Abstract
The K8 gene of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) shares many functional similarities with the BZLF1 gene of Epstein-Barr virus. The protein products of K8 and BZLF1, K-bZIP (RAP, K8) and Zta (BZLF1, ZEBRA, Z) have both been proposed to be members of the bZIP family of transcription factors, forming multimers via a coiled-coil motif termed a leucine zipper. Substantial evidence supporting this model for Zta is published. Here, we demonstrate that the proposed leucine zipper region of K-bZIP (amino acids 182 to 218) is required for multimer formation but that it does not fold as a coiled coil.
Kaposi's sarcoma-associated herpesvirus (KSHV) (or human herpesvirus 8 [HHV8]) is implicated in the development of three potentially fatal human diseases (reviewed in references 6, 7, 11, 20, 30, 37, 38, and 45). It is a member of the human gammaherpesvirus family, and its genome displays sequence homology with the prototypical member of the family, Epstein-Barr virus (EBV) (34). The K8 gene of KSHV encodes K-bZIP (RAP, K8α) which is involved in viral lytic replication (5, 25, 48). K-bZIP interacts with replication structures (4, 47) and with three individual cellular proteins, p53 (31), CBP (19), and C/EBPα (51), and it regulates the expression of the cellular gene encoding p21CIP1 (51). Thus, K-bZIP is a candidate to mediate two important events: viral replication and cell cycle arrest (41).
K8 appears to be a homologue of the EBV gene BZLF1 (14, 26, 32). The protein encoded by BZLF1, Zta (BZLF1, ZEBRA, or Z), plays a key role in the EBV replicative cycle (reviewed in references 12, 29, 39, 41, and 43). Zta also shares with K-bZIP the ability to interact with its origin of lytic replication (35, 36), to promote cell cycle arrest (13), and to interact with the cellular proteins p53, CBP, and C/EBPα (1, 28, 41, 50, 52, 53). Given the functional similarities between these two proteins, it is tempting to speculate that a similar molecular mechanism is exploited by each virus to achieve the same ends.
The overall homology between K-bZIP and Zta is low but rises to 30% similarity (18% identity) within the C-terminal half. This region contains the well-characterized bZIP domain of Zta, which directs the formation of homodimers through the leucine zipper motif and interacts with DNA through the adjacent basic region (15, 16, 29, 39, 41-43). Interestingly, both K-bZIP and Zta form homo-multimers involving the C-terminal half of the protein (14, 26), suggesting that multimerization could be mediated in a similar manner for both viral proteins. In this study, we question whether K-bZIP contains a functional bZIP domain.
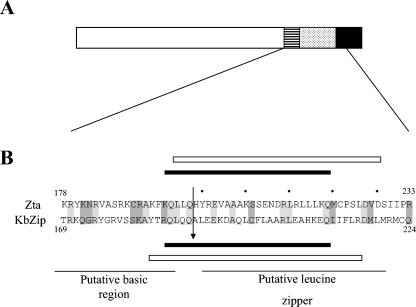
Similarities between Zta and K-bZIP primary and secondary structures are illustrated in Fig. 1. Alignment of the carboxy-terminal halves of both Zta and K-bZIP using ClustalW (17, 46) reveals scattered homology throughout the putative basic and leucine zipper regions. Leucine zipper multimerization domains comprise two strands of α-helix, which fold together through a hydrophobic face on each helix to form a coiled-coil structure (3, 8, 24, 44). Bioinformatics analyses using the program COILS (27) revealed that the proposed leucine zipper region of K-bZIP is predicted to fold as a coiled coil, as Zta does. This prompted us to question experimentally whether K-bZIP is able to undertake the two key functions of a bZIP protein, namely, (i) to interact with DNA or (ii) to form multimers using a coiled-coil folding motif.
FIG. 1.
Schematic diagram of Zta and K-bZIP. A. A schematic diagram of the known features of Zta is shown. The striped box represents the basic region, the stippled box the coiled coil, and the filled box the CT region. B. Analysis of the primary structure of K-bZIP (KbZip) and Zta using the bioinformatics program ClustalW revealed the indicated region of homology. Conserved amino acids are shown in on a light grey background, and conservative substitutions are shown on a dark grey background. The dots above the Zta sequence indicate those amino acids in the leucine zipper at “d” positions in the proposed coiled coil. The propensity of equivalent regions of Zta and K-bZIP to fold as coiled coils were investigated using the predictive program COILS and are shown as black bars below or above each sequence. The extent of the synthetic peptides used for biophysical analyses are indicated as boxes above or below each sequence. The position of the junction within the Z-K hybrid is indicated by an arrow.
In light of the scattered homology between the basic (DNA contact) region of Zta and K-bZIP, we questioned whether K-bZIP is able to interact with an AP1 site. Polyhistidine-tagged versions of both full-length proteins were generated in vitro using a reticulocyte lysate translation system and equal amounts of each protein were added to an in vitro DNA-binding assay with AP1 and a mutant version of that site (as described in reference 15). As expected, only His-Zta showed a clear interaction with the AP1 site (data not shown).
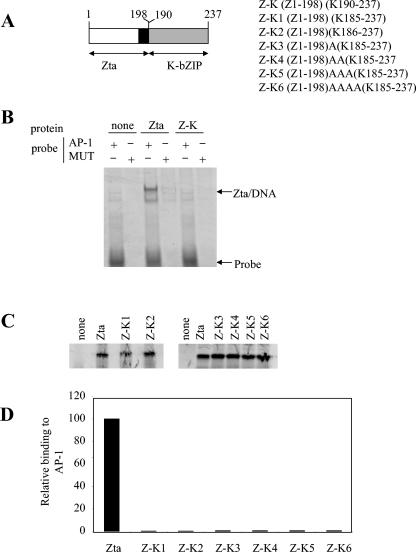
We sought to determine whether K-bZIP encodes a leucine zipper domain by undertaking domain swaps with the bZIP protein Zta. Functional swaps of the leucine zipper have been documented for this protein with leucine zippers of other members of the bZIP family (for an example, see reference 49). Using the alignment shown in Fig. 1, the first domain swap maintained the exact register of the alignment, ending Zta at amino acid Q198 and replacing the remainder of the coding sequence with amino acids A190 to the carboxyl terminus of K-bZIP (Fig. 2). His-Zta and His-K-bZIP proteins were generated in vitro, and their ability to interact with DNA was assessed using a DNA-binding assay. Surprisingly, this exchange did not result in a hybrid protein capable of interacting with AP1. The alignment of the basic and leucine zipper regions is important for the function of b-ZIP proteins (2, 23), so six further domain swaps were generated that contained between zero and four alanine residues between the basic and proposed zipper regions in order to align the proposed zipper in different conformations with respect to the basic region. However, none of the hybrids was able to interact with DNA, suggesting that a functional bZIP domain is not formed by any of the combinations tested. It is possible that this approach omitted a crucial configuration; however, the data shown lend no support to a model in which the putative zipper region of K-bZIP contains a functional leucine zipper.
FIG. 2.
K-bZIP domain swap proteins do not function as bZIPs. A. Vectors encoding hybrid proteins composed of the Zta transactivation and basic domains and the K-bZIP putative zipper domain were generated and are shown schematically here. The Zta basic region is shown in black. The region of K-bZIP from the proposed zipper to the carboxy terminus is shown as a grey box. B. Electrophoretic mobility shift assay reactions were undertaken with equal quantities of the indicated proteins and probes (as described in reference 15). Following electrophoresis, the locations of the probe and DNA complexes were visualized using phosphorimaging. C. Products from the indicated translation reactions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantitated by phosphorimaging. The migration of protein molecular weight markers (in kDa) is indicated on the left. D. Following the electrophoretic mobility shift assay analysis with equal amounts of protein, the specific binding of His-Zta and each hybrid protein to DNA is shown.
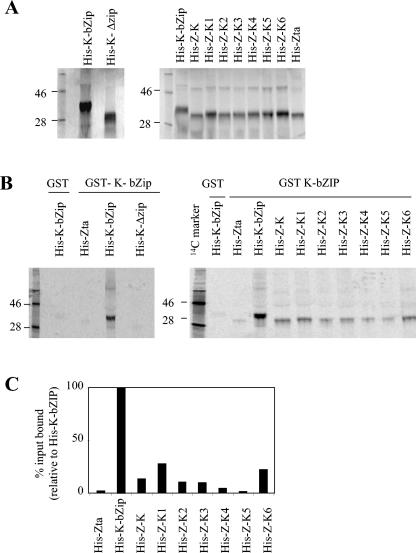
In order to probe whether the predicted coiled coil within the putative bZIP region of K-bZIP is required for multimerization, a deletion mutant, lacking amino acids 182 to 218, was generated by site-directed mutagenesis. The ability of a polyhistidine-tagged version of this protein, His-K-ΔZIP, to form multimers with full-length K-bZIP (14) was assessed using an in vitro association assay with a glutathione S-transferase (GST)-tagged version of K-bZIP (Fig. 3). In conditions where no association between GST and His-K-bZIP or GST-K-bZIP and His-Zta was observed, His-K-bZIP showed a clear association with the full-length GST-K-bZIP. Furthermore, the deletion within His-K-ΔZIP prevented association with GST-K-bZIP, thus demonstrating a role for amino acids 182 to 218 in multimerization. We then explored the ability of the hybrid proteins to form multimers with full-length K-bZIP protein. His-K-bZIP showed a strong association with GST-K-bZIP, whereas His-Zta revealed negligible binding, setting the background level of the assay (2% of the binding of His-K-bZIP). All of the His-Z-K hybrids showed a degree of multimerization, with His-Z-K1 and His-K-6 reaching over 20% of the binding of His-K-bZIP and His-Z-K and with His-Z-K2 and His-Z-K3 clearly also forming multimers. This demonstrates that the region of K-bZIP between 190 and 237 contains a multimerization domain that can transfer its function onto another protein.
FIG. 3.
The formation of multimers of K-bZIP requires amino acids within the proposed leucine zipper region. A. [35S]methionine-labeled His-K-bZIP, His-K-ΔZIP, and the Z-K hybrid series were generated in reticulocyte lysate. Products from the indicated translation reactions were analyzed by SDS-PAGE and quantitated by phosphorimaging. The migration of protein molecular weight markers (in kDa) is indicated on the left of each gel. B. In vitro association assays with GST and GST-K-bZIP agarose beads were undertaken with equal quantities of the indicated proteins. C. The products from the indicated translation reactions were analyzed by SDS-PAGE and quantitated by phosphorimaging.
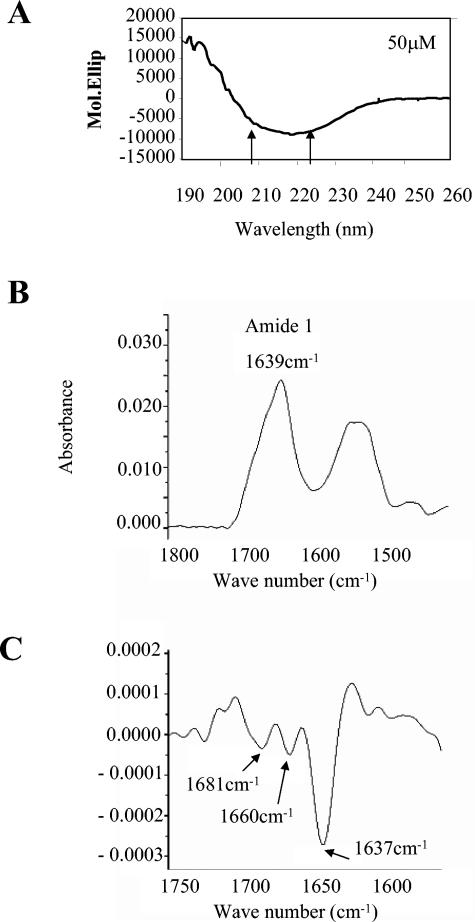
The proposed leucine zipper region of K-bZIP is predicted to have a high probability of folding as a coiled coil, according to the analysis shown in Fig. 1. We questioned whether this occurs by using two biophysical approaches. A synthetic peptide was generated that was equivalent to the region of Zta that we had previously shown to fold as a coiled coil in vitro (16). The propensity of the K-bZIP peptide to fold as an α-helix was assessed first using circular dichroism (CD) spectroscopy. Using this spectroscopic technique, α-helical peptides generate a CD spectrum with two characteristic minima, at 208 and 222 nm. The biophysical properties of the synthetic peptide were analyzed at two concentrations and two pH conditions, pH 7.0 and pH 3.7, where it displayed a greater solubility. All of the data sets generated similar results. A lack of concentration dependence is indicative of this peptide folding as a monomeric species in solution. Surprisingly, none of the data revealed α-helical minima at 222 nm and 208 nm (Fig. 4), suggesting that the peptide did not fold in an α-helical conformation. Indeed, a single trough was observed at 218 nm, which is the characteristic signal generated by peptides folded as β-sheets (21, 22). Further analysis of the folding of the K-bZIP peptide was undertaken using attenuated total reflectance-Fourier transform infrared spectroscopy (ATR-FTIR) (Fig. 4). This analysis revealed a peak for amide I bonds at 1,637 ± 4 cm−1 (characteristic of β-sheet structure). The second derivative of the spectrum of the amide I peaks revealed four components, which all correspond to either β-sheet or β-turn structure (10).
FIG. 4.
The proposed leucine zipper region of K-bZIP does not fold as a coiled coil in vitro. A synthetic peptide corresponding to amino acids 182 to 218 of K-bZIP was synthesized and analyzed by circular dichroism spectroscopy as described in references 15 and 49. A. The normalized spectrum of a 50 μM peptide solution at pH 3.7 is shown. The arrows indicate the two minima, which are characteristic of α-helical secondary structure. The observed minimum, 218 nm, is characteristic of a β-sheet. B. The FTIR-ATR spectrum of peptide is shown. The position of the amide I peak is indicated. C. The second derivative spectrum of the amide I peak revealed three distinct contributions at the indicated wavelengths. The characteristic structure responsible for generating a peak at that wavelength is shown.
Given the functional homology and sequence homology between K-bZIP and Zta, it is tempting to speculate that they contain a common structural element that acts in a similar manner within each protein to achieve their common functions. The obvious candidate for this is the bZIP domain that has been so well characterized for Zta and widely proposed to exist in K-bZIP. Evidence in favor of a bZIP structure for K-bZIP is summarized below.
The initial publications describing the transcript encoding K-bZIP and the primary structure of the protein identified and highlighted the presence of a heptad repeat of leucine residues within the carboxy-terminal half of the protein and termed it a leucine zipper (14, 26, 40, 54). Furthermore, K-bZIP folds as a multimer and the carboxy-terminal half of the protein is required for multimerization (14) (26). Analysis of the primary structure reveals two regions containing clusters of basic amino acid residues, but these do not have direct homology with the motif found in the basic region of bZIP proteins. Indeed, we did not observe any significant interaction between K-bZIP and an AP1 site in in vitro DNA-binding assays. However, it has recently been shown that K-bZIP expressed in vivo associates with DNA, specifically the lytic origin of replication (4, 48, 50). However, whether this occurs by a direct or indirect association and whether it involves the proposed bZIP region remain to be firmly established.
Recently, AuCoin et al. elegantly demonstrated that K-bZIP is required for replication through the lytic origin of the KSHV genome (4) and, furthermore, that this function requires contributions from both the amino- and carboxy-terminal halves of K-bZIP. During their further investigation of the contribution from the carboxy-terminal half of the protein, they generated a series of mutations in the heptad of leucine residues within the proposed leucine zipper region by replacing between one and four leucines in each mutant. Interestingly, two of the resulting mutants retained the ability to replicate through the lytic origin of replication and two lost this function. This study clearly demonstrates that K-bZIP is required for replication through the KSHV lytic origin of replication and, furthermore, it shows a contribution from some of the leucine residues within the proposed zipper to that function. However, no experiments involving multimerization or other biophysical properties were undertaken, and this set of data could be used equally well to support or to detract from a bZIP model for the K-bZIP protein.
Regarding multimerization, we demonstrate that amino acids 190 to 237 of K-bZIP contain a transferable multimerization domain. Although additional contributions from amino acids out of this region cannot be excluded, these amino acids clearly contain sufficient inherent information to fold into a stable multimeric structure. K-bZIP therefore appears to conform to the bZIP model, in terms of the ability to multimerize using the proposed bZIP region; however, the similarity ends there. The proposed zipper region of K-bZIP is not able to confer DNA-binding ability onto the basic region of Zta, despite the ability of several of the hybrid proteins to fold as multimers.
Finally, our biophysical investigations not only fail to support the leucine zipper model but demonstrate that this region preferentially folds into a different secondary structure. The synthetic K-bZIP peptide corresponding to the proposed leucine zipper of K-bZIP appears to fold as a β-sheet rather than as an α-helix in two distinct in vitro assays; CD spectroscopy and FTIR-ATR (Fig. 4). It is worth noting that many sequences that were predicted to fold as α-helices have been shown, using biophysical measurements, to actually fold as β-sheets; these have been termed false zippers (18).
Although β-sheets can form multimeric interfaces in vitro (9, 33), we observed no concentration dependence on the strength of the β-sheet signal observed for the CD spectra, which suggests that the peptide contains insufficient information to fold as multimers. Therefore, despite the clear involvement of the region containing the β-sheet to the multimerization of K-bZIP, it remains to be determined whether multimerization requires a β-sheet interface or whether the β-sheet stabilizes the formation of a different multimerization interface employing amino acids that lie outside residues 182 to 218.
The balance of evidence presented here suggests that the proposed bZIP region within K-bZIP is unlikely to function as a bZIP motif, unlike that of Zta. So, although Zta and K-bZIP hold many functions in common, we conclude that it is unlikely that they exploit similar molecular mechanisms to effect those functions.
Acknowledgments
Henri Gruffat (Lyon) kindly provided a vector encoding K-bZIP, George Papadakis helped to generate the His-tagged version of K-bZIP, Matthew Hicks generated the His-tagged version of Zta, and Derek Woolfson (University of Sussex) advised on biophysical analyses.
This work is supported by grants from the Wellcome trust to A.S. and from the BBSRC (B17958) to Derek Woolfson. S.A.M. was supported by a studentship from the government of the United Arab Emirates.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J. Virol. 73:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agre, P., P. F. Johnson, and S. L. McKnight. 1989. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science 246:922-926. [DOI] [PubMed] [Google Scholar]
- 3.Alber, T. 1992. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2:205-210. [DOI] [PubMed] [Google Scholar]
- 4.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 5.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff, C. 1998. Kaposi's sarcoma-associated herpesvirus: the second human gammaherpesvirus. Epstein-Barr Virus Rep. 5:3-9. [Google Scholar]
- 7.Boshoff, C., and R. A. Weiss. 1998. Kaposi's sarcoma associated herpesvirus. Adv. Cancer Res. 75:57-86. [DOI] [PubMed] [Google Scholar]
- 8.Busch, S. J., and P. Sassone-Corsi. 1990. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6:36-40. [DOI] [PubMed] [Google Scholar]
- 9.Cox, A., M. M. Arroyo, and K. H. Mayo. 2001. Folding of βpep-4 β-sheet sandwich dimers and tetramers is influenced by aliphatic hydrophobic residues at the intersubunit interface. Biochem. J. 357:739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, A., P. Huang, and W. S. Caughey. 1990. Protein secondary structures in water from second-derivative amide I infrared spectra. Biochemistry 29:3303-3308. [DOI] [PubMed] [Google Scholar]
- 11.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruffat, H., S. Portes-Sentis, A. Sergeant, and E. Manet. 1999. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J. Gen. Virol. 80:557-561. [DOI] [PubMed] [Google Scholar]
- 15.Hicks, M. R., S. S. Al-Mehairi, and A. J. Sinclair. 2003. The zipper region of Epstein-Barr virus bZIP transcription factor Zta is necessary but not sufficient to direct DNA binding. J. Virol. 77:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicks, M. R., S. Balesaria, C. Medina-Palazon, M. J. Pandya, D. N. Woolfson, and A. J. Sinclair. 2001. Biophysical analysis of natural variants of the multimerization region of Epstein-Barr virus lytic-switch protein BZLF1. J. Virol. 75:5381-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins, D. G. 1994. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol. Biol. 25:307-318. [DOI] [PubMed] [Google Scholar]
- 18.Hirst, J. D., M. Vieth, J. Skolnick, and C. L. Brooks III. 1996. Predicting leucine zipper structures from sequence. Protein Eng. 9:657-662. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, S., Y. Gwack, H. Byun, C. Lim, and J. Choe. 2001. The Kaposi's sarcoma-associated herpesvirus K8 protein interacts with CREB-binding protein (CBP) and represses CBP-mediated transcription. J. Virol. 75:9509-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 1602:1-22. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, S. M., and N. C. Price. 1997. The application of circular dichroism to studies of protein folding and unfolding. Biochim. Biophys. Acta 1338:161-185. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, S. M., and N. C. Price. 2000. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 1:349-384. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides, T., G. Packham, A. Cook, and P. J. Farrell. 1991. The BZLF1 protein of EBV has a coiled coil dimerization domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene 6:195-204. [PubMed] [Google Scholar]
- 24.Kouzarides, T., and E. Ziff. 1989. Behind the Fos and Jun leucine zipper. Cancer Cells 1:71-76. [PubMed] [Google Scholar]
- 25.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 28.Mauser, A., S. Saito, E. Appella, C. W. Anderson, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503-12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, G. 1989. The switch between EBV latency and replication. Yale J. Biol. Med. 62:205-213. [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, P., and Y. Chang. 1998. Identification of Kaposi's sarcoma-associated sequences of non-human origin. Epstein-Barr Virus Rep. 5:1-2. [Google Scholar]
- 31.Park, J., T. Seo, S. M. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portes-Sentis, S., E. Manet, G. Gourru, A. Sergeant, and H. Gruffat. 2001. Identification of a short amino acid sequence essential for efficient nuclear targeting of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 K8 protein. J. Gen. Virol. 82:507-512. [DOI] [PubMed] [Google Scholar]
- 33.Richardson, J. S., and D. C. Richardson. 2002. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA 99:2754-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schepers, A., D. Pich, and W. Hammerschmidt. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367-376. [DOI] [PubMed] [Google Scholar]
- 36.Schepers, A., D. Pich, and W. Hammerschmidt. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 12:3921-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz, T. F. 1998. Kaposi's sarcoma-associated herpesvirus (HHV-8). J. Gen. Virol. 79:1573-1591. [DOI] [PubMed] [Google Scholar]
- 38.Schultz, T. F. 1998. KSHV—the middle game. Epstein-Barr Virus Rep. 5:101-107. [Google Scholar]
- 39.Schwarzmann, F., M. Jager, N. Prang, and H. Wolf. 1998. The control of lytic replication of Epstein-Barr virus in B lymphocytes. Int. J. Mol. Med. 1:137-142. [DOI] [PubMed] [Google Scholar]
- 40.Seaman, W. T., D. S. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair, A. J. 2003. bZIP proteins of human gamma herpesviruses. J. Gen. Virol. 84:1941-1949. [DOI] [PubMed] [Google Scholar]
- 42.Sinclair, A. J., and P. J. Farrell. 1992. Epstein-Barr virus transcription factors. Cell Growth Differ. 3:557-563. [PubMed] [Google Scholar]
- 43.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 44.Struhl, K. 1989. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem. Sci. 14:137-140. [DOI] [PubMed] [Google Scholar]
- 45.Talbot, S. J., and D. H. Crawford. 2004. Viruses and tumours—an update. Eur. J. Cancer 40:1998-2005. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78:4248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Y., H. Li, M. Y. Chan, F. X. Zhu, D. M. Lukac, and Y. Yuan. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 78:8615-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West, M. J., H. M. Webb, A. J. Sinclair, and D. N. Woolfson. 2004. Biophysical and mutational analysis of the putative bZIP domain of Epstein-Barr virus EBNA 3C. J. Virol. 78:9431-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, F. Y., H. Chen, S. E. Wang, C. M. apRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, F. Y., Q. Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-α. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zerby, D., C. J. Chen, E. Poon, D. Lee, R. Shiekhattar, and P. M. Lieberman. 1999. The amino-terminal C/H1 domain of CREB-binding protein mediates Zta transcriptional activation of latent Epstein-Barr virus. Mol. Cell. Biol. 19:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]