Abstract
Reverse transcriptases (RTs) are found in a wide variety of mobile genetic elements including viruses, retrotransposons, and infectious organellar introns. An invariant triad of aspartates is thought to be required for the catalytic function of RTs. We generated RT mutants in the yeast retrotransposon Ty1, changing each of these active-site aspartates to asparagine or glutamate. All but one of the mutants lacked detectable polymerase activity. The novel exception, D211N, retained near wild-type in vitro polymerase activity within virus-like particles but failed to carry out in vivo transposition. For this mutant, minus-strand synthesis is impaired and formation of the plus-strand strong-stop intermediate is eliminated. Intragenic second-site suppressor mutations of the transposition defect map to the RNase H domain of the enzyme. Our results demonstrate that one of the three active-site aspartates in a retrotransposon RT is not catalytically critical. This implies a basic difference in the polymerase active-site geometry of Ty1 and human immunodeficiency virus RT and shows that subtle mutations in one domain can cause dramatic functional effects on a distant domain of the same enzyme.
Reverse transcriptase (RT), the DNA polymerase that copies RNA templates into DNA, has been identified in numerous biological niches. While vertebrate retrovirus RTs, and in particular human immunodeficiency virus (HIV-1) RT, have been intensively studied (14), families of endogenous RT-encoding retrotransposons are ubiquitous among eukaryotes. Based on the amino acid sequence of their RTs, retrotransposons and retroviruses have been placed into a superfamily of RT-containing genetic elements, where structurally related elements identified in many species cluster with one another (21, 76). Aside from retrotransposons, the superfamily includes classes of RTs found in other viruses (72), bacteria (36), self-splicing introns (78), mitochondrial plasmids (73), and most recently the catalytic component of the nearly universal enzyme telomerase (49). Based on these phylogenetic considerations, RT is thought to be an ancient enzyme that evolved from a primordial RNA-dependent RNA polymerase (25, 55).
Saccharomyces cerevisiae is home to five families of multicopy long terminal repeat (LTR)-containing endogenous retrotransposons, of which Ty1 is the most abundant (40). Studies of Ty1 have demonstrated its essential structural and functional relatedness to vertebrate retroviruses (reviewed in reference 14). The complete element contains two overlapping open reading frames (ORFs), termed TYA (analogous to retroviral GAG) and TYB (analogous to retroviral POL), and is flanked by LTRs made up of distinct U3, R, and U5 regions. Within the second ORF, separate protease, integrase (IN), and RT/RNase H domains have been identified. Ty1 DNA is present on chromosomes and is transcribed and processed using regulatory sequences in its LTR. This RNA is translated and also serves as the genomic RNA for reverse transcription. Ty1 translation products assemble, along with Ty1 RNA and host tRNA, into cytoplasmic virus-like particles (VLPs) in which replication occurs.
The key to replication of both retroviruses and retrotransposons is the RT enzyme, which contains spatially separated amino-terminal polymerase and carboxy-terminal RNase H domains. While the polymerase carries out template- and primer-directed DNA polymerization, using either RNA or DNA as the primer and template, the RNase H degrades RNA within RNA-DNA duplexes (14). Within the context of these basic functions, the two domains of the enzyme are required to carry out specific, sequential, and interrelated reactions in order for the genomic RNA to be successfully copied into an integratable double-stranded cDNA. For example, Ty1 replication is initiated by the polymerase domain of RT copying the 5′ end of Ty1 RNA, using initiator methionine tRNA (IMT) as the minus-strand primer. While polymerase copies the RNA, the associated RNase H activity is presumed to follow and degrade the RNA in the resulting RNA-DNA hybrid. The terminated product is termed the minus-strand strong-stop intermediate (msss). For copying to continue, the RT and its primer end must undergo an RNase H-dependent translocation, termed strand transfer. After transfer, minus-strand synthesis progresses along the RNA template. A short region of RNA just upstream of the 3′ U3, referred to as the polypurine tract (PPT), is specifically spared from RNase H cleavage and is then used by the polymerase as the primer to generate the plus-strand strong-stop intermediate (psss). Subsequent plus-strand transfer allows continued synthesis of both strands. The final product is a double-stranded cDNA which can insert into a new genomic location.
Phylogenetic studies comparing the sequences of RTs from many sources have noted a triad of conserved aspartic acid (D) residues (21, 60, 76). Two of these Ds are part of the YXDD box in motif C, which serves as a signature for RTs, while the other D is ∼75 to 100 amino acids amino terminal to this box in a region referred to as motif A (60) (the equivalent residues are D110, D185, and D186 for HIV-1 RT and D129, D210, and D211 for Ty1). The near invariability of these residues, their alignment with catalytic aspartates from other polymerases (17), and the finding that replacement of any one of these Ds in HIV-1 RT results in the loss of both in vitro RT activity and infectivity (11, 39, 44, 45, 47) have led to the conclusion that these are three critical catalytic residues. Further, multiple crystal structures of the HIV-1 RT (35, 38, 41) and a portion of the Moloney murine leukemia virus RT (33) all place these three residues within the active-site pocket of the enzyme. Within the wider range of sequenced retroelements, however, a few exceptions have been noted. A gene encoding an RT-like protein in the mitochondrial genome of Chlamydomonas reinhardtii has the YXDD box sequence YADN (8). Tas, an Ascaris lumbricoides retrotransposon, contains the corresponding sequence YVDN (27), as do several families of LTR-containing retrotransposons recently identified through the Caenorhabditis elegans sequencing project (9). In none of these cases has the nonstandard RT-like ORF been shown to have biochemical activity, nor have any of these exceptional elements been shown to be transpositionally active. Their existence, however, suggests the possibility that the second aspartate in the YXDD box is not catalytically essential.
We have used yeast Ty1 to look directly at whether all three of the conserved aspartate residues are essential for retrotransposon RT function. We find that unlike all other substitutions examined, substitution of asparagine for aspartate at position 211, the second D in the YXDD box, does not obviously affect the exogenous polymerase activity of the enzyme. However, the mutant Ty1 element is completely incapable of carrying out transposition. Intragenic second-site suppressors mapping to the RNase H domain can restore transposition competence. Our results provide biochemical and genetic evidence that the second aspartate side chain in the YXDD box of retrotransposon RT polymerases is not essential for catalyzing the polymerization reaction but does play critical roles in the replication process, possibly by coordinating RNase H and polymerase activities.
MATERIALS AND METHODS
Strains and culture conditions.
Yeast strain YH50 (MATα ho spt3-202 ura3-167 trp1Δ1 leu2-3 his3Δ200) (20) or the isogenic rad52::LEU2 strain AGY49 (71) were used for genetic assays, while strain YH51 (MATa ura3 his 4-539 lys2-801 spt-202) (29) was used for preparation of VLPs. All strains are spt3, which eliminates endogenous transposition and its potential for complementation of mutant Ty1 elements in trans or background Ty1 RT activity within VLPs (6). Standard synthetic complete (SC) medium with omission of different combinations of amino acids, yeast-peptone-dextrose (YPD) medium and SC medium containing 5-fluoroorotic acid (SC+5FOA) were prepared and used as previously described (5, 66).
Plasmid constructions.
Site-directed mutants were constructed by the method of Kunkel (42). All sequence changes were confirmed.
(i) Mutant versions of pJEF724.
The 1,120-bp KpnI-HindIII fragment from the URA3-marked, 2μm-based, GAL1-driven Ty1-H3 plasmid pJEF724 (4) was ligated with similarly cut M13mp18 replicative-form phage. Recombinant plaques produced single-stranded phage, which was annealed with mutagenic primer AG19 (5′ CAA TAC CAT ATT ATC TAC GAA TAA 3′) or AG20 (5′ CAA TAC CAT TTC ATC TAC GAA TAA 3′) to generate asparagine (D211N) or glutamate (D211E) substitutions, respectively (Ty1-H3, position 4577). The 1,120-bp KpnI-HindIII fragment from mutant replicative-form phages was subcloned back into similarly digested pJEF724. Additional active-site mutants were obtained by the same procedures using mutagenic primers AG37 (5′ TTT TTG CTA AAG AGC ACC ATG TCC TCT ACG AAT AA 3′) and AG38 (5′ CAA TAC CAT GTC GTT AAC GAA TAA ACA 3′) to generate the D210E and D210N substitutions (Ty1-H3 position 4574) and mutagenic primers RAG81 (5′ CGA AGA TAT CTC TAA TTG TG 3′) and RAG82 (5′ TGC CGA AGA AAT ATT TAA TTG TG 3′) to generate the D129E and D129N substitutions (Ty1-H3 position 4357).
(ii) Mutant versions of pX3.
Each of the six pJEF724 mutations was subcloned into pX3 (a derivative of pJEF724 with a TRP1 marker in the Ty1 3′ untranslated region [7]) by ligating the mutant KpnI-HindIII fragment with the largest KpnI-HindIII fragment of pX3, digesting the resulting plasmid with HindIII, ligating it with the 1,093-bp TRP1-containing HindIII fragment from pX3, and subsequently identifying the correctly oriented insert.
(iii) Mutant versions of pGTy1mhis3AI.
Mutant versions of pGTy1mhis3AI were made in several steps. First, pGTyCla (AGE565), which is equivalent to pJEF724 except for the presence of a unique ClaI site in the Ty1 3′ untranslated region (Ty1-H3 position 5561), was constructed by digesting plasmid pBJC8 (15) with ClaI and self-ligating the large fragment. Second, the 4.4-kb XhoI-HindIII Ty1-containing fragment of AGE565 was subcloned into pBSIIKS(+) and the resulting phagemid was used to introduce a new MluI site at Ty1-H3 position 3219 with primer RAG205 (5′ GTT TTA GAA ACG CGT TTC GAA T 3′) (MluI site underlined). Third, the MluI-containing phagemid (AGE1495) was digested with BstEII and HindIII and the resulting 2.8-kb Ty1 fragment was ligated to similarly digested AGE565 to generate plasmid AGE1542. Fourth, AGE1542 was linearized at its unique ClaI site and ligated to the 990-bp his3AI-containing fragment obtained by digesting BJC267 (15) with ClaI. Finally, the resulting plasmid with the correct orientation of his3AI was partially digested with ClaI and the downstream ClaI site was filled in with Klenow polymerase and then religated to generate AGE1589. The D211N version of pGTy1mhis3AI (AGE1603) contains both the D211N mutation and a new adjacent BstBI site. Both were engineered by site-directed mutagenesis of AGE1495, using RAG 204 (5′ ATT TAG ATT TTT CGA AAA CAA TAC CAT ATT ATC TAC GAA T 3′) (the new BstBI site underlined). The resulting phagemid (AGE1532) was digested with BstEII and HindIII and ligated to similarly digested AGE565 to generate AGE1598. AGE1598 was digested with XhoI and ClaI, and the 5328-bp Ty1-containing fragment was ligated to the similarly digested AGE1589.
(iv) pGTy1mhis3AIΔKpnI.
pGTy1mhis3AIΔKpnI (AGE1627) was constructed by digesting AGE1589 with KpnI and self-ligating the largest fragment. This procedure eliminates a 2,600-bp internal fragment of the plasmid containing a portion of IN, the entire RT domain and a portion of the his3AI gene.
(v) Second-site suppressor mutations.
Second-site suppressor mutations were introduced into both wild-type (WT) and D211N Ty1 plasmids. First, the 1,413-bp HindIII-BamHI fragment from pJEF724 was ligated to similarly digested pBSIIKS(+) to create AGE1038. Then AGE1038 was mutagenized with oligonucleotides RAG567 (5′ CAG TAA GTA AGC CGC GGA TAA TTG GTT CTT GTT AAG 3′), RAG568 (5′ CTG ATA CTT CAT CTC TGA GAC CCA TTG CCT TTG 3′), and RAG565 (5′ CCT GAT ACT TCA TCA CGA AGT GAC ATT GCC TTT G 3′) to introduce the K463R, R495G, and R495S substitutions, respectively (Ty1-H3 coordinates 5332, 5434, and 5434). In each case, the resulting mutagenized phagemid was digested at a unique BglII site in the Ty1 3′ untranslated region, filled in with Klenow polymerase, and religated to generate a new, unique ClaI site. The resulting phagemids were transformed into a Dam methyltransferase-defective Escherichia coli strain, repurified, and digested with HindIII and ClaI. The 939-bp Ty-containing fragments were ligated to similarly digested D211-containing AGE565 vectors or to similarly digested D211N- containing AGE1598 vectors. Finally, each of the six resulting plasmids was digested with XhoI and ClaI and the 5,328-bp Ty1-containing fragment was ligated to the similarly digested AGE1589 vector to generate AGE1904 (K463R,), AGE1907 (R495G), AGE1906 (R495S), AGE1898 (D211N+K463R), AGE1902 (D211N+R495G), and AGE1901 (D211N+R495S) respectively.
(vi) D468S RNase H mutant plasmids.
The construction of the D468S RNase H mutant plasmids (AGE1160 without his3AI and AGE1455 with his3AI, corresponding to Ty1-H3 position 5347), was similar to the procedure described above but will be presented in detail elsewhere (E. Mules and A. Gabriel, unpublished data).
Qualitative transposition assay.
Patches from single colonies containing various pGTy1mhis3AI plasmids were grown at 30°C on agar plates containing SC medium lacking uracil plus 2% glucose (SC-Ura+GLU), replica plated to SC medium lacking uracil plus 2% galactose (SC-Ura+GAL), incubated at 22°C for 5 days, replica plated to SC medium lacking uracil and histidine plus 2% glucose (SC-Ura-His+GLU), and observed after 3 days at 30°C.
Determination of transposition frequency.
For initial tests of transposition (see Table 1), yeast strain YH50 containing various mutant versions of pX3 was grown at 30°C as patches on SC-Ura+GLU plates, replica plated to SC-Ura+GAL plates, incubated at 22°C for 5 days, then sequentially replica plated at 30°C to SC-Ura+GLU, YPD, and SC+FOA plates. The cells were scraped from the SC+FOA plates into sterile water, and various dilutions were plated on SC+FOA and SC+FOA-Trp and subsequently counted.
TABLE 1.
Properties of engineered Ty1 RT active-site mutants
| Ty1 RT sequencea
|
60-kDa RT protein by Western blotting | Exogenous polymerase activity via
|
Transpositiond | |||
|---|---|---|---|---|---|---|
| D129 | D210 | D211 | Oligo (dG)-poly(rC)b | Activated calf thymus DNAc | ||
| D | D | D | Present | ++ | ++ | Present |
| D | D | E | Present | − | − | Absent |
| D | D | N | Present | ++ | + | Absent |
| D | E | D | Present | − | NDe | Absent |
| D | N | D | Present | − | ND | Absent |
| E | D | D | Present | − | ND | Absent |
| N | D | D | Present | − | ND | Absent |
The RT amino acid sequence numbering is based on the amino terminus of Ty1 RT at position 3946 in Ty1-H3 (52).
++ indicates >40% of WT polymerase incorporation. Aside from multiple WT and D211N VLP preps, no other VLP source gave detectable polymerase activity.
VLPs were mixed with activated calf thymus DNA, all four dNTPs and either [α-32P] dGTP, [α-32P]dATP, or [α-32P]TTP at a 1:100 molar ratio with unlabeled nucleotide. ++ indicates WT polymerase activity, and + indicates >20% of WT polymerase incorporation. Similar results were obtained for each of the three labeled nucleotides.
Transposition frequencies of the TRP1-marked pX3 derived plasmids were determined as the fraction of cells per milliliter growing on SC+FOA−Trp plates compared to the cells grown on SC+FOA plates. Under the conditions described in Materials and Methods, ∼35% of Ura− WT cells were Trp+, compared to < 0.5 % of Ura− cells with any of the mutant versions of Ty1.
ND, not determined.
Determination of transposition rate.
Ten independent cultures of each strain containing various pGTy1mhis3AI plasmids were grown for 2 days in liquid SC-Ura+raffinose medium, diluted to ∼500 cells per ml of SC-Ura+GAL, and incubated at 22°C until saturation. Dilutions were plated on both SC-Ura+GLU and SC-Ura-His+GLU plates. The frequency of transposition was calculated for each culture by dividing the number of colonies per milliliter of culture growing on SC-Ura-His plates by the number of colonies per milliliter growing on SC-Ura plates. The median frequency for each construct was found, and the transposition rate per cell per division was calculated by the method of Drake (23).
Genetic screen for second-site suppressors.
Random mutations were introduced into the D211N version of the Ty1 RT region by PCR amplification (77) of a ∼3.55-kb segment of Ty1mhis3AI, using primers RAG34 (5′ CAT ATC ATT CGT TCA TTG CGT 3′) (Ty1-H3 positions 3041 to 3061) and RAG450 (5′ TAC CAC CCA TAA TGT AAT AGA TC 3′) (Ty1-H3 positions 5562 to 5584) with AGE1603 as the template. The PCR conditions used are available on request. PCR products were mixed with equimolar amounts of KpnI-linearized pGTy1m his3AIΔKpn (AGE1627) and cotransformed into competent yeast strain YH50 using the high-efficiency TRAFO transformation method (R. Agatep, R. D. Kirkpatrick, D. L. Parchaliuk, R. A. Woods, and R. D. Gietz, Technical Tips Online; http://tto.trends.com). Gap-repaired plasmids were selected by growth on SC-Ura+GLU plates. In control experiments, we found a 19:1 ratio of gap repair to end joining. Since AGE1627 lacks the entire RT domain, its recircularization and transposition competence depends on recombination either with the PCR product or with an intact genomic Ty1 element. A total of 8,500 colonies from among those capable of growth on SC-Ura+GLU plates were patched onto fresh SC-Ura+GLU plates, which then served as masters for duplicate replica plating onto SC-Ura+GAL. Replicas were grown at 22°C for 5 days, replica plated to SC-His+GLU, and inspected after 3 days at 30°C. Patches with lawns of growth on SC-His+GLU or inconsistencies between the duplicates were ignored. A total of 101 patches with multiple papillae were further analyzed. In each case a His+ Ura+ colony was purified, the yeast plasmid was shuttled into E. coli (34), and the purified plasmid was digested with MluI and BstBI to screen for candidates that had not recombined with genomic Ty1 elements and had consequently lost these engineered restriction sites. The RT domains of six candidate plasmids which retained these restriction sites were sequenced to confirm the D211N mutation and to identify suppressor mutations. For each identified suppressor mutation, site-directed mutagenesis was used to recreate the same substitution in a native version of plasmid AGE1603 and confirm its ability to suppress the D211N transposition defect.
VLP and nucleic acid preparation.
Ty1 VLPs were partially purified through sequential sucrose gradients and concentrated as previously described (12, 29, 53). Either these were used directly or nucleic acids were extracted by proteinase digestion (50 μg of proteinase K per ml, 25 mM EDTA [pH 8.0], 0.5% sodium dodecyl sulfate SDS) at 50°C for 1 h, followed by phenol-chloroform extraction and ethanol precipitation. The total protein concentration of VLP fractions was determined by the Bradford method.
Exogenous polymerization reactions.
Partially purified VLPs were used to perform exogenous RT reactions, using either poly(rC)-oligo(dG) as the template and primer as previously described (24, 29, 31) or using 10 μg of activated calf thymus DNA (Sigma) per ml as the random primer and template under the same reaction conditions as for homopolymers, along with 10 μM each unlabeled nucleotide and 100 nM labeled nucleotide.
Endogenous polymerization reactions.
VLPs were incubated with all four nucleotides plus [α-32P]dATP at room temperature for 90 min, as previously described (13); D. Voytas, personal communication). Each reaction mixture was divided between two tubes; one tube was incubated with 0.325 N NaOH at 55°C for 1 h and then 0.325 N HCl was added. Nucleic acids from both tubes were then extracted as above, and resuspended reaction products were separated on an 8% denaturing polyacrylamide gel.
Reiterative primer extension analysis.
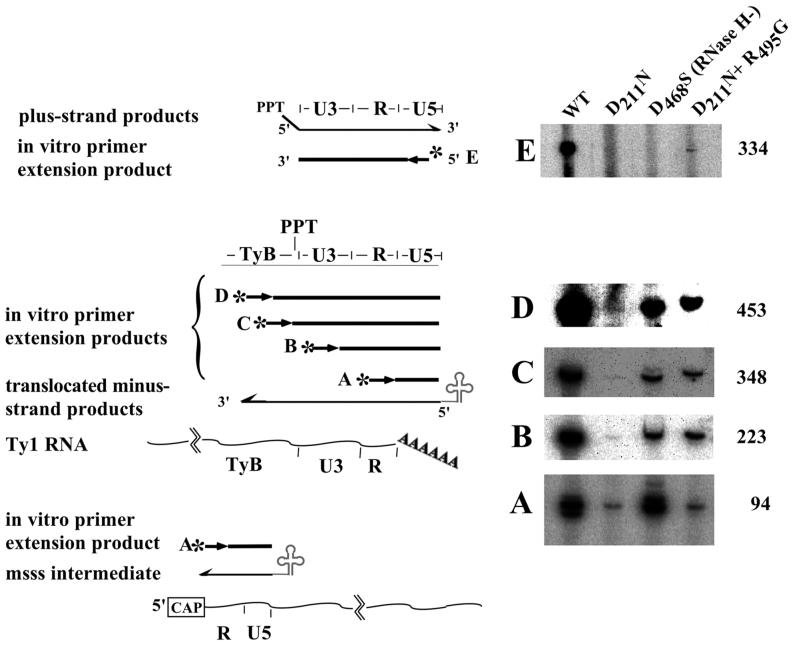
To detect Ty1 replication intermediates, reiterative primer extension was carried out with 5′-end-labeled strand-specific oligonucleotide primers. VLPs (see Fig. 4A) or nucleic acids extracted from VLPs (see Fig. 4B through E) were digested with 0.1 mg of RNase A per ml at 37°C for 10 min and then incubated with labeled primers, using standard PCR buffers in a 20-μl volume, along with 2 U of Taq polymerase, 200 nM labeled primer, and 200 μM each deoxynucleoside triphosphate (dNTP). The PCR program was 30 cycles of 95°C for 30 s, 53°C for 2 min, and 72°C for 3 min. The extension products were separated on an 8% denaturing polyacrylamide gel, dried, and then analyzed by phosphorimaging. Band intensities were quantified using Image-Quant software. The primers used were RAG646 (5′ GGA GAA CTT CTA GTA TAT TCT GTA TAC C 3′) (Ty1-H3 positions 243 to 270 or 5827 to 5854, and primer A in Fig. 4), RAG647 (5′ GTG GAA GCT GAA ACG CAA GG 3′) (Ty1-H3 positions 113 to 132 or 5697 to 5716, and primer B in Fig. 4), RAG743 (5′ CTA TTA ACT AAC AAA TGG ATT CAT TAG 3′) (Ty1-H3 positions 5543 to 5562, and primer C in Fig. 4), RAG266 (5′ CGA GAC CAA GAA GAA CAT TG 3′) (Ty1-H3 positions 5475 to 5494, and primer D in Fig. 4), and RAG322 (5′ AGA ATT GGG TGA ATG TTG AG 3′) (Ty1-H3 positions 332 to 313 or 5915 to 5897, and primer E in Fig. 4).
FIG. 4.
Reiterative primer extension analysis of Ty1 replication intermediates. To determine the presence and abundance of different-sized replication intermediates, 5′-end-labeled primers A through E were annealed directly to VLPs (A) or nucleic acids extracted from VLPs (B to E) and then repeatedly extended with Taq polymerase. Since the absolute amounts of nucleic acids were not comparable in each VLP preparation the relative intensities within each panel cannot be directly compared. Further, the panels represent different exposure times, to accentuate different points. The same amounts of VLP nucleic acids, however, were used in each column, regardless of exposure, so that the relative levels of each cDNA template within a column can be compared.
For direct quantitative comparisons of specific replication intermediates in different VLP preps (see Fig. 4 A and Table 3), the volume of VLPs added to the primer extension analysis mixture was normalized to equal amounts of Ty1 RT protein, based on Western blot analysis. To compare different VLP preparations for the relative amounts of strand-transferred product, a 68-mer oligonucleotide complementary to both primers A and B (RAG822 [5′ TAT GCA ATG CGT CGA GCT CGA GGG TAT ACA GAA TAT ACT AGA AGT TCT CCT TGC GTT TCA GCT TCC AC 3′]) was added to each primer extension reaction mixture at 0.4 nM to serve as an internal control and to normalize results from different reactions.
TABLE 3.
Mutant VLP-associated RT activities relative to WT
| RT source | Exogenous polymerase activity per unit of proteina | Endogenous minus-strand cDNA synthesis per unit of RT proteinb | Relative proportion of minus-strand transfer productc |
|---|---|---|---|
| Ty1 WT | 1.00 | 1.00 | 1.00 |
| Ty1 D211N | 1.00 | 0.06 | 0.19 |
| Ty1 D468S | 1.27 | 1.28 | 0.03 |
| Ty1 D211N+R495G | 0.28 | 0.07 | 0.55 |
Based on poly(rC)-oligo(dG) homopolymer assays, using appropriate volumes of VLPs to give equivalent quantities of Ty1 RT protein (as determined by Western blot analyses) per reaction.
Based on quantitation of the phosphorimager signal from reiterative primer extension shown in Fig. 4A. Direct primer extension of VLPs was performed using appropriate volumes of VLPs to give equivalent quantities of Ty1 RT protein.
Based on quantitation of the phosphorimager signal from reiterative primer extension using purified VLP nucleic acids and primers A and B from Fig. 4. Signal intensities were normalized to a simultaneously extended 68-mer oligonucleotide complementary to primers A and B. The intensity of the normalized signal for primer B compared to primer A was determined, and mutant ratios were compared to WT.
Immunoblot analysis.
Multiple dilutions of VLPs from different sources were denatured and separated through a 10% polyacrylamide–1% SDS denaturing gel, transferred to nitrocellulose membranes, and incubated with polyclonal antiserum TyB8, raised against a fusion protein from the Ty1 RT domain (32), to compare Ty1 RT mutants. Antibody detection was carried out by enhanced chemiluminescence as specified by the manufacturer (Amersham).
RESULTS
An active-site mutant that can polymerize but not transpose.
We generated a series of site-directed mutant versions of the galactose-inducible, plasmid-based Ty1-H3 element, in which each of the three invariant RT active-site aspartates was replaced with either asparagine or glutamate. We transformed these constructs into yeast, induced VLP formation, and collected VLPs for analysis of Ty1 RT function. VLPs made from WT or mutant Ty1 elements all contained similar levels of a ∼60-kDa Ty1 RT protein (Table 1), indicating that these substitutions affect neither the apparent size nor the stability of the protein. We expected the mutants to all be devoid of polymerase activity. However, when each VLP preparation was tested for in vitro polymerase activity, using a standard homopolymer primer-template, we found that the D211N substitution mutant retained near-WT levels of polymerase activity (Table 1; also see Table 3). We confirmed this unexpected result by rescuing the plasmid from yeast back into E. coli and sequencing the RT region to show that the site had not reverted. Further, we recreated the substitution and obtained similar results with VLPs generated from several independent plasmids. This indicated that the D211N Ty1 RT mutant retained significant polymerase activity using an exogenous DNA-RNA homopolymer. Since the homopolymer primer-template is an unnatural substrate, we also examined a random substrate, i.e., activated calf thymus DNA. The WT and D211N mutant enzymes were both able to incorporate dNTPs into this primer-template (Table 1), while the D211E mutant enzyme was not. Thus, only the D211N mutant is capable of exogenous DNA polymerization, utilizing either a DNA or RNA template. Finally, we asked whether a Ty1 element containing an active-site aspartate mutation is still able to transpose. As shown in Table 1, neither the D211N mutant nor any of the other active-site mutants appeared competent for transposition in a standard plasmid-based transposition assay (7).
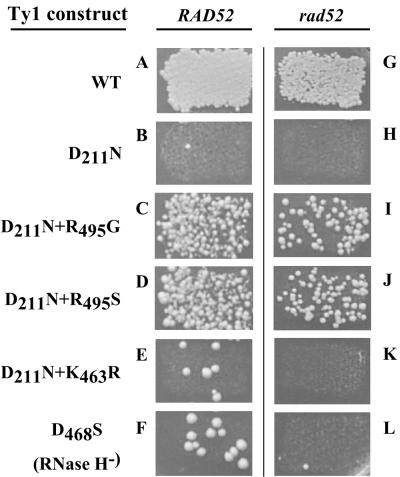
To examine the transposition defect of the D211N mutant in greater detail, we next utilized the “HIS3-artificial intron” (i.e., his3AI) assay for cDNA formation (16). In this assay, an antisense HIS3 gene in the Ty1 3′ untranslated region of a pGALTy1 plasmid contains an artificial intron (AI) inserted in the antisense orientation relative to HIS3. The insertion makes the his3AI gene nonfunctional. The AI can only be spliced out of the Ty1 RNA. If this spliced mRNA is reverse transcribed and the resulting cDNA is inserted back into the genome, by either transposition or recombination, a functional HIS3 gene will be created, resulting in histidine prototrophy. Using this system, expression of a wild-type pGALTy1mhis3AI plasmid leads to high levels of histidine prototrophy (Fig. 1A) while the D211N mutant version is only rarely associated with histidine prototrophy (Fig. 1B). Quantitatively, the frequency of histidine prototrophy for the D211N mutant, in a recombination-proficient strain, is ∼2,000-fold lower than that of WT Ty1 (data not shown). Histidine prototrophy can result from either transposition or cDNA recombination (65). We can distinguish between these two mechanisms by inducing Ty1 expression in a recombination-deficient rad52 yeast strain, since recombination is RAD52 dependent while transposition is RAD52 independent. We found that in a rad52 strain WT Ty1 could still generate histidine prototrophs but the D211N mutant could not (Fig. 1G and H; Table 2). This indicates that the rare prototrophs observed for the D211N mutant in the RAD52 strain result from one or more recombinational process.
FIG. 1.
Qualitative analysis of transposition and recombination using a HIS3 marker gene. Yeast colonies are shown growing on SC−Ura−His+GLU plates. Isogenic RAD52 (YH50) and rad52 (AGY49) yeast strains were transformed with plasmids AGE1589 (A and G), AGE1603 (B and H), AGE1902 (C and I), AGE 1901 (D and J), AGE1898 (E and K), or AGE1455 (F and L) and induced to undergo Ty1 replication before being replica plated to a medium lacking histidine.
TABLE 2.
Determination of the transposition rate for WT and mutant Ty1 elements
| Ty1 constructa | Rate of histidine prototrophy per cell per generation (relative to WT)b |
|---|---|
| WT | 4.77 × 10−3 (1) |
| D211N | NAc |
| D211N + K463R | 4.71 × 10−7 (0.0001) |
| K463R | 1.18 × 10−2 (2.50) |
| D211N + R495G | 2.81 × 10−4 (0.059) |
| R495G | 1.87 × 10−4 (0.039) |
| D211N + R495S | 2.53 × 10−4 (0.053) |
| R495S | 1.47 × 10−3 (0.31) |
Rates were determined in the spt3 rad52 strain AGY49
All determined rates were different from one another at the 5% significance level, using the Mann-Whitney rank sum test, except D211N + R495G versus R495G, D211N + R495G versus D211N + R495S, and R495G versus D211N + R495S.
NA, not applicable. See text for details.
To examine the basis for the rare histidine prototrophs in RAD52 strains carrying the D211N mutant Ty1 element, we isolated 54 individual His+ colonies and rescued the Ty1-containing plasmid back into E. coli. By restriction digestion analysis of the recovered plasmids, we found that 31 (55%) had lost the two restriction sites (MluI and BstBI) that had been engineered into the mutant plasmid to distinguish it from endogenous Ty1 elements (Fig. 2A and data not shown). The loss of these sites most probably represents gene conversion of the plasmid Ty1 by a genomic Ty1 element, resulting in replacement of the mutant asparagine codon with the WT aspartate codon and restoration of WT RT function (51). The remaining 23 plasmids (45%) retained the distinguishing restriction sites and presumably the D211N mutation. In these cells, the mutant enzyme probably carried out sufficient reverse transcription to generate cDNA, which could recombine with homologous plasmid or chromosomal sequences (18, 19). To test this, we replica plated His+ colonies to SC+FOA−His plates. On this medium, absence of growth implies that histidine prototrophy is linked to the URA3-containing plasmid, i.e., the HIS3 cDNA recombined with the his3AI segment on the plasmid. We found this linkage for 16 of the 23 events, while HIS3 was chromosomal in only 7 of 23 events. Thus, despite its near-normal levels of exogenous polymerase activity, the D211N mutant is incapable of transposition and even its ability to generate HIS3 cDNAs, which can function in homologous recombination, is greatly reduced.
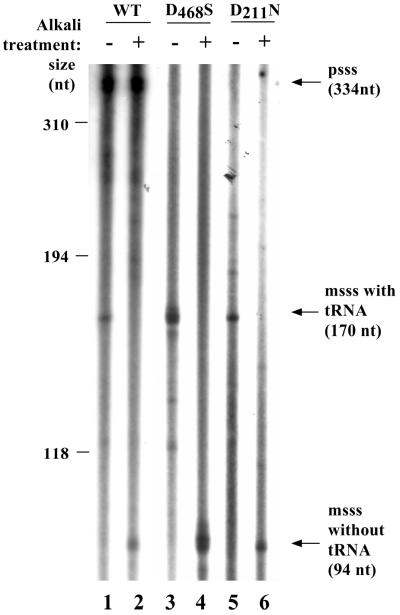
FIG. 2.
Observation of psss and msss intermediates. Endogenously labeled replication intermediates extracted from VLPs derived from WT, the D468S RNase H active-site mutant, or the D211N polymerase mutant versions of Ty1 were either run directly on a denaturing polyacrylamide gel (lanes 1, 3, and 5) or pretreated with alkali to hydrolyze RNA. The expected sizes of the msss intermediate, with and without attached tRNA, and the psss intermediate are shown to the right of the lanes.
The D211N mutant has endogenous polymerase activity.
Since the D211N mutant can polymerize using an exogenous template but cannot transpose, we next tested endogenous polymerase activity by examining labeled intermediates of Ty1 replication. We performed endogenous RT reactions, incubating VLPs derived from WT or D211N mutant Ty1 elements with [α-32P]dATP in the presence of all four nucleotides, then extracted nucleic acids from the VLPs and separated them on polyacrylamide gels. As a control, we also tested VLPs from a Ty1 element that contained a site-directed mutation in one of the presumed active-site aspartates within the RNase H domain (D468S). As shown autoradiographically, a discrete labeled product was observed at ∼170 nucleotides (nt) for WT, the D468S RNase H mutant, and the D211N mutant (Fig. 2, lanes 1, 3, and 5). This is the size expected for the msss intermediate (13). To confirm this supposition, we treated nucleic acids derived from endogenously labeled VLPs with alkali to hydrolyze the RNA components of the labeled products. In each case the ∼170-nt product disappeared and an ∼94-nt product appeared, as would be predicted if the IMT had been degraded, leaving only the labeled cDNA portion (lanes 2, 4, and 6). The D211N mutant RT is therefore capable of endogenous polymerization. It should be noted, however, that the D211N mutant RT generates a much smaller amount of endogenous product than does the WT or the D468S mutant per unit of VLP (Table 3). The three pairs of lanes shown in Fig. 2 represent different exposures and are not directly comparable.
In the WT lanes, a prominent alkali stable band is present at ∼334 nt, the size expected for the psss intermediate (Fig. 2, lanes 1 and 2). Similar-size VLP-associated nucleic acids have been previously noted and attributed to psss (54, 61). The band is absent from in vivo nucleic acids derived from the RNase H mutant VLPs (lanes 3 and 4). This is expected, since RNase H cleavage generates the PPT, which is required to prime plus-strand synthesis. The absence of this band in nucleic acids derived from the D211N mutant (lanes 5 and 6) is not obviously explained but does indicate that this mutant is incapable of carrying out some replication step beyond msss synthesis, that is required for subsequent psss synthesis.
Generation of intragenic second-site suppressors of the D211N transposition defect.
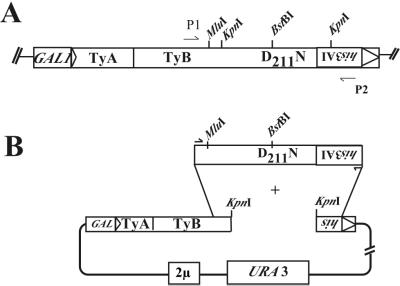
The finding that the D211N mutant is incompetent for transposition suggests that this subtle mutation creates one or more crucial defects in the transposition process that are unrelated to its basic ability to polymerize. To gain insights into the nature of these potential defects, we used the his3AI marker system to carry out a genetic screen for intragenic second-site suppressor mutations that would allow the D211N RT mutant to overcome its transposition defect. As shown in Fig. 3A, we PCR amplified a 3,548-bp portion of the Ty1mhis3AI plasmid, using the D211N mutant version as the PCR template, to randomly generate one or two mutations per PCR product. The PCR fragments were cotransformed into a recombination-proficient yeast strain along with the linearized URA3 marked plasmid pGTymhis3AIΔKpnI (Fig. 3B). This plasmid lacks the entire RT domain and a portion of the his3AI marker but retains ∼500 bp of flanking homology to the PCR product. By selecting for uracil prototrophy after transformation, we enriched the population for plasmids which have been “gap repaired” by recombination with the PCR product. Colonies that could grow on glucose plates lacking uracil were patched and induced to express Ty1 by replica plating to SC−Ura+GAL. Patches were subsequently replica plated onto glucose medium lacking histidine to select for cells which had become histidine prototrophs, signifying that they had regained the ability to synthesize cDNAs (see Materials and Methods for further experimental details).
FIG. 3.
Important landmarks on the modified Ty1 element and scheme for generating intragenic second-site suppressors via gap repair. (A) Scale drawing of the his3AI marked Ty1-H3 element, showing the position of the D211N mutation, and positions of restriction sites (MluI and BstBI) engineered into the construct to distinguish it from endogenous Ty1 elements or used to create the gapped plasmid (KpnI). P1 (RAG34) and P2 (RAG450) refer to the plus- and minus-strand primers, respectively, used to generate a randomly mutagenized 3,548-bp PCR product spanning the carboxy-terminal half of the TyB ORF. (B) Plasmid AGE1627 was linearized with KpnI and cotransformed with the randomly mutagenized PCR fragment into the recombination-proficient yeast strain YH50 to repair the gap and generate a circular plasmid.
By screening 8,500 cotransformants, we obtained 6 candidates with elevated levels of histidine prototrophy in the presence of the D211N mutation. We sequenced the entire RT domain of each of these candidates and observed three classes of suppressor mutations. The first class, obtained independently four times, consisted of a R495G substitution (Fig. 1C). The second class, obtained once, consisted of a K463R substitution (Fig. 1E). Finally, a plasmid with two substitutions, A36V and R495S, was obtained once.
For each mutation, we used site-directed mutagenesis to recreate the substitution in fresh plasmids that contained either D or N at RT position 211. In this way we confirmed the second-site suppressor status of the first two mutations and showed that the A36V mutation was incidental, since suppression required only the R495S substitution (Fig. 1D). Therefore, two different substitutions of the same R495 residue can suppress the transposition defect. Both R495 and K463 are located in the region that would be expected to be within the Ty1 RNase H domain. Our attempt to model either R495 or K463 within the context of the HIV-1 RNase H domain were unrevealing, however, since both of these residues are present in nonconserved locations of the RNase H sequence (A. Gabriel and S. Sarafianos, unpublished observation).
We tested each recreated mutant version of Ty1 for histidine prototrophy in an isogenic rad52 strain to determine whether the second-site suppressors actually restored transposition or, alternatively, just restored the ability to synthesize cDNAs which could recombine into the genome. As shown in Fig. 1G through L, histidine prototroph formation for WT Ty1, as well as the D211N+R495S and the D211N+R495G double mutants, was independent of RAD52, consistent with bona fide transposition. However, histidine prototrophy was RAD52 dependent for D211N, D211N+K463R, and the RNase H mutant, consistent with cDNA recombination.
To quantitatively compare the various Ty1 mutants, we next determined the rate of transposition for each of the second-site suppressors, in the absence or presence of the D211N mutation, and compared this to the rate of WT transposition under the same conditions (Table 2). We used a rad52 strain so that only transposition events would be observed. In this strain, the frequency of histidine prototrophy for the D211N mutant is so low (i.e., 40-fold lower than for the D211N+K463R double mutant) that its transposition rate could not be directly compared with that of the other strains. As shown in Table 2, both the D211N+R495G and D211N+R495S constructs restored transposition to ∼5% of WT levels while the D211N+K463R construct resulted in only 0.01% of WT levels of transposition, in agreement with our qualitative results (Fig. 1G to L). In the context of WT D211, the R495S and R495G mutations showed mild to moderate defects in transposition (31 and 3.9% of WT) whereas the K463R substitution showed a slight (2.5-fold) but significant enhancement of transposition. These results suggest that mutations in R495 or K463 affect different steps in the Ty1 replication process.
The active-site mutant generates reduced levels of smaller endogenous replication intermediates.
The endogenous polymerization assay (Fig. 2) suggested that the D211N mutant RT generates reduced levels of the msss intermediate. To quantify this observation and to determine the presence and relative abundance of other expected replication intermediates in WT and mutant versions of Ty1 VLPs, we carried out a reiterative primer extension analysis (46). As cartooned in Fig. 4, labeled strand-specific primers will anneal to VLP-derived nucleic acids and be subsequently extended only if particular intermediates are present. The intensity of the resulting primer extension signal is proportionate to the abundance of the intermediates. Because the 5′ ends of minus-strand and plus-strand cDNA intermediates are fixed by the IMT and PPT respectively, we can identify discrete labeled extension products distinct from potentially contaminating Ty1 genomic or plasmid DNA that might also anneal to the primers.
We carried out primer extension reactions using WT −, D211N −, D468S−, and D211N+R495G-derived VLPs (Fig. 4). In addition to the plus-strand primer in the R region (Fig. 4A), which will detect all minus-strand cDNAs, we used a plus-strand primer corresponding to the 5′ end of U3, which will anneal and extend only if minus-strand transfer has occurred (Fig. 4B). We used two additional plus-strand primers 27 and 110 bases upstream of the PPT/U3 border (Fig. 4C and D), to determine if longer minus-strand products are synthesized. Furthermore, we used a minus-strand primer corresponding to the 3′ end of U5 to detect the presence of plus-strand products (Fig. 4E). As shown in Fig. 4, the different mutants showed distinct patterns of primer extension products. While high levels of all extension products were detected with WT VLPs, levels of extension products for the D211N mutant were low throughout and progressively diminished from those in panels A through D. This suggests that fewer and shorter in vivo cDNAs are made within D211N VLPs. For the RNase H mutant, the absolute level of msss product was similar to that of the WT, but the proportion of translocated products was only ∼3% of that of the WT (Table 3). The absolute level of minus-strand products was low for the D211N+R495G mutant, but the translocated products were long, similar to WT and the RNase H mutant. With the double mutant, we could also detect a low level of plus-strand intermediates (Fig. 4E).
To quantify the relative amount of minus-strand DNA derived from different mutant RTs, we carried out reiterative primer extension reactions using volumes of VLPs previously determined by Western blot analyses to contain equivalent amounts of RT protein. When normalized in this way, the amount of exogenous polymerase activity was equivalent for WT and D211N VLPs (Table 3), but the amount of minus-strand cDNA present within the D211N VLPs was ∼20-fold smaller than for either WT or the RNase H mutant. Interestingly, in the presence of the transposition-restoring R495G suppressor mutation, the relative amount of minus-strand cDNAs does not dramatically change. These results do not correlate with the in vitro polymerase activities, indicating that the D211N mutant has an in vivo defect in synthesizing minus-strand cDNA. However, this is not the crucial transposition defect for the mutant enzyme, since the transposition-competent double mutant has similarly low levels of minus-strand cDNA.
The observed patterns of intermediates provide clues to the basis for the D211N transposition defect. For the RNase H mutant, the small proportion of translocated products was expected, since RNase H activity has been shown in retroviral systems to be required for strand transfer (14). For the D211N and double mutant, however, the proportions of strand-transferred products were intermediate between those for the RNase H mutant and WT, i.e., ∼19 and 55% relative to WT. This implies that the D211N mutant enzyme retains nonspecific RNase H activity. While we could not accurately quantify the relative levels of long D211N-derived minus-strand products because of the low signal-to-noise ratio (Fig. 4C and D), the progressive decrease of the primer extension signal in these lanes suggests the possibility that the mutant enzyme has a defect in processive synthesis along its natural RNA template. Finally, the absence of plus-strand intermediates in the D211N VLPs, even though some minus-strand synthesis does proceed past the PPT (Fig. 4C and D), is noteworthy. The ability of the double mutant to transpose is correlated with the reappearance of plus-strand products (Fig. 4 panel E). From this we infer that one crucial (and suppressible) defect in the D211N mutant is an inability to generate and/or extend the PPT primer that is required for plus-strand synthesis.
DISCUSSION
In this paper we demonstrate that the second aspartate of the canonical YXDD box is not essential for Ty1 RT to carry out polymerization, a property which distinguishes this enzyme from HIV-1 RT. Given the universal occurrence of adjacent aspartates in biochemically confirmed RTs, it has generally been assumed that these residues, along with one additional invariant aspartate, are critical for catalysis (17, 57). Our work indicates that this is not entirely true. Replacement of Ty1 D211 with an asparagine side chain leads to comparable levels of in vitro polymerase activity, using two different primer-template substrates (Tables 1 and 3), as well as reduced but measurable in vivo polymerase activity (Fig. 2 and 4; Table 3). The loss of transposition, however, and our observations that a D211E mutation does eliminate polymerase activity, indicate that while not critical for catalysis, this residue is intimately involved in the function of the enzyme. Other than for HIV-1, equivalent site-directed mutations of active-site aspartates have not previously been reported for any other RT source. Given our finding, it will be important to experimentally verify whether this aspartate is catalytically nonessential for other retrovirus, retrotransposon, and retroelement RTs.
The alignment of aspartate or glutamate residues in the primary sequences of all RNA- and DNA-dependent polymerases suggests a universal role for negatively charged carboxylates in polymerization (17). The widely accepted “two-metal-ion mechanism” for catalysis of polymerization by phosphoryl transfer proposes that divalent metal ions function to position both the primer terminus and the incoming nucleotide for nucleophilic attack, as well as to stabilize the transition state (2). As determined in the crystal structures of multiple polymerases, carboxylate side chains contribute to coordinating the metal ions in the active site (22, 33, 35, 48, 58). However, structural comparison of the active sites from four different ternary complexes of DNA polymerases with bound primer-template and nucleotide substrates (HIV-1 RT, T7 DNA polymerase, Klentaq DNA polymerase, and DNA polymerase β), demonstrates that only two of the three conserved carboxylate residues are superimposable in all four enzymes. These are the two residues which directly coordinate metal ions (reviewed in reference 63). The positioning of the second aspartate in the YXDD box of RTs, or its equivalent residue in other polymerases, is more variable. Only in the most distantly related polymerase β are all three carboxylate residues clearly seen directly coordinating metals in the ternary complex (58). The fact that only two aspartates are structurally conserved in polymerase active sites has led Steitz to conclude that only two of the three aspartates are actual metal binding residues (68).
Further evidence for the flexibility of the third active-site carboxylate in polymerases comes from phylogenetic and site-directed mutagenesis studies of polymerases other than RTs. In sequence alignments of DNA-directed RNA polymerases, no carboxylates equivalent to the second aspartate in motif C are present (17). Several functional polymerases from minus-strand RNA viruses, including Sendai virus, measles virus, rabies virus, and vesicular stomatitis virus (VSV), encode DN rather than DD within their motifs C (reviewed in reference 60). Substitution of the VSV polymerase DN with DD results in an enzyme with reduced but observable polymerase activity (67). For the equivalent “CGDD” box in hepatitis C virus, site-directed mutation to either CGDN or CGDE results in enzymes with ∼8% of the WT in vitro polymerase activity (50). Thus, for a retrotransposon RT, as well as several distantly related viral RNA-dependent RNA polymerases, the second D in motif C allows limited side chain replacement. Given the belief that RTs are derived from RNA-dependent RNA polymerases, these results suggest that the active-site geometry of retrotransposon polymerases share a flexibility with this class of viral enzymes that may have been lost in the HIV-1 polymerase.
An alternative interpretation of the polymerase activity associated with the D211N mutant form of Ty1 RT is that this residue is particularly prone to hydrolytic deamidation, thereby recreating the wild-type enzyme. Slow spontaneous asparagine deamidation does occur in many proteins as a function of aging, e.g., lens crystallins (3). More interestingly, rapid reversion of a mutant active-site asparagine to WT aspartate has been observed for two dehalogenases, possibly catalyzed by the presence of the reaction product (62, 75). However, it is unlikely that this can explain our data. In particular, the specific activities of the mutant and WT enzymes are similar (Table 3), implying that most of the mutant protein would have to be reverted to the WT form. In that case, the mutant Ty1 element should be competent for transposition. Instead, the mutant is completely dead for transposition, consistent with the mutated side chain being present in the enzyme, blocking one or more step in the transposition process. The presence of different second-site suppressor mutants, which have varied effects on restoring cDNA recombination or transposition, implies complex functional interactions between the active-site mutation and other domains within the enzyme, rather than an effect on deamidation of asparagine to aspartate. Further, D211N VLPs show a distinctive pattern of replication intermediates (Fig. 4), as expected for an enzyme with multiple defects in the polymerization process, rather than simply a reduced level of WT enzyme. Thus, while definitive proof of deamidation will require direct sequence analysis of a purified form of the mutant Ty1 RT, such an explanation is inconsistent with our combined biochemical and genetic data.
The D211N mutant enzyme cannot direct transposition, despite the observed in vitro and in vivo polymerase activity. Our finding that both of the intragenic second-site suppressor mutations map to the RNase H domain raises the possibility that either the original mutation in the polymerase active site causes a defect in RNase H function or the substitutions in the RNase H domain partially restore some defect in polymerase function. A recent study suggests that the Ty1 RT and RNase H active sites are separated by a 14-nt RNA-DNA heteroduplex (74). Work with retroviral systems has shown that the polymerase and RNase H domains of RTs are structurally distinct but interrelated entities. Active isolated RNase H domains can be expressed (26, 64), and pairs of defective MuLV RTs, with active-site mutations in polymerase and RNase H, are capable of in vivo complementation (70). On the other hand, studies have identified mutations in one domain that affect only the activity of the other domain (10, 30, 56, 69). These results have led to the intriguing idea that mutations in one domain can exert their effects over long distances by altering the positioning of the nucleic acid duplex between the active sites, which consequently affect the precise orientation of substrate at the active sites (10, 11, 30).
We have no direct assay to measure Ty1 VLP-associated RNase H nuclease activity, so we can make only indirect inferences about its function in the mutant enzyme. The D211N mutant does not behave like the D468S RNase H active-site mutant during strand transfer, suggesting that nonspecific RNA hydrolysis activity is present in the D211N mutant. The essential replication defect of the D211N mutant could, however, be an indirect consequence of positional changes of the RNA-DNA hybrid on more subtle aspects of RNase H function. For example, the D211N mutant is incapable of plus-strand synthesis, but this defect can be partially suppressed by a second mutation in the RNase H domain at R495. Initiation of plus-strand synthesis requires several specific events. First, minus-strand cDNA synthesis must proceed past the PPT, to generate an RNA-DNA hybrid of the region. Second, RNase H must recognize the PPT and make specific cleavages upstream and downstream of this sequence to generate the PPT primer. Finally, the polymerase active site must specifically bind the PPT primer and minus-strand cDNA template, to initiate plus-strand synthesis. Our data on D211N replication intermediates demonstrate a low level of minus-strand extension beyond the PPT. Therefore we propose that the D211N mutation interferes either with RNase H correctly recognizing and cleaving the PPT substrate or with the polymerase recognizing and extending the PPT primer once it has been generated. According to this scenario, the R495 suppressor mutation causes a reorientation of the nucleic acid substrates that either allows PPT to be generated or partially restores the primer recognition or utilization function of the polymerase.
An additional D211N defect is its significantly lower level of msss synthesis, which is not corrected by the R495G suppressor mutation. The reduced level of msss is not due simply to reduced processive synthesis, since reiterative primer extension using a plus-strand primer in U5, much closer to the IMT primer, gave the same result as in Fig. 4A (data not shown). Further, the D211N+R495G double mutant appears capable of synthesizing long minus-strand products but still has reduced levels of msss. While we can only speculate about the nature of this minus-strand defect, plausible explanations include decreased RT-mediated packaging of the tRNA primer (59), decreased formation of the initiation complex consisting of a tRNA primer annealed to the RNA template in the enzyme active site (1, 28), and even a defect in the chemistry of polymerization initiation, distinct from elongation, which specifically involves interactions between the enzyme and the tRNA primer (37, 43). Distinguishing these possibilities is amenable to existing experimental approaches and is an area of future interest.
What is the mechanistic significance of the functional aspartate substitution in the Ty1 RT polymerase active site? Our findings suggest that the Ty1 D211 side chain is positioned within the active site but that the negatively charged carboxylate of this residue is not essential for the phosphoryl transfer reaction. If D211 does retain a catalytic role, then the retrotransposon enzyme must be able to compensate for the side chain alteration. It is possible that the asparagine can still stabilize the transition state, position the primer end, or even coordinate a metal ion indirectly through an intervening water molecule. Such an enzyme would probably have significant alterations in its kinetic properties compared to WT. Alternatively, the second D in the YXDD box of Ty1 RT may simply play an important structural role without any direct catalytic function. Using our current biochemical assays for VLP-associated polymerase activity, it is impractical to carry out the thorough enzymological studies to distinguish these possibilities. However, a recombinant form of Ty1 RT has recently been reported (74). Future use of this recombinant enzyme should allow us to compare the detailed enzymatic properties of the WT and mutant Ty1 enzymes. Ultimately, understanding the basis for our observations will require analysis of the crystal structure of retrotransposon RTs. With the availability of an active recombinant Ty1 enzyme, this goal may now be feasible.
ACKNOWLEDGMENTS
This work was funded in part by the Charles and Johanna Busch Endowment, the Lucille P. Markey Charitable Trust, and National Institutes of Health grants AI00803 and AI39201.
We thank D. Garfinkel for providing polyclonal antisera TyB8; J. Curcio for providing plasmids; D. Voytas for sharing unpublished protocols; A. Gill, E. Mules, R. Robichaud, and Z. Liu for giving technical assistance; E. Arnold, S. Brill, M. Gartenberg, M. Georgiadis, S. Goff, S. Sarafianos, N. Uzun, and members of the Gabriel laboratory for helpful discussions; and R. Steward, M. Georgiadis, and S. Sarafianos for manuscript review. Special thanks go to Jef Boeke, in whose laboratory this work was initiated.
Footnotes
Dedicated to the memory of Esther M. Gabriel.
REFERENCES
-
1.Arts E J, Ghosh M, Jacques P S, Ehresmann B, Le Grice S F. Restoration of tRNA
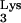 -primed (−)-strand DNA synthesis to an HIV-1 reverse transcriptase mutant with extended tRNAs. Implications for retroviral replication. J Biol Chem. 1996;271:9054–9061. doi: 10.1074/jbc.271.15.9054. [DOI] [PubMed] [Google Scholar]
-primed (−)-strand DNA synthesis to an HIV-1 reverse transcriptase mutant with extended tRNAs. Implications for retroviral replication. J Biol Chem. 1996;271:9054–9061. doi: 10.1074/jbc.271.15.9054. [DOI] [PubMed] [Google Scholar] - 2.Beese L S, Steitz T A. Structural basis for 3′-5′ exonuclease activity of Escherichia coli DNA polymerase 1: a two-metal-ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloemendal H, Berns A J, van der Ouderaa F, de Jong W W. Evidence for a “non-genetic” origin of the A1 chains of alpha-crystallin. Exp Eye Res. 1972;14:80–81. doi: 10.1016/0014-4835(72)90147-9. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Garfinkel D J, Styles C A, Fink G R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine 5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, Styles C A, Fink G R. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol Cell Biol. 1986;6:3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, Xu H, Fink G R. A general method for the chromosomal amplification of genes in yeast. Science. 1988;239:280–282. doi: 10.1126/science.2827308. [DOI] [PubMed] [Google Scholar]
- 8.Boer P H, Gray M W. Genes encoding a subunit of respiratory NADH dehydrogenase (ND1) and a reverse transcriptase-like protein (RTL) are linked to ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. EMBO J. 1988;7:3501–3508. doi: 10.1002/j.1460-2075.1988.tb03226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowen N J, McDonald J F. Genomic analysis of Caenorhabditis elegans reveals ancient families of retroviral-like elements. Genome Res. 1999;9:924–935. doi: 10.1101/gr.9.10.924. [DOI] [PubMed] [Google Scholar]
- 10.Boyer P L, Ferris A L, Clark P, Whitmer J, Frank P, Tantillo C, Arnold E, Hughes S H. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 (HIV-1) reverse transcriptase. J Mol Biol. 1994;243:472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- 11.Boyer P L, Ferris A L, Hughes S H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992;66:1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braiterman L T, Monokian G M, Eichinger D J, Merbs S L, Gabriel A, Boeke J D. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene. 1994;139:19–26. doi: 10.1016/0378-1119(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 13.Chapman K B, Byström A S, Boeke J D. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci USA. 1992;89:3236–3240. doi: 10.1073/pnas.89.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 15.Curcio M J, Garfinkel D J. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics. 1994;136:1245–1259. doi: 10.1093/genetics/136.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curcio M J, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 18.Derr L K, Strathern J N. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- 19.Derr L K, Strathern J N, Garfinkel D J. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- 20.Dombroski B A, Feng Q, Mathias S L, Sassaman D M, Scott A F, Kazazian H H, Boeke J D. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4485–4492. doi: 10.1128/mcb.14.7.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–29. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 22.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 23.Drake J W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eichinger D J, Boeke J D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 25.Eickbush T H. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 26.Evans D B, Brawn K, Deibel M R, Jr, Tarpley W G, Sharma S K. A recombinant ribonuclease H domain of HIV-1 reverse transcriptase that is enzymatically active. J Biol Chem. 1991;266:20583–20585. [PubMed] [Google Scholar]
- 27.Felder H, Herzceg A, de Chastonay Y, Aeby P, Tobler H, Muller F. Tas, a retrotransposon from the parasitic nematode Ascaris lumbricoides. Gene. 1994;149:219–225. doi: 10.1016/0378-1119(94)90153-8. [DOI] [PubMed] [Google Scholar]
-
28.Friant S, Heyman T, Bystrom A S, Wilhelm M, Wilhelm F X. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA
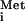 primer are required for initiation of reverse transcription in vivo. Mol Cell Biol. 1998;18:799–806. doi: 10.1128/mcb.18.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
primer are required for initiation of reverse transcription in vivo. Mol Cell Biol. 1998;18:799–806. doi: 10.1128/mcb.18.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar] - 29.Gabriel A, Boeke J D. Reverse transcriptase encoded by a retrotransposon from the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci USA. 1991;88:9794–9798. doi: 10.1073/pnas.88.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao H Q, Boyer P L, Arnold E, Hughes S H. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 31.Garfinkel D J, Boeke J D, Fink G R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 32.Garfinkel D J, Hedge A, Youngren S D, Copeland T D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgiadis M M, Jensen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 35.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 36.Inouye M, Inouye S. Retroelements in bacteria. Trends Biochem Sci. 1991;16:18–21. doi: 10.1016/0968-0004(91)90010-s. [DOI] [PubMed] [Google Scholar]
-
37.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA
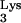 EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar] - 38.Jacobo-Molina A, Ding J, Nanni R G, Clark A D J, Lu X, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaushik N, Rege N, Yadav P N, Sarafianos S G, Modak M J, Pandey V N. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry. 1996;35:11536–11546. doi: 10.1021/bi960364x. [DOI] [PubMed] [Google Scholar]
- 40.Kim J M, Vanguri S, Boeke J D, Gabriel A, Voytas D F. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 41.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 42.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanchy J M, Keith G, Le Grice S F, Ehresmann B, Ehresmann C, Marquet R. Contacts between reverse transcriptase and the primer strand govern the transition from initiation to elongation of HIV-1 reverse transcription. J Biol Chem. 1998;273:24425–24432. doi: 10.1074/jbc.273.38.24425. [DOI] [PubMed] [Google Scholar]
- 44.Larder B A, Kemp S D, Purifoy D J M. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci USA. 1989;86:4803–4807. doi: 10.1073/pnas.86.13.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larder B A, Purifoy D J, Powell K L, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 46.Lauermann V, Boeke J D. Plus strand strong-stop DNA transfer in yeast Ty retrotransposons. EMBO J. 1997;16:6603–6612. doi: 10.1093/emboj/16.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Grice S F J, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lingner J, Hughes T R, Shevchenko M M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 50.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melamed C, Nevo Y, Kupiec M. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1613–1620. doi: 10.1128/mcb.12.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore S P, Garfinkel D J. Expression and partial purification of enzymatically active recombinant Ty1 integrase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:1843–1847. doi: 10.1073/pnas.91.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mules E H, Uzun O, Gabriel A. Replication errors during in vivo Ty1 transposition are linked to heterogeneous RNase H cleavage sites. Mol Cell Biol. 1998;18:1094–1104. doi: 10.1128/mcb.18.2.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Müller F, Laufer W, Pott U, Ciriacy M. Characterization of products of Ty1-mediated reverse transcription in Saccharomyces cerevisiae. Mol Gen Genet. 1991;226:145–153. doi: 10.1007/BF00273598. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura T M, Cech T R. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 56.Palaniappan C, Wisniewski M, Jacques P S, LeGrice S F, Fay P J, Bambara R A. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J Biol Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 57.Patel P H, Jacobo-Molina A, Ding J, Tantillo C, Clark A D, Raag R, Nanni R G, Hughes S H, Arnold E. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995;34:5351–5365. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 58.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 59.Peters G G, Hu J. Reverse transcriptase as the major determinant for selective packaging of tRNAs into avian sarcoma virus particles. J Virol. 1980;36:692–670. doi: 10.1128/jvi.36.3.692-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pochart P, Agoutin B, Rousset S, Chanet R, Doroszkiewicz V, Heyman T. Biochemical and electron microscope analyses of the DNA reverse transcripts present in the virus-like particles of the yeast transposon Ty1. Identification of a second origin of Ty1 DNA plus strand synthesis. Nucleic Acids Res. 1993;21:3513–3520. doi: 10.1093/nar/21.15.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pries F, Kingma J, Janssen D B. Activation of an Asp-124→Asn mutant of haloalkane dehalogenase by hydrolytic deamidation of asparagine. FEBS Lett. 1995;358:171–174. doi: 10.1016/0014-5793(94)01420-6. [DOI] [PubMed] [Google Scholar]
- 63.Sarafianos S G, Das K, Ding J, Boyer P L, Hughes S H, Arnold E. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem Biol. 1999;6:R137–R146. doi: 10.1016/s1074-5521(99)80071-4. [DOI] [PubMed] [Google Scholar]
- 64.Schultz S J, Champoux J J. RNase H domain of Moloney murine leukemia virus reverse transcriptase retains activity but requires the polymerase domain for specificity. J Virol. 1996;70:8630–8638. doi: 10.1128/jvi.70.12.8630-8638.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 67.Sleat D E, Banerjee A K. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steitz T A. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 69.Telesnitsky A, Goff S P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Telesnitsky A, Goff S P. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 1993;12:4433–4438. doi: 10.1002/j.1460-2075.1993.tb06128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teng S-C, Kim B, Gabriel A. Retrotransposon reverse transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 72.Toh H, Hayashida H, Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. Nature. 1983;305:827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Lambowitz A M. The Mauriceville plasmid reverse transcriptase can initiate cDNA synthesis de novo and may be related to reverse transcriptase and DNA polymerase progenitor. Cell. 1993;75:1071–1081. doi: 10.1016/0092-8674(93)90317-j. [DOI] [PubMed] [Google Scholar]
- 74.Wilhelm M, Boutabout M, Wilhelm F-X. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem J. 2000;348:337–342. [PMC free article] [PubMed] [Google Scholar]
- 75.Xiang H, Dong J, Carey P R, Dunaway-Mariano D. Product catalyzes the deamidation of D145N dehalogenase to produce the wild-type enzyme. Biochemistry. 1999;38:4207–4213. doi: 10.1021/bi982670b. [DOI] [PubMed] [Google Scholar]
- 76.Xiong Y, Eickbush T H. Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Y, Zhang X, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimmerly S, Guo H, Perlman P S, Lambowitz A M. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:545–554. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]