Abstract
Efficient replication of murine cytomegalovirus (MCMV) in macrophages is a prerequisite for optimal growth and spread of the virus in its natural host. Simultaneous deletion of US22 gene family members M139, M140, and M141 results in impaired replication of MCMV in macrophages and mice. In this study, we characterized the proteins derived from these three genes and examined the impact of individual gene deletions on viral pathogenesis. The M139, M140, and M141 gene products were identified as early proteins that localize to both the nucleus and cytoplasm in infected cells. Gene M139 encodes two proteins, of 72 and 61 kDa, while M140 and M141 each encode a single protein of 56 (pM140) and 52 (pM141) kDa, respectively. No role for the M139 proteins in MCMV replication in macrophages or mice was determined in these studies. In contrast, deletion of either M140 or M141 resulted in impaired MCMV replication in macrophages and spleen tissue. Replication of the M140 deletion mutant was significantly more impaired than that of the virus lacking M141. Further analyses revealed that the absence of the pM140 adversely affected pM141 levels by rendering the latter protein unstable. Since the replication defect due to deletion of M140 was more profound than could be explained by the reduced half-life of pM141, pM140 must exert an additional, independent function in mediating efficient replication of MCMV in macrophages and spleen tissue. These data indicate that the US22 genes M140 and M141 function both cooperatively and independently to regulate MCMV replication in a cell type-specific manner and, thus, to influence viral pathogenesis.
The US22 genes of cytomegalovirus (CMV) are members of a multigene family unique to the betaherpesviruses. This gene family is characterized by the presence of one, two, three, or four conserved motifs (4, 7, 8, 12, 24, 25, 27). Consensus sequences for motifs I and II have been identified, and they contain short stretches of hydrophobic and charged residues. The less-well-defined motifs III and IV also have stretches of nonpolar residues. At the left end of betaherpesvirus genomes is a cluster of US22 family genes that exhibit homology among human CMV (HCMV) genes (UL23, UL24, UL28, and UL29), murine CMV (MCMV) genes (M23, M24, m25.1, and m25.2), and human herpesvirus 6 (HHV-6) or HHV-7 genes (U2, U3, U7, and U8). Farther downstream are HCMV US22 genes UL36 and UL43, which are respectively homologous to M36 and M43 in MCMV and to U16/17 and U25 in HHV-6 or HHV-7. At the right end of the MCMV genome is US22 gene M128 (ie2), which is homologous to the HHV-6 or HHV-7 U95 gene. Finally, at the far right end of the HCMV and MCMV genomes lies another cluster of US22 genes not present in HHV-6 or HHV-7. They are HCMV genes US22, US23, US24, US26, and IRS1/TRS1 and homologous MCMV genes M139, M140, M141, m142, and m143.
At least two US22 genes within each betaherpesvirus genome appear to function as transcriptional transactivators of heterologous promoters. These include HCMV immediate-early (IE) genes UL36 (5) and IRS1/TRS1 (30), MCMV IE genes M128 (1) and m142 (B. L. Dalton and A. E. Campbell, unpublished data), and HHV-6 or HHV-7 DR7 (11, 31) and U3 (23). Therefore, these genes may regulate either host or betaherpesvirus gene expression. However, their roles during a natural infection in the host are not entirely clear. For example, M128 is dispensable for growth of MCMV in vitro (1, 17) and has no apparent impact on replication in target organs in vivo (1).
There are other members of the US22 gene family that are required for optimal virus replication in specific tissues. Deletion of M43 compromises replication of MCMV specifically in salivary glands (33). The M139, M140, and/or M141 gene functions to regulate MCMV replication in macrophages, a major target cell for both acute and latent MCMV infection within the host. Deletion of M139, M140, and M141 in mutant virus RV10 impairs growth of MCMV in macrophages in vitro and in macrophage-dense organs in vivo (10). In the spleen, where the virus is confronted with an organized network of macrophages, RV10 replicates to titers at least 2 log10 lower than those of wild-type (WT) or revertant virus (10). Interestingly, restoration of RV10 replication in the spleen is achieved when splenic macrophages are depleted prior to mutant-virus infection (10), indicating that replication competency in macrophages is the factor limiting growth of RV10 in the spleen. In SCID mice, RV10 is highly attenuated compared to WT virus with respect to lethality (10). Therefore, these genes likely function to regulate viral replication competence, at least in macrophages, and this level of regulation has profound effects on the pathogenesis of MCMV in its natural host.
MCMV M139, M140, and M141 RNAs are expressed abundantly at early times and throughout the late phase of infection in both fibroblasts and macrophages (9). Transcripts that map to this gene region in MCMV HindIII-I are 3′ coterminal; they are transcribed from right to left and terminate at a poly(A) site within m138 (9). One transcript maps to M140, and two transcripts each map to the M139 and M141 genes. Sequence analysis of the complete open reading frames (ORFs) predicts that all four US22 motifs are present in M139, M140, and M141 (9).
Our long-term goal is to identify the functions of MCMV US22 gene products in regulating MCMV pathogenesis. The goals of the present study were to characterize the protein products of the M139, M140, and M141 genes and to identify which of the gene products function to regulate macrophage tropism and, hence, pathogenesis in the mouse. We considered the possibility that efficient replication in macrophages might be conferred by one of these three genes or, alternatively, by more than one of them functioning either redundantly or cooperatively. Mutant MCMVs with interruptions in M139, M140, or M141 provided a means of assessing the role of each gene in regulating growth of MCMV in macrophages in vitro and in vivo, as well as of genetically verifying the origin of the newly identified protein(s) produced from each ORF. The M139 gene products were dispensable for efficient MCMV replication in macrophages and spleen tissue. In contrast, mutation of either M140 or M141 resulted in impaired viral replication in this cell and tissue type. Both cooperative and independent functions for these two proteins were revealed in these studies.
MATERIALS AND METHODS
Mice.
Six-week-old male BALB/cAnN (Harlan Sprague Dawley, Indianapolis, Ind.) mice were housed in sterile microisolator cages with sterile food, water, and bedding.
Cells.
Murine NIH 3T3 fibroblasts (ATCC CRL-1658; American Type Culture Collection, Manassas, Va.) were propagated in Dulbecco's modified Eagle medium (Mediatech, Herndon, Va.) supplemented with 10% heat-inactivated bovine calf serum (HyClone Laboratories, Logan, Utah) and 1% l-glutamine (Gibco/BRL, Grand Island, N.Y.). IC-21 cells, a simian virus 40-transformed peritoneal macrophage cell line (ATCC TIB 186) (19), were propagated in RPMI 1640 medium (Mediatech) supplemented with 10% heat-inactivated fetal calf serum (Gibco/BRL) and 1% l-glutamine.
Viruses.
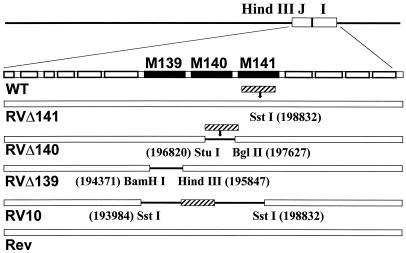
The WT virus in these studies was MCMV Smith strain (ATCC VR 194). Generation and characterization of mutant virus RV10 (from which ORFs M139, M140, and M141 were deleted) and the corresponding revertant virus, RV10Rev, were described previously (10). Viruses with mutations affecting single genes were generated by homologous recombination as previously described (2). The mutant viruses are illustrated in Fig. 1. The plasmid used for generation of RVΔ141 was made by partial digestion of HindIII-I (2) with SstI and insertion of the e1–β-glucuronidase (β-glu) expression cassette. To make RVΔ140, we started with a plasmid, pJI, containing a fragment spanning the HindIII-J and -I junction, from the last BamHI site in HindIII-J (base 194417) to the first PvuII site in HindIII-I (base 199657), inserted between the BglII and EcoRV sites in pcDNA3 (Invitrogen, Carlsbad, Calif.). This plasmid was digested with StuI and EcoRV and then religated, thereby deleting bases 196821 to 196844 and generating a unique BamHI site from the junction. Finally, the plasmid was digested with BamHI and BglII, and the e1–β-glu cassette was inserted, producing a mutant with a deletion from the original StuI site (base 196820) to the BglII site (base 198627). RVΔ139 was generated during the rescue of a mutant virus with deletions in both M139 and M140 and containing WT sequence between the deletions. Mutant RVΔ139 does not contain the e1–β-glu cassette.
FIG. 1.
Map of MCMV mutants. The open boxes indicate WT MCMV sequences within the HindIII-J and -I fragments of the MCMV genome. Solid black lines denote deleted sequences. Black boxes show the location of ORFs M139, M140, and M141. Hatched boxes indicate the locations of the e1–β-glu cassette. The restriction sites and the bases at which the cuts occur are indicated below each mutation site. Rev represents the revertant viruses generated from each mutant, in which the WT sequence was restored.
Revertant viruses were derived from the mutants by homologous recombination as described for RV10Rev, using the same plasmid, pDJI (MCMV bases 191764 to 201966 in plasmid pcDNA3 [10]). The genotype of each mutant and revertant virus was confirmed by Southern blot analysis (data not shown). In addition, all revertant viruses were verified as WT by comparing their replication in fibroblasts and macrophages with that of the WT Smith strain. All virus stocks were prepared in NIH 3T3 cells and quantitated by standard plaque assay. Mock virus preparations were supernatants from uninfected NIH 3T3 cells.
Virion purification by gradient centrifugation.
Mutant MCMVs and their revertants were purified by centrifugation on a 20 to 70% sorbitol gradient according to protocols used for HCMV (6). Briefly, MCMV was grown in 150-ml flasks and the supernatants were harvested when the cytopathic effect reached 100%. Bacitracin (Calbiochem, La Jolla, Calif.) was added to the purified supernatants to a final concentration of 100 μg/ml. The supernatants were layered over cushions of 20% sorbitol in Tris-buffered saline (TBS; 30 mM Tris, 150 mM NaCl, pH 7.5) with bacitracin (100 μg/ml) and centrifuged for 1 h at 26,000 × g and 18°C. The pelleted virus was resuspended in 2 ml of TBS-bacitracin, layered onto a 20 to 70% sorbitol gradient, and centrifuged at 65,000 × g and 18°C for 1 h. Bands were present at all of the interfaces, but the majority of filterable infectious virus was located at the 50–60% interface. Virus was harvested from that interface, diluted 1:10 in TBS-bacitracin, and pelleted over a 20% sorbitol cushion for 1 h at 26,000 × g and 18°C. The virus was resuspended in TBS-bacitracin and frozen prior to quantitation of infectious units by plaque assay.
In vitro growth of MCMV mutants in fibroblasts and macrophages.
The abilities of the mutant and revertant MCMVs to replicate in fibroblast and macrophage cell lines were compared. Approximately 106 NIH 3T3 fibroblasts or IC-21 macrophages in T-25 flasks were infected at a multiplicity of infection (MOI) of 0.1 PFU/cell for multistep growth curves or at 10 PFU/cell for single-step growth curves. After a 2-h incubation to allow virus binding and penetration, the inoculum was removed, the cells were washed with sterile phosphate-buffered saline, and 5 ml of viral growth medium (Dulbecco's modified Eagle medium with 5% serum) was added. At various times postinfection, both intracellular virus and extracellular virus were harvested collectively and titers were determined on NIH 3T3 cells. In one-step growth curves, the hour 0 titer is that of the initial inoculum. For multistep growth curves, samples taken immediately after addition of the medium, subsequent to virus adsorption, were used as day 0 samples. Growth of each virus in the two cell types was quantitated at least twice.
In vivo growth of MCMV mutants.
The abilities of the recombinant and revertant viruses to replicate in mouse spleen and liver tissues were compared. Each mouse received 3 × 105 PFU of tissue culture-passaged virus intravenously (i.v.) in the tail vein. Organs were harvested from individual mice as 20% (wt/vol) homogenates on days 1, 2, and 3 after infection, and titers were determined as described previously (2).
Northern blot analysis.
Northern blot analysis was performed to compare MCMV gene expression by mutant and revertant viruses in IC-21 macrophages. Two million IC-21 macrophages were mock infected or infected with 2 PFU of gradient-purified virus/cell. Infection was allowed to proceed for 2 h, the inoculum was removed, and 10 ml of cell growth medium was added per plate. RNA was harvested 2 h later (at 4 h postinfection) by using TriReagent (Molecular Research Center, Inc., Cincinnati, Ohio) according to the manufacturer's instructions. Northern blot analysis was performed as previously described (10). The probe used for the major IE genes was the HindIII-L fragment. To verify equal loading of RNA samples, the Northern blot was stripped by boiling for 10 min in 0.1% sodium dodecyl sulfate (SDS) and hybridized with a probe specific for the 18S rRNA (EcoRI fragment; ATCC 1227).
Southern blot analysis.
Southern blot analysis was used to confirm the generation of the mutant viruses as described previously (10). Southern blot analysis was also performed to confirm equivalent levels of infection by the mutant viruses in the Northern blot analyses. For this purpose, DNA was harvested both from the above-described TriReagent samples according to the manufacturer's instructions and from replicate plates which had been washed with acid-glycine saline (0.8% NaCl, 0.038% KCl, 0.01% MgCl2, 0.01% CaCl2, 0.7% glycine [pH 3.0]) for 1 min at 4°C. The DNA from the replicate plates was harvested at 4 h postinfection and extracted by using a GenomicPrep DNA isolation kit (Amersham Pharmacia Biotech, Piscataway, N.J.). At this time point, the viral DNA present consisted of unreplicated input genomes. Seven micrograms of total DNA was digested and subjected to Southern blot analysis, performed as described previously (2). The probe used for Southern blot analysis was the HindIII-J fragment.
Antibodies.
Recombinant His-tagged M139, M140, and M141 proteins were produced using the pTrcHis system (Invitrogen). For M139, a HindIII-EcoRI fragment (bases 195847 to 193529; lacking the first 169 bases of M139) was inserted into pTrcHisA. The M140 construct contained a SalI-HindIII fragment (bases 197269 to 195847) lacking 247 bases at the N terminus of the ORF. The M141 construct was generated by insertion of an MscI-XbaI fragment (bases 199159 to 197547) of HindIII-I into EcoRV- and XbaI-digested pTrcHisA, yielding M141 lacking the first 72 bases. His-tagged M139 and M140 were extracted by sonication, while His-tagged M141 was solubilized and extracted with B-PER (Pierce, Rockford, Ill.) and solubilization reagent (Pierce). All of the proteins were purified by using the Xpress system (Invitrogen) under denaturing conditions according to the manufacturer's directions. The recombinant proteins were used to generate rabbit polyclonal antisera (Cocalico, Reamstown, Pa.).
MCMV M44-specific antibody 25G11, a positive control for both viral infection and nuclear localization (15), was a generous gift from Carol Wu and John Shanley (University of Connecticut, Farmington). Protein tyrosine phosphatase (PTP)-PEST-specific antiserum, a positive control for cytoplasmic specificity (3), was the generous gift of Michael Tremblay (McGill University, Montreal, Quebec, Canada).
Western blot analysis of viral gene expression.
Expression of the M139, M140, and M141 proteins in mutant- and revertant-virus-infected cells was monitored by Western blot analysis. Two million NIH 3T3 fibroblasts or IC-21 macrophages were infected with mutant or revertant viruses. Total infected-cell lysates were harvested in lysis buffer (50 mM Tris, 1% SDS, pH 7.5). For time courses, MOIs of 2 and 4 were used for NIH 3T3 fibroblasts and IC-21 macrophages, respectively. Equal amounts of lysate were electrophoresed in the lanes of a 12.5% acrylamide gel for Western blot analysis. For comparisons of mutant viruses, the protein in infected-cell extracts was quantitated using the bicinchoninic acid protein assay reagent (Pierce) according to the manufacturer's instructions. Fifty micrograms of total protein was loaded per lane and electrophoresed on a 12.5% polyacrylamide gel under denaturing conditions for Western blot analysis. All blots were blocked with TBS containing 5% milk and either 0.1% (for PTP-PEST and M141) or 0.5% (for M44, M139, and M140) Tween 20 and washed with TBS containing the same concentration of Tween 20.
For intracellular localization of viral proteins in infected cells, cell fractionation was performed by using an NE-PER kit (Pierce) according to the manufacturer's instructions. For mutant-virus analysis, protease inhibitors aprotinin and phenylmethylsulfonyl fluoride (PMSF; Sigma, St. Louis, Mo.) were added as recommended by Pierce. Volumes of lysate from the cytoplasmic and nuclear fractions representing equivalent numbers of infected cells were electrophoresed on 12.5% acrylamide gels and blotted as described above.
Pulse-chase analysis.
NIH 3T3 fibroblasts were infected with MCMV single-deletion mutants or with RV10Rev at an MOI of 1 PFU/cell. Nineteen hours postinfection, cells were starved of methionine and cysteine for 1 h and then radiolabeled with 450 μCi of [35S]methionine-[35S]cysteine protein labeling mix per ml for 2 h. Labeled proteins were chased with complete medium for various periods of times. The cell lysates were harvested in 1 ml of standard immunoprecipitation lysis buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM PMSF, 10 μg of aprotinin/ml) and clarified from cellular debris. Three hundred microliters of each sample was incubated with 5 μl of polyclonal antiserum overnight at 4°C with constant rocking; then 60 μl of a 50% slurry of protein A-agarose (Roche Molecular Biochemicals, Indianapolis, Ind.) was added to each sample, and the mixtures were further incubated for at least 4 h. Immunoprecipitated complexes were washed three times with SNNTE buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 500 mM NaCl, 5% sucrose, 1% NP-40) and three times with radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1% deoxycholate). Washed immune complexes were resuspended in 50 μl of 1% SDS loading dye, boiled for 5 min, and electrophoresed on 12.5% acrylamide gels. The gels were dried and exposed to X-ray film to obtain autoradiographs. The autoradiographs were scanned with a PowerLook II scanner (UMAX Data Systems, Inc., Hsinchu, Taiwan, Republic of China), and the relative intensities of the bands were quantitated using the Kodak (New Haven, Conn.) Digital Science 1D program.
RESULTS
Multistep growth analysis of MCMV mutant and revertant viruses.
In a previous publication we reported that simultaneous deletion of ORFs M139, M140, and M141 results in a mutant virus (RV10) which displays a cell type-specific replication defect. RV10 exhibits WT replication in fibroblasts but is significantly impaired for replication in differentiated peritoneal macrophages (10). To determine which gene(s) is responsible for mediating efficient replication of MCMV in macrophages, we performed multistep growth analyses using mutant viruses with independent deletions in each of the three ORFs. None of the single-gene deletion mutants was impaired for replication in NIH 3T3 fibroblasts (data not shown), as expected, since RV10 is unimpaired for replication in this cell type (10).
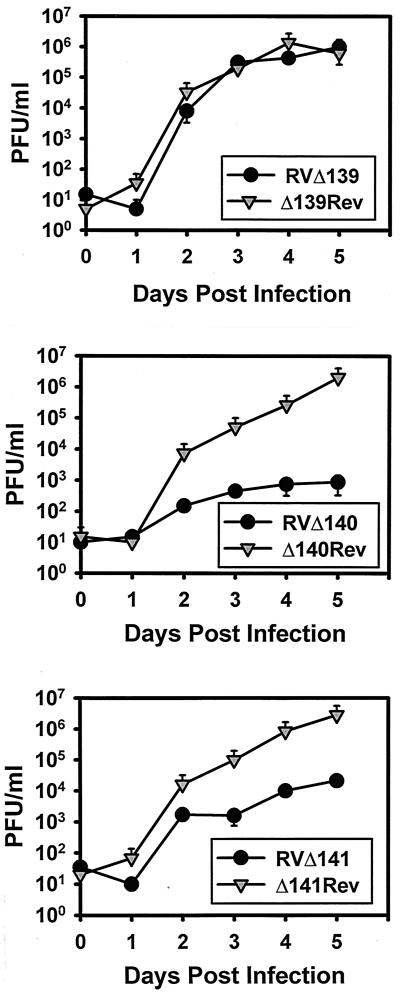
Results of multistep growth analyses of the single-mutant viruses in IC-21 macrophages are shown in Fig. 2. Deletion of M139 alone had no impact on viral replication in IC-21 cells. We therefore ruled out the possibility of an independent role for M139 in optimizing growth of MCMV in macrophages. Deletion of M140, on the other hand, produced a virus with a phenotype nearly identical to that of the triple-deletion mutant RV10 (10). Like RV10, growth of RVΔ140 in IC-21 macrophages was consistently reduced 2 to 3 log10 from that of the WT or revertant virus. Thus, M140 is essential for efficient replication of MCMV in differentiated peritoneal macrophages. Since deletion of M140 reproduced the RV10 phenotype, it was surprising to find that deletion of M141 also resulted in impaired growth of the virus in macrophages. However, at most a 2 log10 difference was consistently evident when comparing RVΔ141 growth to that of its WT revertant. The finding that deletion of a single gene (M140) resulted in the RV10 phenotype while deletion of a second gene (M141) produced an intermediate phenotype was not expected but suggested that these genes participate to different degrees in regulating efficient replication of MCMV in macrophages.
FIG. 2.
Multistep replication of MCMV mutants in IC-21 macrophages. Monolayers of IC-21 macrophages were infected at an MOI of 0.1 PFU/cell. The titers of cell-free and cell-associated viruses were collectively determined on the indicated days postinfection by plaque assay on NIH 3T3 cells. Means and standard deviations for replicates within an experiment are shown. The analysis was repeated at least twice for each mutant.
Single-step growth analysis of MCMV mutant and revertant viruses.
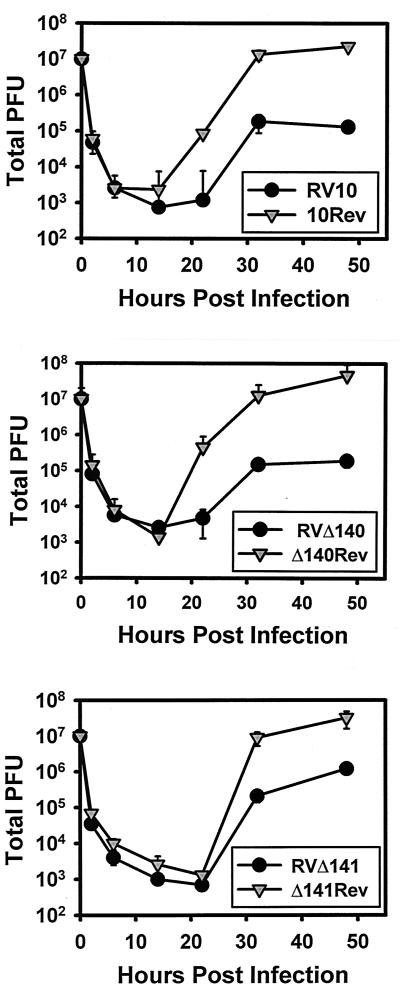
For some mutant CMVs, impairment in replication is overcome by a high MOI (20, 21). Such single-step growth analyses can also differentiate between a defect in virus replication and a defect in virus spread. Thus, such analyses (which had not previously been performed with RV10) may assist in defining the function of these genes. The results of single-step growth analyses of mutant viruses impaired for multistep growth in macrophages (RV10, RVΔ140, and RVΔ141) are shown in Fig. 3. These analyses led to two major findings. First, the defect in replication of these mutant viruses could not be compensated by a high MOI with gradient-purified virions. Second, when comparing results of the multi- and single-step growth curves, it was evident that in macrophages both M140 and M141 function primarily to regulate MCMV replication. In some repetitions of the single-step growth curves for RVΔ140, extracellular and intracellular virus titers of mutant and revertant viruses were compared separately. The magnitude of the impairment of mutant-virus growth in macrophages was the same for both extracellular and intracellular virus (data not shown). These data indicated that there was no obvious defect in release of infectious virus as a consequence of the M140 deletion. Collectively, the data from the single-step growth curves revealed that M140 and M141 gene products regulate one or more stages in replication of MCMV in macrophages.
FIG. 3.
Single-step replication of MCMV mutants in IC-21 macrophages. Monolayers of IC-21 macrophages were infected at an MOI of 10 PFU/cell with gradient-centrifugation-purified mutant or revertant viruses. Titers of cell-free and cell-associated viruses were collectively determined at the indicated times postinfection by plaque assay on NIH 3T3 cells. Means and standard deviations for replicates within an experiment are shown.
In vivo growth of viruses lacking M139, M140, or M141.
We previously reported that impaired replication of mutant MCMV in macrophages correlates with attenuated growth of the virus in vivo (2, 10). When injected i.v. into young-adult BALB/c mice, RV10 failed to replicate to detectable levels in the spleen during a 3-day interval, during which the revertant virus replicated at least 2 log10 (10). Replication of RV10 in the liver, where hepatocytes support growth of MCMV, is unimpaired by the triple deletion in RV10 (10). It was of interest, therefore, to determine if the impaired growth of RVΔ140 and RVΔ141 in macrophages in vitro impacted on replication of these viruses in the spleen in vivo. Replication of RVΔ139 was also assessed to confirm that this mutant virus, exhibiting a WT phenotype with respect to growth in macrophages, replicated like the WT virus in vivo.
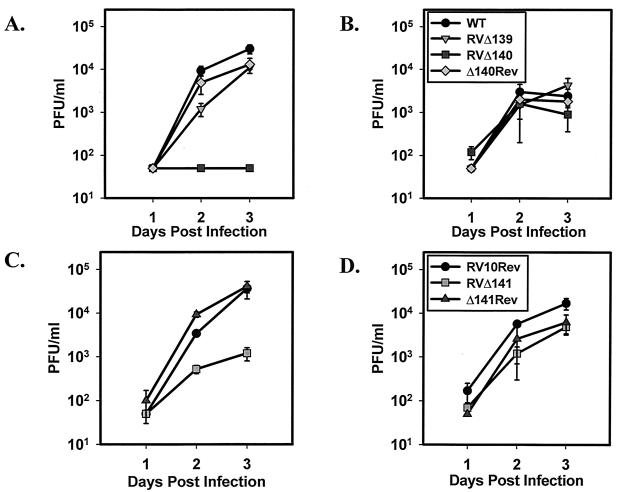
The results of these experiments are shown in Fig. 4. Mutant virus RVΔ140 failed to replicate to detectable levels in the spleen through day 3 postinfection, a time by which WT virus and the RVΔ140 revertant virus reached titers of at least 104 PFU/ml of tissue homogenate (Fig. 4A). RVΔ141 displayed an intermediate growth phenotype in vivo, reaching a mean titer of 1.2 × 103 PFU/ml on day 3, when the mean revertant-virus titer was 4.1 × 104 PFU/ml (Fig. 4C). Both RVΔ140 and RVΔ141 replicated efficiently in liver tissue, reaching peak titers comparable to those of WT or revertant viruses (Fig. 4B and D). Mutant virus RVΔ139 replicated to peak titers equivalent to those of the WT virus by day 3 postinfection in both the spleen and liver (Fig. 4A and B). These data confirmed the previously reported correlation between the capacity of MCMV to replicate in macrophages in vitro and the ability of the virus to replicate in the macrophage-dense environment of the spleen. The data also attested to the fact that deletion of M140 yields a growth phenotype indistinguishable from that of the triple-deletion mutant RV10 (10). Finally, the intermediate role of M141 in dictating macrophage replication in vitro was extended to an intermediate role for this gene product in regulating MCMV growth in the spleen. The in vivo studies therefore supported the in vitro data implicating primarily M140 and secondarily M141 as determinants of MCMV pathogenesis.
FIG. 4.
Growth of MCMV mutant and revertant viruses in spleen and liver tissues. BALB/c mice were inoculated i.v. with 3 × 105 PFU of virus. Spleen and liver tissues were harvested at the indicated times postinoculation. Virus titers in 20% (wt/vol) tissue homogenates were determined by plaque assay on NIH 3T3 cells. Data points are the averages of values for three individual mice, and error bars represent standard deviations. (A and B) Results for the spleen (A) and liver (B) from one experiment in which mice received WT virus, RVΔ139, RVΔ140, or RVΔ140Rev. The key for graphs A and B is shown in panel B. (C and D) Results for the spleen (C) and liver (D) from a second experiment in which mice were injected with RV10Rev, RVΔ141, or RVΔ141Rev. The key for graphs C and D is shown in panel D.
M139, M140, and M141 protein expression from WT and mutant MCMVs.
In characterizations of the single-gene-deletion mutant viruses, each mutant was found to produce transcripts that hybridized to probes corresponding to the neighboring ORFs (data not shown). However, the presence of these transcripts did not guarantee expression of the authentic proteins. We were especially concerned that the intermediate phenotype of RVΔ141 might be due to altered expression of the M140 protein in this mutant virus or that both M140 and M141 were affected by the deletion in RVΔ140. To address these concerns, we generated M139-, M140-, and M141-specific antisera and analyzed first the WT expression of these proteins and then the impact of the mutations on protein expression.
(i) Characterization of M139, M140, and M141 proteins in WT-MCMV-infected NIH 3T3 fibroblasts and IC-21 macrophages.
The M139, M140, and M141 RNAs are all expressed at early times in the MCMV replication cycle (9, 32). In this study, we examined the expression of these proteins by Western blot analysis, assessing both size and kinetics (Fig. 5). As expected from the low level of homology among the proteins, the antisera were not cross-reactive (Fig. 5A). These specific proteins were consistently detected irrespective of both the rabbit from which the antiserum was derived and the WT virus stock used to generate the infected-cell lysates (data not shown).
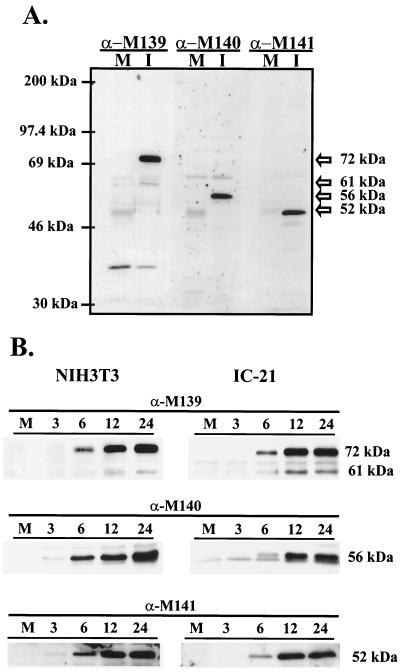
FIG. 5.
Expression of M139, M140, and M141 proteins in NIH 3T3 fibroblasts and IC-21 macrophages infected with WT MCMV. Monolayers of NIH 3T3 fibroblasts or IC-21 macrophages were infected with 2 or 4 PFU of WT virus/cell, respectively. Cell lysates were harvested, and equal volumes of lysates were electrophoresed on a 12.5% acrylamide gel for Western blot analysis using the indicated rabbit polyclonal antisera (α-M139, anti-M139 antibody; α-M140, anti-M140 antibody; α-M141, anti-M141 antibody). M, mock-infected cells; I, infected cells. (A) Lysates from NIH 3T3 fibroblasts 24 h postinfection. (B) Lysates from NIH 3T3 cells or IC-21 macrophages harvested at the indicated hours postinfection.
Two proteins were detected by the M139-specific antiserum, one of 72 kDa (p72M139) and one of 61 kDa (p61M139) (Fig. 5A). Both proteins were detectable by 6 h postinfection in either fibroblasts (longer exposure) or macrophages, and their steady-state levels increased over the course of infection (Fig. 5B). However, p61M139 was consistently less abundant at all times postinfection. The 72-kDa M139 protein closely matched the predicted size of 71.8 kDa (27), suggesting that this protein is a full-length product. The smaller protein may be a processed product derived from p72M139 or may arise from the second transcript.
The M140-specific antiserum recognized a single viral protein of the predicted size of 56 kDa, designated pM140 (Fig. 5A). This protein was detectable by 3 h postinfection in fibroblasts and by 6 h postinfection in IC-21 macrophages (Fig. 5B). As with the M139 proteins, the steady-state level continued to rise over the course of infection.
The M141 antiserum reacted with a viral protein of 52 kDa (Fig. 5A). The 52-kDa M141 protein (pM141) was detectable by 3 h postinfection in fibroblasts and by 6 h postinfection in IC-21 macrophages (Fig. 5B). Therefore, the kinetics of expression of M141 was identical to that of M140.
(ii) Analysis of M139, M140, and M141 protein expression in MCMV mutant-virus-infected cells.
Analysis of expression of the M139, M140, and M141 proteins in macrophages infected with each of the single-deletion mutant viruses individually addressed two objectives: first, it provided a means to confirm the identities of the proteins; and second, it showed whether the mutations altered the steady-state levels of the other proteins encoded in this region. The results are shown in Fig. 6. In this figure, the results for one revertant virus are shown because infection with each revertant virus resulted in a WT pattern of M139, M140, and M141 protein expression.
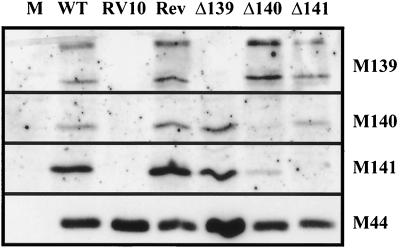
FIG. 6.
Expression of M139, M140, and M141 proteins in IC-21 macrophages infected with MCMV mutants. Monolayers of IC-21 macrophages were infected at an MOI of 4 PFU/cell with the indicated WT and mutant viruses. Fifty micrograms of total protein was loaded per lane, and Western blot analysis was performed as indicated in Materials and Methods and in the legend to Fig. 5. M, mock-infected cells; Rev, lysate from cells infected with RVΔ140 revertant.
Assessment of the proteins expressed by the mutant viruses provided a genetic approach to verifying the identities of the M139, M140, and M141 proteins detected originally in WT-virus-infected cells. Deletion of M139 sequences in RVΔ139 resulted in the loss of both the 72- and 61-kDa bands. This confirmed that both proteins are products of the M139 ORF. Likewise, deletion of M140 sequences in RVΔ140 abolished expression of the 56-kDa protein, confirming its identity as a product of the M140 ORF. Finally, the insertion mutation in RVΔ141 disrupted expression of the 52-kDa protein. An important observation was that the mutations disrupted only the targeted genes and did not prevent synthesis of proteins from the other 3′-coterminal transcripts.
Deletion of M139 had no impact on the level of pM140 or pM141. Similarly, the insertion in M141 did not impact upon M139 or M140 protein levels. In contrast, deletion of M140, while not affecting M139 protein levels, had a dramatic impact on the steady-state levels of pM141. pM141 was barely detectable in RVΔ140-infected cells, in sharp contrast to the abundance of pM141 in cells infected with the WT virus or RVΔ139. This reduction in pM141 levels in RVΔ140-infected cells was highly reproducible among replicate experiments in either fibroblasts or macrophages.
Since deletion of pM141 had no impact on pM140's size or levels, the phenotype of RVΔ141 could not be explained by a simple alteration in pM140. However, the significant reduction in pM141 levels in RVΔ140-infected cells indicates that the mutant phenotype of RVΔ140 may not be due solely to the absence of pM140 but instead may be attributable to a combined effect of the absence of pM140 and the significant reduction of pM141 levels.
Stability of the M141 protein in WT- or mutant-virus-infected cells.
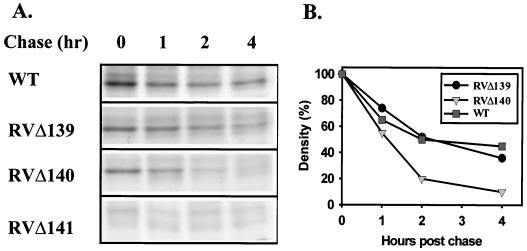
The significantly reduced steady-state levels of pM141 in RVΔ140-infected cells did not correlate with the WT levels of M141 transcripts in cells infected with this mutant virus (data not shown). Therefore, we assessed the stability of pM141 in WT-virus (RV10Rev in this experiment)- and RVΔ140-infected cells by pulse-chase analysis (Fig. 7). The stability of pM141 was significantly compromised by deletion of the M140 gene. The half-life of pM141 was approximately 1 h in RVΔ140-infected cells, compared to 2 h in cells infected with WT virus or RVΔ139. By 4 h postchase, only 10% of the initial pM141 remained in RVΔ140-infected-cell lysates, in contrast to more than 40% of the labeled pM141 remaining in WT- or RVΔ139-infected cells. Thus, pM140 positively influences, either directly or indirectly, the stability of pM141.
FIG. 7.
Stability of pM141 in WT- or mutant-virus-infected cells. Monolayers of NIH 3T3 fibroblasts were infected with the indicated viruses at an MOI of 1 PFU/cell. At 19 h postinfection, the cells were pulsed for 2 h and the chased for the indicated number of hours. (A) Protein precipitating with pM141-specific antiserum was analyzed by polyacrylamide gel electrophoresis. (B) Quantitation of the autoradiographs was performed using Kodak Digital Science software; intensities are expressed relative to the intensity of the pM141 band at time zero.
Intracellular localization of M139, M140, and M141 proteins in infected fibroblasts and macrophages.
Information about the subcellular localization of the M139, M140, and M141 proteins could provide direction for additional studies of the functions mediated by these proteins. M139 and M141 both contain potential nuclear localization signals, while M140 does not. The HCMV homologue of M139, US22, is localized to both the cytoplasm and nucleus of infected cells (22), suggesting that at least M139 may have dual localization. The coordinate regulation of M139, M140, and M141 and the ability of pM140 to stabilize pM141 made the determination of protein colocalization particularly interesting. Unfortunately, standard procedures for fixation and immunostaining of infected cells for visualization by confocal microscopy failed to detect M139, M140, and M141 proteins. Therefore, Western blot analysis of fractionated cells was used to assess localization of each protein in infected cells. The purity of the fractions was confirmed with antibodies specific for known nuclear (MCMV M44 [15]) and cytoplasmic (PTP-PEST [3]) proteins. As with the protein characterization studies, localization experiments were performed first with WT-infected fibroblasts and macrophages and subsequently with mutant-virus-infected cells.
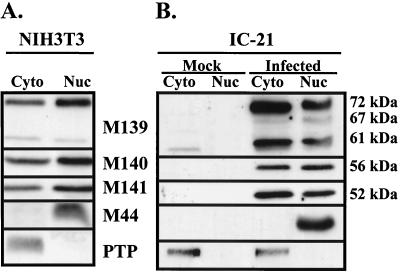
The M139, M140, and M141 proteins were detected in both the nuclear and cytoplasmic fractions in fibroblasts and macrophages at late times after WT virus infection (Fig. 8). Interestingly, an additional low-abundance, 67-kDa protein (p67M139) was detected with M139-specific antiserum in both the cytoplasmic and nuclear fractions of infected macrophages, but not in infected fibroblasts or mock-infected cells. This protein, which was not previously seen in unfractionated lysates from infected cells (Fig. 6), may represent a degradation product of the 72-kDA protein generated during the fractionation procedure. Although Western blot analysis of the cell fractions, as performed here, was not precisely quantitative, the data suggest that M139, M140, and M141 proteins are in general equally distributed between the cytoplasm and nucleus of infected fibroblasts and macrophages.
FIG. 8.
Subcellular localization of M139, M140, and M141 proteins in infected NIH 3T3 fibroblasts and IC-21 macrophages. Monolayers of NIH 3T3 fibroblasts (A) or IC-21 macrophages (B) were infected as described in the legend to Fig. 5. Cells were harvested 24 h postinfection, and the cytoplasmic (Cyto) and nuclear (Nuc) fractions were prepared using an NE-PER kit. Cytoplasmic and nuclear extracts representing equal numbers of cells were electrophoresed on a 12.5% acrylamide gel for Western blot analysis using polyclonal antisera specific for the indicated proteins. Antibodies specific for the nuclear MCMV protein M44 and the cellular cytoplasmic protein PTP-PEST (PTP) were included to demonstrate the purity of the fractions. Mock, extracts from mock-infected cells.
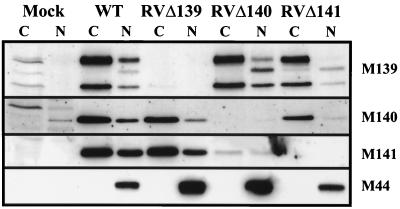
Western blot analyses of cytoplasmic and nuclear fractions of macrophages infected with the various mutant viruses were also performed. The results are shown in Fig. 9. In addition to allowing us to assess the impact of the deletions on neighboring protein localization, this fractionation analysis confirmed the origin of p67M139, since this band was absent when M139 was deleted.
FIG. 9.
Localization of M139, M140, and M141 proteins in IC-21 macrophages infected with MCMV mutants. Monolayers of IC-21 macrophages were infected as described in the legend to Fig. 5. Preparation of the cytoplasmic (C) and nuclear (N) extracts was performed as described for Fig. 8 except that the protease inhibitors aprotinin and PMSF were added to the extraction buffers, and Western blot analysis was performed as described in the legend to Fig. 6. M44 served as a control both for level of viral infection and for purity of the cytoplasmic fraction. Mock, extracts from mock-infected cells.
Deletion of M139 clearly had no impact on the cellular distribution of either pM140 or pM141. Deletion of M140 reduced the relative level of p72M139 in the nucleus compared to the cytoplasmic level. It is unclear whether this was related to the accompanying increase in p67M139. The M141 protein, although expressed in significantly reduced amounts, was detected in both the cytoplasm and nucleus of RVΔ140-infected cells. The disruption of pM141 expression had no impact on localization of pM140. Interestingly, however, p72M139 was undetectable in the nuclei of RVΔ141-infected cells. This exclusion of p72M139 from the nucleus was selective, since p67M139 and p61M139 retained their dual localization. Any impact of this altered localization on replication of MCMV in macrophages is difficult to discern because of the WT phenotype of RVΔ139.
Expression of major IE transcripts in WT- or mutant-MCMV-infected IC-21 macrophages.
Our previous studies using RV10 indicated that the block in replication of this triple-deletion mutant in macrophages correlates with a defect early in the course of infection. In the absence of M139, M140, and M141, expression of IE transcripts in infected macrophages is dramatically reduced (10). We therefore examined whether the block in RVΔ140 replication in macrophages also occurred at the IE phase.
The expression of IE transcripts in IC-21 macrophages infected with each of the single-deletion mutants was assessed by Northern blot analysis (Fig. 10). Surprisingly, we consistently observed that unlike RV10, the RVΔ140 mutant displayed WT levels of IE transcripts in infected macrophages (Fig. 10A). Northern blot analysis of 18S rRNA was performed to confirm that equivalent amounts of RNA were loaded for all samples (Fig. 10B). The block in replication of RVΔ140 in macrophages therefore occurred downstream of IE gene expression.
FIG. 10.
Expression of IE genes in MCMV mutant- and revertant-infected IC-21 macrophages. Monolayers of IC-21 macrophages were infected with gradient-centrifugation-purified MCMV at an MOI of 2 PFU/cell. Five micrograms of total RNA harvested at 4 h after infection was electrophoresed on a formaldehyde gel for Northern blot analyses. (A) The RNA was hybridized to a probe corresponding to the HindIII-L region of MCMV to detect RNA from ie1 (2.7 kb) or ie3 (2.7 kb). (B) The Northern blot was stripped and reprobed with a probe specific for the 18S rRNA to confirm equivalent loading of RNA. (C) Seven micrograms of total DNA harvested from the same cells was digested with BglII and HindIII, and the level of cell-associated viral DNA was assessed by Southern blot analysis. The fragment shown is from the left end of the HindIII-J region (bases 1879444 in M130 to 190639 in M136). M, mock-infected cells.
The reduction in IE gene expression evident in RV10-infected macrophages was not attributable to reduced levels of input genomic DNA (Fig. 10C). The data shown are for DNA isolated from the samples used for RNA extractions with TriReagent, but the results were similar when DNA was isolated from replicate plates which were subjected to acid-glycine saline washes to remove attached but noninternalized virions (data not shown). These results indicated that reduced IE gene expression in RV10-infected cells is not likely attributable to a block in virus entry. These results were in contrast to our previously published data on RV10. We believe that this difference is due to the use of single-capsid or small multicapsid virions selected by gradient centrifugation in the present experiments rather than the unpurified virus preparations employed in the previously published work (10).
Thus, while RVΔ140 exhibited a phenotype indistinguishable from that of RV10 with respect to replication in macrophages and in mice, this single-deletion mutant (as well as RVΔ139 and RVΔ141) did not display the reduction in IE expression evident when all three genes (M139, M140, and M141) were deleted. This suggests that more than one of these genes may independently mediate efficient IE expression. More importantly, these results indicate that the function of pM140 and pM141 necessary for efficient replication in macrophages is unrelated to the IE gene defect seen in RV10 and is exerted at an early or late stage of infection.
DISCUSSION
This study introduced the characterization of the products of M139, M140, and M141 and assessed the role of these US22 family members in MCMV pathogenesis. For clarity, we first discuss the characterization of the proteins and then address their potential roles in viral pathogenesis.
Protein characterization.
The results of this study provide the first characterization of the protein products of MCMV US22 genes M139, M140, and M141. All protein products were initially detected at early times postinfection and were upregulated to abundant levels at late times in the MCMV replication cycle in fibroblasts and macrophages. It appears, therefore, that like that of some other 3′-coterminal CMV genes (13), the expression of their gene products is coordinately regulated, suggesting cooperativity in function. The products of all three genes localized to both the cytoplasm and nucleus of infected cells. This localization in both the nuclear and cytoplasmic fractions of infected cells is certainly not unusual for herpesvirus structural, as well as nonstructural, proteins. A wide range of proteins exhibit this type of localization, including herpes simplex virus type 1 tegument protein UL37 (29) and its varicella-zoster virus homologue derived from gene 21 (16), the Epstein-Barr virus Bcl-2 homologue BHRF1 (14), and herpes simplex virus type 1 ICP22/US1.5-overlapping gene products, which affect the stability of certain viral mRNAs (26). The relative roles of the cytoplasmic and nuclear fractions of M139, M140, and M141 are in question, but it is possible that the proteins provide independent and/or multiple functions at each cellular locale.
(i) M139 proteins.
Two major gene products of the M139 ORF were detected at early and late times in infected fibroblasts and macrophages. These 72- and 61-kDa proteins are likely encoded by the abundant 3.8-kb and less-abundant 3.0-kb transcripts, respectively, which map to the M139 gene (9). Transcript mapping data suggest that this smaller transcript arises from a separate promoter within the larger ORF. There are two potential start sites that would yield proteins of approximately 61 kDa: an ATG at position 195766 and an ATG at position 195667, predicted to yield proteins of 62 and 58 kDa, respectively. The presence of two overlapping potential TATA boxes between bases 195847 and 195831 supports the possibility of a transcript originating in this region. However, since M140, M141, and the larger M139 ORF all have TATA-less promoters, these potential TATA boxes are not necessarily indicative of the origin of the smaller transcript.
Interestingly, the M139-specific antiserum also recognized an additional, 67-kDa protein in extracts of infected macrophages but not in those of fibroblasts. In the absence of cell fractionation to enrich for nuclear proteins, this product was not detected. The 67-kDa protein may be derived from proteolytic processing of the 72-kDa protein in the environment of the macrophage, which contains a plethora of proteolytic enzymes potentially released upon cell lysis. This supposition is based on the correlation between reduced levels of the 72-kDa band and increased levels of the 67-kDa band in some of the mutant-virus-infected cells. Expression of the three M139 proteins was abolished by deletion of the M139 ORF, thus genetically identifying the M139 ORF as the origin of the 72-, 67-, and 61-kDa proteins. Aside from the potential processing of the 72-kDa protein to the 67-kDa protein in macrophages, there appeared to be minimal posttranslational modifications of the 72- and 61-kDa proteins, since the sizes of these proteins match the sizes predicted on the basis of amino acid sequence alone.
While the 67-kDa protein localized preferentially to the nucleus of infected macrophages, the 72- and 61-kDa proteins were found in both the nuclear and cytoplasmic fractions of infected fibroblasts and macrophages. In the absence of pM141, there was appreciably less localization of the 72-kDa protein to the nucleus than occurred in WT-virus-infected cells. Since the M139 proteins are predicted (based on sequence analysis) to contain a nuclear targeting signal, it is unlikely that another viral protein, such as pM141, is needed to transport p72M139 to this locale. However, it is possible that pM141 influences retention of the M139 proteins in the nucleus.
(ii) M140 protein.
The M140 gene encodes a single 56-kDa protein expressed at early and late times postinfection. This protein localized to both the nucleus and cytoplasm in WT- and mutant-virus-infected cells, although the sequence of the M140 gene does not reveal a consensus nuclear targeting signal. Thus, it is likely that either pM140 has an atypical nuclear localization signal or it interacts with one or more other proteins that mediate its transport to the nucleus. However, it is also possible that pM140 enters the nucleus by passive diffusion, since it is smaller than 60 kDa, the maximum size of proteins which enter the nucleus by this process (reviewed in reference 18).
(iii) M141 protein.
The M141 gene encodes a 52-kDa protein from the larger of the two transcripts mapping to this gene (9). This early protein is expressed with kinetics identical to that of pM140 and, likewise, is located in both the cytoplasm and nucleus of infected cells. Unlike M140, the M141 sequence contains a putative nuclear targeting signal. The most striking feature of pM141 is its dependence on the presence of pM140 for stability. It is tempting to speculate that these two proteins form a complex, either by themselves or with other cellular or viral proteins. Data from an independent study support the hypothesis that pM140 and pM141 interact in at least one complex in MCMV-infected cells (Z. Karabekian and A. E. Campbell, unpublished data).
The M141 ORF would be predicted to encode a PEST sequence, a region which is enriched in proline, glutamate, serine, and threonine and which targets a gene product for potential degradation under certain physiological conditions (28). This PEST sequence may therefore contribute to the instability of pM141. Interestingly, however, the M139 gene also contains a putative PEST sequence that apparently does not render the M139 proteins unstable during replication in fibroblasts and macrophages, at least. Explanations for why pM141 is unstable and how pM140 contributes to pM141 stability await further studies.
Pathogenesis.
Previously, we showed that one or more of the US22 gene family members M139, M140, and M141 are involved in regulation of efficient replication in macrophages and in spleen tissue in vivo. In attempts to identify a role for each US22 protein, mutations were made in the individual genes. While the results leave in question the function or role of M139 and M141 in viral pathogenesis, the studies indicated that deletion of M140 alone recapitulated the replication phenotype of the triple-deletion mutant RV10 and revealed that this US22 gene family member mediates more than one function in the regulation of MCMV pathogenesis.
(i) M139 and M141 proteins.
The function of the M139 proteins in viral pathogenesis is not clear. Deletion of this ORF alone had no impact on replication of MCMV either in the differentiated macrophage cell line or in spleen tissue of young-adult BALB/c mice. In a previous study, we found that unlike the triple-deletion mutant, RVΔ139 was lethal for SCID mice, but the time of death was delayed compared to the WT virus (L. K. Hanson, H. W. Virgin IV, and A. E. Campbell, unpublished data). Therefore, the M139 gene products likely influence some aspect of MCMV pathogenesis. Inclusion of a larger repertoire of cell types and mouse organs may be necessary to reveal the function of these proteins.
The M141 gene product, on the other hand, clearly has a role in MCMV pathogenesis. The intermediate phenotype of RVΔ141 compared to that of RV10, RVΔ140, or WT virus is intriguing. In multistep growth curves, RVΔ141 replicated in macrophages to levels approximately 2 log10 lower than its WT revertant, while the level of RVΔ140 was reduced 3 to 4 log10 from the level of the WT virus. In vivo, RVΔ141 replicated in the liver and spleen, but splenic titers were 30-fold lower than those of WT virus on day 3 postinfection. The fact that pM141 is unstable in the absence of pM140 suggests that these two proteins may function cooperatively; however, the mechanism(s) by which pM141 functions and contributes to viral pathogenesis remains to be elucidated.
(ii) M140 protein.
A major role for pM140 in regulating growth of MCMV in macrophages and in the BALB/c mouse spleen was revealed by use of the M140 deletion mutant. This mutant was defective for replication in macrophages, even at a high MOI. In vivo, replication of RVΔ140 was not detected in the spleen through 3 days postinfection, a time by which WT virus had replicated at least 3 log10. Based on our previous studies (10), we speculate that the inability to detect RVΔ140 replication in the spleen was directly related to its retarded growth in splenic macrophages. However, we recognize that deletion of this gene may have multiple effects influencing tropism for a variety of splenic cells. Replication of RVΔ140 in macrophages was hindered at some stage of the replication cycle downstream of ie1 and ie3 expression. Further studies are needed to define the block in replication of this mutant in macrophages and mouse spleen.
The M140 protein clearly exhibits at least two separate roles in MCMV pathogenesis. First, pM140 is needed to stabilize pM141 expression in infected cells. However, if this were the only function of pM140, the phenotypes of RVΔ141 and RVΔ140 would be the same. Since the replication impairment in RVΔ140 is significantly greater than that in RVΔ141, pM140 must have an additional, independent function. Thus, this viral protein appears to function both cooperatively and independently to mediate the same phenotype. It is tempting to speculate that the HCMV homologues of M140 and M141 (US23 and US24, respectively) confer similar cooperative and independent functions in regulating cell or tissue tropism.
ACKNOWLEDGMENTS
We gratefully acknowledge the contributions of and the reagents provided by Timothy Bos, Michael Tremblay, and Carol Wu. We thank Julie Kerry and Mark Birkenbach for critical evaluation of the manuscript.
This work was supported by the National Cancer Institute (PHS grant R01-CA41451) and the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust.
REFERENCES
- 1.Cardin R D, Abenes G B, Stoddart C A, Mocarski E S. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology. 1995;209:236–241. doi: 10.1006/viro.1995.1249. [DOI] [PubMed] [Google Scholar]
- 2.Cavanaugh V J, Stenberg R M, Staley T L, Virgin IV H W, MacDonald M R, Paetzold S, Farrell H E, Rawlinson W D, Campbell A E. Murine cytomegalovirus with a deletion of genes spanning HindIII-J and -I displays altered cell and tissue tropism. J Virol. 1996;70:1365–1374. doi: 10.1128/jvi.70.3.1365-1374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charest A, Wagner J, Shen S H, Tremblay M L. Murine protein tyrosine phosphatase-PEST, a stable cytosolic protein tyrosine phosphatase. Biochem J. 1995;308:425–432. doi: 10.1042/bj3080425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kousarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 5.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36–38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 7.Efstathiou S, Lawrence G L, Brown C M, Barrell B G. Identification of homologues to the human cytomegalovirus US22 gene family in human herpesvirus 6. J Gen Virol. 1992;73:1661–1671. doi: 10.1099/0022-1317-73-7-1661. [DOI] [PubMed] [Google Scholar]
- 8.Gompels U A, Nicholas J, Lawrence G, Jones M, Thompson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 9.Hanson L K, Dalton B L, Karabekian Z, Farrell H E, Rawlinson W D, Stenberg R M, Campbell A E. Transcriptional analysis of the murine cytomegalovirus HindIII-I region: identification of a novel immediate-early gene region. Virology. 1999;260:156–164. doi: 10.1006/viro.1999.9796. [DOI] [PubMed] [Google Scholar]
- 10.Hanson L K, Slater J S, Karabekian Z, Virgin IV H W, Biron C A, Ruzek M C, van Rooijen N, Ciavarra R P, Stenberg R M, Campbell A E. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J Virol. 1999;73:5970–5980. doi: 10.1128/jvi.73.7.5970-5980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashanchi F, Thompson J, Sadaie M R, Doniger J, Duvall J, Brady J N, Rosenthal L J. Transcriptional activation of minimal HIV-1 promoter by ORF-1 protein expressed from the SalI-L fragment of human herpesvirus 6. Virology. 1994;201:95–106. doi: 10.1006/viro.1994.1269. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T, Bankier A T, Satchwell S C, Preddy E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 13.Leatham M P, Witte P R, Stinski M F. Alternate promoter selection within a human cytomegalovirus immediate-early and early transcription unit (UL119–115) defines true late transcripts containing open reading frames for putative viral glycoproteins. J Virol. 1991;65:6144–6153. doi: 10.1128/jvi.65.11.6144-6153.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu M-Y, Shih Y-Y, Li L-Y, Chou S-P, Sheen T-S, Chen C-L, Yang C-S, Chen J-Y. Expression of the Epstein-Barr virus BHRF1 gene, a homologue of Bcl-2, in nasopharyngeal carcinoma tissue. J Med Virol. 2000;61:241–250. doi: 10.1002/(sici)1096-9071(200006)61:2<241::aid-jmv11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Loh L C, Keeler V D, Shanley J D. Sequence requirements for the nuclear localization of the murine cytomegalovirus M44 gene product pp50. Virology. 1999;259:43–59. doi: 10.1006/viro.1999.9700. [DOI] [PubMed] [Google Scholar]
- 16.Mahalingam R, Lasher R, Wellish M, Cohrs R J, Gilden D H. Localization of varicella-zoster virus gene 21 protein in virus-infected cells in culture. J Virol. 1998;72:6832–6837. doi: 10.1128/jvi.72.8.6832-6837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning W C, Mocarski E S. Insertional mutagenesis of the murine cytomegalovirus genome: one prominent alpha gene (ie2) is dispensable for growth. Virology. 1988;167:477–484. [PubMed] [Google Scholar]
- 18.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 19.Mauel J, Defendi V. Infection and transformation of mouse peritoneal macrophages by simian virus 40. J Exp Med. 1971;134:335–350. doi: 10.1084/jem.134.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier J L, Pruessner J A. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J Virol. 2000;74:1602–1613. doi: 10.1128/jvi.74.4.1602-1613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1491aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mocarski E S, Pereira L, McCormick A L. Human cytomegalovirus ICP22, the product of the HWLF1 reading frame, is an early nuclear protein that is released from cells. J Gen Virol. 1988;69:2613–2621. doi: 10.1099/0022-1317-69-10-2613. [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Yagi H, Shimamoto T, Isegawa U, Sunagawa T, Inagi R, Kondo K, Tano Y, Yamanishi K. Analysis of human herpesvirus 6 U3 gene, which is a positional homolog of human cytomegalovirus UL24 gene. Virology. 1998;249:129–139. doi: 10.1006/viro.1998.9305. [DOI] [PubMed] [Google Scholar]
- 24.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas J, Martin M E D. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J Virol. 1994;68:597–610. doi: 10.1128/jvi.68.2.597-610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogle W O, Roizman B. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 29.Schmitz J B, Albright A G, Kinchington P R, Jenkins F J. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology. 1995;206:1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- 30.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the α gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J, Choudhury S, Kashanchi F, Doniger J, Berneman Z, Frenkel N, Rosenthal L J. A transforming fragment within the direct repeat region of human herpesvirus type 6 that transactivates HIV-1. Oncogene. 1994;9:1167–1175. [PubMed] [Google Scholar]
- 32.Vieira J, Farrell H E, Rawlinson W D, Mocarski E S. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with a lacZ/gpt cassette. J Virol. 1994;68:4837–4846. doi: 10.1128/jvi.68.8.4837-4846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao J, Tong T, Zhan X, Haghjoo E, Liu F. In vitro and in vivo characterization of a murine cytomegalovirus with a transposon insertional mutation at open reading frame M43. J Virol. 2000;74:9488–9497. doi: 10.1128/jvi.74.20.9488-9497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]