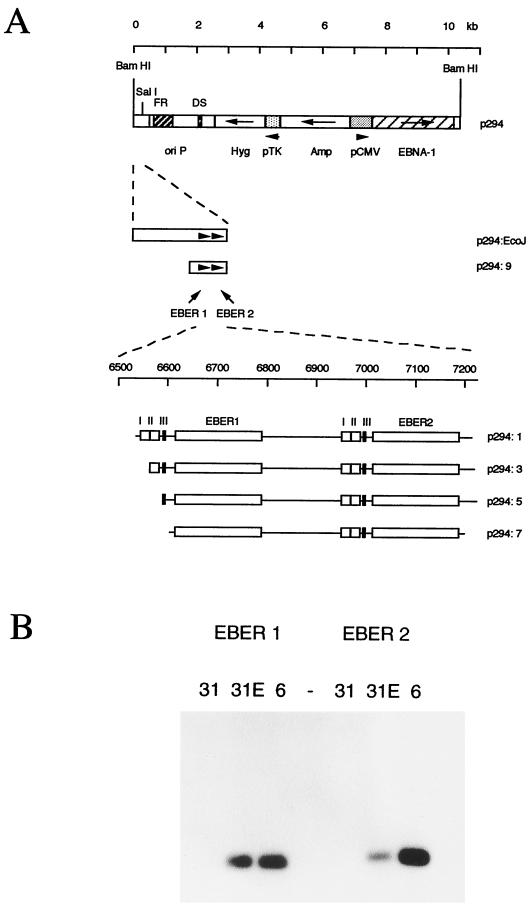
(A) Plasmids used for EBER transfection assays. The vector p294 (also named CMVpEBNA1 in reference 23) was a gift from Bill Sugden. The EcoRI J fragment of B95-8 EBV was subcloned into the EcoRI site of Bluescript II pBSK+, choosing the orientation that places the EBER genes close to the polylinker SalI site. The EBV sequence was then excised with the polylinker enzymes BamHI and SalI and cloned between the BamHI and SalI site of p294, creating the plasmid p294:EcoJ. Deletions 1, 3, 5, 7, and 9 were made by PCR from p294:EcoJ. The PCR products were cloned into the pTA vector and then subcloned into p294. The EBV content of deletion 1 is from position 6551 to 7315, 3 is from 6574 to 7315, 5 is from 6601 to 7315, 7 is from 6607 to 7315, and 9 is from 6297 to 7315. (B) p294:EcoJ was transfected into the EBV-negative Akata 31 cell line (17), and hygromycin-resistant cells were selected (lanes 31E). Cytoplasmic RNA was extracted and compared on Northern blots to RNA from EBV-positive Akata 6 cells (17) (lanes 6) or untransfected Akata 31 cells (lanes 31). Replica blots were probed for EBER1 or EBER2 with probes described previously (8).
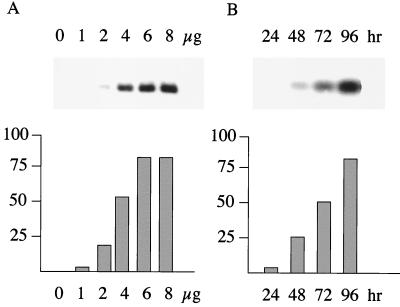
In the p294:EcoJ plasmid (Fig. 3A), the EBER genes are reconstructed close to their normal configuration relative to oriP. EBER1 expression from this construct was compared to that of a series of 5′ deletions upstream of EBER1 (Fig. 3A). EBER RNA expression from the plasmids was measured by transfection of 293 cells and analysis of total cell RNA by Northern blotting with probes specific for EBER1 or EBER2. The amount of transfected plasmid and the time of harvesting the RNA after transfection were titrated so that the subsequent assays would be within the linear range of the assay (Fig. 4A). Assay conditions of 4 μg of transfected DNA (with 0.25 μg of pCMV-βgal) were selected with an assay time of 48 to 50 h (Fig. 4A).
FIG. 4.
(A) Dose response. 293 cells were transfected by the calcium phosphate method with increasing amounts of a p294:EcoJ plasmid, with all transfections also receiving 0.25 μg of pCMV-βgal to monitor transfection efficiency. Forty-eight hours after transfection, the RNA was isolated and analyzed by Northern blotting for EBER1. The phosphorimager data were corrected for transfection efficiency with the corresponding β-galactosidase values and then plotted (bottom) on an arbitrary scale. (B) Time course. 293 cells were transfected as shown in panel A with 4 μg of the same p294:EcoJ plasmid and 0.25 μg of pCMV-βgal. Cells were harvested at the times indicated, the resulting RNA was analyzed by Northern blotting for EBER1, and the EBER1 expression was quantitated.
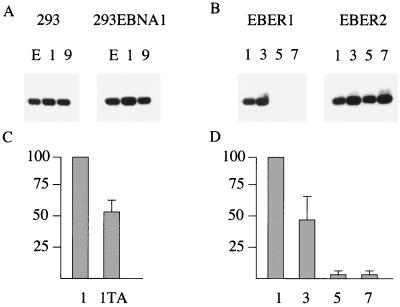
5′ deletion of the p294:EcoJ plasmid through p294:9 or p294:1 made little difference to the expression of EBER1, indicating that sequences upstream of box I were not required for EBER1 expression (Fig. 5A). To ensure that the level of EBNA1 expressed from the p294-transfected vector was sufficient for oriP function, the same plasmids were also transfected into 293 cells constitutively expressing EBNA1; only a small increase was observed, of about 50% (Fig. 5A). Successive deletion of boxes I, II, and III (plasmids p294:3, p294:5, and p294:7) showed a major effect on EBER1 transcription of deleting box II (Fig. 5B). The results of five similar experiments were averaged to give the quantitation of EBER1 expression shown in Fig. 5D. The RNA from the experiment shown in Fig. 5B was also blotted for EBER2 expression (Fig. 5B); this did not vary significantly as EBER1 promoter elements were deleted and confirms the good quality of the RNA (Fig. 5B, lanes 5 and 7) of the EBER1 experiment. Although little additional effect was seen when box III was deleted, this may be because the level of transcription was already very low after deletion of box II. Deletion of box I caused a 50% reduction, on average, of EBER1 expression. This is a much smaller reduction than was observed upon mutation of box I in the EBER2 analysis (14).
FIG. 5.
(A) 293 cells or 293 EBNA1 cells (constitutively expressing EBNA1) were transfected with 4 μg of p294:EcoJ plasmid (lanes E), plasmid 1 or 9, and 0.25 μg of pCMV-βgal. After 48 h, RNA was isolated and analyzed by Northern blotting for EBER1 expression. (B) 293 cells were transfected as in panel A with plasmids p294:1, p294:3, p294:5, and p294:7 and analyzed for expression of EBER1 or EBER2. (C) 293 cells were transfected as described in panel A with p294:1 or pTA:1 and analyzed by Northern blotting for EBER1 expression. The data from five such experiments were quantitated on the phosphorimager, normalized on the values for p294:1, corrected for transfection efficiency using the corresponding β-galactosidase values, and then shown as an average with a standard error bar. (D) The data from Fig. 5B and four similar experiments were quantitated as described in Fig. 5C to give average values for the activities of EBER1 expression and standard error bars.
Some of the EBER promoter mutants were also cloned into the pTA vector, which lacks oriP and EBNA1. The expression of EBER1 from construct 1 from the TA vector in 293 cells was about 50% of that of the corresponding p294 construct (Fig. 5C), suggesting only a modest enhancement of EBER expression by oriP, in contrast to the up to 200-fold increase reported with oriP on the Cp and LMP1 promoters (9, 23).
In summary, we have mapped a region spanning the EBER genes and the FR segment of oriP in which the DNA seems to be more sensitive to micrococcal nuclease than the rest of the EBV episome, not having a standard nucleosomal structure. The sequences around oriP and the EBER genes appear to be exceptionally sensitive to digestion rather than protected from access to the micrococcal nuclease (there is very little signal on the Southern blots with the EBER and oriP probes), so access of micrococcal nuclease to the DNA is not prevented in this region. The region of variant chromatin structure corresponds approximately to the MAR identified previously by different methods (16), consistent with the notion of attachment to the cell chromosomes through this part of the episome. Since chromosome attachment is considered to be via EBNA1 binding to the FR segment of oriP, it was surprising that the region of variant chromatin structure extended so far, particularly on the EBER side of the FR. It might be that the reason for the EBER genes having a variant chromatin structure is more related to their high level of transcription than the MAR function, which was not mapped very precisely in the earlier experiments (16). Since the EBV episomes contain genes transcribed by RNA polymerases II and III and the episomes could potentially be isolated as chromatin, EBV may be a useful system with which to study the relationship between chromatin structure and gene expression.
The sequences regulating EBER2 transcription have been studied previously (14). Because of the presence of similar sequences upstream of EBER1, it was assumed that the regulation of EBER1 was the same. We have now tested this directly and shown that box II plays a major role in regulating EBER1 expression in the assay system we have used. The presence of oriP and EBNA1 increased EBER transcription two- to fourfold but had a much smaller effect than has been observed on the Cp or LMP1 promoters, which are conventional RNA polymerase II promoters of EBV. Whether this reflects the different chromatin states of the EBER genes compared to the polymerase II promoters or the different transcription factors involved is currently unknown. Box II contains a potential ATF binding site (sequence TGACGTAG), with which a standard ATF site competes effectively in a transcription assay (14). We have found that transfection of an ATF1 expression vector caused a modest increase in expression of EBER1 (data not shown), but the ATF transcription factor family is a very complex matrix of protein dimers (12), so it is difficult to know which ATF factor might be the relevant one to study in connection with this site. With run-on assays, EBER transcription has been shown to decline during lytic replication (11). It is remarkable that there is very little or no EBER detectable during the intense EBV lytic replication that is observed in oral hairy leukoplakia lesions (10), bearing in mind the very high abundance of EBERs in most EBV-infected cell lines and tumors and the ubiquitous expression of RNA polymerase III. It would be difficult to determine exactly which forms of ATF are present in oral hairy leukoplakia, since such small amounts of tissue are available from the patients. However, changes in the ATF family profile during epithelial cell differentiation in the squamous epithelium might account for the lack of EBER expression, in the same way that changes in ATF activity have previously been proposed to be related to induction of the lytic cycle of EBV (1, 21).
Acknowledgments
B.W. and A.S. are equal first authors.
REFERENCES
- 1.Adamson A L, Darr D, Holley-Guthrie E, Johnson R A, Mauser A, Swenson J, Kenney S. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J Virol. 2000;74:1224–1233. doi: 10.1128/jvi.74.3.1224-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allday M J, Farrell P J. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol. 1994;68:3491–3498. doi: 10.1128/jvi.68.6.3491-3498.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P, Barrell B. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 4.Clemens M J. Functional significance of the Epstein-Barr virus-encoded small RNAs. Epstein-Barr Virus Rep. 1994;1:107–111. [Google Scholar]
- 5.Davenport M G, Pagano J S. Expression of EBNA-1 mRNA is regulated by cell cycle during Epstein-Barr virus type I latency. J Virol. 1999;73:3154–3161. doi: 10.1128/jvi.73.4.3154-3161.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson P J, Farrell P J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 7.Farrell P. Epstein-Barr virus immortalizing genes. Trends Microbiol. 1995;3:105–109. doi: 10.1016/s0966-842x(00)88891-5. [DOI] [PubMed] [Google Scholar]
- 8.Farrell P J, Hollyoake M, Niedobitek G, Agathanggelou A, Morgan A, Wedderburn N. Direct demonstration of persistent EBV gene expression in peripheral blood of infected common marmosets and analysis of virus infected tissues in vivo. J Gen Virol. 1997;78:1417–1424. doi: 10.1099/0022-1317-78-6-1417. [DOI] [PubMed] [Google Scholar]
- 9.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilligan K, Rajadurai P, Resnick L, Raab-Traub N. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc Natl Acad Sci USA. 1990;87:8790–8794. doi: 10.1073/pnas.87.22.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greifenegger N, Jager M, Kunz-Schughart L A, Wolf H, Schwarzmann F. Epstein-Barr virus small RNA (EBER) genes: differential regulation during lytic viral replication. J Virol. 1998;72:9323–9328. doi: 10.1128/jvi.72.11.9323-9328.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai T W, Liu F, Coukos W J, Green M R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 13.Howe J, Shu M. Upstream basal promoter element important for exclusive RNA polymerase III transcription of the EBER 2 gene. Mol Cell Biol. 1993;13:2655–2665. doi: 10.1128/mcb.13.5.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe J G, Shu M D. Epstein-Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell. 1989;57:825–834. doi: 10.1016/0092-8674(89)90797-6. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh D, Camiolo S, Yates J. Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent replication. EMBO J. 1993;12:4933–4944. doi: 10.1002/j.1460-2075.1993.tb06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jankelevich S, Kolman J L, Bodnar J W, Miller G. A nuclear matrix attachment region organizes the Epstein-Barr viral plasmid in Raji cells into a single DNA domain. EMBO J. 1992;11:1165–1176. doi: 10.1002/j.1460-2075.1992.tb05157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins P, Binné U, Farrell P. Histone acetylation and reactivation of Epstein-Barr virus from latency. J Virol. 2000;74:710–720. doi: 10.1128/jvi.74.2.710-720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 2343–2396. [Google Scholar]
- 19.Komano J, Maruo S, Kurozumi K, Oda T, Takada K. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J Virol. 1999;73:9827–9831. doi: 10.1128/jvi.73.12.9827-9831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larminie C G, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacCallum P, Karimi L, Nicholson L J. Definition of the transcription factors which bind the differentiation responsive element of the Epstein-Barr virus BZLF1 Z promoter in human epithelial cells. J Gen Virol. 1999;80:1501–1512. doi: 10.1099/0022-1317-80-6-1501. [DOI] [PubMed] [Google Scholar]
- 22.Shaw J E, Levinger L F, Carter C., Jr Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979;29:657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]