Abstract
Molecular analysis of a cytopathogenic (cp) bovine viral diarrhea virus (BVDV) isolate (1741) obtained from a case of mucosal disease (MD) led to the identification of five different viral subgenomic RNAs in addition to a noncytopathogenic (noncp) strain (NCP 1741). For each of the subgenomes, a large internal deletion was found together with an inserted sequence encoding part of ribosomal protein S27a fused to an N-terminally truncated ubiquitin monomer. Surprisingly, the two cellular insertions together with flanking viral sequences encoding parts of NS3 and NS4B are >99% identical to the previously described sequence of BVDV vaccine strain RIT (P. Becher, M. Orlich, and H.-J. Thiel, J. Virol. 72:8697–8704, 1998), while the remainder of the subgenomes is derived from the genome of NCP 1741. Further analyses including molecular cloning and nucleotide sequencing of the recombination partners revealed that both homologous and nonhomologous RNA recombination contributed to the generation of the viral subgenomes. Interestingly, for another cp BVDV isolate (CP 4584) from an independent case of MD, again an insertion of a RIT-derived sequence element was detected. In contrast to CP 1741, for CP 4584 a duplication of the genomic region encoding NS3 and parts of NS4A and NS4B was found. Transfection of bovine cells with RNA transcribed from a chimeric cDNA construct showed that the RIT-derived insertion together with the CP 4584-specific duplication of viral sequences represents the genetic basis of cytopathogenicity of CP 4584. Remarkably, passages of the recovered cp virus in cell culture led to emergence of noncp BVDV and a number of viral subgenomes whose genome organization was similar to that in BVDV 1741.
The genera Pestivirus, Flavivirus, and Hepacivirus constitute the family Flaviviridae. The genus Pestivirus is represented by the species Bovine viral diarrhea virus 1 (BVDV-1), BVDV-2, Classical swine fever virus (CSFV), and Border disease virus (18). Pestiviruses have a positive-sense single-stranded RNA genome of about 12.3 kb in length with one large open reading frame (ORF) flanked by 5′ and 3′ nontranslated regions (NTR) (see references 26 and 33 for reviews). This ORF encodes a polyprotein of approximately 3,900 amino acids (aa) which is co- and posttranslationally processed by viral and cellular proteases, leading to the mature viral proteins. The first third of the ORF encodes an autoprotease and four structural proteins, while the 3′ part of the RNA genome codes for the other nonstructural (NS) proteins (see references 26 and 33 for reviews). Based on the effects in tissue culture, two biotypes, cytopathogenic (cp) and noncytopathogenic (noncp), are distinguished (17, 20).
BVDV represents one of the most important pathogens of cattle, causing significant economical losses worldwide (1). Horizontal BVDV infection can have different consequences, such as abortion, diarrhea, hemorrhagic syndrome, and, most frequently, inapparent courses (see references 1 and 33 for reviews). Diaplacental infection with noncp BVDV can result in the birth of persistently infected animals with an acquired immunotolerance to the original BVDV strain. Such persistently infected animals may come down with mucosal disease (MD). In addition to the persisting noncp BVDV, a cp BVDV can always be isolated from animals with MD (12, 26).
Molecular characterization of several BVDV pairs strongly suggested that the cp viruses can evolve from the respective noncp viruses by nonhomologous RNA recombination (see reference 26 for a review). For the cp viruses, various genomic alterations were identified, including insertions of cellular sequences, frequently together with large duplications of viral sequences, and genomic rearrangements with large duplications and deletions (2, 4, 8, 23, 26, 30). One important difference between cp and noncp BVDV is the expression of NS3, which is colinear to the C-terminal part of NS2-3. While NS2-3 is expressed in both cp and noncp BVDV-infected cells, NS3 is found exclusively after infection with cp BVDV. Accordingly, NS3 is regarded as the marker protein for cp BVDV strains.
In this paper, we report the identification of BVDV vaccine strain RIT-derived insertions in the genomes of two cp BVDV isolates obtained from independent cases of MD. The results of this study, including the molecular characterization of the putative recombination partners, strongly suggest that homologous and nonhomologous RNA recombination between persisting noncp BVDV and BVDV vaccine strain RIT can be responsible for induction of fatal MD.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK) cells were obtained from the American Type Culture Collection (Manassas, Va.). Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% horse serum. The cp BVDV isolates 1741 and 4584 were isolated from cattle in Germany in 1996 that came down with MD. BVDV strains CP7 and NCP7 as well as BVDV vaccine strain RIT 4350 (Pfizer, Karlsruhe, Germany) have been described previously (8, 12, 21, 24). Comparative sequence analyses indicate that all virus isolates included in this study are BVDV-1 strains.
Infection of cells.
Supernatants and lysates of infected cells were combined and used for infection of MDBK cells. Material for infection was prepared by freezing and thawing cultures 48 h postinfection and stored at −70°C. Infection with noncp BVDV was detected by immunofluorescence (IF) with monoclonal antibody 8.12.7 (directed against NS3), kindly provided by E. J. Dubovi (Cornell University, Ithaca, N.Y.).
RNA preparation, gel electrophoresis, and Northern (RNA) hybridization.
Preparation of RNA, gel electrophoresis, radioactive labeling of the probes, hybridization, and posthybridization washes were performed as described previously (3). A 2.5-kb NotI-NsiI fragment from the cDNA clone pCP7-5A (7) encompassing the NS3 gene of BVDV CP7 was used as a probe.
Oligonucleotides.
If not otherwise indicated, numbering of nucleotides throughout this work refers to the genomic sequence of BVDV SD-1, the first completely sequenced noncp BVDV strain (13). Oligonucleotides were purchased from MWG Biotech GmbH (Ebersberg, Germany). Oligonucleotides Ol BVDV 7100 (nucleotides [nt] 7313 to 7335; sense), Ol NS3R (nt 5326 to 5343; antisense), Ol 100 (nt 107 to 127; sense), Ol 1400R (nt 1430 to 1448; antisense), Ol Rit-ubi4 (corresponding to part of the ubiquitin coding sequence; sense), and Ol Rit-4AR (nt 7417 to 7434; antisense) have been described previously (5, 6, 8). Primers Ol 5150 (5′-AGRGGGCCWGCCGTGTG-3′; R = A or G, W = A or T; corresponding to nt 5150 to 5166; sense), Ol 7400R (5′-AGTCTCYTTCCCCTCAGTTC-3′; Y = C or T; corresponding to nt 7359 to 7378; antisense), Ol NS3 (5′-CTGGCAGTWGACCTCCTAG-3′; W = A or T; corresponding to nt 7109 to 7127; sense), and Ol 8600R (5′-GCYTTCATCTCATARCCRCA-3′; Y = C or T, R = A or G; corresponding to nt 8609 to 8628; antisense) were designed by using published sequences of BVDV-1 strains NADL, SD-1, Osloss, and CP7. The sequences of the following primers were derived from the obtained sequences of CP 1741 and CP 4584: Ol 1741-AgeIR (5′-CCAACCGGTTTCCAATCCCCTCCTCACCTTTAGCAATGCTG-3′, antisense) and Ol 4584-MluI (5′-TTACGCGTCCGGGGGATTGGATTTC-3′, sense).
RT-PCR and molecular cloning.
Reverse transcription-PCR (RT-PCR) of approximately 500 ng of heat-denatured RNA was carried out as described previously (5). The cDNA fragments obtained after RT-PCR were separated by agarose gel electrophoresis and purified by using a QIAEX DNA purification kit (Qiagen, Hilden, Germany). The respective cDNA fragments were cloned by using a TA cloning kit (Invitrogen, De Schelp, The Netherlands).
For NCP 1741, part of the 5′ NTR together with the genomic region encoding Npro, C, and part of Ems was amplified by RT-PCR with primers Ol 100 and Ol 1400R. For NCP 1741 and NCP 4584, the genomic region encoding NS3, NS4A, NS4B, and part of NS5A was amplified by using primer pairs Ol 5150-Ol 7400R and Ol NS3-Ol 8600R, respectively. For amplification of the genomic regions encoding NS3, NS4A, and part of NS4B of CP 1741 and CP 4584, primers Ol Rit-ubi4 and Ol Rit-4AR were used.
Nucleotide sequencing and sequence analysis.
Nucleotide sequences were determined by cycle sequencing with a Thermo Sequenase kit (Amersham Buchler, Braunschweig, Germany) and Li-Cor 4000 L DNA sequencer (MWG Biotech). All sequences were determined by sequencing both complementary strands of at least three independent cDNA clones. Computer analysis of sequence data was performed with HUSAR (DKFZ, Heidelberg, Germany), which provides the Genetics Computer Group software package (14).
Construction of BVDV full-length cDNA clone.
The noncp BVDV full-length cDNA clones pNCP7-5A and pNCP7-5A-(AgeI-) have been described previously (2). These plasmids are derivatives of the cp BVDV infectious full-length cDNA clone pCP7-5A (7). Numbering of nucleotides throughout this paragraph refers to the NCP7-5A sequence. Construction of p7/4584 was based on pNCP7-5A-(AgeI-), which differs from pNCP7-5A by the absence of the single AgeI site (nt 5309 to 5314). A CP 4584-specific MluI/AgeI fragment was obtained by PCR with primer Ol 4584-MluI (encompassing an MluI site which corresponds to nt 7434 to 7439) and primer Ol 1741-AgeIR, using the CP 4584 -derived cDNA as template, and cloned into pCR2.1. The resulting plasmid was termed p4584-MluI. Next, the SacI (nt 5842 to 5847)/MluI (nt 7434 to 7439) fragment of pNCP7-5A was cloned into p4584-MluI precut with SacI (located in the polylinker) and MluI. Addition of the pNCP7-5A-derived AgeI/SalI (nt 7716 to 7721) fragment completed the cloning of the SacI/SalI fragment, which was then introduced into pNCP7-5A-(AgeI-) precut with SacI and SalI. The genome organization of the resulting chimeric cDNA construct p7/4584, the genomic region derived from CP 4584, and positions of the AgeI and MluI sites are indicated in Fig. 4A. According to comparative analysis of the NS3 coding sequences, CP 4584 and NCP7 are about 94% identical.
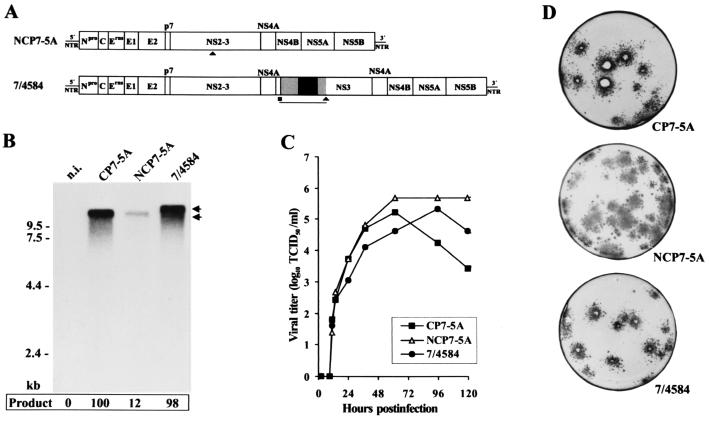
FIG. 4.
Transfection experiments with RNA transcribed from infectious cDNA clones pNCP7-5A, pCP7-5A, and p7/4584. (A) Genome organization of noncp BVDV strain NCP7-5A and the chimeric cDNA construct p7/4584 that mirrors the genome structure of CP 4584. Generation of p7/4584 was based on cDNA clone pNCP7-5A-(AgeI-), which differs from the infectious noncp cDNA clone pNCP7-5A by the absence of a single AgeI site. For 7/4584, the CP 4584-specific part of the genome encompassing part of the CP RIT-derived insertion (gray and black boxes) and positions of the MluI (■) and AgeI (▴) sites used for cDNA cloning are indicated below the bars. The remaining part of p7/4584 was derived from pNCP7-5A-(AgeI-). The lengths of the bars are not drawn to scale. (B) Northern blot analysis of total RNA from MDBK cells infected with the indicated viruses at an MOI of 0.05. The infected cells were processed in parallel with those used to determine the growth rates (C). RNAs were extracted at 24 h after infection. Northern blotting was performed as described in the legend for Fig. 1A. Numbers refer to RNA ladder sizes. Migration positions of the viral genomic RNAs are marked with arrows. The intensity of bands was determined with a phosphorimager. The relative amounts of viral genomic RNAs are indicated below the blot (percentage of CP7-5A value [100%]) n.i., RNA from noninfected MDBK cells. (C) Growth curves of the recovered BVDV CP7-5A, NCP7-5A, and 7/4584 determined on MDBK cells infected at an MOI of 0.05. The titers of released virus were determined over a 5-day period. (D) Plaques produced by BVDV CP7-5A and 7/4584 and foci produced by BVDV NCP7-5A at 4 days after infection of MDBK cells. For infection, dilutions of the supernatants from cells transfected with CP7-5A RNA, NCP7-5A RNA, and 7/4584 RNA were used. Infection of cells was visualized by immunostaining.
In vitro transcription and transfection of RNA.
In vitro synthesis of RNA and transfection of MDBK cells were carried out as described elsewhere (7). About 2 μg of RNA was used for each transfection. For electroporation (one pulse, 950 μF and 180 V), a Gene Pulser II (Bio-Rad, Munich, Germany) was used.
Plaque assay.
MDBK cells were infected with 10-fold serial dilutions of supernatants from transfected cells. After incubation at 37°C for 4 h, the attached cells were overlaid with semisolid medium containing 0.6% low-melting-point agarose (Gibco-BRL) and 5% horse serum. After 4 days of incubation at 37°C, the agarose overlays were removed, and the cells were washed with phosphate-buffered saline and then fixed with acetone-methanol (1:1) for 1 h at −20°C. Immunostaining of cells using the BVDV E2-specific monoclonal antibody D5 (kindly provided by E. Weiland, Tübingen, Germany) was carried out as described elsewhere (7).
Determination of growth kinetics.
MDBK cells (106 in a six-well dish) were infected with transcript-derived virus at a multiplicity of infection (MOI) of 0.05. After adsorption for 1 h at 37°C, the cells were washed six times with phosphate-buffered saline, overlaid with medium containing 10% horse serum, and then incubated for 5 days. After the indicated time intervals, aliquots (200 μl) of the cell culture supernatant were removed and used for titration on MDBK cells. The viral yields were determined as the titer of 50% tissue culture infectious doses (TCID50) per milliliter.
Analysis of viral RNA synthesis by Northern blotting.
MDBK cells (106) were infected with transcript-derived virus at an MOI of 0.05 and processed parallel to cells used for determination of the growth kinetics. After 1 and 2 days of incubation at 37°C, RNA was prepared using an RNeasy total RNA kit (Qiagen). Five micrograms of glyoxylated RNA was subjected to Northern blot analysis. The viral RNAs were detected by autoradiography, and the intensity of bands was determined with a phosphorimager (Fujik BAS 1000; Fuji).
Nucleotide sequence accession numbers.
Sequence data from this study have been deposited in the EMBL and GenBank data libraries and assigned accession numbers AF321450 to AF321458.
RESULTS
Characterization of BVDV 1741 by hybridization.
BVDV 1741 was obtained from a bovine that died of MD, the fatal form of a BVDV infection. In MDBK cells, this virus causes a cytopathic effect. Limiting dilution of BVDV 1741 from the third cell passage allowed isolation of an accompanying noncp virus, termed NCP 1741. However, several attempts failed to biologically clone the cp virus by plaque purification. In the text that follows, cp BVDV 1741 is termed CP 1741.
Previous studies have demonstrated that most cp BVDV isolates contain RNA genomes significantly larger or shorter than those of noncp BVDV; the alterations are due to large duplications of viral sequences or deletions, respectively (2, 4, 8, 23, 26). To investigate whether similar genomic differences are present in CP 1741, a Northern blot analysis was performed. Total RNA from MDBK cells infected with either CP 1741 or NCP 1741 was hybridized with a BVDV-specific cDNA probe (Fig. 1A). Viral genomic RNA with a size of about 12.3 kb was detected in cells infected with CP 1741 and NCP 1741. In addition, subgenomic RNA with a size of about 8 kb was identified after infection with CP 1741 but not NCP 1741. Accordingly, cytopathogenicity of CP 1741 correlates with the presence of viral subgenomic RNA.
FIG. 1.
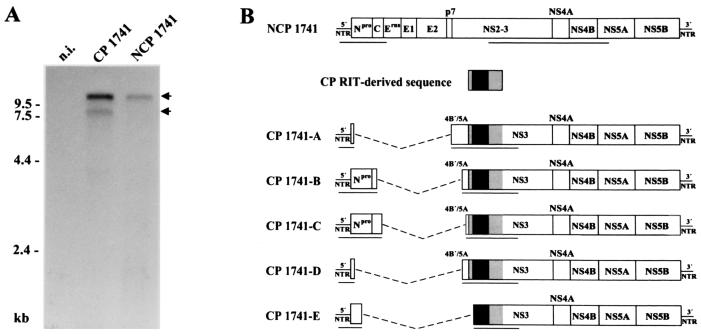
Analysis of BVDV 1741. (A) Northern blot analysis of total RNA from MDBK cells infected with CP 1741, NCP 1741, and noninfected MDBK cells (n.i.). RNA was separated by denaturing agarose gel electrophoresis, blotted onto a nylon membrane and hybridized with a 2.5-kb NotI-NsiI fragment from the cDNA clone pCP7-5A (7). Numbers refer to RNA ladder sizes. Migration positions of the viral genomic and subgenomic RNAs are marked with arrows. (B) Genome organization of NCP 1741 and the CP 1741 subgenomic RNAs. All subgenomic RNAs contain an insertion of CP RIT-derived sequences which encode part of S27a (S27a∗) fused to an N-terminally truncated ubiquitin (ubi∗) (black box) as well as flanking viral sequences (gray boxes). The deletions are indicated by dashed lines. The lengths of the bars are not drawn to scale. The underlined parts of the (sub)genomes have been sequenced. With respect to the analyzed regions, all subgenomes maintain one large ORF.
RT-PCR analysis.
So far, subgenomic RNAs have been described for two cp BVDV isolates (19, 32) as well as for four closely related cp BVDV isolates obtained during an outbreak of MD on a single farm (4). A common feature of these subgenomes is the deletion of all or almost all of the genomic region encoding the structural proteins, as well as the nonstructural proteins p7 and NS2. To determine the deletion which led to the subgenomic RNA of CP 1741, we performed an RT-PCR analysis using antisense primer Ol NS3R, located in the NS3 coding region, and sense primer Ol 100, located in the 5′ NTR. Interestingly, five specific products ranging in size from 0.9 to 1.7 kb were generated (data not shown). Changing the conditions of RT-PCR to include elongation times of up to 300 s did not result in detection of additional cDNA fragments. Under the chosen conditions, cDNA to be obtained from viral RNA without deletion with an expected size of about 5.2 kb was not amplified. We did not investigate whether this variety of cDNA fragments changed with culture conditions.
Genome organization of the subgenomes and identification of CP RIT-derived sequences.
For further characterization, the amplified cDNA fragments were cloned and subjected to nucleotide sequence analysis. To allow comparison with the corresponding sequences of NCP 1741, part of the 5′ NTR together with the genomic regions encoding Npro and C as well as NS3, NS4A, NS4B, and part of NS5A of NCP 1741 were amplified by RT-PCR, cloned, and sequenced. Analysis of the CP 1741-derived cDNA fragments resulted in identification of five subgenomes differing in size, termed CP 1741-A, -B, -C, -D, and -E. To our knowledge, there is only one other report on cp BVDV isolates consisting of such a variety of viral subgenomes in addition to noncp BVDV (4). In contrast, all other described cp BVDV isolates comprise only one cp (sub) genome. Comparative sequence analyses revealed that each of the subgenomes carries two cellular insertions which encode part of ribosomal protein S27a (S27a∗) fused to a truncated ubiquitin monomer lacking the N-terminal 3 aa (ubi∗). Interestingly, these two cellular insertions have been previously found in the genome of BVDV CP RIT, a temperature-sensitive strain widely used for vaccination (8). Moreover, further analyses of the CP 1741 sequences demonstrated that the two cellular insertions as well as flanking viral sequences encoding part of NS4B and the N-terminal region of NS3 are >99% identical to the CP RIT sequence, while the identity between these viral sequences flanking the insertions and the corresponding sequences of NCP 1741 is <93% (Fig. 1B). In contrast, the region upstream of the RIT-specific sequence element is >99% identical to the corresponding sequence of NCP 1741 but <91% identical to the CP RIT sequence. These results strongly suggest that the CP 1741 subgenomes were generated by RNA recombination between NCP 1741 and vaccine strain CP RIT.
The genome organizations of the five identified subgenomes of CP 1741 are similar to each other. The following features are common to all subgenomes described here: (i) deletion of either all or most of the genomic region encoding the structural proteins, p7, and NS2 and (ii) presence of the same host-specific insertion directly upstream of the NS3 gene. Remarkably, all deletions and insertions occurred in frame. Subgenomes CP 1741-A, -B, -C, and -D each carry an additional NCP 1741-derived viral insertion encoding various C-terminal parts of NS4B together with 3 aa of the NS5A N terminus directly upstream of the RIT-specific sequence. This duplication of viral sequences is not present in subgenome CP 1741-E, which also lacks 9 nt from the 5′ end of the S27a∗ coding region compared to the other subgenomes and CP Rit (Fig. 1B).
Comparison of NS3 coding sequences.
Our analysis of the CP 1741 subgenomic RNAs indicated that the 5′ regions of the subgenomes were derived from NCP 1741, while the two cellular insertions together with flanking viral sequences were obtained from CP RIT. On the basis of these data, however, we did not know whether the entire 3′ region or only a part of it was derived from CP RIT. To address this question, the genomic region encoding NS3, NS4A, and part of NS4B of CP 1741 was amplified by RT-PCR (using a sense primer encompassing part of the ubiquitin coding sequence and an antisense primer corresponding to the NS4B coding region), cloned, and subjected to nucleotide sequence analysis. Interestingly, the 5′ terminal 280 nt of the NS3 gene of CP 1741 were >99.5% identical to the sequence of CP RIT but <92% identical to the NCP 1741 sequence. Analysis of the remaining part of the NS3 gene of CP 1741 revealed only 93% identity to CP RIT, while 99.5% of nucleotides were identical to the sequence of NCP 1741. Moreover, RT-PCR using an antisense primer corresponding to nt 553 to 572 of the NS3 gene and a set of sense primers each spanning the internal deletion of the individual subgenomes was performed. Sequence analysis of the obtained cDNA fragments demonstrated for each of the subgenomes that the genomic region encompassing the 5′-terminal 280 nt of the NS3 gene was derived from CP RIT, while the remaining part of the NS3 coding region was derived from NCP 1741. Interestingly, the junction of the NS3 coding sequences occurred without deletion or insertion of nucleotides at the cross over site and thus resulted from precise homologous recombination. In contrast, the internal deletions and the 5′ junction between the NCP 1741-specific sequences and the CP RIT-derived sequences (see above) are due to nonhomologous recombination. Taken together, the results indicate that both nonhomologous and homologous recombination contributed to the generation of the viral subgenomes. Comparative analysis of the recombination junctions in the subgenomes with the corresponding regions of the recombination partners revealed short regions of sequence similarity at several cross over sites (data not shown), suggesting that base pairing between the nascent RNA and the acceptor template may facilitate template switching of the RNA-dependent RNA polymerase and thereby contributes to nonhomologous RNA recombination. The presence of such regions of sequence similarity at cross over sites has recently been reported for another set of BVDV subgenomes (4).
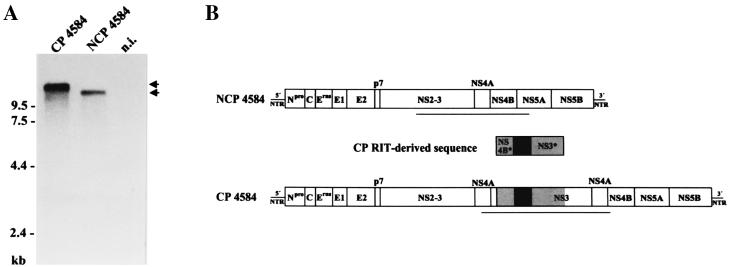
Characterization of BVDV 4584.
BVDV 4584 was also isolated from an animal that died of MD. After two passages in tissue culture cells, cp and noncp BVDV were biologically cloned and termed CP 4584 and NCP 4584, respectively. Northern blot analysis revealed that the genome of CP 4584 is about 15.2 kb long, or about 2.9 kb longer than the genomes of NCP 4584 and other noncp BVDV strains (Fig. 2A). Molecular cloning of parts of the genome of CP 4584 by RT-PCR using primers Ol 7100 and Ol NS3R and subsequent nucleotide sequencing indicated the presence of duplicated viral sequences; these encode NS3, NS4A, and aa 1 to 29 and 52 to 132 of NS4B. In addition, two cellular insertions encoding parts of ribosomal protein S27a and ubiquitin were found (Fig. 2B). Comparative analyses of the genomic region encoding NS3, NS4A, and part of NS4B of CP 4584, NCP 4584, and vaccine strain RIT demonstrated that the part of the CP 4584 sequence which encodes the two cellular insertions (S27a∗-ubi∗) as well as flanking viral sequences encoding aa 52 to 132 of NS4B (NS4B∗) and aa 1 to 371 of NS3 (NS3∗) are >99% identical to the published sequence of BVDV RIT (8), while NS4B∗ and NS3∗ are only 93 and 95%, respectively, identical to the NCP 4584 sequence. In contrast, the remaining part of the determined CP 4584 sequence encoding NS4A, aa 1 to 29 of NS4B, as well as aa 372 to 683 of NS3 is >99% identical to the sequence of NCP 4584 but <95% identical to the RIT sequence (Fig. 2B). The results of our analysis strongly suggest that CP 4584 evolved by RNA recombination between NCP 4584 and BVDV vaccine strain RIT. RNA recombination between the NS3 coding sequences of BVDV RIT and NCP 4584 occurred without deletion or insertion of nucleotides and is therefore homologous in nature, while recombination between the NS4B coding sequences of these strains resulted from a nonhomologous reaction. It should be noted that the RIT-derived insertion of CP 4584 differs from that identified in the subgenomes of CP 1741 with respect to the 5′ and 3′ borders of the insertions (Fig. 3).
FIG. 2.
Analysis of cp and noncp BVDV 4584. (A) Northern blot analysis of total RNA from MDBK cells infected with BVDV CP 4584, NCP 4584, and noninfected MDBK cells (n.i.). RNA ladder sizes are indicated on the left. Migration positions of the viral genomic RNAs are marked with arrows. (B) Genome organization of NCP 4584 and CP 4584. The genome of CP 4584 contains a duplication of viral sequences encoding NS3, NS4A, and part of NS4B together with an insertion of CP RIT-derived sequences which encode S27a∗ and ubi∗ (black box) as well as flanking viral sequences (NS4B∗, NS3∗; gray boxes). The lengths of the bars are not drawn to scale. The underlined parts of the genomes have been sequenced.
FIG. 3.
Schematic representation of part of the genome of BVDV vaccine strain RIT (top) and the RIT-derived sequence elements identified in the subgenomes A to D of CP 1741 and in the genome of CP 4584. The RIT-derived sequence elements encode S27a∗ and ubi∗ (black box) as well as flanking viral sequences (NS4B∗, NS3∗; gray boxes). The lengths of the bars are not drawn to scale. The numbers of amino acids corresponding to the indicated proteins or parts thereof are shown below the bars.
Infectious BVDV cDNA clone with S27a∗ and ubi∗ coding insertions and duplicated NS3 gene.
The subgenomes of CP 1741 and the genome of CP 4584 were partially sequenced. Accordingly, it cannot be excluded that other genetic changes are present in these viral (sub)genomes in addition to the identified insertions and deletions or duplications. To investigate whether the S27a∗-ubi∗ coding insertion together with the identified duplication of viral sequences represents the genetic basis of cytopathogenicity of CP 4584, we generated the chimeric cDNA construct p7/4584, which resembles the genome of CP 4584; this full-length clone is based on the full-length cDNA clone pNCP7-5A-(AgeI-) and contains the S27a∗-ubi∗ coding insertion together with a duplication of viral sequences encoding NS3, NS4A, and part of NS4B (Fig. 4A).
Northern blot analysis of cells infected with BVDV 7/4584 led to identification of genomic RNA exhibiting the expected size of about 15 kb (Fig. 4B). In contrast, the RNAs of CP7-5A and NCP7-5A are 12.3 kb long. For analysis of viral RNA synthesis, cells were infected at an MOI of 0.05 and processed in parallel with cells used for determination of growth kinetics. Total cellular RNA was prepared from cells at 24 h after infection and used for Northern blot analysis; after this incubation time, the titers of infectious virus were similar for NCP7-5A, CP7-5A, and 7/4584 (see below). Interestingly, determination of the relative amounts of viral genomic RNA by phosporimager analysis revealed that the amounts of viral RNAs from cells infected with the cp viruses CP7-5A and 7/4584 were about eight times higher than that produced by noncp BVDV NCP-5A (Fig. 4B). The results of our analysis indicate that viral RNA synthesis is apparently not significantly affected by the different lengths of the genomes of CP7-5A and 7/4584. Similar differences in viral RNA synthesis have recently been reported for another isogenic BVDV pair consisting of cp and noncp BVDV derived from cDNA constructs as well as three additional BVDV pairs (22, 35).
Cell lysis was observed 2 days after transfection of bovine cells with 7/4584 RNA. To characterize the recovered cp virus 7/4584, a plaque assay was performed on MDBK cells infected with supernatants from transfected cells (Fig. 4D). For comparison, plaque and focus-forming assays were also performed with supernatants from MDBK cells transfected with CP7-5A RNA and NCP7-5A RNA, respectively. The plaques generated by 7/4584 were slightly smaller than the ones produced by CP7-5A. As expected, no signs of cytopathology were observed in NCP7-5A-infected cells. For further characterization, the growth kinetics of 7/4584, CP7-5A, and NCP7-5A were determined. MDBK cells were infected with supernatants from cells transfected with the various RNAs at an MOI of 0.05. After incubation at 37°C, virus released into the medium was titrated over a 5-day period. The chimeric virus 7/4584 grew slightly more slowly than the other two viruses (Fig. 4C). Each of the three viruses reached titers of >105 TCID50/ml.
Generation of noncp BVDV and subgenomic RNAs after passaging of BVDV 7/4584.
In a recent study, we demonstrated that replication of a cp BVDV strain carrying a cellular NEDD8 coding insertion together with duplicated viral sequences rapidly resulted in the emergence of noncp BVDV (2). It was therefore of interest to investigate whether passages of 7/4584 in MDBK cells would also lead to generation of noncp BVDV. The possible emergence of noncp BVDV was monitored by IF of infected cells and Northern blot analysis. IF analysis actually indicated the presence of noncp BVDV in cells infected with 7/4584. With respect to the fifth and all subsequent passages of 7/4584, viral RNA with a size of 12.3 kb was detected in addition to the larger viral RNA of 7/4584 (Fig. 5A). For further analysis, the emerged noncp BVDV was biologically cloned using serial dilutions of supernatants from cells infected with 7/4584. To characterize this noncp BVDV, part of the NS2-3 coding region was cloned and sequenced; within this region, BVDV strain NCP7 and the engineered virus NCP7-5A carry an AgeI site (corresponding to positions 5309 to 5314 of the NCP7-5A sequence) which is absent in the genome of 7/4584. Sequence analysis revealed the absence of the AgeI site for the emerged noncp BVDV, which allows a clear distinction from NCP7 and NCP7-5A. To address the possibility that the emergence of noncp deletion genomes resulted from (i) the presence of low-level deletion templates in the plasmid DNA used for in vitro transcription or (ii) low-level deletion transcript RNAs generated by the SP6 RNA polymerase, a plaque assay using dilutions of cells transfected with 7/4584 RNA was performed directly after transfection; after 5 days a single plaque was harvested from a six-well dish with a total of seven plaques and used for further propagation. After three passages in MDBK cells noncp BVDV emerged which was biologically cloned and shown to contain the above-described genetic marker. Taken together, the results of our analyses demonstrate that the noncp BVDV evolved from 7/4584 by RNA recombination during replication in tissue culture cells.
FIG. 5.
Emergence of noncp BVDV and subgenomic RNAs after passages of cp BVDV 7/4584 in MDBK cells. (A) Northern blot of total RNA from MDBK cells infected with the indicated passage number (passage 1 [P1] and P5 to P10) of 7/4584 at an MOI of about 0.1. RNAs were extracted at 48 h after infection. Northern blotting was performed as described in the legend for Fig. 1A. Numbers refer to RNA ladder sizes. Migration positions of the genomic 7/4584 RNA (upper arrow), the genomic RNA of the emerged noncp BVDV (middle arrow), and the emerged subgenomic RNAs (lower arrow) are indicated on the right. (B) Genome organizations of BVDV 7/4584 and the subgenomic RNAs 7/4584-A (B to F) which evolved after 8 and 10 passages of 7/4584 in MDBK cells. The CP RIT-derived sequences are marked by gray and black boxes. Deletions are indicated by dashed lines. The lengths of the bars are not drawn to scale. The sequenced regions of the subgenomes are indicated by a line below the bar at the bottom. With respect to the analyzed regions, all subgenomes maintain one large ORF. The genome organization of the emerged noncp BVDV is not included.
Surprisingly, Northern blot analysis of viral RNA from cells infected with the 8th, 9th, and 10th passages of 7/4584 led to identification of subgenomic RNAs in addition to the genomic RNAs of 7/4584 and the emerged noncp BVDV (Fig. 5A). For characterization of these subgenomes, the RNAs obtained after 8 and 10 passages of 7/4584 were subjected to RT-PCR analysis using primers Ol NS3R and Ol 100. For both RNAs obtained after 8 and 10 passages, a number of cDNA fragments ranging in size between 0.8 and 1.3 kb were generated. With respect to the eighth passage of 7/4584, molecular cloning and nucleotide sequencing of the obtained fragments resulted in identification of four different subgenomic RNAs (7/4584-A, -B, -C, and -D), two of which were also found after 10 passages (Fig. 5). Furthermore, two additional subgenomes (7/4584-E, and -F) were detected after 10 passages of 7/4584 in MDBK cells. The genome organizations of the 7/4584-derived subgenomes are very similar to each other, both exhibiting (i) a large internal deletion of viral sequences and (ii) the presence of S27a∗-ubi∗ coding sequences directly upstream of the NS3 gene. Remarkably, all deletions resulting in generation of the subgenomes occurred in frame. To our knowledge, this is the first report on the emergence of noncp BVDV and a number of subgenomic RNAs during propagation of cp BVDV in tissue culture cells.
DISCUSSION
One particularly interesting aspect of pestivirology concerns the existence of two biotypes, noncp and cp viruses. In the BVDV system, the occurrence of cp BVDV in cattle persistently infected (p.i.) with noncp BVDV is directly linked to induction of lethal MD. In p.i. animals, cp BVDV can be generated in the absence of exogenous BVDV and thus by endogenous evolution (see reference 26 for a review). Moreover, superinfection of p.i. animals with cp BVDV strains may result in this fatal disease (10, 11, 15, 28). In this context, it has been speculated that vaccination with live attenuated cp BVDV may cause MD (9, 16, 29). In the present study, two cp BVDV isolates (1741 and 4584) obtained from independent cases of MD were characterized at the molecular level. The results of our analyses strongly suggest that RNA recombination between persisting noncp BVDV and cp BVDV vaccine strain RIT resulted in generation of cp BVDV and induction of MD. This conclusion is consistent with the fact that the animals had been vaccinated with BVDV RIT prior to onset of disease. In Germany, several hundred thousand bovines are vaccinated with BVDV RIT per year. In this context, it is noteworthy that molecular analyses of three additional cp BVDV isolates from animals with MD housed on different farms from distinct geographic areas in Germany also led to detection of BVDV RIT-derived sequences within the genomes of the viruses (P. Becher, unpublished data). The RIT-derived insertions of CP 1741, CP 4584, and the other three cp BVDV isolates encompass sequences encoding parts of ribosomal protein S27a and ubiquitin (S27a∗-ubi∗) together with flanking viral sequences of different lengths. Apart from the host-specific S27a∗-ubi∗ coding sequences, only short viral sequences derived from CP RIT are present in the genomes of the recombinant cp viruses (Fig. 1B, 2B, and 3). It appears reasonable to assume that the acquired immunotolerance of the p.i. animals selected against the presence of larger genomic regions derived from vaccine strain RIT with regard to both structural and nonstructural proteins. To our knowledge, there is only one previous report which suggested recombination between noncp BVDV and a BVDV vaccine strain and where sequences from cp BVDV-1 vaccine strain NADL were found in the genome of a BVDV-2 strain (31). Taking into account the latter study and our data from five cp BVDV strains, it can be concluded that RNA recombination between persisting noncp BVDV and cp BVDV vaccine strains can lead to fatal MD in cattle.
Molecular characterization of CP 1741 and CP 4584 revealed the presence of S27a∗-ubi∗ coding sequences upstream of the genomic region encoding NS3. In a previous study, it was demonstrated that S27a∗-ubi∗ serves as a processing signal to yield NS3 (8). Transfection experiments with RNAs transcribed from infectious cDNA clone p7/4584 showed that the insertion of S27a∗-ubi∗ coding sequences directly upstream of the NS3 coding region also represents the genetic basis of cytopathogenicity of CP 4584 (Fig. 4). Furthermore, transfection experiments with engineered subgenomic RNAs carrying insertions of S27a∗-ubi∗ coding sequences revealed that these subgenomes are capable of autonomous replication and induction of cytopathogenicity (data not shown).
Molecular characterization of noncp and cp pestiviruses has revealed that the genomes of cp viruses comprise a broad range of genomic alterations which in general are not present in the genomes of noncp viruses (26). The genetic alterations of most cp pestiviruses resulted from RNA recombination (4, 26). With respect to integration of cellular sequences as well as duplication and deletion of viral sequences, the nature of recombination is nonhomologous. In contrast to other positive-stranded RNA viruses, nonhomologous RNA recombination has been observed in the BVDV system far more frequently than homologous reactions. In this context, it is noteworthy that both nonhomologous and homologous RNA recombination between CP RIT and persisting BVDV contributed to generation of BVDV CP 1741 and CP 4584.
For CP 1741, five different viral subgenomic RNAs were identified, each comprising a unique deletion together with an insertion of S27a∗-ubi∗ coding sequences. Moreover, a common crossover site within the NS3 gene was detected for all of these subgenomes. It is thus considered unlikely that the different subgenomes were generated by independent RNA recombination events. Alternatively, the various subgenomes may have developed in the course of two separate recombination processes. In a first step, integration of S27a∗-ubi∗ coding sequences together with flanking viral sequences derived from CP RIT into the genome of NCP 1741 resulted in a hypothetical precursor virus with a duplication of viral sequences encoding NS3, NS4A, and part of NS4B. In a second step, several different deletions resulted in emergence of the various subgenomes. Interestingly, our analysis of 7/4584 demonstrated that replication of a virus carrying S27a∗-ubi∗ coding sequences together with duplicated viral sequences actually led to generation of a number of different subgenomes (Fig. 5). The observed emergence of 7/4584-derived subgenomes supports the hypothesis that the CP 1741 subgenomes evolved from a common precursor with a genomic structure similar to that of 7/4584.
Tissue culture passages of cp BVDV 7/4584 carrying a large duplication of viral sequences resulted in the evolution of noncp BVDV and at least six different subgenomic RNAs (Fig. 5). To our knowledge, this is the first report about emergence of a variety of unique viral subgenomes during replication in tissue culture cells. Apart from cp CSFV subgenomes generated by internal deletion from the genomes of noncp CSFV (25, 27, 34), there are no reports about emergence of cp pestiviruses during replication of noncp pestiviruses in cell culture. Similar to the observed generation of noncp BVDV after passages of cp BVDV 7/4584, a switch from cp to noncp biotype has been demonstrated for another recently described chimeric cp BVDV strain with duplicated viral sequences (2). There is so far no published evidence that replication of naturally occurring cp BVDV strains in tissue culture cells leads to generation of noncp BVDV. In this context, it is interesting to mention that 10 passages of the biologically cloned CP 4584 in bovine cells did not result in noncp BVDV. It is therefore remarkable that tissue culture passages of two different cp BVD viruses derived from cDNA constructs with duplications of viral sequences led to deletion of part of the genomes and thereby to the emergence of noncp BVDV. Moreover, it can be speculated that evolution of noncp BVDV from cp BVDV genomes also occurs during replication of cp BVDV in the animal. Animal experiments, in particular with cp pestiviruses derived from cDNA constructs, will show whether noncp BVDV actually emerge during replication of cp BVDV in the animal. Such studies will provide further insight into the fascinating changes from noncp to cp biotype and vice versa.
ACKNOWLEDGMENTS
We thank M. Baroth for help with cDNA cloning and nucleotide sequencing.
This study was supported by SFB 535 “Invasionsmechanismen and Replikationsstrategien von Krankheitserregern” from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Baker J C. Bovine viral diarrhea virus: a review. J Am Vet Med Assoc. 1987;190:1449–1458. [PubMed] [Google Scholar]
- 2.Baroth M, Orlich M, Thiel H-J, Becher P. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology. 2000;278:456–466. doi: 10.1006/viro.2000.0644. [DOI] [PubMed] [Google Scholar]
- 3.Becher P, Meyers G, Shannon A D, Thiel H-J. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J Virol. 1996;70:2992–2998. doi: 10.1128/jvi.70.5.2992-2998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher P, Orlich M, König M, Thiel H-J. Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease. J Virol. 1999;73:5646–5653. doi: 10.1128/jvi.73.7.5646-5653.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becher P, Orlich M, Shannon A D, Horner G, König M, Thiel H-J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 1997;78:1357–1366. doi: 10.1099/0022-1317-78-6-1357. [DOI] [PubMed] [Google Scholar]
- 6.Becher P, Orlich M, Thiel H-J. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher P, Orlich M, Thiel H-J. Mutations in the 5′ nontranslated region of bovine viral diarrhea virus result in altered growth characteristics. J Virol. 2000;74:7884–7894. doi: 10.1128/jvi.74.17.7884-7894.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becher P, Orlich M, Thiel H-J. Ribosomal S27a-coding sequences upstream of ubiquitin-coding sequences in the genome of a pestivirus. J Virol. 1998;72:8697–8704. doi: 10.1128/jvi.72.11.8697-8704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bittle J L, House J A. Comments on bovine viral diarrhea vaccine reactions. J Am Vet Med Assoc. 1973;163:879. [Google Scholar]
- 10.Bolin S R, McClurkin A W, Cutlip R C, Coria M F. Severe clinical disease induced in cattle persistently infected with noncytopathogenic bovine viral diarrhea virus by superinfection with cytopathogenic bovine viral diarrhea virus. Am J Vet Res. 1985;46:573–576. [PubMed] [Google Scholar]
- 11.Brownlie J, Clarke M C, Howard C J. Experimental production of fatal mucosal disease in cattle. Vet Rec. 1984;114:535–536. doi: 10.1136/vr.114.22.535. [DOI] [PubMed] [Google Scholar]
- 12.Corapi W V, Donis R O, Dubovi E J. Monoclonal antibody analyses of cytopathic and noncytopathic viruses from fatal bovine viral diarrhea infections. J Virol. 1988;62:2823–2827. doi: 10.1128/jvi.62.8.2823-2827.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng R, Brock K V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathogenic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–879. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O A. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritzemeier J, Greiser-Wilke I, Haas L, Pituco E, Moennig V, Liess B. Experimentally induced “late-onset” mucosal disease—characterization of the cytopathogenic viruses isolated. Vet Microbiol. 1995;46:285–294. doi: 10.1016/0378-1135(95)00093-p. [DOI] [PubMed] [Google Scholar]
- 16.Fuller D A. When to vaccinate for IBR-VD. Mod Vet Pract. 1965;46:40–43. [Google Scholar]
- 17.Gillespie J H, Baker J A, McEntee K. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 1960;50:73–79. [PubMed] [Google Scholar]
- 18.Heinz F X, Collett M S, Purcell R H, Gould E A, Howard C R, Houghton M, Moormann R J M, Rice C M, Thiel H-J. Family Flaviviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 2000. pp. 859–878. [Google Scholar]
- 19.Kupfermann H, Thiel H-J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K M, Gillespie J H. Propagation of virus diarrhea virus of cattle in tissue culture. Am J Vet Res. 1957;18:953. [PubMed] [Google Scholar]
- 21.Lobmann M, Charlier P, Florent G, Zygraich N. Clinical evaluation of a temperature-sensitive bovine viral diarrhea vaccine strain. Am J Vet Res. 1984;45:2498–2503. [PubMed] [Google Scholar]
- 22.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyers G, Stoll D, Gunn M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J Virol. 1998;72:4139–4148. doi: 10.1128/jvi.72.5.4139-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers G, Thiel H-J. Cytopathogenicity of classical swine fever virus caused by defective interfering particles. J Virol. 1995;69:3683–3689. doi: 10.1128/jvi.69.6.3683-3689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 27.Mittelholzer C, Moser C, Tratschin J-D, Hofmann M A. Generation of cytopathogenic subgenomic RNA of classical swine fever virus in persistently infected porcine cell lines. Virus Res. 1997;51:125–137. doi: 10.1016/S0168-1702(97)00081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moennig V, Frey H-R, Liebler E, Polenz P, Liess B. Reproduction of mucosal disease with cytopathogenic bovine viral diarrhoea virus seleced in vitro. Vet Rec. 1990;127:200–203. [PubMed] [Google Scholar]
- 29.Peter C P, Tyler D E, Ramsey F K. Characterization of a condition following vaccination with bovine virus diarrhea vaccine. J Am Vet Med Assoc. 1967;150:46–52. [Google Scholar]
- 30.Qi F, Ridpath J F, Berry E S. Insertion of a bovine SMT3B gene in NS4B and duplication of NS3 in a bovine viral diarrhea virus genome correlate with the cytopathogenicity of the virus. Virus Res. 1998;57:1–9. doi: 10.1016/s0168-1702(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 31.Ridpath J F, Bolin S R. Delayed onset postvaccinal mucosal disease as a result of genetic recombination between genotype 1 and genotype 2 BVDV. Virology. 1995;212:259–262. doi: 10.1006/viro.1995.1480. [DOI] [PubMed] [Google Scholar]
- 32.Tautz N, Thiel H-J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1059–1073. [Google Scholar]
- 34.Tratschin J-D, Moser C, Ruggli N, Hofmann M A. Classical swine fever virus leader proteinase Npro is not required for viral replication in cell culture. J Virol. 1998;72:7681–7684. doi: 10.1128/jvi.72.9.7681-7684.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassilev V B, Donis R O. Bovine viral diarrhea virus induced apoptosis correlates with increased intracellular viral RNA accumulation. Virus Res. 2000;69:95–107. doi: 10.1016/s0168-1702(00)00176-3. [DOI] [PubMed] [Google Scholar]