Abstract
A series of novel thiazole-based chalcones were evaluated for their anticancer activity as potential tubulin polymerization inhibitors. In vitro anticancer screening for the thiazole derivatives 2a–2p exhibited broad-spectrum antitumor activity against various cancer cell lines particularly Ovar-3 and MDA-MB-468 cells with a GI50 range from 1.55 to 2.95 μΜ, respectively. Compound 2e demonstrated significant inhibition of tubulin polymerization, with an IC50 value of 7.78 μM compared to Combretastatin-A4 (CA-4), with an IC50 value of 4.93 μM. Molecular docking studies of compounds 2e, 2g, and 2h into tubulin further supported these findings, revealing that they bind effectively to the colchicine binding site, mirroring key interactions exhibited by CA-4. Computational predictions suggested favorable oral bioavailability and drug-likeness for these compounds, highlighting their potential for further development as chemotherapeutic agents.
Keywords: thiazole chalcones, anticancer, tubulin inhibitors, colchicine binding site
1. Introduction
Microtubules are dynamic cytoskeletal elements in human cells, involved in cellular activities throughout cell division [1]. The highly dynamic behavior of microtubules can be successfully targeted to combat rapidly replicating cancer cells [1,2]. Tumor cells possess unique traits such as unlimited growth, angiogenesis, adaptability, and effortless spread throughout the body [3,4]. These features strongly rely on the involvement of microtubules, making microtubules an essential target for treating cancer [5,6]. Antimitotic drugs are a variety of cyclic compounds that interfere with cell division [7,8] polymerization binding. Traditional antimitotic drugs directly bind to tubulin to stabilize formed microtubules or prevent tubulin from polymerizing to form microtubules. These agents create abnormalities in the mitotic spindle, leading to an extended pause in mitosis that initiates apoptosis [7,9]. The effectiveness of antimitotic therapies indicates that focusing on mitosis is a promising strategy for creating novel anticancer medications [10].
Antimitotic drugs that target tubulin bind at four distinct binding sites, taxanes, vinca alkaloids, colchicine, and laulimalide sites [11,12]. Tubulin inhibitors that target vinca alkaloids and taxane sites, like paclitaxel, vinblastine, and ixabepilone, have been commonly used in medical practice for years [13,14]. Nevertheless, due to their limited water solubility, narrow therapeutic range, and the development of drug resistance, there is a push to find safer and more potent antimitotic drugs [15,16,17].
Colchicine, a natural product, binds to a different location on tubulin and successfully prevents tubulin assembly [14]. Colchicine is not utilized clinically due to its significant toxicity [18]. Additionally, there are presently no FDA-approved tubulin inhibitors that target the colchicine site [19]. Hence, it is crucial to create new antimitotic drugs that target the colchicine binding site [20]. There is a growing interest in antimitotic agents that interact with the colchicine binding site because they are simple molecules with enhanced solubility in water and a wide therapeutic range [13,21]. Compared to other binding sites, targeting the colchicine binding site is reported to cause a rapid disruption of existing tumor vasculature and decrease multidrug resistance [21,22]. In addition, CA-4 is a potent antimitotic agent that binds to the colchicine binding site and suppresses tubulin assembly [23,24]. Nevertheless, the in vivo efficacy of CA-4 is limited due to its unfavorable pharmacokinetics, which is caused by its high hydrophobicity, low aqueous solubility, and isomerism to a less active E-isomer [11,17].
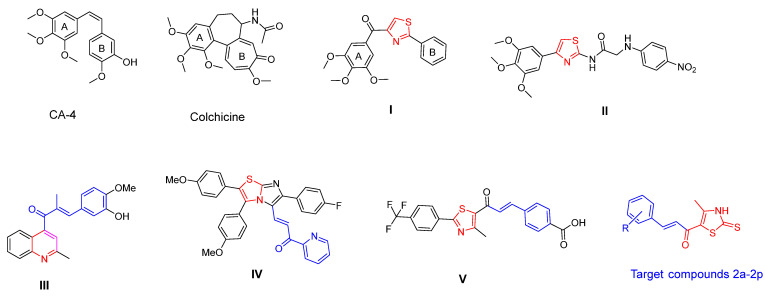
Chalcones are an important group of flavonoid compounds with unique structures in medicinal chemistry [23,25]. Due to their uncomplicated structure and anticancer characteristics, chalcones can be easily hybridized with other anticancer pharmacophores, creating several bioactive derivatives [15,25]. Hundreds of chalcone derivatives were synthesized and evaluated as tubulin inhibitors [26]. Thiazole-linked chalcone V revealed remarkable anticancer activity against the colorectal cancer cells, HT-29, HCT-116, and Lovo, with IC50 values of 7.94, 3.12, and 2.21 μM, respectively [27]. On the other hand, thiazole was incorporated into many chemotherapeutic agents due to its favorable pharmacokinetic and pharmacodynamic characteristics [3,28]. Various clinically used anticancer drugs contain thiazole rings like bleomycin, ixabepilone, and dasatinib [29]. Bioactive thiazole enhances the binding to target, molecular conformation, water solubility, physicochemical properties, and pharmacokinetic properties [3,30]. Many CA-4 analogs such as tubulin polymerization inhibitors have been reported as replacing the double bond of CA-4 with a thiazole ring to maintain the cis-conformation, for example, compounds I and II [31,32]. Several research teams have designed heterocycle–chalcone hybrids to improve both the pharmacokinetics and pharmacodynamics of chalcones as antimitotic agents [33,34]. Li and coworkers incorporated a quinoline moiety in chalcone to improve physicochemical properties as compound III [21]. Conversely, Kamal and his team designed imidazothiazole-chalcone IV that showed enhanced binding interactions with the colchicine binding region in the tubulin dimer, compared to CA-4 (Figure 1) [27,32,35].
Figure 1.
Some reported heterocyclic-chalcones and thiazole derivatives targeting colchicine binding site on tubulin.
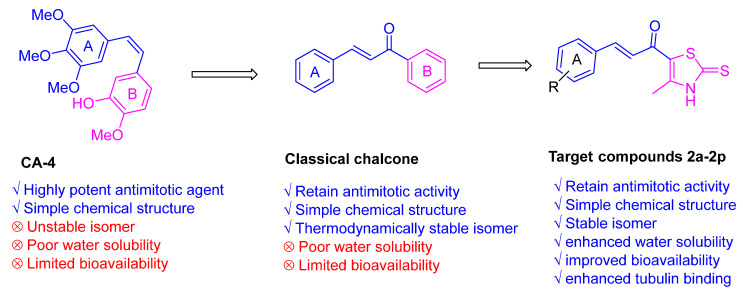
Motivated by these findings and continuing our endeavors to create novel derivatives with anticancer activity, we designed and synthesized a range of thiazole–chalcone derivatives as potential antimitotic agents (Figure 2). Subsequently, we assessed the effectiveness of these derivatives in inhibiting the growth of different types of human cancer cells. To investigate the mechanism by which these derivatives exhibit their anticancer activities, their impact on tubulin polymerization was evaluated.
Figure 2.
Design of novel thiazole-chalcones targeting colchicine binding site.
2. Results and Discussion
2.1. Chemistry
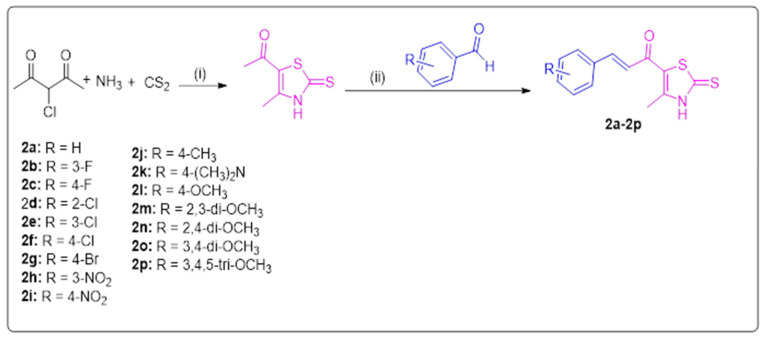
Both thiazole chalcones 2a–2p and their intermediate 1 were prepared as illustrated in Scheme 1. 1-(4-methyl-2-thioxo-2,3-dihydrothiazol-5-yl)ethan-1-one was synthesized following the previously published procedure [36]. The thiazole derivatives 2a–2p were synthesized in ethanol by condensation of intermediate 1 with the appropriate aromatic aldehyde in the presence of sodium hydroxide [2]. The structures of target compounds 2a–2p were elucidated using 1H NMR, 13 C NMR, elemental analysis, and mass spectrometry. The 1H NMR spectra of chalcones derivatives 2a–2p showed a singlet at δ = 2.50–2.60 ppm due to the methyl group of the thiazole ring. Also, two doublets appeared at δ = 7.10–7.80 ppm due to chalcone protons. One singlet for the NH appeared at δ 13.63–13.70 ppm. Moreover, 13 C NMR showed characteristic signals at δ = 188.91–189.44 ppm, 179.87–180.24 ppm, and 14.14–15.02 ppm, which corresponds to C=S, C=O, and the methyl group of the chalcone scaffold, respectively.
Scheme 1.
Synthesis of thiazole-based chalcones 2a–2p. Reagents and conditions: (i) ethanol, 20 °C, 6 h.; (ii) appropriate aromatic aldehyde, 60% NaOH, ethanol, 0 °C, 18 h.
2.2. Biological Investigation
2.2.1. In Vitro Screening of Anticancer Activity of Thiazole Derivatives 2a–2p at 10 μM
According to the protocol of the drug evaluation, all thiazole chalcones 2a–2p were selected by the National Cancer Institute (NCI) for in vitro screening of their anticancer activities at a single dose of 10 μM against nine tumor subpanels, including leukemia, CNS, melanoma, colon, lung, breast, ovarian, renal, and prostate cancer (PC) cell lines (Table 1). From the results in Table 1, it is clear that all tested thiazole derivatives displayed a broad range of antiproliferative and cytotoxic activity against most of the tested cell lines, with mean growth percentages ranging from −21.75 to 77.71. Chalcone derivatives 2c, 2e, 2f, 2g, 2h, 2i, and 2p showed remarkable anticancer activity against most of the tested cell lines with mean growth percentages equal 36.74, 22.13, 23.72, 34.25, −21.75, 25.23, and 14.89, respectively. Among the tested derivatives, compound 2h was the most potent derivative, with broad cytotoxic effect (negative value) against LOX IMVI, RXF 393, UO-31, HCC-2998, SF-539, MDA-MB-468, OVCAR-3, KM12, U251, NCI-H23, SK-MEL-5, NCI-H522, ACHN, UACC-62, T-47D, CAKI-1, SK-MEL-28, MCF7, MALME-3M, NCI-H226, SW-620, OVCAR-5, MDA-MB-435, HCT-15, NCI-H460, SN12C, COLO 205, MDA-MB-231/ATCC, BT-549, SR, 786-0, and HCT-116 cancer cells with growth percentages of −98.99, −91.92, −89.63, −86.72, −85.88, −82.86, −79.48, −78.4, −75.99, −72.77, −68.8, −58.81, −58.35, −56.71, −56.63, −56.46, −47.02, −45.42, −42.94, −41.9, −37.8, −37.4, −36.95, −36.42, −30.68, −25.64, −24.88, −21.75, −17.58, −14.03, −10.99, −10.85, and −6.25, respectively. Also, it displayed remarkable cytostatic action (positive value) against EKVX, HOP-62, M14, SF-295, HL-60(TB), NCI/ADR-RES, PC-3, DU-145, RPMI-8226, UACC-257, IGROV1, SNB-19, OVCAR-4, MOLT-4, OVCAR-8, K-562, CCRF-CEM, SK-MEL-2, A549/ATCC, HT29, HS 578T, SF-268, NCI-H322M, SK-OV-3, and A498 cancer cells with growth inhibition percentages of 0.65, 0.68, 1.86, 4.14, 5.25, 8.19, 8.43, 8.55, 9.39, 9.4, 10.16, 10.38, 13.05, 13.31, 13.44, 13.77, 13.78, 16.88, 19.51, 20.35, 24.47, 27.46, 28.92, 44.61, and 8.87, respectively. Thiazole chalcones 2c, 2e, 2f, 2g, 2i, and 2p exhibited broad and cytotoxic effects against most of the tested cells with growth percentages of −13.63 to 119.09, −26.09 to 98.01, −48.15 to 105.85, −34.08 to 108.90, −86.21 to 107.77, and −64.57 to 96.50, respectively. Thiazole derivatives 2a, 2b, 2d, 2j, 2k, 2l, 2m, 2n, and 2o showed moderate potency against most of the tested cancer cell lines with growth percentages of 14.13 to 128.73, −22.69 to 106.03, 14.10 to 116.44, 11.77 to 130.77, 35.38 to 129.94, 15.11 to 120.21, 5.15 to 108.77, 19.19 to 122.75, −0.42 to 125.58, respectively. The data presented in Table 1 showed that substituting the phenyl ring of thiazole chalcones greatly impacts the potency against various cancer cell lines. The presence of an electron-withdrawing group increases cytotoxic activity. In contrast, an electron-donating group decreases the anticancer activity of thiazole derivatives.
Table 1.
Illustration of the in vitro screening results of the anticancer activity of thiazole derivatives 2a–2p at a dose of 10 μM.
| Cell Line Panel/Cell Line Name | Growth Percentage of Thiazole Chalcones 2a–2p | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2a | 2b | 2c | 2d | 2e | 2f | 2g | 2h | 2i | 2j | 2k | 2l | 2m | 2n | 2o | 2p | ||
| Leukemia | CCRF-CEM | 37.22 | 15.42 | −0.21 | 46.79 | 15.19 | 3.18 | −11.02 | 13.78 | −7.25 | 24.6 | 68.52 | 58.98 | 12.41 | 53.72 | 15.53 | 4.92 |
| HL-60(TB) | 49.59 | 12.04 | 12.98 | 53.03 | 25.32 | −0.98 | 13.16 | 5.25 | −5.39 | 61 | 87.05 | 58.48 | 12.74 | 55.84 | 35.6 | 3.95 | |
| K-562 | 50.96 | 22.81 | 20.43 | 47.41 | 16.76 | 7.94 | 13.64 | 13.77 | 12.28 | 34.16 | 47.03 | 56.62 | 27.51 | 48.87 | 32.43 | 8.21 | |
| MOLT-4 | 74.19 | 43.64 | 58.71 | 84.22 | 19.78 | 10.18 | 30.17 | 13.31 | 14.2 | 68.76 | 62.1 | 81.55 | 36.78 | 72.27 | 73.82 | 9.11 | |
| RPMI-8226 | 14.13 | 15.24 | 2.32 | 28.65 | 10.37 | 13.82 | 1.4 | 9.39 | −7.9 | 18.86 | 41.48 | 19.63 | 9.09 | 35.64 | 18.58 | 5.31 | |
| SR | 32.39 | 6.82 | 18.33 | 14.1 | 3.42 | ND | 8.46 | −10.99 | −4.81 | 45.55 | 57.41 | 43.69 | 5.15 | ND | 32.51 | ND | |
| NSCLC | A549/ATCC | 82.31 | 76.38 | 55.96 | 87.57 | 26.49 | 61.14 | 64.07 | 19.51 | 75.71 | 74.27 | 68.06 | 82.86 | 43.09 | 90 | 64.57 | 26.57 |
| EKVX | 73.11 | 68.61 | 51.04 | 35.62 | 34.72 | 40.96 | 62.9 | 0.65 | 74.66 | 55.52 | 67.01 | 84.75 | 54.52 | 56.24 | 54.16 | 23.4 | |
| HOP-62 | ND | 34.77 | ND | 91.65 | 10.45 | −2.73 | 11.94 | 0.68 | 21.63 | 54.1 | 86.25 | ND | 91.75 | 88.82 | 34.07 | 4.61 | |
| HOP-92 | 89.61 | ND | 45.42 | ND | ND | 34.78 | 67.12 | ND | 57.63 | 98.01 | 118.71 | 100.67 | 61.54 | 103.21 | 83.73 | 16.45 | |
| NCI-H226 | 96.48 | 85.65 | 67.1 | 89.61 | 32.73 | 20.49 | 54.51 | −41.9 | 91.38 | 93.76 | 88.28 | 93.62 | 90.78 | 79.73 | 80.5 | 16.43 | |
| NCI-H23 | 77.52 | 71.09 | 62.66 | 91.77 | 18 | 36.43 | 65.01 | −72.77 | 49.2 | 68.54 | 71.13 | 81.76 | 86.47 | 72.87 | 65.13 | 7.27 | |
| NCI-H322M | 66.51 | 84.46 | 63.98 | 106.22 | 50.15 | ND | 44.13 | 28.92 | 50.72 | 62.15 | 81.14 | 92.4 | 62.76 | ND | 63.43 | ND | |
| NCI-H460 | 56.7 | 43.22 | 39.31 | 82.17 | 12.05 | 82.56 | 39.06 | −30.68 | 11.33 | 53.09 | 85.67 | 85.62 | 40.11 | 89.77 | 40.01 | −1.34 | |
| NCI-H522 | 71.74 | 63.86 | 44.19 | 90.13 | 21.99 | 13.54 | 21.1 | −58.81 | 33 | 60.24 | 58.91 | 61.35 | 32.87 | 73.46 | 56.8 | 16.3 | |
| Colon Cancer | COLO 205 | ND | 94.51 | ND | 117.87 | 70.71 | 78.15 | 89.96 | −24.88 | 91.53 | 121.66 | 115.46 | ND | 102.52 | 122.75 | 115.96 | 81.66 |
| HCC-2998 | 91.33 | 58.22 | 27.06 | 132.42 | 13.88 | −21.28 | 3.23 | −86.72 | −11.87 | 77.15 | 95.3 | 100.59 | 79.76 | 80.73 | 41.71 | 19.01 | |
| HCT-116 | 49.38 | 21.2 | 23.71 | 44.58 | 3.28 | 6.98 | 16.57 | −6.25 | −47.76 | 46.74 | 72.22 | 75.95 | 46.54 | 62.3 | 34.55 | 6.81 | |
| HCT-15 | 35 | 19.06 | 14.71 | 57.4 | −22.01 | −19.78 | 5.48 | −36.42 | −62.1 | 38.58 | 64.44 | 50.75 | 32.68 | 45.78 | 31.33 | −3.26 | |
| HT29 | 99.67 | 47.06 | 73.33 | 122.14 | 29.92 | 31.17 | 76.26 | 20.35 | 62.83 | 98.71 | 94.03 | 96.79 | ND | 114.99 | 97.72 | 40.09 | |
| KM12 | 29.04 | 11.91 | 6.85 | 63.81 | −30.72 | 6.77 | 1.17 | −78.4 | −72.47 | 29.81 | 62.65 | 42.17 | 25.26 | 73.4 | 42.25 | 9.52 | |
| SW-620 | 69.11 | 20.94 | 13.76 | 82.04 | 8.6 | 8.02 | 5.7 | −37.8 | −44.79 | 35.87 | 89.86 | 75.28 | 47.89 | 84.93 | 34.92 | 6.92 | |
| CNS Cancer | SF-268 | 69.81 | 82.33 | 58.62 | 80.38 | 51.84 | 67.94 | 46.1 | 27.46 | 47.36 | 86.89 | 99.28 | 81.05 | 54.04 | 89.73 | 67.22 | 18.5 |
| SF-295 | 88.86 | 38.42 | 60.61 | 82.55 | 41.84 | 42.62 | 90.37 | 4.14 | 86.08 | 83.38 | 79.76 | 83.49 | 26.31 | 72.5 | 66.5 | 42.28 | |
| SF-539 | 41.72 | 6.17 | −4.61 | 93.59 | −10.65 | −30.44 | −25.34 | −85.88 | −25.22 | 42.96 | 88.7 | 69.82 | 22.73 | 87.29 | 29.82 | −13.9 | |
| SNB-19 | 66.75 | 44.03 | 33.66 | 85.22 | ND | 15.84 | 32.68 | 10.38 | 41.18 | 65.54 | 71.2 | 75.59 | 37.21 | 77.37 | 61.91 | 19.56 | |
| SNB-75 | 92.71 | ND | 36.49 | ND | 8.74 | ND | 2.52 | ND | 35.98 | 78.06 | 96 | 98.01 | 50.41 | ND | 58.12 | ND | |
| U251 | 56.91 | 17.63 | 14.09 | 63.31 | 12.07 | 11.35 | 15.53 | −75.99 | 13.65 | 60.37 | 67.11 | 62.29 | 20.33 | 68.6 | 33.72 | 14.98 | |
| Melanoma | LOX IMVI | 31.05 | 13.83 | 8.91 | 57.46 | −26.09 | −48.15 | −5.17 | −98.99 | −86.21 | 31.22 | 60.24 | 44.25 | 21.68 | 57.81 | 26.92 | 17.13 |
| MALME-3M | 87.78 | 101.24 | 85.79 | 104.51 | 86.93 | ND | 72.94 | −42.94 | 75.16 | 84.65 | 79.42 | 90.04 | 97.13 | ND | 89.57 | ND | |
| M14 | 77.06 | 78.45 | 49.58 | 82.29 | 40.25 | 34.39 | 74.48 | 1.86 | 69.52 | 83.02 | 83.18 | 87.51 | 53.99 | 75.84 | 82.23 | 15.43 | |
| MDA-MB-435 | 50.1 | 35.94 | 4.67 | 80.75 | 17.26 | 22.51 | 8.26 | −36.95 | 3.76 | 39.01 | 58.35 | 63.89 | 51.86 | 84.73 | 38.89 | 10.98 | |
| SK-MEL-2 | 68.19 | 81.22 | 56.59 | 84.85 | 32.1 | 58.38 | 62.33 | 16.88 | 49.55 | 89.83 | 88.89 | 80.29 | 94.22 | 64.72 | 68.6 | 34.94 | |
| SK-MEL-28 | 85.25 | 93.67 | 50.57 | 101.08 | 44.69 | 45.71 | 64.25 | −47.02 | 23.87 | 92.85 | 92.66 | 101.91 | 70 | 92.8 | 83.19 | 24.24 | |
| SK-MEL-5 | 68.82 | 62.16 | 42.56 | 68.46 | 16.05 | 27.99 | 48.84 | −68.8 | 53.78 | 56.13 | 51.26 | 55.87 | 23.46 | 37.84 | 63.12 | −64.57 | |
| UACC-257 | 76.15 | 83.72 | 64.09 | 87.25 | 52.71 | 66.8 | 69.1 | 9.4 | 63.26 | 80.08 | 64.13 | 67.97 | 35.39 | 84.44 | 71.4 | 42.08 | |
| UACC-62 | 65.26 | 58.32 | 47.27 | 72.88 | 29.97 | 33.88 | 46.84 | −56.71 | 47.53 | 59.43 | 58.71 | 71.85 | 45.46 | 65.16 | 63.85 | −3.11 | |
| Ovarian Cancer | IGROV1 | 81.82 | 32.3 | 51.24 | 91.91 | 33.98 | ND | 37.11 | 10.16 | 16.06 | 83.42 | 82.27 | 89.07 | 73.86 | ND | 69.47 | ND |
| OVCAR-3 | 42.69 | −1.17 | −10.94 | 77.36 | 8.26 | −24.25 | −12.3 | −79.48 | −6 | 14.99 | 82.2 | 66.86 | 24.27 | 91.16 | 3.22 | −25.23 | |
| OVCAR-4 | 76.39 | 79.55 | 48.43 | 90.32 | 31.68 | 76.31 | 49.51 | 13.05 | 49.16 | 59.22 | 51.23 | 72.33 | 62.2 | 52.05 | 47.63 | 15.6 | |
| OVCAR-5 | 128.73 | 88.15 | 119.09 | 116.44 | 36.37 | 50.81 | 115.99 | −37.4 | 109.56 | 138.42 | 142.09 | 144.69 | 81.68 | 120.39 | 131.45 | 6.56 | |
| OVCAR-8 | 65.76 | 60.46 | −13.63 | 85.43 | 10.39 | 4.96 | −34.08 | 13.44 | 7.06 | 17.26 | 83.37 | 76.93 | 44.61 | 81.51 | 34.3 | 4.32 | |
| NCI/ADR-RES | 64.78 | 10.81 | −10.78 | 88.8 | −5.11 | −8.39 | −13.89 | 8.19 | −4.45 | 31 | 56.92 | 76.38 | 30.95 | 48.05 | 26.85 | 0.58 | |
| SK-OV-3 | ND | 110.63 | ND | 118.47 | 59.91 | 60.83 | 96.66 | 44.61 | 107.77 | 102.36 | 92.59 | ND | 85.62 | 100.52 | 76.93 | 31.19 | |
| Renal Cancer | 786-0 | 68.1 | 46.91 | 37.27 | 93.1 | 13.35 | 16.59 | 25.03 | −10.85 | 25.64 | 70.71 | 91.67 | 78.11 | ND | 85.94 | 60 | 21.09 |
| A498 | 105.99 | 106.3 | 115 | 116 | 98.01 | 105.85 | 108.9 | 98.87 | 104.64 | 130.77 | 129.94 | 120.21 | 108.77 | 100.71 | 125.58 | 96.5 | |
| ACHN | 71.77 | 55.96 | 47.17 | 93.42 | 22.9 | 34.61 | 55.01 | −58.35 | 3.88 | 71.22 | 75.67 | 85.64 | 52.53 | 76.96 | 64.88 | 19.58 | |
| CAKI-1 | 52.53 | 37.69 | 31.6 | 68.49 | 23.85 | 53.21 | 32.76 | −56.46 | 31.07 | 43.41 | 68.64 | 59.04 | 41.93 | 82.85 | 42.04 | 28.19 | |
| RXF 393 | 46.35 | 23.17 | 23.24 | 93.84 | 10.66 | 9.78 | 15.96 | −91.92 | −4.35 | 55.42 | 90.79 | 77.53 | 52.45 | 68.86 | 48.08 | 30.28 | |
| SN12C | 66.07 | 32.09 | 24.05 | 80.04 | 12 | 4.62 | 19.22 | −25.64 | 18.63 | 62.33 | 75.73 | 74.87 | 44.99 | 80.95 | 45.37 | 7.3 | |
| TK-10 | 80.33 | ND | 40.37 | ND | ND | 40.37 | 37.82 | ND | 12.9 | 79.78 | 86.81 | 80.89 | 64.47 | 94.1 | 75.43 | 52.99 | |
| UO-31 | 39.33 | 44.34 | 28.13 | 73.69 | 34.33 | 30.44 | 22.38 | −89.63 | −41.32 | 46.79 | 48.26 | 49 | 53.74 | ND | 49.17 | ND | |
| PC | PC-3 | 77.6 | 54.71 | 43.41 | 71.78 | 31.83 | ND | 48.79 | 8.43 | 51.28 | 74.84 | 71.73 | 82.15 | 41.63 | 75.69 | 68.88 | 35.71 |
| DU-145 | 27.95 | 12.86 | 20.32 | 75.95 | 16.1 | 7.29 | 23.9 | 8.55 | 21.39 | 54.64 | 92.79 | 58.53 | 41.98 | 86.87 | 48.1 | 12.86 | |
| Prostate Cancer | MCF7 | 19.79 | 7.97 | 9.48 | 28.93 | 7.9 | 4.86 | 10.52 | −45.42 | 5.51 | 28.33 | 59.99 | 30.84 | 16.65 | 45.98 | 28.86 | 9.35 |
| MDA-MB-231/ATCC | 70.05 | 67.38 | 29.72 | 101.38 | −0.43 | 39.29 | 9.49 | −17.58 | 10.75 | 71.69 | 90.28 | 89.32 | 68.26 | 98.69 | 58.36 | 2.51 | |
| HS 578T | 90.12 | 79.81 | 95.29 | 92.76 | 47.21 | 47.98 | 74.6 | 24.47 | 68.11 | 101.32 | 118 | 96.5 | 49.25 | 89.49 | 84.71 | 3.94 | |
| BT-549 | 68.85 | 65.65 | 23.21 | 76.82 | 2.28 | −14.41 | 19.5 | −14.03 | 20.09 | 56.13 | 82.22 | 75.04 | 70.95 | 59.75 | 18.94 | −11.78 | |
| T-47D | ND | 0.69 | ND | 47.34 | 4.54 | 1.38 | 29.03 | −56.63 | 44.32 | 38.31 | 32.35 | ND | 34.48 | 30.94 | 44.15 | −1.59 | |
| MDA-MB-468 | 20.79 | −22.69 | −4.51 | 30.83 | −7.12 | −21.19 | −4.75 | −82.86 | −18.95 | 11.77 | 35.38 | 15.11 | 41.07 | 19.19 | −0.42 | 3.79 | |
| Mean growth percentage | 64.97 | 47.5 | 36.74 | 79.42 | 22.13 | 23.72 | 34.25 | −21.75 | 25.23 | 63.31 | 77.71 | 74.58 | 49.88 | 75.09 | 55.11 | 14.89 | |
ND = not determined.
2.2.2. Screening of the Anticancer Activity at Five Doses
The findings from the five-dose experiments (100 μM, 10 μM, 1 μM, 0.1 μM, and 0.01 μM) conducted on compounds 2c, 2e, 2f, 2g, 2h, 2i, and 2p indicate that these compounds had significant and wide-ranging antitumor activity toward the cancer cell line panels that were evaluated (Table 2). Moreover, the IC50 of compounds 2e, 2f, 2h, and 2p were evaluated. Compound 2c (R = 4-F) had significant efficacy against all cancer cell lines tested, with GI50 Value (growth inhibition 50%) ranging from 1.93 μM (against MDA-MB-468) to 16.7 μM (against MOLT-4). Compound 2e (R = 3-Cl) exhibited significant efficacy against all selected cell lines, especially against HCT-116, LOX IMVI, and cell lines (IC50 2.95, 2.88, 2.88 μM, respectively). Compared to 2e, compound 2f (R = 4-Cl) showed its highest inhibitory activity against different cell lines namely, CCRF-CEM, RPMI-8226, OVCAR-3, and MDA-MB-468 (IC50 2.88, 2.40, 2.82, 2.51 μM, respectively). Compound 2g (R = 4-Br) had significant efficacy against all cancer cell lines tested, with GI50 ranging from 1.79 μM (against MDA-MB-468) to 15.40 μM (against OVCAR-5). On the other hand, compound 2h (R = 3-NO2) exhibited broad activity against the tested cell lines with remarkable inhibition against both U251 and LOX IMVI cell lines (IC50 3.09 and 2.75 μM, respectively). Compound 2i (R = 4-NO2) showed significant inhibition against all tested cancer cell lines, with GI50 ranging from 1.85 μM (against LOX IMVI) to 18.6 μM (against SK-OV-3). Finally, compound 2p (R = 3,4,5-tri-OCH3) showed anticancer activity against CCRF-CEM, NCI-H460, SK-MEL-5, and OVCAR-8 (IC50 3.55, 3.31, 3.31, and 3.55 μM, respectively).
Table 2.
Five dose results of compounds 2c, 2e, 2f, 2g, 2h, 2i, 2p, and CA-4 in μM.
| Cell Line Panel/Cell Line NAME | 2c | 2e | 2f | 2g | 2h | 2i | 2p | CA-4 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI50 | LC50 | TGI | GI50 | IC50 | LC50 | TGI | GI50 | IC50 | LC50 | TGI | GI50 | LC50 | TGI | GI50 | IC50 | LC50 | TGI | GI50 | LC50 | TGI | GI50 | IC50 | LC50 | TGI | GI50 | LC50 | TGI | ||
| Leukemia | CCRF-CEM | 3.68 | >100 | >100 | 2.45 | 3.72 | >100 | 7.94 | 2.88 | 3.98 | >100 | 11.22 | 3.06 | >100 | 61.5 | 2.14 | 3.39 | >100 | 6.61 | 4.56 | >100 | >100 | 2.45 | 3.55 | >100 | 8.71 | 0.10 | 89.95 | 5.43 |
| HL-60(TB) | 2.83 | >100 | 32.2 | 2.24 | 4.27 | >100 | 7.08 | 3.55 | 5.89 | >100 | 38.02 | 1.94 | >100 | 9.92 | 1.95 | 3.31 | >100 | 5.75 | 3.05 | >100 | 16.9 | 2.57 | 3.89 | >100 | 11.22 | 0.03 | 59.02 | 0.06 | |
| K-562 | 3.25 | >100 | 48.8 | 3.39 | 4.27 | >100 | >100 | 3.89 | 4.68 | >100 | >100 | 2.73 | >100 | >100 | 3.09 | 3.72 | >100 | >100 | 3.5 | >100 | >100 | 3.02 | 3.72 | >100 | >100 | 0.03 | >100 | 2.09 | |
| MOLT-4 | 16.7 | >100 | 64.3 | 2.82 | 4.27 | >100 | >100 | 14.45 | 24.55 | >100 | 79.43 | 3.57 | >100 | 36.6 | 3.47 | 5.01 | >100 | 30.2 | 3.79 | >100 | >100 | 2.88 | 4.17 | >100 | 22.91 | 0.10 | 78.34 | 3.72 | |
| RPMI-8226 | 2.85 | >100 | >100 | 2.04 | 3.72 | >100 | 5.5 | 2.4 | 4.68 | >100 | 8.32 | 2.29 | >100 | >100 | 2.04 | 3.72 | >100 | 6.31 | 3.55 | >100 | >100 | 2.19 | 4.47 | >100 | 9.12 | 0.15 | 96.61 | 6.50 | |
| SR | 3.19 | >100 | >100 | 2.63 | 3.89 | >100 | >100 | 3.55 | 5.37 | >100 | 56.23 | 2.83 | >100 | 23.2 | 2.75 | 3.89 | >100 | >100 | 2.86 | >100 | >100 | 2.34 | 3.8 | >100 | 16.22 | 0.11 | >100 | 75.34 | |
| NSCLC | A549/ATCC | 12.4 | 57.7 | 26.8 | 4.79 | 7.41 | >100 | 47.86 | 10.72 | 16.22 | 52.48 | 23.99 | 11.7 | 55.3 | 25.4 | 3.24 | 4.47 | 54.95 | 12.02 | 17 | 69 | 34.2 | 3.55 | 5.01 | 44.67 | 13.8 | 0.07 | 96.38 | 76.56 |
| EKVX | 12.8 | 64.6 | 28.7 | 10.47 | 23.99 | 95.5 | 31.62 | 12.88 | 23.99 | 52.48 | 26.3 | 10.6 | 51 | 23.3 | 5.75 | 15.14 | 46.77 | 19.5 | 16.4 | 58.1 | 30.9 | 4.07 | 10.96 | >100 | 22.91 | 0.36 | >100 | 82.04 | |
| HOP-62 | 3.83 | 48.7 | 16.2 | 2.57 | 4.68 | 74.13 | 6.31 | 5.37 | 12.02 | 72.44 | 22.91 | 3.94 | 61.2 | 14 | 3.98 | 6.76 | 40.74 | 12.3 | 6.43 | 67.7 | 23.2 | 3.16 | 5.37 | >100 | 16.22 | 0.18 | 89.13 | 2.53 | |
| HOP-92 | 3.35 | 56 | 15.8 | 3.55 | 79.43 | >100 | 30.9 | 11.48 | 30.2 | 63.1 | 26.92 | 6.44 | 58.8 | 22.5 | 3.89 | 22.39 | 44.67 | 16.6 | 12.2 | 64.4 | 28 | 2.95 | 44.67 | >100 | 19.95 | 0.22 | >100 | 36.31 | |
| NCI-H226 | 5.27 | >100 | 27.6 | 4.37 | 29.51 | >100 | 38.9 | 13.8 | 36.31 | >100 | 38.02 | 4.13 | >100 | 25.4 | 11.22 | 33.11 | >100 | 38.02 | 15.6 | >100 | 42.1 | 3.63 | 32.36 | >100 | 36.31 | 0.67 | 96.16 | 48.98 | |
| NCI-H23 | 10.6 | 49.9 | 23 | 2.09 | 3.63 | 12.88 | 4.68 | 14.79 | 26.92 | 58.88 | 29.51 | 8.23 | 48 | 21.3 | 5.37 | 10 | 45.71 | 18.2 | 12.2 | 52.6 | 25.3 | 2.95 | 5.25 | 44.67 | 10.47 | 0.02 | 96.16 | 0.40 | |
| NCI-H322M | 4.1 | 44.8 | 17.9 | 6.03 | 16.22 | 85.11 | 26.3 | 10.23 | 19.95 | 51.29 | 22.91 | 3.76 | 41.3 | 15.2 | 8.71 | 17.38 | 50.12 | 21.88 | 11 | 54.6 | 24.5 | 4.57 | 14.13 | 51.29 | 18.62 | 0.07 | >100 | 74.82 | |
| NCI-H460 | 15.2 | 83.3 | 35.6 | 2.75 | 3.55 | 45.71 | 7.59 | 12.02 | 19.05 | 69.18 | 28.84 | 14.3 | 80.9 | 34.1 | 3.63 | 4.27 | 54.95 | 13.8 | 15.9 | 72.5 | 34 | 2.14 | 3.31 | 44.67 | 5.62 | 0.03 | >100 | 66.83 | |
| NCI-H522 | 8.63 | 46.8 | 21.2 | 4.57 | 11.48 | 45.71 | 17.38 | 11.75 | 22.39 | 52.48 | 25.12 | 3.37 | 41.2 | 14.3 | 5.01 | 11.75 | 46.77 | 17.78 | 9.37 | 51.5 | 22.5 | 3.16 | 7.59 | 40.74 | 12.3 | 0.03 | 88.51 | 3.46 | |
| Colon Cancer | COLO 205 | 15.4 | 80.4 | 35.2 | 3.55 | 5.25 | 51.29 | 14.45 | 19.05 | 33.88 | 74.13 | 38.02 | 13.9 | 80 | 33.3 | 5.75 | 8.71 | 51.29 | 19.05 | 15.9 | >100 | 41.1 | 16.22 | 28.84 | 70.79 | 33.88 | 2.49 | >100 | 43.25 |
| HCC-2998 | 3.46 | 36.5 | 13.2 | 2.57 | 4.37 | 28.18 | 7.76 | ND | ND | ND | ND | 2.72 | 31.6 | 9.62 | 2 | 3.63 | 11.75 | 4.57 | 9.37 | 47 | 21.5 | 5.13 | 11.75 | 44.67 | 17.38 | 0.05 | 26.85 | 1.22 | |
| HCT-116 | 3.99 | 42.5 | 15.2 | 1.82 | 2.95 | 7.41 | 3.63 | 6.61 | 7.94 | 74.13 | 24.55 | 3.82 | 40.9 | 13.1 | 2.88 | 3.31 | >100 | >100 | 2.78 | 44.3 | 7.89 | 3.16 | 3.63 | 69.18 | 14.45 | 0.03 | >100 | 0.22 | |
| HCT-15 | 3.27 | 43.7 | 14.2 | 2.09 | 3.55 | 46.77 | 5.89 | 4.07 | 5.37 | 48.98 | 16.6 | 2.66 | 36.4 | 10 | 2.14 | 3.47 | 31.62 | 7.76 | 2.46 | 33.7 | 7.86 | 3.24 | 4.17 | 74.13 | 15.49 | 0.03 | >100 | 9.51 | |
| HT29 | 10.6 | 52.4 | 23.6 | 2.24 | 3.55 | 12.59 | 4.9 | 6.76 | 9.55 | 53.7 | 20.89 | 5.09 | 57.8 | 19.3 | 5.5 | 7.24 | 60.26 | 19.95 | 10.7 | 68.7 | 27.1 | 3.47 | 4.57 | >100 | 13.49 | 0.66 | 38.73 | 5.51 | |
| KM12 | 4.28 | 41 | 15.4 | 1.95 | 3.39 | 7.76 | 3.89 | 4.17 | 8.13 | 45.71 | 16.6 | 3.27 | 35.9 | 11 | 3.63 | 5.37 | 42.66 | 13.18 | 3.8 | 40 | 13.6 | 3.24 | 6.46 | 42.66 | 13.8 | 0.05 | 91.41 | 0.76 | |
| SW-620 | 3.37 | 49.3 | 15 | 1.91 | 3.16 | 7.59 | 3.8 | 3.09 | 4.37 | 38.9 | 11.22 | 2.82 | 41.6 | 11.7 | 3.39 | 4.07 | 45.71 | 10.96 | 3.59 | 44.2 | 13.9 | 2.57 | 3.72 | 46.77 | 9.77 | 0.03 | >100 | 86.30 | |
| CNS Cancer | SF-268 | 13 | 63.1 | 28.7 | 3.72 | 7.59 | 97.72 | 16.98 | 12.02 | 24.55 | 64.57 | 28.18 | 13.5 | 64.2 | 29.5 | 10.47 | 19.05 | 57.54 | 24.55 | 15.6 | 67.3 | 32.4 | 2.63 | 6.46 | 54.95 | 11.22 | 0.13 | 91.62 | 49.66 |
| SF-295 | 12.5 | 52.9 | 25.7 | 4.37 | 11.48 | 43.65 | 16.98 | 15.85 | 28.84 | 54.95 | 29.51 | 12.7 | 52.7 | 25.8 | 3.63 | 8.71 | 39.81 | 14.79 | 16.1 | 56.5 | 30.2 | 5.5 | 14.45 | 66.07 | 21.38 | 0.07 | >100 | 1.28 | |
| SF-539 | 2.94 | 35.5 | 10.8 | 2 | 3.63 | 8.32 | 4.07 | 6.92 | 16.22 | 47.86 | 20.42 | 2 | 12.8 | 4.67 | 2.19 | 3.98 | 12.88 | 4.9 | 2.31 | 27.2 | 7.02 | 2.19 | 4.47 | 20.89 | 5.5 | 0.04 | 86.90 | 0.20 | |
| SNB-19 | 4.87 | 43.1 | 17.8 | 7.76 | 16.98 | 48.98 | 21.38 | 12.02 | 22.39 | 50.12 | 24.55 | 3.47 | 37.7 | 13.4 | 4.9 | 11.75 | 41.69 | 16.98 | 5.42 | 43.2 | 18.3 | 5.01 | 13.18 | 43.65 | 17.78 | 0.04 | >100 | 15.85 | |
| SNB-75 | ND | ND | ND | 1.66 | 3.8 | 7.76 | 3.63 | 2.19 | 8.51 | 28.84 | 6.76 | ND | ND | ND | 2.29 | 8.51 | 27.54 | 7.41 | ND | ND | ND | 2.19 | 7.59 | 28.84 | 6.17 | 0.83 | 82.41 | 24.72 | |
| U251 | 3.63 | 41.9 | 15.1 | 1.91 | 3.31 | 7.24 | 3.72 | 3.39 | 4.79 | 40.74 | 13.18 | 2.87 | 39.7 | 11.7 | 1.74 | 3.09 | 5.89 | 3.24 | 4.1 | 53 | 17 | 3.02 | 4.17 | 43.65 | 12.02 | 0.04 | 98.86 | 13.21 | |
| Melanoma | LOX IMVI | 3.26 | 36.6 | 12.7 | 1.66 | 2.88 | 5.75 | 3.09 | 3.39 | 4.47 | 38.02 | 12.02 | 2.8 | 33.7 | 10.2 | 1.58 | 2.75 | 6.03 | 3.09 | 1.85 | 8.91 | 4.06 | 3.24 | 4.47 | 38.9 | 12.02 | 0.01 | >100 | 12.74 |
| MALME-3M | 12.4 | 60.9 | 27.5 | 2.24 | 5.13 | 50.12 | 5.75 | 14.45 | 30.9 | 75.86 | 33.11 | 12.5 | 67.2 | 29 | 12.3 | 24.55 | 54.95 | 26.3 | 13.7 | 70.8 | 31.1 | 5.75 | 17.78 | 54.95 | 20.42 | 0.44 | >100 | 70.31 | |
| M14 | 6.97 | 56.7 | 22.2 | 2.88 | 4.17 | >100 | 9.55 | 14.13 | 24.55 | 72.44 | 32.36 | 9.02 | 54.4 | 22.9 | 7.08 | 11.75 | 83.18 | 25.7 | 12.2 | 66.3 | 28.4 | 3.24 | 5.13 | 72.44 | 13.18 | 0.14 | 88.31 | 0.24 | |
| MDA-MB-435 | 4.24 | 48.7 | 17.5 | 2.09 | 3.55 | 16.98 | 5.01 | 5.5 | 10.96 | 48.98 | 19.05 | 4.12 | 45.1 | 16.7 | 4.57 | 7.41 | 44.67 | 16.98 | 7.39 | 50.9 | 21.1 | 2.57 | 4.47 | >100 | 10.47 | 0.03 | 95.06 | 0.08 | |
| SK-MEL-2 | 11.6 | 50.1 | 24.1 | 10.47 | 29.51 | 70.79 | 27.54 | 13.8 | 27.54 | 56.23 | 28.18 | 5.87 | 47.9 | 19.5 | 12.59 | 26.92 | 53.7 | 26.3 | 13.6 | 62 | 29 | 3.09 | 10.96 | 42.66 | 11.48 | 8.20 | >100 | 60.53 | |
| SK-MEL-28 | 11.8 | 51.4 | 24.7 | 2.14 | 4.07 | 13.49 | 4.9 | 14.45 | 26.3 | 53.7 | 28.18 | 11.9 | 50.2 | 24.5 | 5.5 | 13.18 | 44.67 | 18.2 | 13.7 | 52.5 | 26.8 | 4.07 | 9.12 | 57.54 | 17.38 | 3.16 | 90.78 | 82.41 | |
| SK-MEL-5 | 6.77 | 44.2 | 19.5 | 4.79 | 9.12 | 41.69 | 16.98 | 11.22 | 20.42 | 48.98 | 23.44 | 5.82 | 43 | 18.4 | 13.18 | 22.91 | 51.29 | 26.3 | 12 | 49.7 | 24.4 | 1.58 | 3.31 | 10 | 3.98 | 0.01 | 94.19 | 53.70 | |
| UACC-257 | 12.6 | 56.6 | 26.7 | 3.63 | 9.77 | 63.1 | 14.79 | 13.8 | 26.92 | 56.23 | 27.54 | 11.3 | 51.9 | 24.2 | 3.39 | 10 | 38.02 | 13.49 | 15.1 | 61.4 | 30.4 | 5.13 | 18.62 | 75.86 | 21.88 | 4.95 | 1.97 | 0.01 | |
| UACC-62 | 5.3 | 45.7 | 19.3 | 2.34 | 4.17 | 34.67 | 8.71 | 9.77 | 16.98 | 48.98 | 22.39 | 5.17 | 44.2 | 18.6 | 3.55 | 7.94 | 41.69 | 15.85 | 10.5 | 48.1 | 22.4 | 2.82 | 5.37 | 42.66 | 12.3 | 0.01 | >100 | 80.54 | |
| Ovarian Cancer | IGROV1 | 4.93 | 62.3 | 19.7 | 2.45 | 4.17 | >100 | 6.46 | 13.18 | 25.7 | 93.33 | 34.67 | 3.29 | 53.4 | 11.9 | 3.02 | 4.9 | 47.86 | 12.3 | 4.43 | 50.8 | 14.9 | 3.31 | 5.75 | >100 | 13.8 | 0.06 | >100 | 63.53 |
| OVCAR-3 | 2.42 | 24.5 | 6.81 | 2.29 | 4.17 | 22.91 | 6.03 | 2.82 | 6.46 | 33.11 | 10.72 | 1.88 | 9.7 | 4.27 | 2.4 | 4.37 | 23.99 | 6.46 | 2.22 | 16.6 | 5.28 | 2.45 | 5.25 | 25.7 | 6.92 | 0.04 | 93.54 | 61.66 | |
| OVCAR-4 | 6.03 | 44.7 | 19.6 | 4.47 | 12.88 | >100 | 41.69 | 13.8 | 25.12 | 54.95 | 27.54 | 10.7 | 48.5 | 22.8 | 10.96 | 20.42 | 51.29 | 23.99 | 14.9 | 53.8 | 28.3 | 2.82 | 6.61 | 51.29 | 12.59 | 0.26 | 20.23 | 0.16 | |
| OVCAR-5 | 15.6 | 57.7 | 30 | 5.25 | 15.49 | 57.54 | 19.95 | 17.78 | 32.36 | 60.26 | 33.11 | 15.4 | 57.3 | 29.7 | 3.63 | 9.12 | 38.9 | 13.8 | 17.8 | 58.2 | 32.2 | 3.8 | 12.3 | 43.65 | 15.14 | 0.18 | >100 | 83.18 | |
| OVCAR-8 | 3.99 | 50.9 | 16.2 | 2.69 | 3.89 | >100 | 7.59 | 8.51 | 14.45 | 54.95 | 22.91 | 2.86 | 37.1 | 8.77 | 3.31 | 4.57 | 38.02 | 11.75 | 7.73 | 65.2 | 23.9 | 2.14 | 3.55 | 45.71 | 5.5 | 0.03 | 90.78 | 78.52 | |
| NCI/ADR-RES | 5.92 | 85 | 24.9 | 2.88 | 5.01 | >100 | >100 | 3.72 | 7.08 | 77.62 | 17.38 | 6.04 | 73.4 | 23.4 | 2.51 | 4.27 | >100 | 5.75 | 14.9 | 82.8 | 35.2 | 2.4 | 4.37 | >100 | 7.41 | 0.06 | 90.36 | 39.17 | |
| SK-OV-3 | 11 | 61.8 | 26.1 | 17.38 | 51.29 | >100 | >100 | 15.85 | 30.2 | 66.07 | 32.36 | 13.8 | 74.9 | 32.2 | 5.89 | 15.14 | 45.71 | 18.62 | 18.6 | 76.9 | 37.8 | 7.41 | 46.77 | >100 | >100 | 28.44 | 89.54 | 0.36 | |
| Renal Cancer | 786-0 | 4.59 | 47.1 | 17.6 | 2.51 | 4.07 | 37.15 | 6.31 | 4.68 | 7.76 | 58.88 | 20.42 | 3.33 | 41.8 | 13.3 | 3.98 | 6.31 | 50.12 | 15.85 | 3.99 | 54 | 17.6 | 4.57 | 6.61 | 97.72 | 20.89 | 0.02 | >100 | >100 |
| A498 | 13.6 | 52.6 | 26.8 | 19.5 | 47.86 | 91.2 | 42.66 | 15.85 | 30.9 | 56.23 | 29.51 | 13.5 | 51.8 | 26.5 | 14.79 | 29.51 | 54.95 | 28.18 | 15.5 | 55.3 | 29.3 | 11.48 | 43.65 | >100 | 34.67 | 0.05 | >100 | 25.47 | |
| ACHN | 7.02 | 44.3 | 19.6 | 2.29 | 3.72 | 21.38 | 6.03 | 13.8 | 23.99 | 51.29 | 26.92 | 7.67 | 45.4 | 20.3 | 4.07 | 6.17 | 38.9 | 14.79 | 11.3 | 48.4 | 23.3 | 3.89 | 6.46 | 54.95 | 16.6 | 0.01 | >100 | 0.72 | |
| CAKI-1 | 6.89 | 45.4 | 20.2 | 4.68 | 15.49 | 69.18 | 20.89 | 4.47 | 8.71 | 43.65 | 17.38 | 10 | 47.2 | 21.8 | 10.47 | 20.42 | 51.29 | 22.91 | 13.9 | 52.7 | 27.1 | 2.57 | 4.68 | 45.71 | 13.18 | 0.04 | 97.05 | 8.22 | |
| RXF 393 | 3.02 | 37.2 | 13 | 1.91 | 5.13 | 10 | 4.37 | 5.89 | 23.99 | 48.98 | 19.95 | 2.58 | 33 | 9.43 | 10.23 | 24.55 | 47.86 | 22.39 | 4.88 | 44.8 | 18.6 | 2.51 | 11.22 | 30.9 | 7.08 | 0.89 | >100 | 13.87 | |
| SN12C | 3.95 | 46.7 | 17 | 2 | 3.55 | 8.91 | 4.27 | 4.9 | 10.72 | 43.65 | 18.2 | 3.15 | 36.8 | 13.1 | 2.69 | 4.37 | 33.11 | 10.23 | 6.06 | 44.7 | 19.3 | 2.29 | 3.89 | 46.77 | 9.55 | 0.01 | 91.62 | 82.04 | |
| TK-10 | 12.5 | 50.7 | 25.2 | 19.5 | 45.71 | >100 | 47.86 | 15.14 | 28.18 | 54.95 | 28.84 | 3.04 | 35.2 | 11.8 | 11.22 | 22.91 | 50.12 | 23.44 | 12.4 | 51.3 | 25.3 | 5.01 | 16.22 | 47.86 | 17.78 | 0.57 | >100 | 11.51 | |
| UO-31 | 2.88 | 41.2 | 15.7 | 1.55 | 2.88 | 6.03 | 3.09 | 4.07 | 11.22 | 47.86 | 18.2 | 2.46 | 39.7 | 13.5 | 3.89 | 10.72 | 43.65 | 16.98 | 4.86 | 46.3 | 18.8 | 3.02 | 7.76 | 46.77 | 15.49 | 0.19 | >100 | 4.16 | |
| PC | PC-3 | 5.82 | >100 | 29.7 | 2.88 | 4.9 | >100 | 13.8 | 5.37 | 9.77 | 81.28 | 23.44 | 4.68 | >100 | 26 | 4.07 | 8.13 | 56.23 | 17.78 | 10.3 | >100 | 37.3 | 5.01 | 8.91 | >100 | 41.69 | 0.03 | >100 | 51.40 |
| DU-145 | 4.32 | 39.9 | 15.7 | 3.24 | 4.79 | 42.66 | 11.48 | 4.9 | 8.91 | 42.66 | 17.38 | 3.52 | 35.3 | 12 | 2.4 | 3.89 | 23.44 | 6.17 | 3.41 | 34.5 | 11 | 3.24 | 5.75 | 69.18 | 15.49 | 0.01 | >100 | 78.89 | |
| Breast Cancer | MCF7 | 2.65 | 37 | 11.4 | 3.31 | 8.13 | 37.15 | 12.3 | 3.16 | 4.68 | 43.65 | 13.18 | 2.57 | 38.7 | 10.6 | 2.75 | 6.31 | 38.02 | 9.77 | 2.67 | 51.3 | 11.7 | 3.02 | 4.27 | 45.71 | 10.72 | 0.04 | >100 | >100 |
| MDA-MB-231/ATCC | 15.7 | 66.1 | 32.2 | 2.45 | 6.17 | 39.81 | 6.92 | 16.22 | 30.9 | 61.66 | 31.62 | 10.9 | 50.9 | 23.5 | 5.25 | 16.22 | 44.67 | 17.78 | 12 | 51.5 | 24.8 | 3.24 | 10 | >100 | 13.18 | 0.01 | 91.62 | 73.62 | |
| HS 578T | 13.2 | >100 | 40.2 | 3.09 | >100 | >100 | 85.11 | 21.38 | >100 | >100 | 77.62 | 10.4 | >100 | 33 | 17.38 | 67.61 | >100 | 52.48 | 13 | 80.9 | 32.4 | 2.88 | >100 | >100 | 39.81 | 0.15 | >100 | 0.22 | |
| BT-549 | 2.34 | 33.1 | 8.27 | 2.24 | 5.89 | 28.18 | 6.17 | 5.75 | 20.89 | 57.54 | 20.42 | 2.6 | 30.6 | 7.28 | 3.8 | 13.49 | 43.65 | 14.79 | 5.04 | 48.5 | 19.3 | 2.95 | 11.75 | 50.12 | 10.72 | 0.01 | 99.54 | 8.09 | |
| T-47D | 3.94 | >100 | 28.8 | 3.39 | 7.94 | >100 | 20.89 | 3.47 | 14.13 | 81.28 | 19.05 | 2.77 | 61.7 | 11.8 | 2 | 4.9 | 66.07 | 7.08 | 4.96 | >100 | 25.7 | 2.19 | 7.24 | >100 | 13.18 | 47.97 | >100 | 2.04 | |
| MDA-MB-468 | 1.93 | 25.3 | 6.46 | 3.8 | 12.59 | 46.77 | 15.85 | 2.51 | 12.02 | 30.2 | 8.32 | 1.79 | 27.2 | 6.77 | 2.95 | 7.76 | 40.74 | 10.72 | 2.2 | 31.4 | 8 | 1.55 | 5.75 | 17.78 | 4.47 | ND | ND | ND | |
ND: not determined.
From the data in Table 2, it can be concluded that an electron-withdrawing group on the phenyl ring is crucial for the anticancer activity. The meta position is optimal for anticancer activity (thiazole chalcones 2h (R = 3-NO2) and 2e (R = 3-Cl)). Shifting the substituent from meta to para slightly reduces the antitumor activity (thiazole derivatives 2f (R = 4-Cl) and 2i (R = 4-NO2)). On the other hand, the introduction of an electron donating group markedly decreases the anticancer activity (thiazole derivatives 2j (R = 4-CH3), 2k (R = 4-(CH3)2N), and 2l (R = 4-OCH3)). Thiazole chalcone with trimethoxy phenyl moiety displayed remarkable anticancer activity.
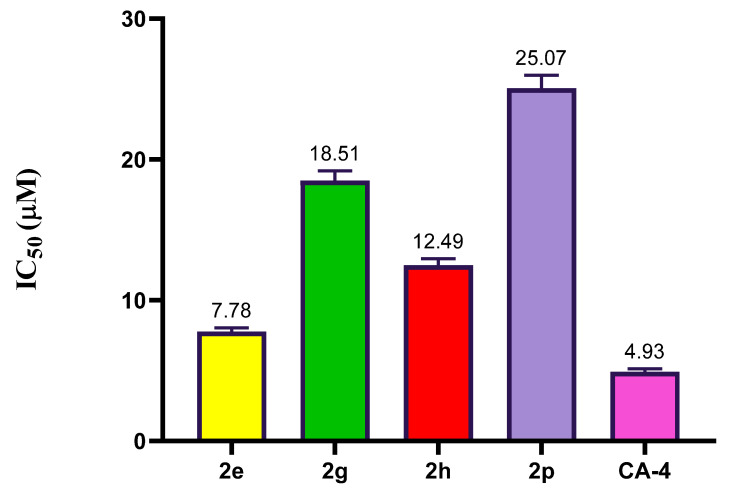
2.2.3. Effect of Compound 2e, 2g, 2h, and 2p on Tubulin Polymerization
To investigate how thiazole derivatives exert their cytotoxic effects, particularly the most potent ones, the effects of compounds 2e, 2g, 2h, and 2p on tubulin polymerization were examined compared to the positive control CA-4. Both CA-4 and 2e demonstrated significant inhibition of tubulin polymerization, with IC50 values of 7.78 μM and 4.93 μM, respectively. Conversely, compounds 2g, 2h, and 2p displayed moderate inhibition of tubulin polymerization compared to CA-4, with IC50 values of 18.51 μM, 12.49 μM, and 25.07 μM, respectively, as shown in Figure 3. These findings suggest that thiazole derivative 2e may disrupt tubulin polymerization.
Figure 3.
The inhibitory activity of thiazole derivatives 2e, 2g, 2h, 2j, 2q, and CA-4 on tubulin polymerization.
2.3. In Silico Studies
2.3.1. Molecular Docking Studies
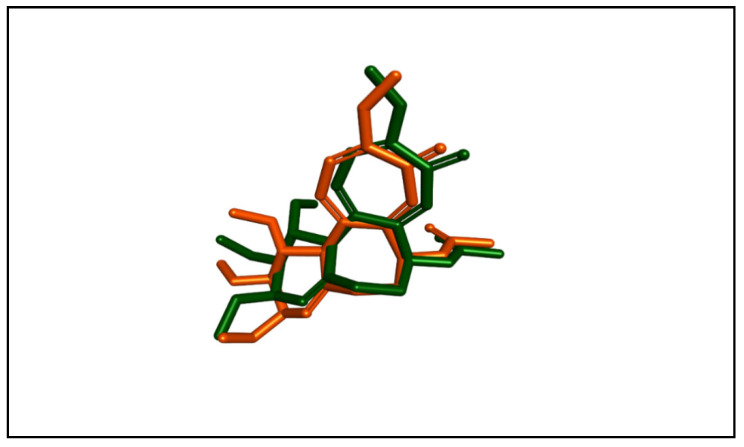
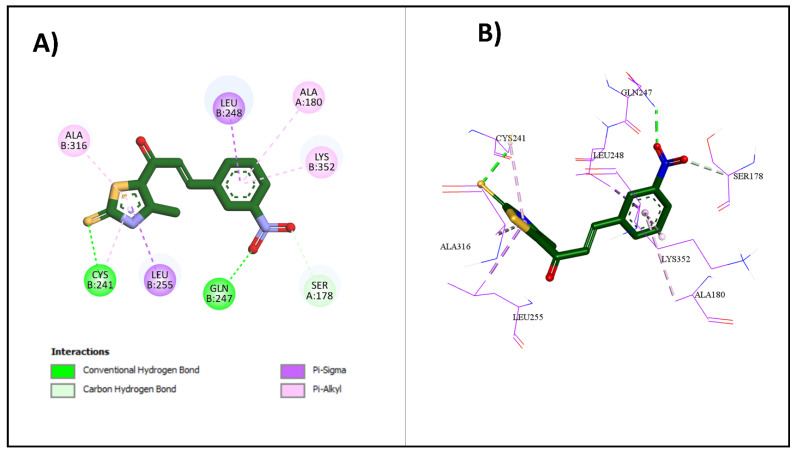
Molecular docking studies were performed to assess the binding abilities and modes of the most potent compounds at the colchicine binding site of tubulin. These studies sought to compare the interactions of these compounds with the established CA-4 and to elucidate their binding mechanisms. Autodock vina was used for the docking simulations. A crystal structure of the tubulin–colchicine complex was used (PDB ID: 4O2B) [37]. To validate the molecular docking method, we performed a redocking procedure of colchicine into its binding site. Results indicated that the redocked colchicine exhibited an affinity of −8.7 kcal/mol. The redocked ligand had a root mean square deviation (RMSD) value of 1.0090 Å compared to the co-crystallized pose. It established most of the binding interactions exhibited by the native ligand. These results confirm the accuracy and reliability of our docking protocol for evaluating the binding interactions of the newly synthesized compounds. The superimposition of both the redocked and co-crystallized poses of colchicine is depicted in Figure 4.
Figure 4.
The superimposition of the redocked (orange) and co-crystallized (green) poses of colchicine (RMSD = 1.0090 Å).
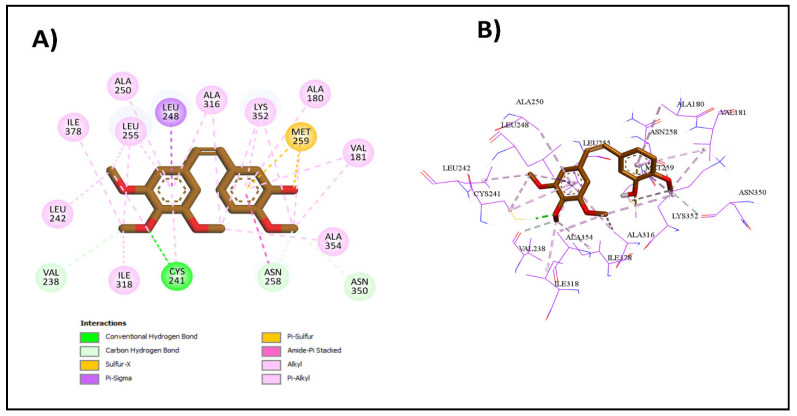
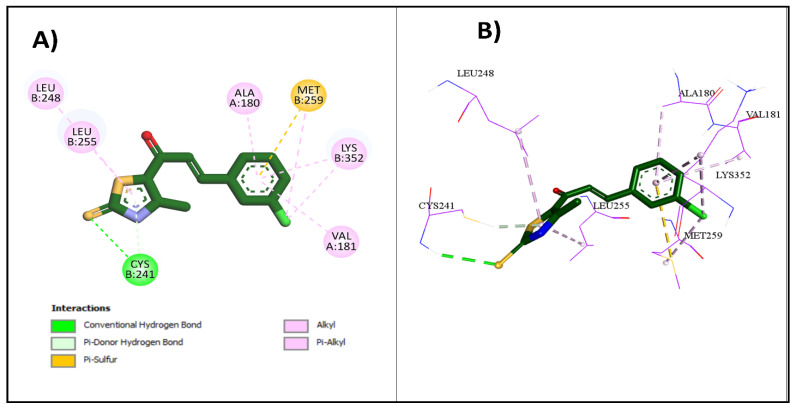
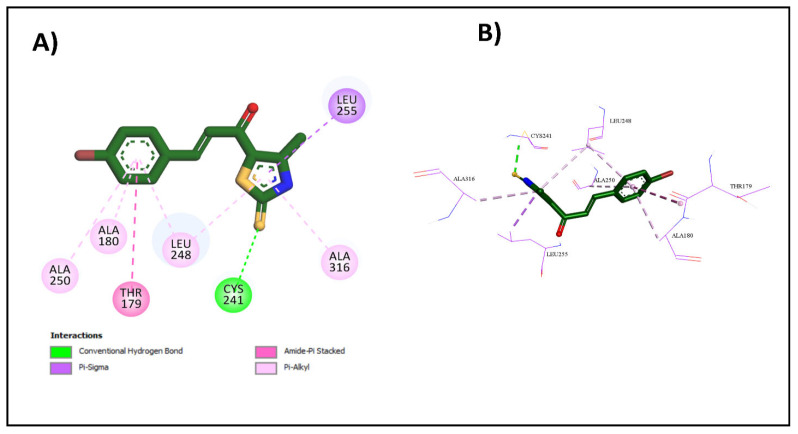
Upon docking of the most potent compounds into the colchicine binding site, the binding affinities of the docked compounds 2e, 2g, and 2h ranged from −7.3 to −8.9 kcal/mol, which was nearly comparable to the affinity of CA-4 at −9.2 kcal/mol (Table 3). Additionally, by examining the best docking poses for these compounds, they seemed to share a similar ligand–receptor interaction profile where they were able to establish key hydrogen bonding with Cys241 through its thiocarbonyl moiety, highlighted, which is pivotal in facilitating the tight binding of CA-4 and colchicine with β-tubulin as highlighted by Gracheva et al. (Figure 5) [38]. Additional non-classical hydrogen bonding between the thiazole ring and Cys241 in compounds 2e and 2h could explain their higher potency than compound 2g. The thiazole ring also formed several hydrophobic interactions with amino acids Leu255, Ala316, and Leu248. The incorporation of the chalcone moiety also contributed to further anchoring these compounds into the pocket by forming more hydrophobic interactions through the phenyl ring with several amino acids, such as Ala180 and Lys352. Both the 2D and 3D diagrams of the best docking poses and their interactions are illustrated in Figure 6, Figure 7 and Figure 8. The docking results presented here align well with the in vitro findings, further substantiating the potential of these newly synthesized compounds as promising candidates for further investigation as tubulin polymerization inhibitors.
Table 3.
The binding affinities and interactions of 2e, 2g, 2h, and CA-4.
| Compound | Binding Affinity (kcal/mol) |
Classical Hydrogen Bonding | Non-Classical Hydrogen Bonding | Hydrophobic Interactions |
|---|---|---|---|---|
| CA4 | −9.2 | Cys241 | Asn258, Asn350, Val238 | Met259, Ala316, Ala354, Val181, Lys352, Cys241, Leu242, Leu255, Ile318, Ile378, Ala180, Leu248, Ala250, Asn258 |
| 2e | −8.6 | Cys241 | Cys241 | Met259, Lys352, Leu248, Leu255, Ala180, Val181, Lys352 |
| 2g | −7.3 | Cys241 | N/A | Thr179, Ala180, Leu248, Leu255, Ala316, Ala250 |
| 2h | −8.9 | Cys241, Gln247 | Ser178 | Cys241, Leu255, Ala316, Ala180, Leu248, Lys352 |
Figure 5.
Interactions of CA-4 with colchicine binding site: (A) the 2D binding interactions; (B) the 3D binding interactions.
Figure 6.
Interactions of 2e with colchicine binding site: (A) the 2D binding interactions; (B) the 3D binding interactions.
Figure 7.
Interactions of 2g with colchicine binding site: (A) the 2D binding interactions; (B) the 3D binding interactions.
Figure 8.
Interactions of 2h with colchicine binding site: (A) the 2D binding interactions; (B) the 3D binding interactions.
2.3.2. Physicochemical and ADME Prediction
In the drug design journey, it is necessary to optimize the pharmacodynamics and pharmacokinetics of potential drug candidates in a parallel way. Therefore, we investigated the physicochemical and pharmacokinetic characteristics of the designed thiazole chalcones 2a–2p; computational calculations were conducted using the SwissADME website to determine the physicochemical and ADME parameters. The Supplementary Materials section displays comprehensive results of the in silico studies (Tables S1–S5).
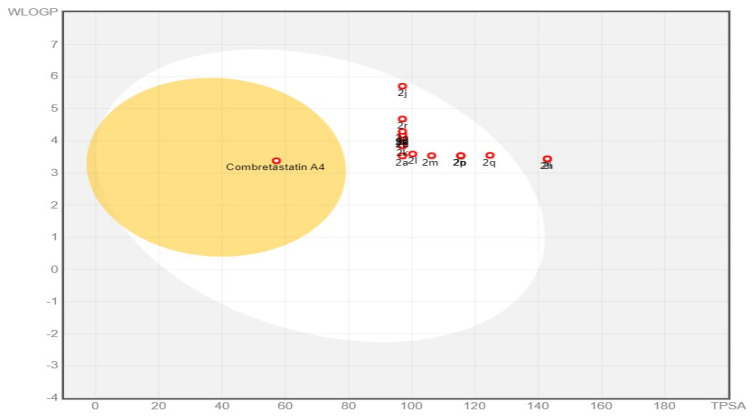
The BOILED Egg approach is a reliable model that precisely predicts the absorption of drug candidates in the gastrointestinal tract and their accessibility via BBB. It achieves this by estimating their lipophilicity (measured in WLOGP) against their polarity (measured in TPSA) (Figure 9). All designed compounds except 2h and 2i appeared in a white zone, indicating a significant gastrointestinal absorption level. This can be attributed to a balance between their lipophilicity (WLOGP 3.38–5.7) and their polarity (TPSA 97.00–142.82 Å2) (Supplementary Materials Tables S2 and S3). Compared to CA-4, all target compounds 2a–2p are predicted not to cross the BBB, which confirms their favorable CNS safety profile. All designed compounds appeared as red points in the BOILED Egg plot, which means they are predicted not to be p-gp substrates, which enhances gastrointestinal absorption and overcomes one of the drug resistance mechanisms [39,40].
Figure 9.
BOILED Egg plot of target compounds 2a–2p and CA-4.
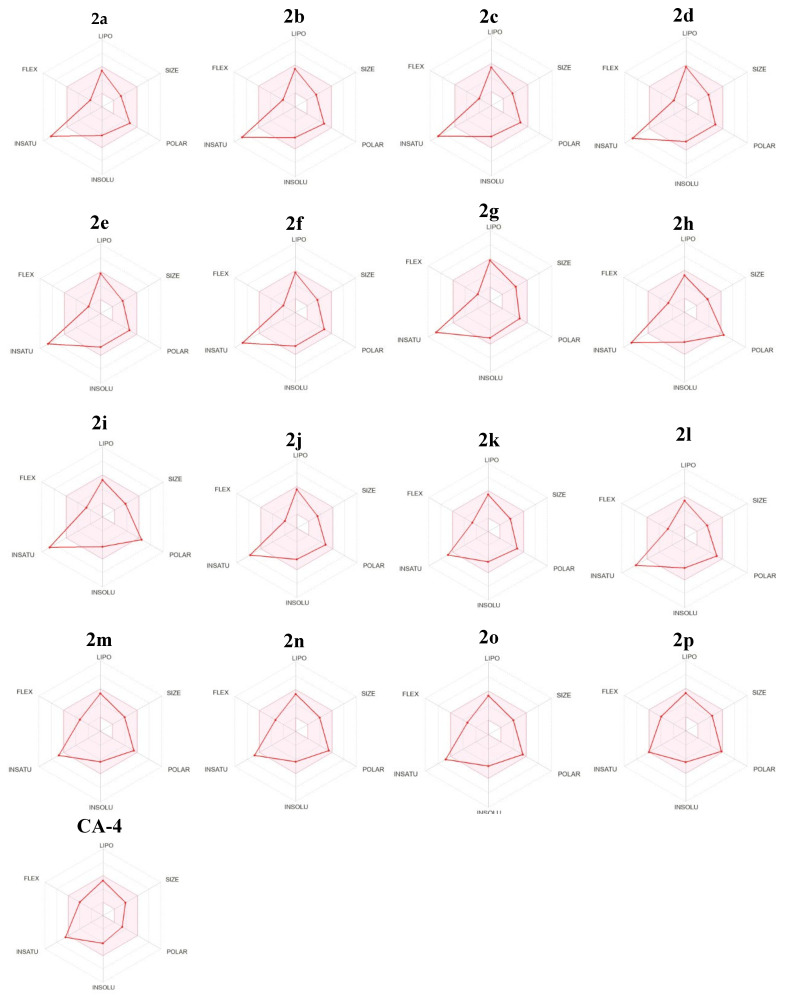
Most target compounds demonstrate favorable anticipated physicochemical properties that make them suitable for oral bioavailability. The bioavailability radar can serve as a convenient means of representing this concept (Figure 10). In the bioavailability radar plot, the pink zone represents the best range for six physicochemical parameters: size, solubility, lipophilicity, polarity, saturation, and flexibility. These properties are considered optimal for achieving optimal oral bioavailability. The majority of the thiazole chalcones are concentrated in the pink region. However, there is a little deviation in the degree of saturation from the pink region due to the presence of less than 0.25 sp3 hybridized carbons [3].
Figure 10.
Rader model for target compounds 2a–2p and Combretastatin A4.
It is worth noting that all target compounds 2a–2p satisfy the drug-likeness criteria of Lipinski’s rule [41]. Additionally, all target compounds satisfy the Ghose filter [42]. Except for 2h and 2i, all target compounds satisfy the Veber rule [43] and the Egan filter [44]. Finally, all target compounds satisfy Muegge’s filter (Supplementary Materials Table S5) [45].
Based on the findings of the in silico ADME prediction studies, it can be inferred that the designed thiazole chalcones exhibit substantial cytotoxic and tubulin polymerization inhibitory properties, as well as favorable physicochemical, pharmacokinetic, and drug-likeness characteristics. These attributes make them suitable for further optimization as potential chemotherapeutic agents.
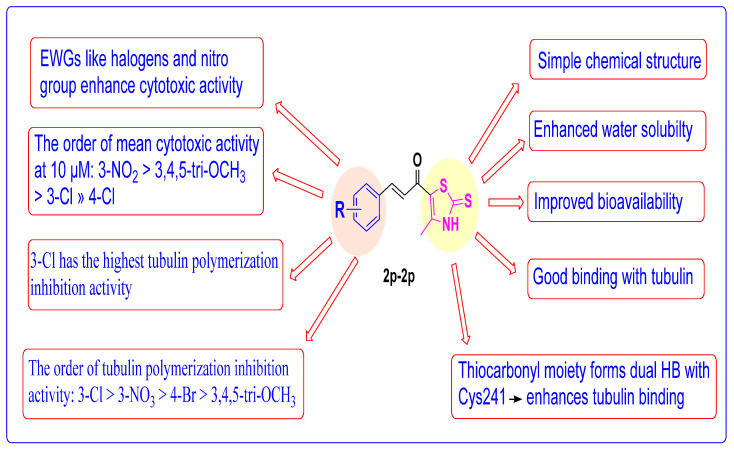
2.4. Structure Activity Relationship (SAR) Studies
SAR studies revealed that substituting the phenyl ring of thiazole chalcones 2a–2p greatly impacts the potency against various cancer cell lines. SAR findings re-garding the anticancer activity, tubulin polymerization inhibitory activity, molecular docking, and ADME Studies are summarized in Figure 11.
Figure 11.
Structure–activity relationship of thiazole–chalcone derivatives.
3. Experimental Section
3.1. Chemistry
Thin-layer chromatography (TLC), using a Merck Grade-9385 precoated aluminum TLC plate with silica gel 60, measuring 5*20 cm, and having a thickness of 0.2 mm, was employed to monitor the progress of the chemical reaction. To detect the spots, the plates were exposed to ultraviolet (UV) light with a wavelength of 254 nm. The Stuart Electrothermal, Melting Point Apparatus was also utilized to determine the melting points without correcting the values. Furthermore, NMR spectra were obtained using a Bruker 400 MHz spectrometer operating at 100 MHz for 13C and 400 MHz for 1H. The solvent used was DMSO-d6, and tetramethylsilane served as the internal standard. In this study, the chemical shifts (δ) and coupling constants (J) were reported in parts per million (ppm) and hertz (Hz), respectively. The following abbreviations were employed to describe the diversity of NMR peaks: singlet (s), doublet (d), doublet of doublets (dd), triplet (t), quartet (q), multiplet (m), and broad signal (brs). Elemental analyses were performed using Shimadzu’s GC/MS-QP5050A instrument at the Regional Centre for Mycology and Biotechnology, Al-Azhar University, Cairo, Egypt. Low-resolution mass analyses were performed at Agilent Pharmaceuticals Inc. (Canada). The spectra were acquired using an Agilent 6400 LC/TQ spectrometer in the negative/positive mode of electrospray ionization (ESI). The intermediate 1 was prepared according to the reported method [36].
3.1.1. General Procedures for the Synthesis of Derivatives 2a–2p
An equimolar amount of thiazole derivative 1 (173 mg, 1 mmol) and the appropriate aromatic aldehyde (1 mmol) were dissolved in ethanol, and aqueous NaOH (140 mg, 3.5 mmol 60%) was added dropwise [24].The reaction mixture was stirred in an ice bath for 2 h, then at rt for 18–20 h. The reaction mixture was acidified by diluted acetic acid. The formed precipitate was filtered off and washed with distilled water, then recrystallized from ethanol.
(E)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)-3-Phenylprop-2-En-1-One 2a
Yellow powder; 0.227 g, 87% yield; mp 245–247 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.63 (1H, s, N-H), 7.79–7.80 (2H, m, Ar-H), 7.66 (1H, d, Jtrans = 16 Hz, =CH), 7.45–7.47 (3H, m, Ar-H), 7.24 (1H, d, Jtrans = 16 Hz, =CH), 2.57 (3H, s, CH3); 13C NMR (100 MHz, DMSO-d6) δ 189.2, 180.2, 147.4, 144.1, 134.7, 131.3, 129.5, 129.3, 124.3, 123.8, 14.9; ESI-MS (m/z): Calcd. 261.03, found 260.56 [M-H]−; Anal. Calcd. For C13H11NOS2: C, 59.74%; H, 4.24%; N, 5.36%. Found: C, 59.52%; H, 4.33%; N, 5.45%.
(E)-3-(3-Fluorophenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2b
Yellow powder; 0.203 g, 73% yield; mp 234–236 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.70 (1H, s, N-H), 7.73 (1H, d, J = 8 Hz, Ar-H), 7.66–7.62 (2H, m, Ar-H and =CH), 7.49 (1H, td, J = 8 Hz, Ar-H), 7.31–7.27 (2H, m, Ar-H and =CH), 2.57 (3H, s, CH3). 13C NMR (100 MHz, DMSO) δ 189.3, 180.1, 162.9 (C-3, d, 1JCFipso = 244.01 Hz), 147.8, 142.6, 137.2 (C-1, d, 3JCFmeta = 8.14 Hz), 131.4 (C-5, d, 3JCFmeta = 8.32 Hz), 125.8, 125.1, 124.2, 117.9 (C-2, d, 2JCFortho = 21.43 Hz), 115.3 (C-4, d, 2JCFortho = 21.86 Hz), 14.9; ESI-MS (m/z): Calcd. 279.02, found 278.58 [M-H]−; Anal. Calcd. For C13H10FNOS2: C, 55.90%; H, 3.61%; N, 5.01%. Found: C, 56.04%; H, 3.85%; N, 5.10%.
(E)-3-(4-Fluorophenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2c
Yellow powder; 0.220 g, 79% yield; mp 262–264 °C; 1H NMR (400 MHz, DMSO-d6) 13.63 (1H, s, N-H), 7.89 (2H, dd, J = 4 Hz, Ar-H), 7.66 (1H, d, Jtrans = 16 Hz, =CH), 7.29 (2H, t, JHF = 8 Hz, Ar-H), 7.19 (1H, d, Jtrans = 16 Hz, =CH), δ 2.56 (3H, s, CH3); 13C NMR (100 MHz, DMSO-d6) δ 189.2, 180.2, 164.0 (C-4, d, 1JCFipso = 249.12 Hz), 147.4, 142.9, 131.7 (C-2 and C-6, d, 3JCFmeta = 8.81 Hz), 131.3, 124.2, 123.7, 116.5 (C-3 and C-5, d, 2JCFortho = 21.70 Hz), 14.9; ESI-MS (m/z): Calcd. 279.02, found 278.53 [M-H]−; Anal. Calcd. For C13H10FNOS2: C, 55.90%; H, 3.61%; N, 5.01%. Found: C, 56.03%; H, 3.51%; N, 5.12%.
(E)-3-(2-Chlorophenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2d
Yellow powder; 0.192 g, 65% yield; mp 240–242 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.67 (1H, s, N-H), 8.03 (1H, d, J = 8 Hz, Ar-H), 7.92 (1H, d, Jtrans = 16 Hz, =CH), 7.56 (1H, d, J = 4 Hz, Ar-H), 7.50–7.42 (2H, m, Ar-H), 7.29 (1H, d, Jtrans = 16 Hz, =CH), 2.58 (3H, s, CH3). 13C NMR (100 MHz, DMSO) δ 189.3, 179.8, 148.1, 139.3, 137.8, 134.8, 132.3, 131.3, 129.1, 127.5, 125.7, 123.9, 14.3; ESI-MS (m/z): Calcd. 294.99, found 294.51 [M-H]−; Anal. Calcd. For C13H10ClNOS2: C, 52.79%; H, 3.41%; N, 4.74%. Found: C, 52.91%; H, 3.37%; N, 4.58%.
(E)-3-(3-Chlorophenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2e
Yellow powder; 0.251 g, 85% yield; mp 246–248 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.69 (1H, s, N-H), 7.94 (1H, s, Ar-H), 7.76 (1H, d, J = 8 Hz, Ar-H), 7.62 (1H, d, Jtrans = 16 Hz, =CH), 7.52–7.45 (2H, m, Ar-H), 7.30 (1H, d, Jtrans = 12 Hz, =CH), 2.57 (3H, s, CH3); 13C NMR (100 MHz, DMSO-d6) δ 189.2, 180.1, 147.9, 142.4, 136.9, 134.3, 131.2, 130.9, 128.7, 128.1, 124.2, 124.2, 14.9; ESI-MS (m/z): Calcd. 294.99, found 294.50 [M-H]−; Anal. Calcd. For C13H10ClNOS2: C, 52.79%; H, 3.41%; N, 4.74%. Found: C, 53.01%; H, 3.48%; N, 4.62%.
(E)-3-(4-Chlorophenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2f
Yellow powder; 0.260 g, 88% yield; mp 250–252 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.67 (1H, s, N-H), 7.76 (2H, d, J = 8 Hz, Ar-H), 7.56 (1H, d, Jtrans = 16 Hz, =CH), 7.43 (2H, d, J = 8 Hz, Ar-H), 7.17 (1H, d, Jtrans = 16 Hz, =CH), 2.44 (3H, s, CH3). 13C NMR (100 MHz, DMSO) δ 189.2, 180.0, 147.8, 142.6, 135.8, 133.6, 131.0, 129.5, 124.4, 124.3, 39.9, 14.9; ESI-MS (m/z): Calcd. 294.99, found 294.61 [M-H]−; Anal. Calcd. For C13H10ClNOS2: C, 52.79%; H, 3.41%; N, 4.74%. Found: C, 52.85%; H, 3.64%; N, 4.81%.
(E)-3-(4-Bromophenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2g
Yellow powder; 0.275 g, 80.82% yield; mp 264–266 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.64 (1H, s, N-H), 7,76 (2H, d, J = 8 Hz, Ar-H), 7.65 (2H, d, J = 8 Hz, Ar-H), 7.62 (1H, d, Jtrans = 16 Hz, =CH), 7.26 (1H, d, Jtrans = 16 Hz, =CH), 2.57 (3H, s, CH3); 13C NMR (100 MHz, DMSO) 189.2, 180.0, 147.7, 143.4, 133.3, 131.5, 130.3, 125.4, 124.7, 123.8, 14.3; ESI-MS (m/z): Calcd. 338.94, found 340.50 [M+2-H]−; Anal. Calcd. For C13H10BrNOS2: C, 45.89%; H, 2.96%; N, 4.12%. Found: C, 45.97%; H, 2.78%; N, 4.25%.
(E)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)-3-(3-Nitrophenyl)Prop-2-En-1-One 2h
Yellow powder; 0.226 g, 74% yield; mp 260–262 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.63 (1H, s, N-H), 8.57 (1H, s, Ar-H), 8.19 (2H, d, J = 8 Hz, Ar-H), 7.71–7.64 (2H, m, Ar-H and =CH), 7.35 (1H, d, Jtrans = 16 Hz, =CH), 2.43 (3H, s, CH3); 13C NMR (100 MHz, DMSO-d6) δ 189.3, 180.0, 148.8, 148.2, 141.5, 136.5, 135.1, 130.8, 126.5, 125.4, 123.9, 123.8, 14.9; ESI-MS (m/z): Calcd. 306.01, found 305.51 [M-H]−; Anal. calcd. for C13H10N2O3S2: C, 50.97%; H, 3.29%; N, 9.14%. Found: C, 50.93%; H, 3.14%; N, 8.97%.
(E)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)-3-(4-Nitrophenyl)Prop-2-En-1-One 2i
Yellow powder; 0.196 g, 64% yield; mp 255–257 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.59 (1H, s, N-H), 8.27 (2H, d, J = 8 Hz, Ar-H), 8.07 (2H, d, J = 8 Hz, Ar-H), 7.72 (1H, d, Jtrans = 16 Hz, =CH), 7.41 (1H, d, Jtrans = 16 Hz, =CH), 2.58 (3H, s, CH3); 13C NMR (100 MHz, DMSO) δ 189.4, 179.9, 148.7, 148.4, 141.1, 130.3, 128.0, 127.8, 124.4, 124.01, 15.1; ESI-MS (m/z): Calcd. 306.01, found 305.52 [M-H]−; Anal. calcd. for C13H10N2O3S2: C, 50.97%; H, 3.29%; N, 9.14%. Found: C, 51.15%; H, 3.13%; N, 9.38%
(E)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)-3-(p-Tolyl)Prop-2-En-1-One 2j
Yellow powder; 0.247 g, 90% yield; mp 241–243 °C; 1H-NMR (400 MHz, DMSO-d6) δ 13.60 (1H, s, N-H), 7.68 (2H, d, J = 8 Hz, Ar-H), 7.63 (1H, d, Jtrans = 16 Hz, =CH), 7.27 (2H, d, J = 8 Hz, Ar-H), 7.16 (1H, d, Jtrans = 16 Hz, =CH), 2.56 (3H, s, CH3), 2.35 (3H, s, Ar-CH3); 13C NMR (100 MHz, DMSO) δ 189.1, 180.2, 147.1, 144.9, 141.5, 131.9, 130.9, 128.5, 124.3, 121.9, 20.9, 14.9; ESI-MS (m/z): Calcd. 275.04, found 274.59 [M-H]−; Anal. Calcd. For C14H13NOS2: C, 61.06%; H, 4.76%; N, 5.09%. Found: C, 61.14%; H, 4.67%; N, 5.24%.
(E)-3-(4-(Dimethylamino)Phenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2k
Pale Red powder; 0.222 g, 73% yield; mp 249–251 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.61 (1H, s, N-H), 7.62–7.57 (3H, m, Ar-H and =CH), 6.91 (1H, d, Jtrans = 12 Hz, =CH), 6.74 (2H, d, J = 8 Hz, Ar-H), 3.01(6H, s, N(CH3)2), 2.54 (3H, s, CH3); 13C NMR (100 MHz, DMSO) δ 189.4, 179.9, 152.8, 148.4, 145.5, 131.3, 129.4, 121.9, 117.7, 112.3, 40.7, 14.9; ESI-MS (m/z): Calcd. 304.07, found 303.7 [M-H]−; Anal. Calcd. For C15H16N2OS2: C, 59.18%; H, 5.30%; N, 9.20%. Found: C, 59.02%; H, 5.48%; N, 9.36%.
(E)-3-(4-Methoxyphenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2l
Yellow powder; 0.227 g, 78% yield; mp 246–248 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.59 (1H, s, N-H), 7.76 (2H, d, J = 8 Hz, Ar-H), 7.63 (1H, d, Jtrans = 12 Hz, =CH), 7.08 (1H, d, Jtrans = 16 Hz, =CH), 7.01 (2H, d, J = 8 Hz, Ar-H), 3.83 (3H, s, OCH3), 2.56 (3H, s, CH3); 13C NMR (100 MHz, DMSO) δ 13C NMR (100 MHz, DMSO) δ 189.0, 180.1, 162.1, 146.9, 143.4, 130.4, 127.2, 124.4, 120.4, 114.2, 55.2, 14.1; ESI-MS (m/z): Calcd. 291.04, found 290.57 [M+H]+; Anal. Calcd. For C14H13NO2S2: C, 57.71%; H, 4.50%; N, 4.81%. Found: C, 57.83%; H, 4.65%; N, 4.67%.
(E)-3-(2,3-Dimethoxyphenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2m
Yellow powder; 0.215 g, 67% yield; mp 239–240 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.67 (1H, s, N-H), 7.84 (1H, d, Jtrans = 16 Hz, =CH), 7.43 (1H, s, Ar-H), 7.27 (1H, d, Jtrans = 16 Hz, =CH), 7.17–7.15 (2H, m, Ar-H), 3.84 (3H, s, OCH3), 3.79 (3H, s, OCH3), 2.57 (3H, s, CH3); 13C NMR (100 MHz, DMSO) δ 189.2, 180.2, 153.4, 147.6, 139.3, 137.6, 128.1, 124.3, 124.0, 121.1, 119.1, 115.1, 57.1, 55.2, 14.2; ESI-MS (m/z): Calcd. 321.05, found 320.60 [M-H]−; Anal. Calcd. For C15H15NO3S2: C, 56.05%; H, 4.70%; N, 4.36%. Found: C, 56.14%; H, 4.57%; N, 4.53%.
(E)-3-(2,4-Dimethoxyphenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2n
Yellow powder; 0.221 g, 69% yield; mp 255–257 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.56 (1H, s, N-H), 7.76 (1H, d, Jtrans = 16 Hz, =CH), 7.67 (1H, d, J = 12 Hz, Ar-H), 7.13 (1H, d, Jtrans = 16 Hz, =CH), 7.60–7.56 (2H, m, Ar-H), 3.86 (3H, s, OCH3), 3.80 (3H, s, OCH3), 2.46 (3H, s, CH3); 13C NMR (100 MHz, DMSO) δ 13C NMR (100 MHz, DMSO) δ 188.9, 180.2, 163.8, 160.8, 146.7, 139.6, 131.9, 124.8, 121.0, 115.9, 106.9, 98.8, 56.4, 56.1, 40.4, 14.9; ESI-MS (m/z): Calcd. 321.05, found 320.63 [M-H]−; Anal. Calcd. For C15H15NO3S2: C, 56.05%; H, 4.70%; N, 4.36%. Found: C, 55.90%; H, 4.69%; N, 4.54%.
(E)-3-(3,4-Dimethoxyphenyl)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)Prop-2-En-1-One 2o
Yellow powder; 0.228 g, 71% yield; mp 245–247 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.70 (1H, s, N-H), 7.61 (1H, d, Jtrans = 16 Hz, =CH), 7.40–7.34 (2H, m, Ar-H), 7.10 (1H, d, Jtrans = 16 Hz, =CH), 7.01 (1H, d, J = 12 Hz, Ar-H), 3.83 (3H, s, OCH3), 3.81 (3H, s, OCH3), 2.55 (3H, s, CH3) 13C NMR (100 MHz, DMSO) δ 188.9, 180.2, 152.2, 149.6, 147.0, 143.8, 127.5, 124.2, 120.7, 117.3, 113.2, 111.5, 56.9, 55.4, 14.2. ESI-MS (m/z): Calcd. 321.05, found 320.61 [M+H]+; Anal. Calcd. For C15H15NO3S2: C, 56.05%; H, 4.70%; N, 4.36%. Found: C, 55.91%; H, 4.81%; N, 4.56%.
(E)-1-(4-Methyl-2-Thioxo-2,3-Dihydrothiazol-5-yl)-3-(3,4,5-Trimethoxyphenyl)Prop-2-En-1-One 2p
Yellow powder: 0.312 g, 89% yield; mp 248–250 °C; 1H NMR (400 MHz, DMSO-d6) δ 13.63 (1H, s, N-H), 7.60 (1H, d, Jtrans = 16 Hz, =CH), 7.13–7.18 (3H, m, 2Ar-H and =CH), 3.85(6H, s, 2OCH3), 3.72(3H, s, OCH3), 2.56 (3H, s, CH3). 13C NMR (100 MHz, DMSO) δ 189.1, 180.4, 153.6, 144.6, 140.7, 130.2, 124.0, 123.3, 107.5, 107.2, 60.6, 56.7, 14.9; ESI-MS (m/z): Calcd. 351.06, found 350.80 [M-H]−; Anal. calcd. for C16H17NO4S2: C, 54.68%; H, 4.88%; N, 3.99%. Found: C, 54.61%; H, 4.77%; N, 3.90%.
3.2. Biological Evaluation and In Silico Studies Methodology
3.2.1. Screening of the Anticancer Activity against a Panel of 60 Cell Lines
The methodology of the NCI anticancer screening has been described in detail elsewhere (http://www.dtp.nci.nih.gov) [2]. For detailed information, see Appendix A in the Supplementary Materials.
3.2.2. In Vitro Tubulin Polymerization Inhibition Assay
In vitro determination of the interaction of thiazole derivatives 2e, 2g, 2h, 2p, and the reference drug CA-4 with the microtubule system was carried out according to the reported protocol [32]. See Appendix A in the Supplementary Materials.
3.2.3. In Silico Studies
Molecular Docking
Autodock vina v1.2.0 was used for molecular docking and the best docking poses were visualized using Discovery Studio Visualizer v24.1.0.23298 [46]. Detailed information is provided in Appendix A in the Supplementary Materials.
In Silico Physicochemical and Pharmacokinetic Properties
The physicochemical and pharmacokinetic parameters for 2a–2p were predicted using the SwissADME tool (http://www.swissadme.ch/index.php) [40]. See Appendix A in the Supplementary Materials.
4. Conclusions
In summary, our research has led to the synthesis and evaluation of a series of novel thiazole-privileged chalcones as tubulin polymerization inhibitors with potential anticancer activities. Thiazole derivatives 2c, 2e, 2f, 2g, 2h, 2i, and 2p revealed broad in vitro cytotoxic activity against various cancer cells, specifically leukemia, colon, renal, and breast cancer cells. Thiazole derivative 2e displayed remarkable antitumor activity against UO-31, SNB-75, LOX IMVI, HCT-116, SW-620, U251, RXF 393, and KM12. Also, compound 2e has demonstrated the ability to inhibit tubulin polymerization in vitro. The use of the thiazole ring, instead of the phenyl ring of classical chalcone, has significantly enhanced both pharmacodynamics and pharmacokinetics. Incorporation of the thiazole moiety in these compounds improves their aqueous solubility, bioavailability, and potentiates binding interaction with the colchicine binding site by forming a dual hydrogen bond with Cys241. These findings underscore the potential of thiazole-privileged chalcones 2a–2p as a promising new class of tubulin-inhibiting molecules for further investigation as a potential anti-cancer therapeutics.
Acknowledgments
The authors extend their profound gratitude to the Swenam College Research Office for their unwavering support and invaluable contributions, which have been instrumental in successfully completing this work.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph17091154/s1, Figures S1–S48: NMR and Mass data; Table S1: Physicochemical properties of target compounds 2a–p and combretastatin CA4; Table S2: Lipophilicity parameters of target compounds 2a–p and combretastatin CA4; Table S3: Water solubility parameters of target compounds 2a–p and combretastatin CA4; Table S4: Pharmacokinetics of target compounds 2a–p and combretastatin CA4; Table S5: Drug likeness parameters of target compounds 2a–r and combretastatin CA4.
Author Contributions
Conceptualization, H.H. and S.M.R.; methodology, W.M.A. and A.S.A.-S.; software, A.G.K.H., A.M.E. and A.S.A.-S.; validation, M.A.A.A.-A., A.E.Z., I.T.R. and S.B.; formal analysis, H.H.; investigation, A.M.E. and A.S.A.-S.; resources, W.M.A. and M.A.A.A.-A.; data curation, A.E.Z., and I.T.R.; writing—original draft preparation, H.H., A.H. and S.B.; writing—review and editing, H.H., A.H. and A.M.E.; visualization, A.G.K.H. and A.M.E.; supervision, H.H. and S.M.R.; project administration, H.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare that this study received No funding from Apogee Pharmaceuticals Inc. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Malik H.S., Bilal A., Ullah R., Iqbal M., Khan S., Ahmed I., Krohn K., Saleem R.S.Z., Hussain H., Faisal A. Natural and Semisynthetic Chalcones as Dual FLT3 and Microtubule Polymerization Inhibitors. J. Nat. Prod. 2020;83:3111–3121. doi: 10.1021/acs.jnatprod.0c00699. [DOI] [PubMed] [Google Scholar]
- 2.Mohammed H.H.H., El-Hafeez A.A.A., Ebeid K., Mekkawy A.I., Abourehab M.A.S., Wafa E.I., Alhaj-Suliman S.O., Salem A.K., Ghosh P., Abuo-Rahma G.E.-D.A., et al. New 1,2,3-triazole linked ciprofloxacin-chalcones induce DNA damage by inhibiting human topoisomerase I&II and tu-bulin polymerization. J. Enzym. Inhib. Med. Chem. 2022;37:1346–1363. doi: 10.1080/14756366.2022.2072308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan A., Badr M., Hassan H.A., Abdelhamid D., Abuo-Rahma G.E.A. Novel 4-(piperazin-1-yl)quinolin-2(1H)-one bearing thiazoles with antiproliferative activity through VEGFR-2-TK inhibition. Bioorg. Med. Chem. 2021;40:116168. doi: 10.1016/j.bmc.2021.116168. [DOI] [PubMed] [Google Scholar]
- 4.Mohammed H.H.H., El-Hafeez A.A.A., Abbas S.H., Abdelhafez E.-S.M.N., Abuo-Rahma G.E.-D.A. New antiproliferative 7-(4-(N-substituted carbamoylmethyl)piperazin-1-yl) derivatives of ciprofloxacin induce cell cycle arrest at G2/M phase. Bioorg. Med. Chem. 2016;24:4636–4646. doi: 10.1016/j.bmc.2016.07.070. [DOI] [PubMed] [Google Scholar]
- 5.Yan J., Xu Y., Jin X., Zhang Q., Ouyang F., Han L., Zhan M., Li X., Liang B., Huang X. Structure modification and biological evaluation of indole-chalcone derivatives as anti-tumor agents through dual targeting tubulin and TrxR. Eur. J. Med. Chem. 2022;227:113897. doi: 10.1016/j.ejmech.2021.113897. [DOI] [PubMed] [Google Scholar]
- 6.Al-Ostoot F.H., Salah S., Khamees H.A., Khanum S.A. Tumor angiogenesis: Current challenges and therapeutic opportunities. Cancer Treat. Res. Commun. 2021;28:100422. doi: 10.1016/j.ctarc.2021.100422. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hamashi A.A., Koranne R., Dlamini S., Alqahtani A., Karaj E., Rashid M.S., Knoff J.R., Dunworth M., Pflum M.K.H., Casero R.A., et al. A new class of cytotoxic agents targets tubulin and disrupts microtubule dynamics. Bioorg. Chem. 2021;116:105297. doi: 10.1016/j.bioorg.2021.105297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchner S., Pianowski Z. Photopharmacology of Antimitotic Agents. Int. J. Mol. Sci. 2022;23:5657. doi: 10.3390/ijms23105657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul R., Risinger A.L., Mooberry S.L. Microtubule-Targeting Drugs: More than Antimitotics. J. Nat. Prod. 2019;82:680–685. doi: 10.1021/acs.jnatprod.9b00105. [DOI] [PubMed] [Google Scholar]
- 10.Henriques A.C., Ribeiro D., Pedrosa J., Sarmento B., Silva P.M.A., Bousbaa H. Mitosis inhibitors in anticancer therapy: When blocking the exit becomes a solution. Cancer Lett. 2019;440–441:64–81. doi: 10.1016/j.canlet.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Guo K., Ma X., Li J., Zhang C., Wu L. Recent advances in combretastatin A-4 codrugs for cancer therapy. Eur. J. Med. Chem. 2022;241:114660. doi: 10.1016/j.ejmech.2022.114660. [DOI] [PubMed] [Google Scholar]
- 12.Sun K., Sun Z., Zhao F., Shan G., Meng Q. Recent advances in research of colchicine binding site inhibitors and their interaction modes with tubulin. Future Med. Chem. 2021;13:839–858. doi: 10.4155/fmc-2020-0376. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Miller D.D., Li W. Molecular interactions at the colchicine binding site in tubulin: An X-ray crystallography perspective. Drug Discov. Today. 2022;27:759–776. doi: 10.1016/j.drudis.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y., Chen J., Xiao M., Li W., Miller D.D. An Overview of Tubulin Inhibitors That Interact with the Colchicine Binding Site. Pharm. Res. 2012;29:2943–2971. doi: 10.1007/s11095-012-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., Jin J., Wu Y., Du B., Zhang L., Lu D., Liu Y., Chen X., Lin J., Chen H., et al. Fluoroindole chalcone analogues targeting the colchicine binding site of tubulin for colorectal oncotherapy. Eur. J. Med. Chem. 2023;257:115540. doi: 10.1016/j.ejmech.2023.115540. [DOI] [PubMed] [Google Scholar]
- 16.Fang Y., Wu Z., Xiao M., Wei L., Li K., Tang Y., Ye J., Xiang J., Hu A. Design, synthesis, and evaluation of new 2-oxoquinoline arylaminothiazole derivatives as potential anticancer agents. Bioorg. Chem. 2021;106:104469. doi: 10.1016/j.bioorg.2020.104469. [DOI] [PubMed] [Google Scholar]
- 17.Sun M., Yuan M., Kang Y., Qin J., Zhang Y., Duan Y., Wang L., Yao Y. Identification of novel non-toxic and anti-angiogenic α-fluorinated chalcones as potent colchicine binding site inhibitors. J. Enzym. Inhib. Med. Chem. 2022;37:339–354. doi: 10.1080/14756366.2021.2014831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X.-F., Wang S.-B., Ohkoshi E., Wang L.-T., Hamel E., Qian K., Morris-Natschke S.L., Lee K.-H., Xie L. N-Aryl-6-methoxy-1,2,3,4-tetrahydroquinolines: A novel class of antitumor agents targeting the colchicine site on tubulin. Eur. J. Med. Chem. 2013;67:196–207. doi: 10.1016/j.ejmech.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Jiang S., Li X., Liu Y., Su J., Chen J. Recent advances in trimethoxyphenyl (TMP) based tubulin inhibitors targeting the colchicine binding site. Eur. J. Med. Chem. 2018;151:482–494. doi: 10.1016/j.ejmech.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Fu D.-J., Liu S.-M., Li F.-H., Yang J.-J., Li J. Antiproliferative benzothiazoles incorporating a trimethoxyphenyl scaffold as novel colchicine site tubulin polymerisation inhibitors. J. Enzym. Inhib. Med. Chem. 2020;35:1050–1059. doi: 10.1080/14756366.2020.1753721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W., Xu F., Shuai W., Sun H., Yao H., Ma C., Xu S., Yao H., Zhu Z., Yang D.-H., et al. Discovery of Novel Quinoline–Chalcone Derivatives as Potent Antitumor Agents with Microtubule Polymerization Inhibitory Activity. J. Med. Chem. 2019;62:993–1013. doi: 10.1021/acs.jmedchem.8b01755. [DOI] [PubMed] [Google Scholar]
- 22.McLoughlin E.C., O’Boyle N.M. Colchicine-Binding Site Inhibitors from Chemistry to Clinic: A Review. Pharmaceuticals. 2020;13:8. doi: 10.3390/ph13010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammed H.H.H., Abbas S.H., Hayallah A.M., Abuo-Rahma G.E.-D.A., Mostafa Y.A. Novel urea linked ciprofloxacin-chalcone hybrids having antiproliferative topoisomerases I/II inhibitory activities and caspases-mediated apoptosis. Bioorg. Chem. 2021;106:104422. doi: 10.1016/j.bioorg.2020.104422. [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Lv J., Cheng S., Jing T., Meng T., Huo D., Ma X., Wen R. Recent Progresses in Chalcone Derivatives as Potential Anticancer Agents. Anticancer Agents Med. Chem. 2023;23:1265–1283. doi: 10.2174/1871520623666230223112530. [DOI] [PubMed] [Google Scholar]
- 25.Rudrapal M., Khan J., Dukhyil A.A.B., Alarousy R.M.I.I., Attah E.I., Sharma T., Khairnar S.J., Bendale A.R. Chalcone Scaf-folds, Bioprecursors of Flavonoids: Chemistry, Bioactivities, and Pharmacokinetics. Molecules. 2021;26:7177. doi: 10.3390/molecules26237177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzaei H., Emami S. Recent advances of cytotoxic chalconoids targeting tubulin polymerization: Synthesis and biological activity. Eur. J. Med. Chem. 2016;121:610–639. doi: 10.1016/j.ejmech.2016.05.067. [DOI] [PubMed] [Google Scholar]
- 27.Kesari C., Rama K., Sedighi K., Stenvang J., Björkling F., Kankala S., Thota N. Synthesis of thiazole linked chalcones and their pyrimidine analogues as anticancer agents. Synth. Commun. 2021;51:1406–1416. doi: 10.1080/00397911.2021.1884262. [DOI] [Google Scholar]
- 28.Rana R., Kumar N., Gulati H.K., Sharma A., Khanna A., Pooja R., Badhwar R., Dhir M., Jyoti P.M.S., Singh J.V., et al. A comprehensive review on thiazole based conjugates as anti-cancer agents. J. Mol. Struct. 2023;1292:136194. doi: 10.1016/j.molstruc.2023.136194. [DOI] [Google Scholar]
- 29.Kassem A.F., Althomali R.H., Anwar M.M., El-Sofany W.I. Thiazole moiety: A promising scaffold for anticancer drug discovery. J. Mol. Struct. 2024;1303:137510. doi: 10.1016/j.molstruc.2024.137510. [DOI] [Google Scholar]
- 30.Gümüş M., Yakan M., Koca İ. Recent advances of thiazole hybrids in biological applications. Future Med. Chem. 2019;11:1979–1998. doi: 10.4155/fmc-2018-0196. [DOI] [PubMed] [Google Scholar]
- 31.Sun M., Xu Q., Xu J., Wu Y., Wang Y., Zuo D., Guan Q., Bao K., Wang J., Wu Y., et al. Synthesis and bioevaluation of N,4-diaryl-1,3-thiazole-2-amines as tubulin inhibitors with potent antiproliferative activity. PLoS ONE. 2017;12:e0174006. doi: 10.1371/journal.pone.0174006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Abd A.O., Bayomi S.M., El-Damasy A.K., Mansour B., Abdel-Aziz N.I., El-Sherbeny M.A. Synthesis and Molecular Docking Study of New Thiazole Derivatives as Potential Tubulin Polymerization Inhibitors. ACS Omega. 2022;7:33599–33613. doi: 10.1021/acsomega.2c05077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mezgebe K., Melaku Y., Mulugeta E. Synthesis and Pharmacological Activities of Chalcone and Its Derivatives Bearing N-Heterocyclic Scaffolds: A Review. ACS Omega. 2023;8:19194–19211. doi: 10.1021/acsomega.3c01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallia A., Sloop J. Advances in the Synthesis of Heteroaromatic Hybrid Chalcones. Molecules. 2023;28:3201. doi: 10.3390/molecules28073201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamal A., Balakrishna M., Nayak V.L., Shaik T.B., Faazil S., Nimbarte V.D. Design and synthesis of imidazo[2,1-b]thiazole-chalcone conjugates: Microtubule-destabilizing agents. ChemMedChem. 2014;9:2766–2780. doi: 10.1002/cmdc.201402310. [DOI] [PubMed] [Google Scholar]
- 36.Mabkhot Y.N., Algarni H., Alsayari A., Muhsinah A.B., Kheder N.A., Almarhoon Z.M., Al-aizari F.A. Synthesis, X-ray Analysis, Biological Evaluation and Molecular Docking Study of New Thiazoline Deriva-tives. Molecules. 2019;24:1654. doi: 10.3390/molecules24091654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prota A.E., Danel F., Bachmann F., Bargsten K., Buey R.M., Pohlmann J., Reinelt S., Lane H., Steinmetz M.O. The Novel Microtubule-Destabilizing Drug BAL27862 Binds to the Colchicine Site of Tubulin with Distinct Ef-fects on Microtubule Organization. J. Mol. Biol. 2014;426:1848–1860. doi: 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Gracheva I.A., Shchegravina E.S., Schmalz H.-G., Beletskaya I.P., Fedorov A.Y. Colchicine Alkaloids and Synthetic Analogues: Current Progress and Perspectives. J. Med. Chem. 2020;63:10618–10651. doi: 10.1021/acs.jmedchem.0c00222. [DOI] [PubMed] [Google Scholar]
- 39.Hassan A., Mubarak F.A.F., Shehadi I.A., Mosallam A.M., Temairk H., Badr M., Abdelmonsef A.H. Design and biological evaluation of 3-substituted quinazoline-2,4(1 H, 3 H)-dione derivatives as dual c-Met/VEGFR-2-TK inhibitors. J. Enzym. Inhib. Med. Chem. 2023;38:2189578. doi: 10.1080/14756366.2023.2189578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan A., Badr M., Abdelhamid D., Hassan H.A., Abourehab M.A.S., Abuo-Rahma G.E.A. Design, synthesis, in vitro antiproliferative evaluation and in silico studies of new VEGFR-2 inhibitors based on 4-piperazinylquinolin-2(1H)-one scaffold. Bioorg. Chem. 2022;120:105631. doi: 10.1016/j.bioorg.2022.105631. [DOI] [PubMed] [Google Scholar]
- 41.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 42.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 43.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 44.Egan W.J., Merz K.M., Baldwin J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 45.Hassan A., Mosallam A.M., Ibrahim A.O.A., Badr M., Abdelmonsef A.H. Novel 3-phenylquinazolin-2,4(1H,3H)-diones as dual VEGFR-2/c-Met-TK inhibitors: Design, synthesis, and biological evaluation. Sci. Rep. 2023;13:18567. doi: 10.1038/s41598-023-45687-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Hakkani M.F., Ahmed N., Abbas A.A., Hassan M.H.A., Aziz H.A., Elshamsy A.M., Khalifa H.O., Abdelshakour M.A., Saddik M.S., Elsayed M.M.A., et al. Synthesis, Physicochemical Characterization using a Facile Validated HPLC Quantitation Analysis Method of 4-Chloro-phenylcarbamoyl-methyl Ciprofloxacin and Its Biological Investigations. Int. J. Mol. Sci. 2023;24:14818. doi: 10.3390/ijms241914818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article; further inquiries can be directed to the corresponding author.