Abstract
Trypanosoma cruzi is the etiologic agent of Chagas disease, an infection that can lead to the development of cardiac fibrosis, which is characterized by the deposition of extracellular matrix (ECM) components in the interstitial region of the myocardium. The parasite itself can induce myofibroblast differentiation of cardiac fibroblast in vitro, leading to increased expression of ECM. Inorganic polyphosphate (polyP) is a linear polymer of orthophosphate that can also induce myofibroblast differentiation and deposition of ECM components and is highly abundant in T. cruzi. PolyP can modify proteins post-translationally by non-enzymatic polyphosphorylation of lysine residues of poly-acidic, serine-(S) and lysine (K)-rich (PASK) motifs. In this work, we used a bioinformatics screen and identified the presence of PASK domains in several surface proteins of T. cruzi. We also detected polyP in the external surface of its different life cycle stages and confirmed the stimulation of host cell fibrosis by trypomastigote infection. However, we were not able to detect significant secretion of the polymer or activation of transforming growth factor beta (TGF-β), an important factor for the generation of fibrosis by inorganic polyP- or trypomastigote-conditioned medium.
Keywords: cardiac fibrosis, Chagas disease, transforming growth factor beta, polyphosphate, Trypanosoma cruzi
1. Introduction
Chagas disease is the most prevalent parasitic disease in the Americas, affecting 6 to 7 million people, with 100 million people at risk worldwide [1,2]. In the U.S., it is estimated that 300,000 individuals are infected and unknowingly expose others to infection through blood and tissue donation [3]. No vaccines are available against Chagas disease, despite recent efforts [4,5,6], and several challenges remain with the current anti-parasitic drug treatment including partial or lack of activity in the acute or chronic stage of the disease, respectively, and unwanted side effects [7,8]. Chagas cardiopathy is the most frequent clinical presentation of the chronic disease, which develops in about 30% of the infected patients [9]. Acute myocarditis can occur in about 1% of infected patients [10]. Interstitial fibrosis occurs in both acute and chronic heart disease and is characterized by deposition of extracellular matrix (ECM) components by cardiac myofibroblasts in the interstitial region of the myocardium [10]. The mechanism and the molecular signaling pathways responsible for the deposition of ECM resulting from the infection with T. cruzi are not known, although transforming growth factor beta (TGF-β) was proposed to mediate cardiac fibrosis [11].
Fibrosis results from excessive accumulation of ECM by terminally differentiated fibroblasts (myofibroblasts) in response to injury or illness and leads to organ disfunction and failure. Myofibroblasts are the main cellular effectors in cardiac fibrosis and the main source of matrix proteins [12]. The most abundant ECM components are the collagens, elastins, fibronectins, and proteoglycans [13]. TGF-β is the best-characterized fibrogenic growth factor [14]. It is secreted by many cell types in a latent form and needs to be activated through proteolysis or interaction with ECM proteins to perform its biological role [15]. Once active, TGF-β binds to membrane receptors (type I and type II) and stimulates phosphorylation of proteins via the Smad2/3 proteins or via alternative pathways, MAPK, JNK, p38, and PI3K, inducing a variety of cellular responses including the regulation of more than 500 genes [15]. TGF-β induction and activation has been demonstrated in several models of cardiac fibrosis and in human hearts with fibrotic cardiomyopathic changes [16]. Chagas cardiac fibrosis was proposed to be mediated by TGF-β, as shown by its involvement in the production of several ECM components [11].
Acute Chagas myocarditis’ residual effects include interstitial fibrosis and myofiber hypertrophy, with minimal inflammation, while the chronic phase of Chagas cardiomyopathy includes low-grade myocarditis with myofiber hypertrophy and interstitial fibrosis that develops over years to decades [10]. Deposition of ECM components (fibronectin, laminin, and collagen) in the infected heart was demonstrated in three-dimensional cardiomyocyte cultures [17] and in cardiac myofibroblasts [18] infected with the parasite. T. cruzi was possibly responsible as no inflammatory cells were present, and treatment with posaconazole reverted this effect [19].
Suess et al. [20] demonstrated that polyphosphate (polyP) is a potent inducer of fibroblast chemotaxis, myofibroblast differentiation, and production of ECM components, such as a-smooth muscle actin (α-SMA), stress fibers, and collagen. PolyP is a polymer of three to hundreds of orthophosphate units bound by high-energy phosphoanhydride bonds and is highly abundant in T. cruzi [21]. We have found that polyP is released by cells such as platelets [22], and mast cells [23]. Platelet-released polyP is a potent procoagulant [24] and proinflammatory [25] agent, and acts as a chemotactic agent for neutrophils, and a stimulant of fibroblast differentiation to myofibroblasts [20]. PolyP present in extracellular vesicles derived from prostate cancer cells (prostasomes) activates coagulation Factor XII [26]. PolyP is also present in extracellular vesicles from different cancer cells and mediates binding of Factor XII, contact activation, and thrombosis [27].
PolyP has been implicated in many processes from serving as an energy source to regulating gene expression [28,29]. In T. cruzi, polyP was first found in acidocalcisomes [21]. However, using a combination of biotinylated polyP and a bioinformatics approach, several glycolytic/gluconeogenic proteins, ribosomal proteins, and other nucleolar proteins that putatively interact with this polymer were identified, and polyP was detected in the glycosomes and nucleoli of these parasites using the polyP-binding domain of E. coli exopolyphosphatase (PPBD) coupled to Alexa 488 [30].
Azevedo et al. [31] first reported that polyP can be attached to lysine residues of proteins within PASK domains and used the term polyphosphorylation to define this non-enzymatic post-translational modification. This modification was found initially in two yeast proteins, nuclear signal recognition 1 (Nsr1), and its interacting partner topoisomerase 1 (Top1). Polyphosphorylation downregulated their interaction and inhibited Top1 enzymatic activity [31]. Further work found evidence that polyphosphorylation can occur in several other Saccharomyces cerevisiae [32,33,34], as well as in human [32] enzymes.
In this work, we used a bioinformatics screen developed by Bentley-DeSousa et al. [32] to search the proteome of T. cruzi for proteins harboring putative PASK-like domains. Interestingly, we found many surface proteins containing this domain and that polyP was present on the surface of T. cruzi at different life stages. Since it has been reported that a trypomastigote invasion of host cells leads to fibrosis and polyP has been shown to induce fibrosis of mammalian cells, we investigated whether polyP was released from the trypomastigote surface and stimulated fibrosis of host cells through TGF-β activation.
2. Materials and Methods
2.1. Chemicals and Reagents
BCA Protein Assay Kit was from Thermo Fisher Scientific Inc. (Waltham, MA, USA) PureView prestained protein ladder was from Azura Genomics (Raynham, MA, USA). Alexa Fluor-conjugated secondary antibodies were purchased from Life Technologies (Carlsbad, CA, USA). Fluoromount-G® was from SouthernBiotech (Birmingham, AL, USA). Nitrocellulose membranes were from Bio-Rad (Hercules, CA, USA). Recombinant human latent TGF-β 1 protein was from R&D Systems (Minneapolis, MN, USA). PolyP60 was a gift from Dr. Toshikazu Shiba (Kitasato University). All other reagents of analytical grade were from Sigma (St. Louis, MO, USA).
2.2. Cell Cultures
T. cruzi Y strain epimastigotes were cultured in liver infusion tryptose (LIT) medium containing 10% heat-inactivated newborn calf serum at 28 °C [35]. Tissue culture cell-derived trypomastigotes [36] and amastigotes [37] were obtained from infected Vero cells and were collected from the culture medium of infected host cells as described in the references. Vero cells were grown in RPMI supplemented with 10% fetal bovine serum and maintained at 37 °C with 5% CO2.
2.3. PASK Domain Screen
The proteomes of T. cruzi, T. brucei, and L. major were screened for putative PASK domains using a custom script designed by Bentley-Souza et al. [32]. In short, parasite proteomes were retrieved from Uniprot database and searched for proteins containing at least 20 amino acid clusters of ≥75% S/E/D/K with at least one K residue. The identified proteins were analyzed for gene ontology (GO) term enrichment using TritrypDB (VEuPathDB.org accessed on 9 May 2024).
2.4. Polyphosphate Surface Staining
Detection of polyphosphate on the cell surface was performed using the polyP-binding domain (PPBD) from E. coli PPX [38] conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, OR, USA) as described before [30]. Cells were harvested, washed 2× with PBS, and fixed for 15 min with 4% paraformaldehyde in 1× PBS pH 7.4. Cells were allowed to settle on poly-L-lysine-coated cover slips for 30 min and blocked with 5% goat serum, 3% BSA, and 1% fish gelatin for 1 h, at RT. The coverslips were incubated with 8 μg/mL PPBD-Alexa fluor 488 and 5 μg/mL DAPI for 1 h, at RT in the dark. Following incubation, the coverslips were washed 3× with PBS and mounted on slides using fluoromount G. The slides were imaged on a Deltavision deconvolution microscope (GE) equipped with a 100× objective NA 1.35.
2.5. In Vitro Fibroblast Infection Assay
NIH-3T3 cells cultured in DMEM containing high glucose, pyruvate (Thermo Scientific 11995065, Boston, MA, USA), 1% penicillin and streptomycin, 10% heat-inactivated newborn calf serum, and 10 mM Hepes were seeded at 5 × 104 per well in a 6-well dish with glass coverslips, then allowed to incubate for 24 h. Fibroblasts were then rinsed with Dulbecco’s Hanks’ solution, and tissue culture-derived T. cruzi trypomastigotes (Y strain) were used to infect at a 20:1 ratio of parasites to host cells. At 8 h post-infection, coverslips were washed with Dulbecco’s Hanks’ solution to remove extracellular parasites, and the media was replaced. At 24, 48, and 72 h post initiation of infection, coverslips were removed from wells and immediately fixed in 4% paraformaldehyde in PBS, pH 7.4, at room temperature for 15 min. Coverslips were washed once with PBS pH 7.4, then stored in PBS at 4 °C prior to immunofluorescence assay. Uninfected controls were treated in the same manner, without the addition of parasites.
2.6. Immunofluorescence Analyses
To evaluate fibroblast differentiation as evidenced by relative levels of α-SMA, fixed coverslips were permeabilized for 5 min with 0.1% Triton X-100. The cells were then blocked overnight at 4 °C with PBS containing 50 mM NH4Cl, 3% BSA, 5% goat serum, and 1% fish gelatin. Next, the coverslips were incubated with 1:200 monoclonal rabbit anti-α-SMA antibody (ABclonal A17910; Woburn, MA, USA) diluted in 1% BSA in PBS, pH 8.0, for 1 h at room temperature. The coverslips were washed three times with 1% BSA in PBS, pH 8.0, then incubated with 1:1000 TRITC-conjugated goat-anti-rabbit antibodies for 1 h in the dark at room temperature. The coverslips were then washed twice with 1% BSA in PBS, pH 8.0, followed by a final wash with PBS, pH 8.0. Then, the coverslips were mounted on slides using Fluoromount-G mounting medium containing 5 μg/mL of DAPI to stain DNA. The fluorescence images were captured using the DeltaVision II microscope system (Applied Precision, Inc., Issaquah, WA, USA), with a 60× objective using equivalent parameters for each image capture. The images were converted to gray scale and opened in FIJI. Using a pre-assigned circular ROI of equal size and a mean gray area of the TRITC channel for one background region, three randomly chosen regions per cell in frame were measured. Five images were analyzed for each coverslip, with three biological replicas. The mean gray area of the background was subtracted from the mean gray area of cells for each image, giving a normalized measurement of fluorescence in relative quantification units. Data were then compiled, graphed, and analyzed using GraphPad Prism software version 9 (GraphPad, La Jolla, CA, USA). The reported values are means ± standard deviation from 3 biological experiments. The level of significance was evaluated by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test.
2.7. Secretion and Extracellular Vesicle Isolation
Cell culture-derived trypomastigotes were collected, washed, and resuspended at 1 × 108 parasites per milliliter in 10 mL of buffer A with glucose, pH 5 (BAG; 116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 50 mM Hepes, 5.5 mM glucose). Parasites were then incubated for 24 h at 37 °C and 5% CO2 to allow for secretion of molecules and release of extracellular vesicles. At the endpoint of incubation, parasites were resuspended homogenously and counted to determine the terminal cell counts. The total conditioned media and parasites were collected and centrifuged at 3000× g for 10 min to pellet cells. After collecting the supernatant for the next step, the cell pellet was washed with fresh, sterile BAG, pH 7.0, then rapidly frozen using liquid nitrogen and stored at −80 °C. The supernatant was then passed through a 0.45 μm syringe filter to remove any remaining cellular debris. An aliquot of the conditioned media filtrate was then rapidly frozen and stored at −80 °C to represent “total secretion”. The remaining filtrate was then centrifuged at 100,000× g and 4 °C for 2 h to pellet large extracellular vesicles (V2) followed by a 16 h spin to pellet smaller extracellular vesicles (V16) [39]. Vesicle pellets were resuspended in 50 μL of fresh, sterile BAG, pH 7.0, then rapidly frozen in liquid nitrogen and stored at −80 °C. An aliquot of the final EV-depleted supernatant was then collected, rapidly frozen, and stored at −80 °C. Extraction and measurement of short chain polyP was conducted as previously reported [21]. Briefly, samples were resuspended in 1 M perchloric acid and sonicated on ice. Cellular debris were pelleted by centrifuging at 18,000× g for 5 min at 4 °C. The cleared supernatant was then added to 4 mg of washed TiO2 beads and allowed to incubate for 20 min at 4 °C with rotation. The TiO2 beads were then washed twice with 1 M perchloric acid and pelleted. Bound polyphosphates were then eluted by addition of 400 µL total of cold 2.8% ammonium hydroxide in two parts. Sample volumes were reduced using a centrifugal evaporator until the pH of the sample was 7 to 8. Extracted polyP samples were then rapidly frozen and stored at −80 °C for analysis.
2.8. TGF-β Activation
Latent TGF-β 1 (10 ng/mL) was incubated with a DMEM medium containing 1.5% bovine calf serum or with a trypomastigote-conditioned medium in the presence or absence of plasmin (2 U/mL) and in the presence or absence of 5 µM polyP60 (a gift from Toshikazu Shiba, Kitasato University) for 2 h at 37 °C. To obtain the conditioned media from trypomastigotes, the cell-derived trypomastigotes were incubated in phosphate-free buffer A with glucose (BAG, 116 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 50 mM Hepes pH 7.4, and 5.5-mM glucose), at a concentration of 1 × 108 parasites per ml at 37 °C in 5% CO2 for 24 h. This conditioned medium was clarified by centrifugation at 2000× g for 10 min and filtered through a 0.45 µm membrane.
2.9. Western Blot Analyses
The samples were mixed with a 4× Laemmli sample buffer (125 mM Tris-HCl, pH 7, 20% (v/v) glycerol, 4.0% (w/v) SDS, 4.0% (w/v) bromophenol blue) before application to 10% SDS-polyacrylamide gels. The separated proteins were transferred onto nitrocellulose membranes with a Bio-Rad Trans-blot apparatus. The membranes were blocked with 5% nonfat dried skim milk in PBS-T (PBS containing 0.1% v/v Tween 20) overnight at 4 °C. Next, the membranes were incubated for 1 h at room temperature with the primary antibody TGF beta 1 Rabbit from ABclonal (1:2000). After three washes with PBS-T, the blots were incubated with the secondary antibody IRDye 680RD-conjugated goat anti-rabbit IgG (1:10,000) for 1 h at room temperature in the dark. The blots were washed three times with PBS-T, and the Western blot images were obtained and processed with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA). The densities of the Western blot bands were quantified with the ImageStudioLite software v.6 (LI-COR Biosciences).
3. Results
3.1. Bioinformatics Screen of Trypanosomatid Proteins
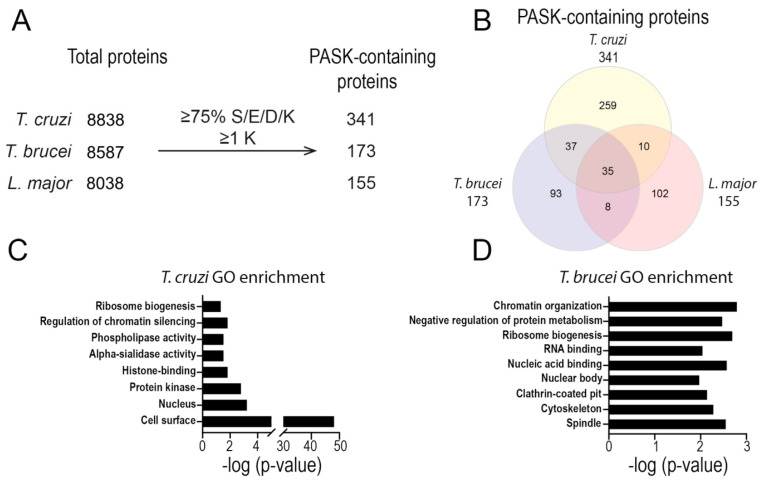
Using a bioinformatics screen developed before [32], we searched the proteomes of T. cruzi, T. brucei, and L. major for proteins possessing putative PASK domains. In total, we found 341 T. cruzi proteins, 173 T. brucei proteins, and 155 L. major proteins that possess regions of 20 amino acid residues containing at least 75% serine (S)/glutamic acid (E)/aspartic acid (D)/lysine (K) residues with at least 1 lysine (Figure 1A and Table S1). Of these proteins, 35 have orthologs present in each kinetoplastid parasite species (Figure 1B). To determine the function of these putative PASK domain-containing proteins, we performed gene ontology (GO) term analysis using TriTrypDB.org. We observed enrichment for proteins involved in processes such as ribosome biogenesis, histone binding, RNA binding, and chromatin organization in both T. cruzi and T. brucei (Figure 1C,D). Unexpectedly, we also saw strong enrichment for proteins annotated to be in the cell surface, especially in T. cruzi (Figure 1C). In total, 9 trans-sialidases, 2 TcMUC II, and 103 mucin-associated surface proteins [MASP] were found to have a PASK domain (Table S1).
Figure 1.
Proteins with a PASK domain in trypanosomatids. (A,B) Number of proteins identified as possessing a PASK domain in the proteomes of T. cruzi, T. brucei, and L. major. (C,D) Analysis of the proteins from T. cruzi (C) or T. brucei (D) according to molecular function. GO terms found for proteins possessing a putative PASK domain are indicated.
3.2. Polyphosphate Localization in the Outer Surface of T. cruzi
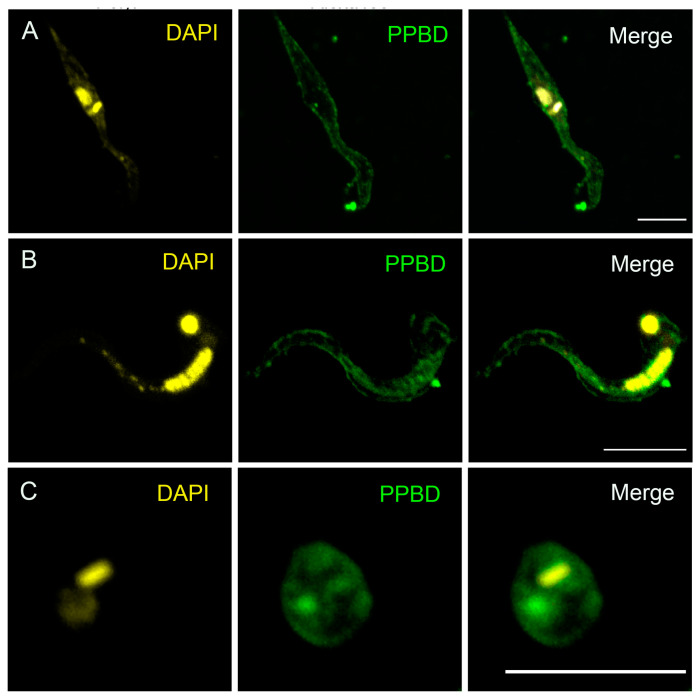
In previous work, we used Alexa Fluor 488-labeled polyP-binding domain (PPBD) of E. coli exopolyphosphatase to investigate the presence of polyP in glycosomes and nucleoli of permeabilized T. cruzi and T. brucei different stages [30]. Because we found that several outer-surface proteins of T. cruzi have PASK domains (Figure 1), we used Alexa fluor 488-labeled PPBD in non-permeabilized trypanosomes, and we detected labeling of polyP at the outer surface of the plasma membrane of epimastigotes (Figure 2A), a localization that has also been reported in fungi [40]. DAPI, which is permeable, labeled the kinetoplast and nuclear DNA. Importantly, we also found surface membrane expression of polyP in T. cruzi trypomastigotes (Figure 2B), and amastigotes (Figure 2C), the infective stages. No surface labeling was found in the permeabilized cells, as reported before [30].
Figure 2.
Presence of surface polyP in T. cruzi different stages. (A) Fluorescence analysis of non-permeabilized T. cruzi wild-type epimastigotes stained with DAPI (left) or PPBD-Alexa488 (middle) and merged images (right) showing the K-DNA and nuclei in yellow and surface expression of polyP in green, respectively. Scale bar = 5 µm. (B,C) Fluorescence analysis of non-permeabilized trypomastigotes (B) and amastigotes (C) stained with DAPI (left), and PPBD-Alexa 488 (PPBD) (middle) and merged images (right). Note that DAPI is cell permeable and strongly labels the nucleus, and k-DNA. Scale bars = 5 µm.
3.3. T. cruzi Infection Induces Myofibroblast Differentiation in NIH-3T3 Cells
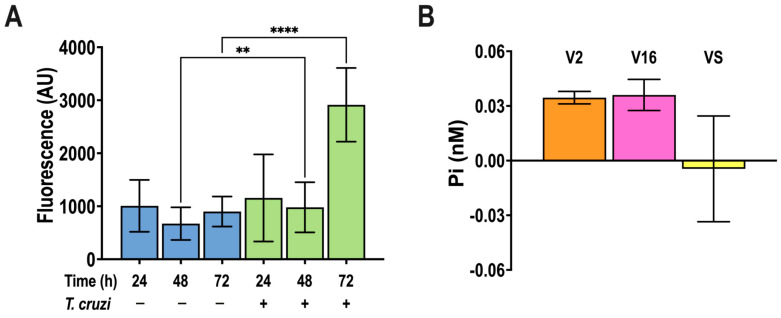
α-Smooth muscle actin (α-SMA) is a cytoskeletal protein that is highly expressed in activated myofibroblasts [41]. By assessing the changes in α-SMA expression, it has been shown that T. cruzi infection activates the differentiation of cardiac fibroblast into myofibroblasts [18]. In agreement with those results, the infection of NIH-3T3 fibroblasts with T. cruzi trypomastigotes showed a time-dependent increase in α-SMA expression, as detected by immunofluorescence analysis using specific antibodies (Figure 3A).
Figure 3.
α-SMA production by T. cruzi-infected NIH-3T3 cells, and polyP presence in trypomastigote extracellular vesicles. (A) NIH-3T3 cells were infected by T. cruzi cell-derived trypomastigotes (ratio 20:1) and α-SMA antibody (1:200) staining was quantified by fluorescence microscopy as a marker of myofibroblast differentiation at different times post-infection. Values are means ± S.D., n = 3, ** p < 0.01, **** p < 0.0001, ANOVA with Turkey’s multiple comparisons test. (B) PolyP extracted from large (V2) and small (V16) extracellular vesicles, and vesicle free supernatant (VS) of 1 × 108/mL cell-derived trypomastigotes incubated for 18 h, at pH = 5.0, detected by the PPX method, and expressed in Pi units.
3.4. Presence of PolyP in Extracellular Vesicles of T. cruzi
T. cruzi trypomastigotes have been shown to release proteins associated with vesicles obtained by fractionation of conditioned cultured supernatant [39]. We applied this fractionation technique and measured their polyP content, as described in Materials and Methods but found only nanomolar levels of polyP (expressed as Pi units) in large (V2) and small (V16) extracellular vesicles and negligible levels in the vesicle supernatant (VS) (Figure 3B).
3.5. Lack of Activation of TGF-β by PolyP- or T. cruzi Trypomastigotes-Conditioned Medium
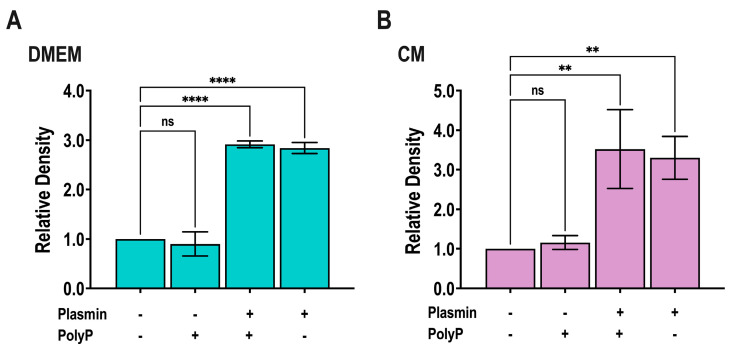
We investigated whether TGF-β is activated by polyP- or T. cruzi trypomastigotes conditioned medium. Plasmin, a known TGF-β activator [42], was used as positive control. Briefly, latent TGF-β was incubated with plasmin (2 U/mL) in a DMEM- or T. cruzi-conditioned medium for 2 h at 37 °C, in the absence or presence of 5 µM polyP60 (the polyP size and conditions used in [20] to demonstrate fibrosis stimulation by polyP). Triplicate samples were prepared for PAGE followed by immunoblot analysis to detect the 13 kDa mature TGF-β. While plasmin was able to activate TGF-β, conditioned medium with or without polyP was not able to activate it and polyP did not potentiate the plasmin effect (Figure 4 and Figure S1).
Figure 4.
Activation of latent TGF-β by plasmin and conditioned medium with and without polyP60. (A,B) Latent TGF-β was incubated with or without plasmin (2 U/mL) in DMEM with 1.5% bovine calf serum (A) or in conditioned medium (B) for 2 h at 37 °C, in the absence or presence of 5 µM polyP60. Samples were prepared for PAGE analysis followed by immunoblot analysis to detect the 13 kDa mature TGF-β, and band density was measured. Values are means ± SD (n = 3). ** p < 0.01 and **** p < 0.0001; n.s., no significant, by one-way ANOVA with Dunnett’s multiple comparison test.
4. Discussion
The use of the polyP-binding domain from E. coli PPX conjugated with Alexa Fluor 488 allowed us to demonstrate the presence of polyP on the external surface of T. cruzi different life cycle stages. These results are in agreement with the presence of PASK domain-containing proteins in several surface proteins of the parasite, suggesting that these proteins could be polyphosphorylated. We confirmed the ability of T. cruzi infective forms to induce myofibroblast differentiation, but we found little evidence that this was caused by secreted polyP or that a trypomastigote-conditioned media were able to induce activation of the fibrosis-inducing TGF-β. However, we cannot rule out the possibility that bound polyP is involved in inducing myofibroblast differentiation.
Our bioinformatics screen of PASK domain-containing proteins identified a lower number of proteins in trypanosomatids than in yeast, but gene ontology-term analysis indicated enrichment in similar functions related to ribosome biogenesis and rRNA processing and proteins with nuclear and nucleolar localizations. Unexpectedly, several of the identified proteins have an external surface localization (mucin-associated surface protein (MASP), trans-sialidase, mucin TcMUCII, and phosphoinositide phospholipase C (PI-PLC) in T. cruzi; variant surface glycoprotein (VSG) in T. brucei, and amastin-like proteins in L. major) (Table S1). Accordingly, polyP was localized to the external surface of T. cruzi different life cycle stages, and it is tempting to speculate that it could be attached to these surface proteins by polyphosphorylation. Further work will be required to demonstrate that this is the case. PolyP has been linked to the persistence of T. cruzi infection [43] and it is possible that this surface localization could contribute to this property.
Until now, only a few proteins have been reported as susceptible to polyphosphorylation in yeast and humans. After the initial description of polyphosphorylation of Nsr1 and Top1 [31], the use of yeast in the bioinformatics screen that we used here led to the selection of a group of 90 potential targets, of which 23 were confirmed as susceptible to polyphosphorylation by the protein mobility shift assay [32,34]. A first group of 15 protein targets included several proteins implicated in ribosome biogenesis [32] while a second group of 8 proteins were shown to interact with previous targets implicated in ribosome biogenesis or those that consisted of vacuolar proteins [34]. Six human proteins were found to be polyphosphorylated following expression of Escherichia coli polyP kinase (PPK) in HEK293T cells, some of them homologs to the yeast targets [32]. Screening of a human protein array with biotinylated polyP identified eight additional human proteins susceptible to polyphosphorylation [33]. Early work showed that polyphosphorylation could affect protein–protein interaction, subcellular localization, or enzymatic activity of the targeted proteins [31].
T. cruzi, like many other cells, has been demonstrated to release extracellular vesicles [39]. The surface localization of polyP suggested that this polymer could possibly be detected in these vesicles and be involved in inducing myofibroblast differentiation and deposition of ECM components. However, only nanomolar levels of polyP could be detected in small and large extracellular vesicles released by infective trypomastigotes while micromolar amounts of polyP were shown to be needed to stimulate ECM components deposition in NIH-3T3 fibroblasts [20]. Neither the T. cruzi trypomastigote-conditioned medium nor synthetic polyP were able to induce TGF-β activation, a reported mediator of cardiac fibrosis [14].
In summary, our work identified the surface localization of polyP and confirmed the myofibroblast deposition of ECM components upon T. cruzi infection of mammalian cells, but did not find significant secretion of polyP or stimulation of TGF-β activation by a polyP- or trypomastigote-conditioned medium.
Acknowledgments
We thank Michael Downey (University of Ottawa, ON, USA) for the provision of the bioinformatics screen to detect PASK domain-containing proteins, James Morrissey (University of Michigan, MI, USA) for the NIH-3T3 fibroblasts, Toshikazu Shiba (Kitasato University) for polyP60, and Muthugapatti Kandasamy and the Biomedical Microscopy Core of the University of Georgia for the use of microscopes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13090776/s1, Figure S1: Latent TGF-β activation; Table S1: T. cruzi, T. brucei, and L. major proteins possessing putative PASK domains.
Author Contributions
Conceptualization, L.P.C., A.G., M.S.B. and R.D.; methodology, L.P.C., A.G., M.S.B. and R.D.; software, L.P.C.; validation, L.P.C., A.G., M.S.B. and R.D.; formal analysis, L.P.C., A.G., M.S.B. and R.D.; investigation, L.P.C., A.G. and M.S.B.; resources, R.D.; data curation, L.P.C., A.G. and M.S.B.; writing—original draft preparation, R.D.; writing—review and editing, L.P.C., A.G., M.S.B. and R.D.; visualization, L.P.C.; supervision, R.D.; project administration, R.D.; funding acquisition, R.D. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Data are contained within the article and Supplementary Information.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This work was funded by the U.S. National Institutes of Health (Grant AI173402) to R.D. L.P.C. and A.G. were supported by the U.S. National Institutes of Health (T32 training grant AI060546). M.S.B. was supported by an American Heart Association postdoctoral fellowship (24POST1196440).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Biolo A., Ribeiro A.L., Clausell N. Chagas cardiomyopathy–where do we stand after a hundred years? Prog. Cardiovasc. Dis. 2010;52:300–316. doi: 10.1016/j.pcad.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Gomez-Ochoa S.A., Rojas L.Z., Echeverria L.E., Muka T., Franco O.H. Global, Regional, and National Trends of Chagas Disease from 1990 to 2019: Comprehensive Analysis of the Global Burden of Disease Study. Glob. Heart. 2022;17:59. doi: 10.5334/gh.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bern C., Montgomery S.P. An estimate of the burden of Chagas disease in the United States. Clin. Infect. Dis. 2009;49:e52–e54. doi: 10.1086/605091. [DOI] [PubMed] [Google Scholar]
- 4.Jones K.M., Poveda C., Versteeg L., Bottazzi M.E., Hotez P.J. Preclinical advances and the immunophysiology of a new therapeutic Chagas disease vaccine. Expert. Rev. Vaccines. 2022;21:1185–1203. doi: 10.1080/14760584.2022.2093721. [DOI] [PubMed] [Google Scholar]
- 5.Dumonteil E., Herrera C., Buekens P. A therapeutic preconceptional vaccine against Chagas disease: A novel indication that could reduce congenital transmission and accelerate vaccine development. PLoS Negl. Trop. Dis. 2019;13:e0006985. doi: 10.1371/journal.pntd.0006985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumonteil E., Herrera C. The Case for the Development of a Chagas Disease Vaccine: Why? How? When? Trop. Med. Infect. Dis. 2021;6:16. doi: 10.3390/tropicalmed6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urbina J.A., Docampo R. Specific chemotherapy of Chagas disease: Controversies and advances. Trends Parasitol. 2003;19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Martinez S.J., Romano P.S., Engman D.M. Precision Health for Chagas Disease: Integrating Parasite and Host Factors to Predict Outcome of Infection and Response to Therapy. Front. Cell Infect. Microbiol. 2020;10:210. doi: 10.3389/fcimb.2020.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saraiva R.M., Mediano M.F.F., Mendes F.S., Sperandio da Silva G.M., Veloso H.H., Sangenis L.H.C., da Silva P.S., Mazzoli-Rocha F., Sousa A.S., Holanda M.T., et al. Chagas heart disease: An overview of diagnosis, manifestations, treatment, and care. World J. Cardiol. 2021;13:654–675. doi: 10.4330/wjc.v13.i12.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonney K.M., Luthringer D.J., Kim S.A., Garg N.J., Engman D.M. Pathology and Pathogenesis of Chagas Heart Disease. Annu. Rev. Pathol. 2019;14:421–447. doi: 10.1146/annurev-pathol-020117-043711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira R.R., Waghabi M.C., Bailly S., Feige J.J., Hasslocher-Moreno A.M., Saraiva R.M., Araujo-Jorge T.C. The Search for Biomarkers and Treatments in Chagas Disease: Insights From TGF-β Studies and Immunogenetics. Front. Cell Infect. Microbiol. 2021;11:767576. doi: 10.3389/fcimb.2021.767576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinz B., Phan S.H., Thannickal V.J., Galli A., Bochaton-Piallat M.L., Gabbiani G. The myofibroblast: One function, multiple origins. Am. J. Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaauboer M.E., Boeijen F.R., Emson C.L., Turner S.M., Zandieh-Doulabi B., Hanemaaijer R., Smit T.H., Stoop R., Everts V. Extracellular matrix proteins: A positive feedback loop in lung fibrosis? Matrix Biol. 2014;34:170–178. doi: 10.1016/j.matbio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Frangogiannis N.G. Cardiac fibrosis. Cardiovasc. Res. 2021;117:1450–1488. doi: 10.1093/cvr/cvaa324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frangogiannis N.G. Transforming growth factor-beta in myocardial disease. Nat. Rev. Cardiol. 2022;19:435–455. doi: 10.1038/s41569-021-00646-w. [DOI] [PubMed] [Google Scholar]
- 17.Garzoni L.R., Adesse D., Soares M.J., Rossi M.I., Borojevic R., de Meirelles Mde N. Fibrosis and hypertrophy induced by Trypanosoma cruzi in a three-dimensional cardiomyocyte-culture system. J. Infect. Dis. 2008;197:906–915. doi: 10.1086/528373. [DOI] [PubMed] [Google Scholar]
- 18.Coelho L.L., Pereira I.R., Pereira M.C.S., Mesquita L., Lannes-Vieira J., Adesse D., Garzoni L.R. Trypanosoma cruzi activates mouse cardiac fibroblasts in vitro leading to fibroblast-myofibroblast transition and increase in expression of extracellular matrix proteins. Parasit. Vectors. 2018;11:72. doi: 10.1186/s13071-018-2614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nisimura L.M., Ferrao P.M., Nogueira A.D.R., Waghabi M.C., Meuser-Batista M., Moreira O.C., Urbina J.A., Garzoni L.R. Effect of Posaconazole in an in vitro model of cardiac fibrosis induced by Trypanosoma cruzi. Mol. Biochem. Parasitol. 2020;238:111283. doi: 10.1016/j.molbiopara.2020.111283. [DOI] [PubMed] [Google Scholar]
- 20.Suess P.M., Smith S.A., Morrissey J.H. Platelet polyphosphate induces fibroblast chemotaxis and myofibroblast differentiation. J. Thromb. Haemost. 2020;18:3043–3052. doi: 10.1111/jth.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz F.A., Rodrigues C.O., Docampo R. Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 2001;276:26114–26121. doi: 10.1074/jbc.M102402200. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz F.A., Lea C.R., Oldfield E., Docampo R. Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 2004;279:44250–44257. doi: 10.1074/jbc.M406261200. [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Sanchez D., Hernandez-Ruiz L., Ruiz F.A., Docampo R. Polyphosphate is a novel pro-inflammatory regulator of mast cells and is located in acidocalcisomes. J. Biol. Chem. 2012;287:28435–28444. doi: 10.1074/jbc.M112.385823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith S.A., Mutch N.J., Baskar D., Rohloff P., Docampo R., Morrissey J.H. Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. USA. 2006;103:903–908. doi: 10.1073/pnas.0507195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller F., Mutch N.J., Schenk W.A., Smith S.A., Esterl L., Spronk H.M., Schmidbauer S., Gahl W.A., Morrissey J.H., Renne T. Platelet polyphosphates are proinflammatory and procoagulant mediators In Vivo. Cell. 2009;139:1143–1156. doi: 10.1016/j.cell.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickel K.F., Ronquist G., Langer F., Labberton L., Fuchs T.A., Bokemeyer C., Sauter G., Graefen M., Mackman N., Stavrou E.X., et al. The polyphosphate-factor XII pathway drives coagulation in prostate cancer-associated thrombosis. Blood. 2015;126:1379–1389. doi: 10.1182/blood-2015-01-622811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim Y.J., Chatterjee V., Swaidani S., Alluri R.K., Kundu S., Merkulova A., Angelini D., You D., Whitney S.A., Feener E.P., et al. Polyphosphate expression by cancer cell extracellular vesicles mediates binding of factor XII and contact activation. Blood Adv. 2021;5:4741–4751. doi: 10.1182/bloodadvances.2021005116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno S.N., Docampo R. Polyphosphate and its diverse functions in host cells and pathogens. PLoS Pathog. 2013;9:e1003230. doi: 10.1371/journal.ppat.1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao N.N., Gomez-Garcia M.R., Kornberg A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 30.Negreiros R.S., Lander N., Huang G., Cordeiro C.D., Smith S.A., Morrissey J.H., Docampo R. Inorganic polyphosphate interacts with nucleolar and glycosomal proteins in trypanosomatids. Mol. Microbiol. 2018;110:973–994. doi: 10.1111/mmi.14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azevedo C., Livermore T., Saiardi A. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell. 2015;58:71–82. doi: 10.1016/j.molcel.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Bentley-DeSousa A., Holinier C., Moteshareie H., Tseng Y.C., Kajjo S., Nwosu C., Amodeo G.F., Bondy-Chorney E., Sai Y., Rudner A., et al. A Screen for Candidate Targets of Lysine Polyphosphorylation Uncovers a Conserved Network Implicated in Ribosome Biogenesis. Cell Rep. 2018;22:3427–3439. doi: 10.1016/j.celrep.2018.02.104. [DOI] [PubMed] [Google Scholar]
- 33.Azevedo C., Singh J., Steck N., Hofer A., Ruiz F.A., Singh T., Jessen H.J., Saiardi A. Screening a Protein Array with Synthetic Biotinylated Inorganic Polyphosphate To Define the Human PolyP-ome. ACS Chem. Biol. 2018;13:1958–1963. doi: 10.1021/acschembio.8b00357. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy L., Bentley-DeSousa A., Denoncourt A., Tseng Y.C., Gabriel M., Downey M. Proteins required for vacuolar function are targets of lysine polyphosphorylation in yeast. FEBS Lett. 2020;594:21–30. doi: 10.1002/1873-3468.13588. [DOI] [PubMed] [Google Scholar]
- 35.Bone G.J., Steinert M. Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature. 1956;178:308–309. doi: 10.1038/178308a0. [DOI] [PubMed] [Google Scholar]
- 36.Docampo R., Moreno S.N., Vercesi A.E. Effect of thapsigargin on calcium homeostasis in Trypanosoma cruzi trypomastigotes and epimastigotes. Mol. Biochem. Parasitol. 1993;59:305–313. doi: 10.1016/0166-6851(93)90228-P. [DOI] [PubMed] [Google Scholar]
- 37.Moreno S.N., Vercesi A.E., Pignataro O.P., Docampo R. Calcium homeostasis in Trypanosoma cruzi amastigotes: Presence of inositol phosphates and lack of an inositol 1,4,5-trisphosphate-sensitive calcium pool. Mol. Biochem. Parasitol. 1992;52:251–261. doi: 10.1016/0166-6851(92)90057-Q. [DOI] [PubMed] [Google Scholar]
- 38.Saito K., Ohtomo R., Kuga-Uetake Y., Aono T., Saito M. Direct labeling of polyphosphate at the ultrastructural level in Saccharomyces cerevisiae by using the affinity of the polyphosphate binding domain of Escherichia coli exopolyphosphatase. Appl. Env. Microbiol. 2005;71:5692–5701. doi: 10.1128/AEM.71.10.5692-5701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayer-Santos E., Aguilar-Bonavides C., Rodrigues S.P., Cordero E.M., Marques A.F., Varela-Ramirez A., Choi H., Yoshida N., da Silveira J.F., Almeida I.C. Proteomic analysis of Trypanosoma cruzi secretome: Characterization of two populations of extracellular vesicles and soluble proteins. J. Proteome Res. 2013;12:883–897. doi: 10.1021/pr300947g. [DOI] [PubMed] [Google Scholar]
- 40.Shari’a A.E.N., Nascimento A.E., Lima M.A.B., Campos-Takaki G.M., de Souza W. Polyphosphate in zygomycetes: A cytochemical study. Braz. J. Microbiol. 2002;33:119–126. doi: 10.1590/S1517-83822002000200004. [DOI] [Google Scholar]
- 41.Asazuma-Nakamura Y., Dai P., Harada Y., Jiang Y., Hamaoka K., Takamatsu T. Cx43 contributes to TGF-β signaling to regulate differentiation of cardiac fibroblasts into myofibroblasts. Exp. Cell Res. 2009;315:1190–1199. doi: 10.1016/j.yexcr.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 42.Lyons R.M., Gentry L.E., Purchio A.F., Moses H.L. Mechanism of activation of latent recombinant transforming growth factor beta 1 by plasmin. J. Cell Biol. 1990;110:1361–1367. doi: 10.1083/jcb.110.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galizzi M., Bustamante J.M., Fang J., Miranda K., Soares Medeiros L.C., Tarleton R.L., Docampo R. Evidence for the role of vacuolar soluble pyrophosphatase and inorganic polyphosphate in Trypanosoma cruzi persistence. Mol. Microbiol. 2013;90:699–715. doi: 10.1111/mmi.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Information.