Abstract
The specificity of usage of promoters for replication and transcription by the pneumoviruses human respiratory syncytial virus (HRSV) and avian pneumovirus (APV) was studied using minigenomes containing a reporter gene. When infectious HRSV or APV was used as helper virus, replication could occur only if both the leader and trailer regions (containing the replicative and transcriptional promoters) were derived from the helper virus. In contrast, when the HRSV replication complex was supplied from cDNA plasmids, a minigenome containing either the APV leader or trailer was recognized and substantial levels of replication and transcription occurred. These data suggest that in pneumovirus-infected cells, helper virus functions can discriminate between genomes on the basis of the terminal sequences and that there is an association between the leader and trailer required for productive replication. This association is required only in virus-infected cells, not when replication and transcription are mediated by plasmid-directed expression of the component proteins required for replication and transcription. The possible implications of this are discussed.
The recent development of reverse genetics systems for nonsegmented negative-strand RNA viruses of the family Paramyxoviridae has allowed investigations into the roles of cis-acting RNA sequences in the replication and transcription of these viruses (18, 26). These systems typically use synthetic genome analogues from which all the viral coding sequences have been replaced by one or more reporter genes. The replication and transcription of these genome analogues, usually referred to as minigenomes, is studied by transfecting RNA transcripts into cells along with helper virus or by expressing minigenomes in cells together with plasmids encoding the viral RNA synthetic proteins. The former method was initially developed for Sendai virus (21) and has been used for a number of other paramyxoviruses, including human respiratory syncytial virus (HRSV) (4). This method has not been successful for the rhabdoviruses (10). The plasmid system typically uses T7 RNA polymerase to drive the expression of the minigenome and the viral polymerase proteins from T7 promoters, and the T7 RNA polymerase is usually provided by a recombinant vaccinia virus (VV-T7). This system was first used for the rhabdovirus vesicular stomatitis virus (22) and has been used for a range of paramyxoviruses and filoviruses, again including HRSV (30). For HRSV in particular, the RNA-helper virus system has been used to analyze transcriptional start and end signals and intergenic regions (14–16). The VV-T7 system has been used with HRSV, initially to show that N, P, and L proteins are the minimal set of proteins required to support replication (30) and then to demonstrate the effects on RNA synthesis of the M2 (3, 8) and NS1 proteins (2) and investigate particle assembly (27), the M2-L gene overlap region (7), the trailer region (23), and mutations in the P (19, 28) and L (13) genes. The effects of varying the relative and absolute amounts of the N and P proteins of HRSV were also studied using the VV-T7 system (6).
An RNA minigenome-helper virus system has also been developed for avian pneumovirus (APV), the causative agent of turkey rhinotracheitis (25). APV shares several properties with other members of the Pneumovirinae, such as possession of SH and M2 genes, but differs from members of the Pneumovirus genus, such as HRSV, in lacking the NS1 and NS2 genes and in having an altered gene order (17, 25, 29). On this basis it has been classified into a separate genus, Metapneumovirus (24).
It was previously shown that a synthetic minigenome based on APV sequences was unable to be rescued (i.e., to produce detectable reporter protein) by HRSV, and a minigenome based on HRSV sequences could not be rescued by APV (25). Rescue of a bovine RSV-derived minigenome by HRSV and ovine RSV has recently been reported (31); however, these viruses are more closely related to HRSV than is APV. For example, the leader sequence of HRSV matches that of bovine RSV for 24 of the first 25 nucleotides (nt) but matches the leader of APV for only 10 of the first 11 nt. The genomic termini (3′ leader and 5′ trailer) of each of these viruses also show complementarity to each other and are presumed to contain the promoters for synthesis of genome, antigenome, and transcriptional initiation of the first gene (9). Between them, these regions must also contain signals for encapsidation by the nucleoprotein, N, and for packaging into virus-like particles.
In this report, we show that the RNA synthetic proteins of HRSV are able to recognize both the replicative and transcriptional promoters of APV in a plasmid-based minigenome system but that for minigenome replication by whole infectious HRSV, both leader and trailer sequences must be derived from HRSV, and the APV sequences are not recognized.
MATERIALS AND METHODS
Plasmid constructs.
The APV minigenome has been described elsewhere (25). The HRSV minigenome was constructed in a similar manner and contains the T7 promoter; trailer and gene end sequence from HRSV strain RSS-2 (nt 15,025 to 15,190; numbering as in GenBank sequence U39662); a complete copy of the chloramphenicol acetyltransferase (CAT) gene open reading frame as a NdeI-to-XbaI fragment, in the opposite sense to the T7 transcript; RSS-2 gene start and leader (nt 1 to 54, nt 4 being altered from C to G); and the hepatitis delta virus antigenomic ribozyme. The C-to-G substitution at nt 4 has been reported to enhance replication in the minigenome system (6, 9) but occurs in some natural HRSV isolates and has no biological effect in full-length virus (10a).
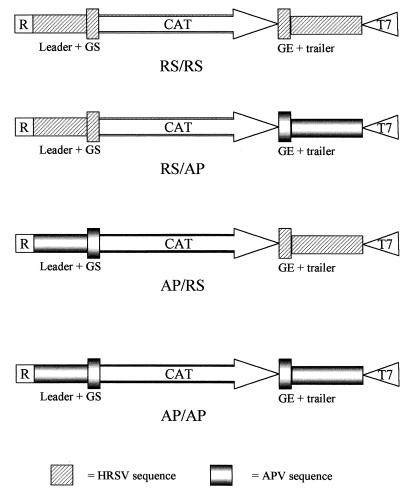
Chimeric minigenomes were constructed by replacing the leader/gene start sequence of the HRSV construct with that of APV and by replacing the trailer/gene end sequence of the HRSV construct with that of APV. This was achieved by inserting the ribozyme, APV leader and gene start, and part of the CAT gene as a BamHI-to-NcoI fragment into the HRSV minigenome from which the corresponding BamHI-to-NcoI fragment had been removed. Similarly, the CAT gene (from the ATG start codon), APV gene end and trailer, T7 promoter, and a portion of the plasmid backbone were excised from the APV minigenome as an NdeI-to-AflIII fragment, which was used to replace the corresponding NdeI-to-AflIII fragment of the HRSV minigenome. Complete nucleotide sequences of each plasmid were determined by cycle sequencing using dye-labeled terminators. The set of four minigenomes were named based on the leader/trailer identity for each, i.e., AP/AP, RS/RS, AP/RS, and RS/AP, where AP refers to APV and RS refers to HRSV. Structures of the minigenomes used are summarized in Fig. 1. The construction of plasmids containing the HRSV strain RSS-2 L, N, P, and M2 genes under the control of T7 promoters has been described elsewhere (19).
FIG. 1.
Diagram of the structures of the minigenomes used in this study. T7, T7 RNA polymerase promoter; R, ribozyme sequence; CAT, CAT reporter gene; GS, gene start signal; GE, gene end signal.
Cells and transfections.
For infectious virus-based rescues, RNA transcripts were synthesized in vitro and transfected into subconfluent monolayers of Vero cells, which had been infected for 1 h with APV strain CVL14/1 or HRSV strain RSS-2, as described previously (25). Alternatively, 2 μg of DNA was transfected using 4 μl of Lipofectace (Gibco-BRL) into virus-infected BHK-T7 cells. These are BHK-21 cells which express T7 RNA polymerase from a defective Sindbis virus replicon, SINrep19/T7pol (1). For plasmid-based rescues, 0.4 μg of minigenome plasmid was transfected into HEp-2 cells, previously infected with recombinant vaccinia virus vTF7-3 (1 PFU per cell), which expresses T7 RNA polymerase (12), along with plasmids encoding HRSV L, N, P, and M2 genes as described previously (19). Alternatively, the same five plasmids were transfected into BHK-T7 cells in the absence of any other virus.
Reporter gene assay and RNA analysis.
Cells were harvested 2 or 3 days (for RNA transfections of Vero cells) posttransfection, and cell lysates were made. CAT enzyme-linked immunosorbent assays, RNA fractionation, micrococcal nuclease digestion, and Northern blotting have been described elsewhere (19). For Northern blots, digoxigenin-labeled riboprobes were transcribed as a 2.6-kb negative-sense copy of the CAT- and Luc-containing minigenome and as a 0.90-kb positive-sense copy of the CAT-containing RS/RS minigenome. Chromatography on oligo(dT)-cellulose was used to isolate mRNA. Micrococcal nuclease digestion was performed on cell lysates prior to RNA extraction, using an established protocol (6); degradation of rRNA was monitored by ethidium bromide staining of RNA gels prior to blotting, and the extent of digestion was confirmed to remove all unencapsidated minigenome RNA (see Results and Fig. 7).
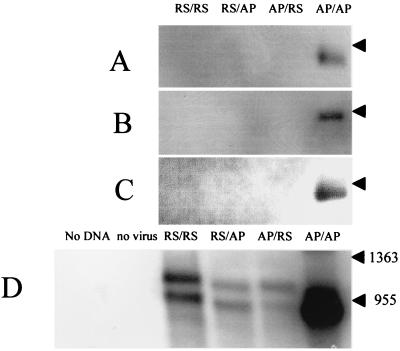
FIG. 7.
Northern blot analysis of RNA from transfected BHK-T7 cells infected with APV helper virus. The minigenome in each lane is indicated at the top. The arrowhead on each panel indicates the position of the 955-nt RNA size marker. (A) Poly(A)+ RNA from cells infected with APV, probed with negative-sense riboprobe. (B) Positive-sense RNA following micrococcal nuclease digestion, from cells infected with APV, probed with negative-sense riboprobe. (C) Total RNA from cells infected with supernatant from transfected, APV-infected cells, probed with negative-sense riboprobe. (D) Negative-sense RNA following micrococcal nuclease digestion, from cells infected with APV, probed with positive-sense riboprobe. The extra lanes show the effect of omitting minigenome DNA from the transfection (no DNA), or transfecting AP/AP minigenome into uninfected BHK-T7 cells (no virus).
RESULTS
Rescue of minigenomes by virus in Vero cells.
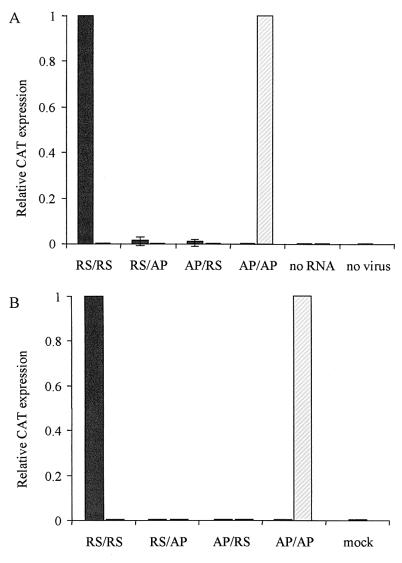
Minigenomes were constructed so as to contain the CAT reporter gene flanked by the leader and trailer regions of HRSV (RS/RS) or of APV (AP/AP) or by the leader of one virus and the trailer of the other (RS/AP and AP/RS chimeric minigenomes [Fig. 1]). Negative-sense RNA transcripts synthesized in vitro were transfected into pneumovirus-infected cells. CAT reporter protein expression was detected in significant amounts only from the minigenome RS/RS in HRSV-infected cells and from the minigenome AP/AP in APV-infected cells (Fig. 2A). Typical values were 900 to 7,200 pg of CAT per 106 cells. Expression of CAT from chimeric or AP/AP minigenomes was at least 50-fold lower than that from the RS/RS minigenome when cells were infected with HRSV, and levels of CAT were at the lowest limits of detection when cells were infected with APV, except when transfected with the AP/AP minigenome. Passage of the supernatant fluid, containing helper virus and any packaged minigenomes, onto fresh cells again produced CAT protein only from the combinations HRSV-RS/RS and APV-AP/AP (Fig. 2B; typical values of >20,000 pg of CAT per 106 cells).
FIG. 2.
CAT protein expression following transfection of RNA minigenomes into Vero cells infected with HRSV (solid bars) or APV (hatched bars). CAT protein was quantified in cell lysates 72 h posttransfection by enzyme-linked immunosorbent assay ELISA. In each case, expression is normalized to the homologous control. Each bar represents the mean of four to six replicates. Error bars show ±1 standard deviation. (A) Lysates of transfected cells. “no RNA” indicates samples infected with virus but not transfected; “no virus” represents uninfected Vero cells transfected with AP/AP RNA. (B) Lysates assayed 72 h after passage of supernatants from transfected cells onto fresh Vero cells. Samples are labeled according to the minigenome used for the primary transfection; “mock” denotes passage of medium from untransfected cells.
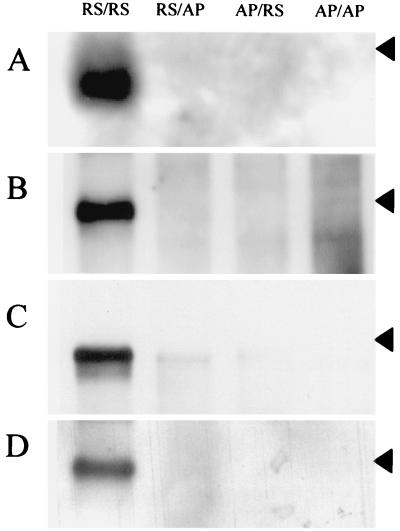
To determine whether the block to reporter gene expression was at the level of transcription or replication, the RNA species produced were analyzed by Northern blotting. As expected, poly(A)+ mRNA was produced only by the homologous virus-RNA combinations which were noted above as producing CAT protein (Fig. 3A for HRSV). Poly(A)− RNA and total cell RNA were not amenable to Northern blot analysis in transfected cells due to the presence of the residual transfected negative-sense RNA. The proportion of negative-sense RNA entering the replication cycle is small (23) and so cannot be resolved from input negative-sense RNA without further analysis. Replication products were therefore identified by micrococcal nuclease digestion of cell lysates prior to Northern blotting, which identified RNA fully encapsidated into nucleocapsid structures. All RNA gels were stained with ethidium bromide before Northern blotting, to ensure that the nuclease digests had proceeded equally for each sample. Control samples in which the helper virus had been omitted showed that the nuclease digest was sufficient to remove all of the nonencapsidated input RNA (an example is shown in Fig. 7D). As shown in Fig. 3B and C, encapsidated positive- and negative-sense RNAs, respectively, were clearly detected only in the HRSV-RS/RS RNA combination but not for the AP/RS or RS/AP minigenomes, suggesting that both leader and trailer sequences are required for productive replication by HRSV. Faint bands visible in the RS/AP and AP/RS tracks of the negative-sense RNA blot (Fig. 3C) probably represent encapsidated input RNA (23). Similar bands are not seen in the blot of positive-sense RNA, suggesting that the level of replication is below the limits of detection (Fig. 3B). Particles released into the medium from transfected cells were passaged onto fresh monolayers of Vero cells, and total RNA was extracted for Northern blotting (Fig. 3D). Antiminigenome RNA was detected only in the HRSV-RS/RS RNA combination, again suggesting that only this combination was able to replicate and then package into infectious particles. The data shown in Fig. 3 are from HRSV as helper virus; analogous results were obtained using APV as helper virus, as expected from the CAT expression data shown in Fig. 2 (data not shown).
FIG. 3.
Northern blot analysis of RNA from transfected cells infected with HRSV helper virus. The minigenome in each lane is indicated at the top. The arrowhead on each panel indicates the position of the 955-nt RNA size marker. (A) Poly(A)+ RNA from transfected cells infected with HRSV, probed with negative-sense riboprobe. (B) RNA following micrococcal nuclease digestion, from transfected cells infected with HRSV, probed with negative-sense riboprobe. (C) RNA following micrococcal nuclease digestion, from transfected cells infected with HRSV, probed with positive-sense riboprobe. (D) Total RNA from cells infected with supernatant from transfected, HRSV-infected cells, probed with negative-sense riboprobe.
Rescue of minigenomes by plasmids in HEp-2 cells.
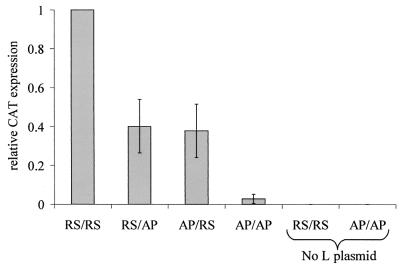
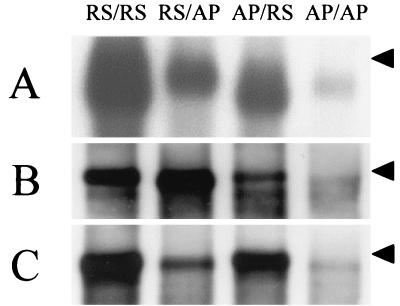
In the plasmid-based rescue system, the minigenome RNA and mRNAs for the HRSV N, P, L, and M2 proteins are all synthesized as T7 transcripts, using T7 RNA polymerase provided by the VV-T7 recombinant. In contrast to the RNA transfection system described above, both RS/AP and AP/RS chimeras produced significant amounts of CAT protein, up to approximately 40% of the level produced by the RS/RS minigenome. Typical values of CAT expression from the RS/RS mingenome were 9,000 to 25,000 pg per 106 cells. The AP/AP minigenome also produced detectable levels of CAT protein in the presence of HRSV N, P, L and M2 proteins, clearly above the background level when no L protein-encoding plasmid was added, but only about 3% of the level produced by the RS/RS minigenome (Fig. 4). The levels of mRNA produced in cells transfected with the plasmid-based rescue system (Fig. 5A) were consistent with the observed CAT protein values (Fig. 4). In addition, RNA replication was observed for all four minigenome constructs, and it was clear that the largest amounts of encapsidated positive-sense RNA were produced from RS/RS and RS/AP minigenomes (Fig. 5B), as would be expected due to the presence of the homologous HRSV leader sequence. In contrast, the largest amounts of encapsidated negative-sense RNA were produced from RS/RS and AP/RS minigenomes (Fig. 5C), correlating with the presence of the HRSV trailer sequence, presumed to contain the promoter for synthesis of genome-sense RNA from an antigenome template. The AP/AP construct was replicated by the HRSV proteins, but to a considerably lower level than the control.
FIG. 4.
CAT protein expression following transfection of plasmid minigenomes along with plasmids encoding the HRSV N, P, L, and M2 proteins into VV-T7-infected HEp-2 cells. Expression is normalized to the level of the RS/RS minigenome. Each bar represents the mean of four replicates. Error bars show standard deviations. For the two tracks labeled “no L plasmid,” the plasmid encoding HRSV L protein was replaced by empty vector.
FIG. 5.
Northern blot analysis of RNAs from VV-T7-infected cells transfected with plasmids, as in Fig. 4. The minigenome in each lane is indicated at the top. The arrowhead on each panel indicates the position of the 955-nt RNA size marker. (A) Poly(A)+ RNA, probed with negative-sense riboprobe. (B) Positive-sense RNA following micrococcal nuclease digestion, probed with negative-sense riboprobe. (C) Negative-sense RNA following micrococcal nuclease digestion, probed with positive-sense riboprobe.
These data clearly demonstrate that the HRSV replicative proteins, expressed in the absence of all other viral proteins, can recognize the APV leader and trailer sequences and utilize them to direct synthesis of RNA. However, this does not happen in the RNA-infectious virus rescue system. A number of possibilities arise to explain this observation: (i) promoter recognition is reduced in specificity by the presence of the VV-T7; (ii) the pool of N protein available for encapsidation is limiting in the helper virus system, such that the heterologous end-containing minigenomes are not able to compete with the helper virus for encapsidation; (iii) the effect is due to continual synthesis of a pool of negative-sense T7 RNA transcripts within the cell, as opposed to a single RNA transfection event; (iv) the reduction in specificity in promoter recognition is due to the host cell environment, i.e., human laryngeal (HEp-2) as opposed to monkey kidney (Vero) cells; and (v) the full specificity of promoter recognition requires an additional virus gene product (other than N, P, L, and M2) or a host protein which is induced by pneumovirus infection. To distinguish between these possibilities, we used a third host cell type, BHK-T7 cells.
Rescue of minigenomes by plasmids in BHK-T7 cells.
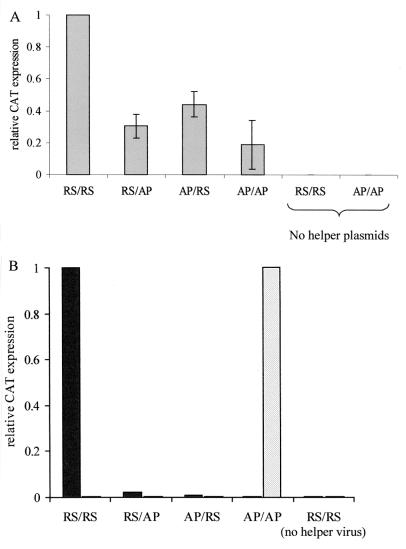
In this case, T7 RNA polymerase is supplied by a defective Sindbis virus replicon which is unable to induce cytopathic effect in BHK cells and is stably transmitted through cell passage in the presence of the selective antibiotic puromycin (1). The relative levels of CAT protein produced from the four minigenomes in BHK-T7 cells in the presence of HRSV N, P, L, and M2 plasmids (Fig. 6A) were not significantly different from the values seen in the VV-T7 system (Fig. 4). Typical values for the RS/RS minigenome were 600 to 900 pg of CAT per 106 cells. The AP/AP minigenome produced over 20% of the CAT levels of the RS/RS minigenome in some experiments. This shows that the reduction in specificity of promoter recognition is not due to the VV-T7 or to the use of HEp-2 cells.
FIG. 6.
CAT protein expression in transfected BHK-T7 cells. (A) Minigenome plasmids cotransfected with plasmids encoding HRSV N, P, L, and M2 proteins. Expression is normalized to the RS/RS control. Each bar represents the mean of four replicates. Error bars show standard deviations. For the tracks labeled “no helper plasmids,” the plasmids encoding HRSV L, N, P, and M2 proteins were replaced by empty vector. (B) Minigenome plasmids transfected into BHK-T7 cells infected with HRSV (solid bars) or APV (hatched bars) helper viruses. Each bar represents the mean of two replicates.
Rescue of minigenomes by helper virus in BHK-T7 cells.
In this case, plasmid DNA encoding the minigenome was transfected into HRSV or APV-infected cells, and negative-sense T7 RNA transcripts were synthesized in the cytoplasm. The pattern of CAT protein production (Fig. 6B) was essentially the same as for the RNA-Vero cell system (Fig. 2A), i.e., very low or no reporter production from either chimera or the heterologous construct. Typical values of CAT expression from the homologous combinations of virus and mingenome were 900 to 7,500 pg per 106 cells. These data were confirmed by Northern blot analysis of the RNA produced (Fig. 7A, B, and D for APV as helper virus; HRSV data not shown). As in the RNA-Vero cell system, mRNA levels were consistent with the amount of CAT protein expressed, and encapsidated RNAs were detected only in the homologous virus-RNA combinations. Hence, the specificity observed is not a function of whether RNA is synthesized in vitro or continuously in the cytoplasm and is not a host cell-dependent phenomenon. Taken together, the data from BHK-T7 cells rule out possibilities i, iii, and iv described above. Full packaging of the AP/AP minigenome by APV helper virus in BHK-T7 cells was confirmed by passage of the supernatant fluid onto fresh uninfected monolayers of Vero cells, which produced high levels of CAT protein (data not shown) and AP/AP RNA (Fig. 7C).
Figure 7D shows nuclease-resistant negative-sense RNAs from APV-infected BHK-T7 cells and has been overexposed to clearly show faint bands. The AP/AP minigenome produced a strong band, confirming that this was the only replicated minigenome. No bands were detected in the absence of minigenome DNA, showing that the probe is minigenome specific, nor was any signal detected in the absence of APV helper virus, confirming that the nuclease completely digests nonencapsidated primary transcripts from the minigenome DNA. The bands seen with minigenomes RS/RS, RS/AP, and AP/AP therefore represent encapsidated negative-sense RNA which has not been replicated. In each lane, the slower-migrating band represents the uncleaved primary transcript, i.e., with the ribozyme sequence still present at the 3′ end, and the faster-migrating band represents the cleaved minigenome, which is predicted from the plasmid sequence to be 200 nt smaller. The uncleaved band must represent primary transcripts from the input plasmid DNA, since any replication by the virus would result in the removal of the ribozyme sequence. Since the cleaved band is approximately the same intensity as the uncleaved band, this indicates that levels of pneumovirus-directed replication in these samples were negligible. The observation that uncleaved primary transcripts were encapsidated (Fig. 7D) also demonstrates that encapsidation does not require an authentic viral sequence at the 3′ end of the RNA. It was already known that an authentic viral 5′ end is not required for encapsidation (23).
These data show that minigenomes containing sequences from HRSV, including RS/RS, which contains no APV sequences, can be encapsidated by the APV N protein in the presence of APV helper virus, suggesting that the helper virus is not outcompeting the minigenomes for a limited pool of N protein but that the block to replication is at a step following primary encapsidation. Analogous data were obtained with HRSV as helper virus (data not shown).
DISCUSSION
Analysis of the cis-acting signals important in replication and transcription of the HRSV genome has been conducted using the minigenome reverse genetics system, as described in the introduction, either using HRSV helper virus to rescue transfected RNA or using a recombinant vaccinia virus (VV-T7) to drive expression of minigenome, and viral N, P, L, and M2 proteins, from T7 promoters. We have shown that the results obtained can depend on the system used, suggesting that the specificity of promoter usage by HRSV is enhanced by a virus-induced product not present in the plasmid-only systems and that in the plasmid-only systems there is less discrimination between the leader and trailer regions of related viruses.
It was previously observed that heterologous rescue between APV or HRSV and the respective minigenomes did not occur (25). This has now been extended to show that both leader and trailer regions of HRSV are required for replication by infectious HRSV, and both leader and trailer regions of APV are required for replication by APV. Encapsidation by N protein, by contrast, does not require either of the genomic ends to be derived from the homologous virus. It has previously been shown that minigenomes based on HRSV, which lack the entire trailer region, are still encapsidated (23). The leader sequences of HRSV and APV are identical at 10 of the first 11 nt from the viral 3′ end (nt 4 is G in RS/RS and A in AP/AP), suggesting that promoter specificity in virus-infected cells is determined by recognition of a sequence or RNA structure located between nt 12 and the first gene start signal at nt 42 or 45 for HRSV and APV, respectively. Initiation of RNA synthesis during replication is, of course, at nt 1 of the leader region. The difference between HRSV and APV at position 4 is unlikely to be involved in promoter specificity: a G4-to-A mutation (i.e., from the HRSV to the APV sequence) has no effect on HRSV minigenome CAT expression (3a), and an A4-to-G mutation in the APV minigenome has little effect on replication or transcription (J. M. Smith and A. J. Easton, unpublished observations). Detailed analysis of the region between nt 12 and 45 in APV is under way. The trailer sequences of APV and HRSV are identical for the first 12 nt from the viral 5′ end, again suggesting that promoter specificity involves recognition downstream of this sequence.
Use of the plasmid-based rescue system gave a similar result regardless of whether the T7 RNA polymerase was supplied by VV-T7 or by SINrep19/T7pol. Expression of the CAT reporter gene from the chimeric constructs RS/AP and AP/RS demonstrated that the HRSV replication complex was able to recognize the APV trailer and leader, respectively, sufficiently well to generate high levels of mRNA and of encapsidated positive- or negative-sense minigenome. This process must have included encapsidation by the HRSV N protein, since both chimeric minigenomes, as well as a small amount of the AP/AP minigenome, were protected against micrococcal nuclease digestion. Significant amounts of CAT expression were obtained from the AP/AP minigenome, and this expression was not observed in the absence of HRSV L protein, demonstrating that the AP/AP RNA was being transcribed, and presumably replicated, albeit at a lower level than the other minigenome constructs.
The similar levels of mRNA produced by the RS/AP and AP/RS chimeric minigenomes in the plasmid-only system, to about 40% of the RS/RS level, can be explained if it is assumed that the transcriptional promoter on the AP/RS negative-sense RNA is recognized at a lower level than the HRSV version of the transcriptional promoter. Mutagenesis of the HRSV gene start signal, showed that the substitution C4→U in the consensus sequence 3′ CCCCGUUUAU 5′ (genome sense) did not affect transcription, whereas U8→C reduced transcription by about 40% (14). These two mutations together produce a sequence very similar to the APV consensus gene start signal, 3′ CCCUGUUCA 5′. It is likely that the wild-type level of negative-sense RNA produced from AP/RS is then transcribed by the HRSV polymerase at reduced efficiency from the APV gene start, whereas the reduced level of negative-sense RNA produced from RS/AP, presumably due to less efficient initiation of negative-sense RNA synthesis from the APV trailer sequence, is then transcribed efficiently from the HRSV gene start, resulting in roughly equal amounts of mRNA from the two constructs. It is likely that the HRSV and APV gene end sequences were utilized with equal efficiency by the HRSV transcriptase complex. This is based on the observations that the HRSV L gene end and APV F gene end both have the sequence 3′ UCAAUAAA(U)4 5′, the HRSV N and M gene ends and the APV L gene end all have the sequence 3′ UCAAUUA(U)5 5′, and the HRSV gene ends are utilized with equal efficiency, with the exception of the NS1 and NS2 gene ends (14).
The difference in promoter specificity seen when N, P, L, and M2 plasmids rather than helper virus were used was independent of cell type, and of virus or replicon used to supply the T7 RNA polymerase, and was not due to intracellular RNA synthesis rather than RNA transfection. We conclude that the specificity is the function of one or more of the remaining viral proteins or of a cellular protein(s) whose synthesis is induced by pneumovirus infection (but not by poxvirus infection). Of the remaining viral proteins, if NS1 or NS2 provide this function for HRSV, a different protein or combination of proteins must do so for APV, which lacks genes for NS1 and NS2 proteins. The remaining proteins for APV comprise M and the membrane glycoproteins F, G, and SH. The further possibility remains that the specificity is conferred by fine-tuning of the N/ P/L/M2 ratio in virus-infected cells which was not achieved when these proteins were derived from plasmids. It has been observed by immunoblotting that steady-state levels of the N and P proteins are approximately the same at the time of harvest between HRSV-infected cells and cells from the plasmid–VV-T7 system (data not shown).
It is possible that L protein is limiting in the helper virus system but not in the plasmid-based system, so that in the former L protein is preferentially bound to the homologous promoter of the helper virus, and in effect the helper virus genome interferes with the replication of the nonhomologous minigenome. A prediction of this possibility would be that when HRSV is used as helper virus, the RS/AP chimera should still be able to synthesize a small amount of positive-sense minigenome and mRNA from the encapsidated, but unreplicated, negative-sense strand. The AP/RS chimera, in contrast, would be predicted not to produce any positive-sense RNA or mRNA from the lower-affinity APV leader promoter. While trace amounts of encapsidated negative-sense RNA were observed from all of RS/AP, AP/RS, and AP/AP (Fig. 3C), no positive-sense RNA was detected from RS/AP (Fig. 3B), nor was any mRNA detected (Fig. 3A). CAT protein levels were not significantly different between RS/AP and AP/RS, either in Vero cells (Fig. 2A) or in BHK-T7 cells (Fig. 6B).
Another possibility is that N protein is limiting in the helper virus system and that the helper virus interferes with the replication of nonhomologous minigenomes by competing for free N protein and preventing the minigenomes from becoming encapsidated. The accepted model of replication for viruses of the Paramyxoviridae requires that only encapsidated RNA be functional in replication. However, we have shown that nonhomologous minigenomes are encapsidated, although not replicated, in the presence of helper virus. This demonstrates that the block to replication is at the stage of initiation or elongation by the replicase complex.
Regardless of the precise protein(s) involved, it was seen that in pneumovirus-infected cells both leader and trailer sequences from the homologous virus were required to be present in the minigenome to enable replication. Since the first round of replication had to initiate on a newly encapsidated negative-sense transcript, the viral polymerase must first recognize a sequence in the leader region of that transcript; however, since only RS/RS was replicated by HRSV and not RS/AP (and conversely, only AP/AP was replicated by APV and not AP/RS), the trailer sequence must also be involved in this recognition step, possibly by directly interacting with the leader in a manner analogous to that proposed for the replication of influenza A virus (11). Alternatively, the polymerase complex may interact with both leader and trailer at different sites, possibly with the involvement of one or more nonviral proteins. This interaction is apparently not necessary when the replication proteins are supplied in a pneumovirus-free context.
The current model of RNA synthesis by the nonsegmented negative-strand viruses suggests that all of the sequences required for synthesis of mRNA and antigenome are contained in the leader and nearby downstream sequences, at the 3′ end of the genome (5). Our data indicate that the situation for pneumoviruses may be more complicated than that. By studying the effects of trailer mutations on production of mRNA and positive-sense minigenome, Peeples and Collins deduced that the leader and trailer of HRSV did not interact, and minigenomes from which the trailer had been deleted were still able to direct synthesis of positive-sense RNA (23). However, the VV-T7–plasmid system was used, and so the study agrees with our data suggesting a lack of any interaction of leader and trailer in the plasmid-based system, and the effect that we describe for the helper virus system would not have been observed. In a set of experiments involving chimeric minigenomes of the filoviruses Marburg (MBG) and Ebola (EBO) viruses, it was found that neither virus replicated the minigenome derived from the other virus (20), as we have described for HRSV and APV. The chimeric minigenome MBG/EBO also failed to be replicated by either virus, but the EBO/MBG chimera was replicated and transcribed by both helper viruses. Similar results were observed using the VV-T7–plasmid system as well as the RNA-helper virus system. The different spectrum of polymerase specificity exhibited by the filoviruses, compared to the pneumoviruses, suggests that promoter specificity may be determined by different strategies between different groups of nonsegmented negative-strand viruses.
We have shown that the outcome of a minigenome replication-transcription experiment can depend on whether the replicative proteins are supplied on plasmids or from whole virus. This should be borne in mind when interpreting the results of minigenome rescue experiments, which have been used widely for many nonsegmented negative-strand viruses. In some cases the whole virus cannot be used; examples include the Rhabdoviridae systems where RNA-helper virus systems have not been successfully developed (10) or a situation in which the system is being used to introduce potentially lethal mutations into the replicative proteins themselves.
ACKNOWLEDGMENTS
We thank C. Rice for supplying the SINrep19/T7pol plasmid and protocols and S. Wilson for technical assistance.
We thank the Biotechnology and Biological Sciences Research Council for award of a studentship to J.M.S. This work was supported by a project grant from the Medical Research Council.
REFERENCES
- 1.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atreya P L, Peeples M E, Collins P L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Collins P L, McIntosh K, Chanock R M. Respiratory syncytial virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1313–1351. [Google Scholar]
- 4.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogues of respiratory syncytial virus genomic RNA and effect of truncation and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conzelmann K-K. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet. 1998;32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 6.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 7.Fearns R, Collins P L. Model for polymerase access to the overlapped L gene of respiratory syncytial virus. J Virol. 1999;73:388–397. doi: 10.1128/jvi.73.1.388-397.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fearns R, Collins P L. Role of the M2–1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol. 1999;73:5852–5864. doi: 10.1128/jvi.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearns R, Collins P L, Peeples M E. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J Virol. 2000;74:6006–6014. doi: 10.1128/jvi.74.13.6006-6014.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finke S, Conzelmann K-K. Virus promoters determine interference by defective RNAs: selective amplification of mini-RNA vectors and rescue from cDNA by a copy-back ambisense rabies virus. J Virol. 1999;73:3818–3825. doi: 10.1128/jvi.73.5.3818-3825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Firestone C-Y, Whitehead S S, Collins P L, Murphy B R, Crowe J E., Jr Nucleotide sequence analysis of the respiratory syncytial virus subgroup A cold-passaged (cp) temperature sensitive (ts) cpts-248/404 live attenuated virus vaccine candidate. Virology. 1996;225:419–422. doi: 10.1006/viro.1996.0618. [DOI] [PubMed] [Google Scholar]
- 11.Fodor E, Pritlove D C, Brownlee G G. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juhasz K, Murphy B R, Collins P L. The major attenuating mutations of the respiratory syncytial virus vaccine candidate cpts530/1009 specify temperature-sensitive defects in transcription and replication and a non-temperature-sensitive alteration in mRNA termination. J Virol. 1999;73:5176–5180. doi: 10.1128/jvi.73.6.5176-5180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo L, Fearns R, Collins P L. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effect of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling R, Easton A J, Pringle C R. Sequence analysis of the 22K, SH and G genes of turkey rhinotracheitis virus and their intergenic regions reveals a gene order different from that of other pneumoviruses. J Gen Virol. 1992;73:1709–1715. doi: 10.1099/0022-1317-73-7-1709. [DOI] [PubMed] [Google Scholar]
- 18.Marriott A C, Easton A J. Reverse genetics of the Paramyxoviridae. Adv Virus Res. 1999;53:321–340. [PubMed] [Google Scholar]
- 19.Marriott A C, Wilson S D, Randhawa J S, Easton A J. A single amino acid substitution in the phosphoprotein of respiratory syncytial virus confers thermosensitivity in a reconstituted RNA polymerase system. J Virol. 1999;73:5162–5165. doi: 10.1128/jvi.73.6.5162-5165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhlberger E, Weik M, Volchkov V E, Klenk H-D, Becker S. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park K H, Huang T H, Correia F F, Krystal M. Rescue of a foreign gene by Sendai virus. Proc Natl Acad Sci USA. 1991;88:5537–5541. doi: 10.1073/pnas.88.13.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattnaik A K, Ball L A, Legrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 23.Peeples M E, Collins P L. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol. 2000;74:146–155. doi: 10.1128/jvi.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pringle C R. Virus taxonomy—San Diego 1998. Arch Virol. 1998;143:1457–1459. doi: 10.1007/s007050050389. [DOI] [PubMed] [Google Scholar]
- 25.Randhawa J S, Marriott A C, Pringle C R, Easton A J. Rescue of synthetic minireplicons establishes the absence of the NS1 and NS2 genes from avian pneumovirus. J Virol. 1997;71:9849–9854. doi: 10.1128/jvi.71.12.9849-9854.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts A, Rose J K. Recovery of negative-strand RNA viruses from plasmid DNAs: a positive approach revitalizes a negative field. Virology. 1998;247:1–6. doi: 10.1006/viro.1998.9250. [DOI] [PubMed] [Google Scholar]
- 27.Teng M N, Collins P L. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villanueva N, Hardy R, Asenjo A, Yu Q, Wertz G. The bulk of the phosphorylation of human respiratory syncytial virus phosphoprotein is not essential but modulates viral RNA transcription and replication. J Gen Virol. 2000;81:129–133. doi: 10.1099/0022-1317-81-1-129. [DOI] [PubMed] [Google Scholar]
- 29.Yu Q, Davis P J, Li J, Cavanagh D. Cloning and sequencing of the matrix protein (M) gene of turkey rhinotracheitis virus reveal a gene order different from that of respiratory syncytial virus. Virology. 1992;186:426–434. doi: 10.1016/0042-6822(92)90007-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Q, Hardy R W, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yunus A S, Krishnamurthy S, Pastey M K, Huang Z, Khattar S K, Collins P L, Samal S K. Rescue of a bovine respiratory syncytial virus genomic RNA analog by bovine human and ovine respiratory syncytial viruses confirms the “functional integrity” and “cross-recognition” of BRSV cis-acting elements by HRSV and ORSV. Arch Virol. 1999;144:1977–1990. doi: 10.1007/s007050050719. [DOI] [PubMed] [Google Scholar]