Abstract
A variety of pathogens use CD46, a ubiquitously expressed membrane protein that regulates complement activation, as a cellular attachment receptor. While the CD46 binding sites of several pathogens, including measles virus, Neisseria gonorrhea, and human herpesvirus 6, have been described, the region of CD46 responsible for adenovirus binding has not been determined. In this study, we used competition experiments with known CD46 ligands, CD46-specific antibodies, and a set of CD46 mutants to localize the binding domain for the group B adenovirus serotype 35 (Ad35). Our results show that Ad35 competes with measles virus for binding to CD46 but not with complement protein C3b. We further show that this interaction is a protein-protein interaction and that N glycosylations do not critically contribute to infection with Ad35 fiber-containing Ad vectors. Our data demonstrate that the native conformation of the CCP2 domain is crucial for Ad35 binding and that the substitution of amino acids at positions 130 to 135 or 152 to 156 completely abolishes the receptor function of CD46. These regions localize to the same planar face of CD46 and likely form an extended adenovirus binding surface, since no single amino acid substitution within these areas eliminates virus binding. Finally, we demonstrate that the infection with a virus possessing human group B serotype Ad11 fibers is also mediated by the CCP2 domain. This information is important to better characterize the mechanisms of the receptor recognition by adenovirus relative to other pathogens that interact with CD46, and it may help in the design of antiviral therapeutics against adenovirus serotypes that use CD46 as a primary cellular attachment receptor.
Understanding how a pathogen interacts with its cellular receptor is a fundamental aspect of virus-host interactions. Delineating these interactions can help to understand viral tropism and can provide novel targets for antiviral therapeutics. CD46, or membrane cofactor protein, is a ubiquitously expressed transmembrane protein belonging to the regulators of complement activation family (24, 41). In addition to its role in preventing complement damage to normal cells, this protein has been described as a cellular receptor for a variety of pathogens, including Neisseria and Streptococcus strains, as well as measles virus (especially the Edmonston strain), human herpesvirus 6, bovine viral diarrhea virus, and different adenovirus serotypes (9, 18, 29, 33, 38, 40, 48, 55). For several of these pathogens the binding sites within CD46 have been described; however, it is still unknown how adenovirus interacts with CD46.
CD46, a membrane-linked glycoprotein, was first described for its complement binding and regulatory properties (reviewed in reference 24). In humans, there are four major isoforms of CD46 (BC1, BC2, C1, and C2), depending on the alternative splicing of a region encoding an extracellular domain and the choice between one or two cytoplasmic tails, Cyt1 and Cyt2 (36). CD46 attaches to complement fragments C3b and C4b that are bound to host cells and then serves as a cofactor for their targeted destruction by the plasma serine protease, factor I (42). This system protects normal cells from the damaging effects of inadvertent complement activation. Human malignant cells significantly up-regulate CD46 expression (14, 19, 32, 52). Therefore, high levels of CD46 on the cell surface may represent a mechanism for tumor cell survival in the host. Four modules, termed complement control protein (CCP) domains CCP1, CCP2, CCP2, and CCP4, form the major part of the extracellular domain of CD46. CCPs contain three N-linked glycosylation sites. Following the CCPs is an area enriched in serines, threonines, and prolines (STP domain) that is a site for O glycosylation. This region undergoes alternative splicing, as does the cytoplasmic tail domain. These splicing events result in four CD46 isoforms, ranging in molecular mass from 55 to 65 kDa. The domains of CD46 important for C3b/C4b binding and cofactor activity have been mapped to CCP2, CCP3, and CCP4 (25). CCP1 and CCP2 interact with measles virus hemagglutinin (H) (3, 15, 27, 28). Human herpesvirus 6 binds to CCP2 and CCP3, while Neisseria gonorrhoeae has been shown to require CCP3 and the STP domain for CD46 binding (13, 17). Binding of Streptococcus pyogenes to human epithelial cells requires CCP2 and CCP3 (10).
We and others have recently shown that various serotypes of adenovirus can use CD46 as a cellular receptor (9, 40, 48, 55). Adenovirus serotypes are divided into six groups (A to F), and all serotypes except those from group B have been shown to use the coxsackievirus and adenovirus receptor (CAR) as a primary cellular attachment receptor. This occurs via interactions of CAR with the trimeric viral fiber knob domain (1, 2, 35, 46). CD46 appears to be a major cellular receptor for all group B adenoviruses as well as for the group D serotype 37. However, the manner in which these adenoviruses interact with CD46 is still unknown. Segerman et al. (40) found that adenovirus serotype 11 (Ad11) infection of CD46-expressing cells could be partially blocked by antibodies against CCP3 and CCP4, while Wu et al. (55) described Ad37 binding to be localized to CCP1 and CCP2. In this study, we employed a panel of CD46 mutants to localize the adenovirus binding domain on CD46, using Ad35 as a representative group B Ad.
MATERIALS AND METHODS
Viruses and cell lines.
Chinese hamster ovary K-1 (CHO) and 293T cells were purchased from the American Type Culture Collection. CHO cells stably expressing the C2 isoform of CD46 (CHO-C2) were described previously (23). Chimeric adenoviruses, based on serotype 5 but possessing the fiber shaft and knob domains of serotype 35 (Ad5/35) or serotype 11-Slobitski strain (Ad5/11), were previously constructed and contain green fluorescent protein (GFP) as a transgene under the control of the cytomegalovirus promoter (45, 49).
Competition experiments.
For C3b competition experiments, CHO-K1 and CHO-C2 cells were seeded in 12-well plates, and on the following day cells were preincubated with various concentrations (1 to 50 μg/ml) of human purified C3b fragment (Antigen Site, San Diego, CA) in 150 μl phosphate-buffered saline (PBS) for 10 min at room temperature. Ad5/35GFP virus was added at a multiplicity of infection (MOI) of 25 PFU/cell in a volume of 150 μl of F12K growth medium and incubated at room temperature for 30 min. Cells were subsequently washed and then incubated overnight at 37°C. The following day, cells were analyzed for GFP expression by flow cytometry using a FACScan instrument (Becton Dickinson, San Jose, CA).
For measles virus hemagglutinin competition, a baculovirus expressing β-galactosidase and a membrane-anchored measles virus hemagglutinin protein were used (gift of Eric Hsu). High-Five cells (gift of Yasuko Mori) were infected with the baculovirus and allowed to express the transgenes for 48 h. Following this incubation, the insect cells were added to confluent monolayers of CHO-K1 or CHO-C2 cells that had been preincubated with recombinant Ad35 or Ad5 fiber knob domain at a concentration of 100 μg/ml. After incubation for 30 min at room temperature, the supernatant containing unbound High-Five cells was removed and the monolayers were washed with PBS twice. A set of cultures were then fixed with 2% glutaraldehyde and stained using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Roche, Alameda, CA). Another set of cultures were lysed, and β-galactosidase activity was measured using a reporter kit (Roche, Alameda, CA).
For antibody competition experiments, 1.5 × 105 CHO-C2 cells were preincubated with either Tra2.10, GB24, or M75 anti-CD46 antibodies (25) at 5 μg/ml in 200 μl PBS for 30 min at room temperature. Ad5/35GFP was added at an MOI of 5 PFU/cell in 200 μl PBS and left for 30 min at room temperature, after which the cells were washed once and incubated overnight at 37°C. GFP expression was measured 24 h later using fluorescence-activated cell sorting.
CD46 deglycosylation.
To determine optimal concentrations of tunicamycin (Sigma, St. Louis, MO), CHO-C2 cells were plated on 12-well plates and incubated with various concentrations of tunicamycin from 0.01 to 0.50 μg/ml for 48 h. Cells were then collected, and lysates were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were blotted and probed with a polyclonal rabbit antibody against CD46. Control recombinant CD46 protein was produced using baculovirus and contains the extracellular domain of CD46 only (31). For infection experiments, optimal concentrations of tunicamycin were used on cells for 48 h, after which cells were infected at an MOI of 10 with Ad5/35GFP for 1 hour at 37°C and then washed and incubated overnight. GFP expression was measured by fluorescence-activated cell sorting analysis.
Studies with mutant CD46 constructs.
For stably transfected cell lines deleted of CCP1 or CCP2, stably transfected clones were isolated from a population of transfected cells by selection using G418 (Invitrogen, Carlsbad, CA) at 0.5 mg/ml. Individual clones were then plated at limiting dilutions and assessed for CD46 expression using CD46-specific antibodies. Isolated clones with expression levels equal to CHO-C2 cells were chosen and plated in 12-well plates. Cells were infected with Ad5/35GFP or Ad5/11GFP at an MOI of 5 PFU/cell for 30 min at 37°C and assessed for GFP expression 24 h postinfection.
Plasmids containing the coding regions for CD46 mutants deleted of CCP2, -3 and -4 (pI/4); CCP1, -3, and -4 (pII/4); or CCP3 and -4 (pI+II/4) under the control of the T7 phage promoter were kindly provided by R. Cataneo (Mayo Clinic, Rochester, MN) (4). For transient expression of mutated CD46, these plasmids were transfected into CHO cells using Fugene reagent (Roche, Alameda, CA), followed by infection with vaccinia virus expressing T7 RNA polymerase as described in detailed in (4). Twenty-four hours after plasmid transfection, CHO cells were infected with Ad5/35GFP vector at an MOI of 10 PFU/cell. Virus was removed 2 hours later, and cells were washed and incubated for another 24 h. The percentage of GFP-expressing cells and the mean GFP fluorescence were determined by flow cytometry.
Constructs with substitutions in CCP2 were transfected into CHO cells by using Lipofectamine. Briefly, 0.85 μg DNA was added to 3 μl Lipofectamine (Invitrogen, Carlsbad, CA), and the mixture was added to 2 × 105 CHO-K1 cells. (The transfection efficiency was routinely between 35 and 40% for CHO cells and all derivatives of this cell line.) To assess CD46 expression levels, at 48 h following transfection, cells were removed from wells by incubation with Versen solution (Earle’s balanced salt solution plus 2 mM EDTA). Cells were then incubated for 1 h with a 1:500 dilution of J4-48 monoclonal antibody against CCP1 of CD46 (Research Diagnostics, Flanders, NJ). Cells were washed twice and incubated for 30 min with a fluorescein isothiocyanate-conjugated anti-mouse secondary antibody (Molecular Probes, Eugene, OR) and then analyzed using flow cytometry. For infection analysis, at 48 h posttransfection, cells were infected with Ad5/35 at an MOI of 10 PFU/cell for 1 h at 37°C. Cells were washed, incubated overnight at 37°C, and assessed for GFP expression the following day by flow cytometry.
Single point mutation constructs were also assessed in a similar manner. CHO-K1 cells were transfected in 12-well plates with 1 μg DNA mixed with 4 μl Fugene 6 (Roche, Alameda, CA) diluted in serum-containing medium. (The transfection efficiency with Fugene was about 60%.) Twenty-four hours after transfection, cells were assessed for CD46 expression as described above and were infected and analyzed for GFP expression.
CD46 modeling.
Crystal structure data were modeled using RasMol (39) from protein data bank files of the crystal structures of CCP1 and CCP2 of CD46 (5).
RESULTS
Virus with Ad35 fiber knob domain competes with measles virus hemagglutinin but not C3b for infection.
To localize the binding site of Ad35 within CD46, we employed known ligands of CD46 as competitors for Ad35 binding. For adenovirus infectivity studies, we used a chimeric adenovirus based on serotype 5 but displaying the receptor-interacting knob domain of serotype 35 and expressing GFP as a reporter gene (Ad5/35GFP). Our earlier studies showed that this chimeric virus has a tropism that mimics that of wild-type Ad35 (45).
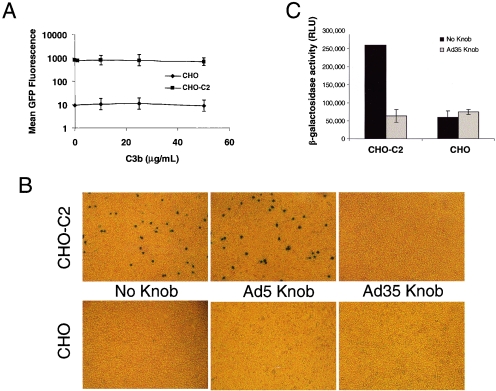
To assess competition with complement fragments (that bind to CCP3 and CCP4 domains of CD46), we infected either CHO cells or CHO cells stably expressing the C2 isoform of CD46 (CHO-C2) with Ad5/35GFP in the presence of increasing amounts of complement factor C3b. As expected, CD46 expression conferred efficient infection of CHO cells by Ad5/35GFP. However, even at a concentration of 50 μg/ml, C3b did not inhibit infection of Ad5/35 as determined by mean GFP fluorescence (Fig. 1A).
FIG. 1.
Ad5/35GFP competes with measles virus but not C3b for binding to CD46. (A) Mean GFP fluorescence of CHO or CHO-C2 cells following infection with Ad5/35GFP in the presence of increasing concentrations of purified human C3b. (B) Visualization of β-galactosidase-producing insect cells attached to either CHO or CHO-C2 cells following preincubation with either saline (no knob), recombinant Ad5 knob, or recombinant Ad35 knob. (C) β-Galactosidase activity of cell lysates from monolayers of either CHO or CHO-C2 cells preincubated with saline (black bars) or Ad35 knob (gray bars) and then incubated with insect cells expressing measles virus hemagglutinin protein and β-galactosidase. RLU, relative light units. Error bars indicate standard deviations.
Measles virus interacts with CD46 through its hemagglutinin protein. Several studies have localized this binding to CCP1 and CCP2 of CD46 (3, 15, 28). In order to determine whether measles virus and adenovirus compete for binding on CD46, we used a convenient binding assay developed by Hsu et al. (15). We used a baculovirus expressing both β-galactosidase and a cell surface-localized measles virus H protein. This system allowed us to track and quantify (via β-galactosidase activity) binding of infected insect cells to CD46-expressing CHO-C2 cells. We preincubated either CHO or CHO-C2 cells with saline, recombinant Ad5, or recombinant Ad35 fiber knob domain. We have previously shown that these fiber knob domains bind to cell surface receptors CAR and CD46, respectively, and efficiently block Ad infection (43). Next we applied baculovirus-infected insect cells to CHO or CHO-C2 monolayers of cells to determine how preincubation with the fiber knob domain affected measles virus H-dependent binding. While the addition of non-CD46 binding Ad5 fiber knob did not affect binding of infected insect cells to CHO-C2 cells (Fig. 1B), Ad35 fiber knob abolished measles virus H binding to background levels seen on CHO cells (Fig. 1B and C).
Since measles virus H protein, but not C3b, efficiently competes with Ad35 fiber knob domain for binding to CD46, our data indicate that the first two CCP domains (CCP1 and CCP2) are likely required for interaction with Ad fiber knob domain.
Adenovirus binding to CD46 does not require glycosylation of CD46.
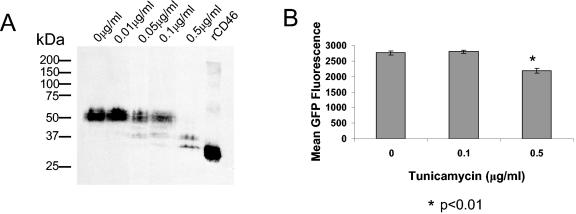
For measles virus, the presence of N-linked glycosylations in CCP1 and CCP2 is critical for a productive binding of the virus to CD46 (26). While measles virus H protein does not directly bind to the glycosylation sites, their presence is required for the proper sterical configuration needed for virus-CD46 interactions. To test whether N-linked glycosylation is important for binding of adenovirus to CD46, we treated CHO-C2 cells with tunicamycin, a bacterial protein that inhibits N-linked glycosylation. We first determined the concentration of tunicamycin that would inhibit glycosylation of CD46. Unglycosylated forms of CD46 were detected based on altered electrophoretic mobility of cellular CD46 in polyacrylamide gels (Fig. 2A). While CD46 N glycosylation was partially blocked after tunicamycin treatment at a concentration of 0.1 μg/ml, pretreatment at a 0.5 μg/ml achieved significant deglycosylation. Pretreatment with 0.1 μg/ml tunicamycin did not reduce GFP expression upon Ad5/35 infection (Fig. 2B). Although the percentage of GFP-expressing cells in those treated with 0.5 μg/ml tunicamycin was comparable to that in untreated cells (data not shown), the level of GFP expression (mean fluorescence) was slightly reduced upon treatment at this concentration (Fig. 2B). We attributed this decrease in GFP expression to cytotoxic effects of tunicamycin. Together, these data imply that unlike the case for measles virus, the interaction of Ad35 with CD46 is not dependent on glycosylation.
FIG. 2.
Glycosylation of CD46 is not essential for Ad35 fiber knob binding. (A) Western blot analysis for CD46 expression in CHO-C2 cells 48 h following treatment with different concentrations of tunicamycin, showing conversion of full-length CD46 to partially and fully deglycosylated CD46. The two bands with lower molecular masses in samples treated with 0.5 μg/ml tunicamycin reflect partially and completely deglycosylated CD46. Recombinant CD46 (rCD46) is shown as a positive control (see Materials and Methods). (B) Mean GFP fluorescence of tunicamycin-treated C2 cells following infection by Ad5/35GFP. Notably, control recombinant CD46 protein was produced using baculovirus and contains the extracellular domain of CD46 only (31), which accounts for the higher electrophoretic mobility. Error bars indicate standard deviations.
N-terminal CCP1 and CCP2 domains of CD46 are important for Ad35 infection.
To evaluate the role of each individual domain of CD46 in mediating Ad35 binding, we used a variety of CD46 mutants and CD46-specific antibodies. First, we used a panel of antibodies specific to different regions of CD46 in Ad infection-blocking experiments. CHO-C2 cells were preincubated with CCP1-specific (Tra2.10), CCP2-specific (M75), or CCP3/4-specific (GB24) antibodies followed by infection with Ad5/35GFP. Ad transduction was analyzed by flow cytometry for GFP expression. These studies revealed that preincubation of CHO-C2 cells with each of the antibodies reduced the efficiency of Ad5/35GFP infection compared to that for control cells preincubated with medium only. However, while reduction of Ad5/35GFP vector infectivity was only modest after preincubation of cells with CCP1- or CCP3/4-specific Tra2.10 or GB24 antibodies (Fig. 3), preincubation of cells with CCP2-specific M75 antibody significantly blocked Ad infection (P < 0.01).
FIG. 3.
Blocking Ad5/35GFP infection of CHO-C2 cells with CD46-specific antibodies. (A) Percentage of cells transduced and (B) mean GFP fluorescence of CHO-C2 cells infected with Ad5/35GFP after preincubation of cells with CCP1-specific Tra2.10, CCP2-specific M75, and CCP3/4-specific GB24 monoclonal antibodies. In control settings (Control), prior to virus infection cells were incubated with medium only. n.s., not statistically significant. *, statistically significant difference (P < 0.01). Error bars indicate standard deviations.
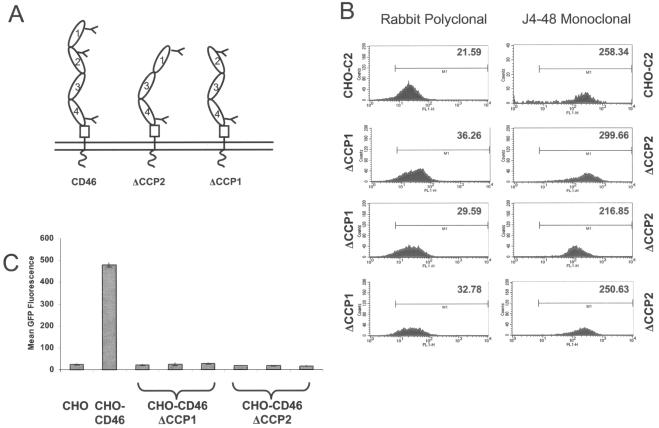
To further analyze the contribution of each individual domain of CD46 in mediating Ad infection, we used cell lines stably expressing membrane-anchored mutants of the BC1 isoform of CD46. The CD46 mutants used in these experiments were deleted for either CCP1 or CCP2 (Fig. 4A). We isolated several clones of each cell line with expression levels of CD46 similar to that of CHO-C2 cells (Fig. 4B). Despite having high levels of mutated CD46 expression, these cells were not susceptible to Ad5/35GFP infection (Fig. 4C). In contrast, CHO-C2 cells, expressing the full-length CD46 molecule, allowed for efficient Ad infection under the same experimental conditions. These results indicate that the presence of intact CCP1 and CCP2 domains, but not C-terminal CCP3 and CCP4 domains, is important for Ad5/35 infection.
FIG. 4.
N-terminal CCP1 and CCP2 are required for adenovirus infection of cells. (A) Schematic of membrane-bound CD46 mutants used for the experiments. (B) Expression levels of CD46 mutants deleted for either CCP1 (ΔCCP1) or CCP2 (ΔCCP2) in three different clonal cell lines, compared to CD46 expression levels of CHO-C2 cells. Mean fluorescence is given in the upper right corner of the histogram plot. ΔCCP1 is recognized with a polyclonal antibody, and ΔCCP2 is recognized by a monoclonal antibody directed against CCP1. (C) Mean GFP fluorescence after infection of CHO, CHO-C2, and three clones each of CHO cells expressing CD46 deleted for either CCP1 or CCP2 with Ad5/35GFP virus. Error bars indicate standard deviations; n = 5.
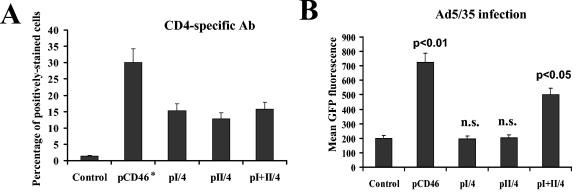
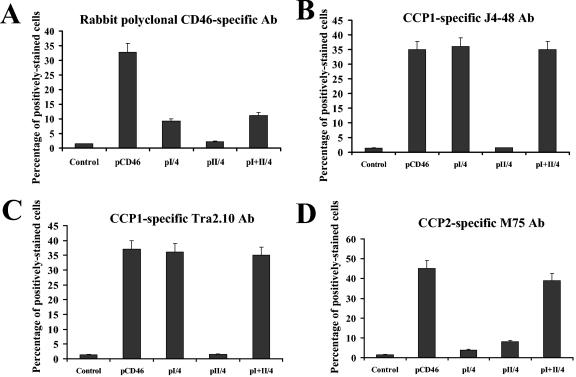
To delineate the contribution of the two distal CCPs in Ad binding and to ultimately exclude the involvement of CCP3 and CCP4 domains of CD46 in mediating Ad infection, we transfected CHO cells with plasmids expressing CCP1 (pI/4), CCP2 (pII/4), or the CCP1 and CCP2 domains together (pI+II/4) (4). In these constructs, the CD46 domains are fused to an immunoglobulin G-like domain of human CD4 that facilitated the detection of CD46-CD4 hybrid expression in transfected cells. A plasmid expressing the complete CD46 molecule (pCD46) was included as a control. Twenty-four hours after transfection, one set of cells was stained with polyclonal anti-CD4 antibody and analyzed by flow cytometry; another set was infected with Ad5/35GFP. Staining of transfected cells with CD4-specific antibody revealed that all cells expressed similar levels of truncated CD46 proteins (Fig. 5A). In contrast, infection of cells with Ad5/35GFP showed that only the full-length CD46 (pCD46) and the truncated CD46 containing both the CCP1 and CCP2 domains (pI+II/4) were able to support Ad5/35GFP infection (Fig. 5B). Taken together, these data confirm that the Ad35 fiber knob recognition site is localized within the N-terminal CCP1 and/or CCP2 domains of CD46.
FIG. 5.
Ad5/35GFP infection of CHO cells expressing truncated forms of CD46. (A) CHO cells were transfected with the indicated plasmids expressing either CCP1 (pI/4), CCP2 (pII/4), or the CCP1 and CCP2 domains together (pI+II/4), joined to a single immunoglobulin G-like domain of human CD4 protein. Transfected cells were stained with CD4-specific polyclonal antibody to detect levels of truncated CD46 protein expression. As a positive control, cells were transfected with full-length CD46-expressing plasmid (pCD46), and the levels of CD46 expression were determined using a rabbit polyclonal antibody (Ab) (indicated by the asterisk). Cells transfected with an unrelated plasmid were used as a control. (B) Efficiency of Ad5/35GFP infection of CHO cells transfected with the plasmids described for panel A. Note that only pCD46 and pI+II/4 plasmid transfection rendered CHO cells susceptible to virus infection. P values demonstrating statistically significant differences between levels of adenovirus infection of control cells and cells transfected with pCD46 or pI+II/4 plasmids are shown above the corresponding bars. n.s., not statistically significant. Error bars indicate standard deviations; n = 4.
The Ad35 binding site is localized within conformationally sensitive areas of the CD46 CCP2 domain.
Since a CCP2-specific monoclonal antibody efficiently blocked Ad5/35GFP infection of CHO-C2 cells (Fig. 3) and a CCP1-specific antibody blocked virus infection at a level comparable to that of CCP3/4-specific antibody, it was somewhat surprising that deletion of either of the N-terminal CD46 domains, CCP1 or CCP2, had equally deleterious effects on the efficiency of Ad5/35GFP infection. One potential explanation of the observed data might be that the CCP1 and CCP2 domains are functionally and structurally interdependent. Therefore, the deletion of one of these two domains would affect the conformation of the other, thus preventing the binding of functional ligands. One of the approaches to detect the changes in conformation of the CCP1 and CCP2 domains is to analyze the efficiency of polyclonal and monoclonal antibodies in binding to cells expressing truncated forms of CD46. The approach of using antibody binding is widely used in studies aimed to detect conformational changes in proteins, including those that occur upon virus-host cell interactions (22).
To analyze whether the conformation of CCP1 and CCP2 domains, when expressed as individual domains, differs from their conformation in wild-type full-length CD46 molecules, we transfected CHO cells with the plasmids expressing N-terminal domains of CD46 described above (Fig. 5) and 24 h later stained them with either rabbit polyclonal anti-CD46 antibody, mouse monoclonal CCP1-specific Tra2.10 and J4-48 antibodies, or a monoclonal CCP2-specific antibody, M74, that efficiently blocked Ad5/35GFP infection as shown above (Fig. 2). All transfected cells were also stained with CD4-specific polyclonal antibody to detect levels of truncated CD46 protein expression. Similarly to experiments described above (Fig. 5A), staining of transfected cells with CD4-specific antibody confirmed that all truncated CD46 forms were expressed at comparable efficiencies (data not shown). We also found that full-length CD46 protein was efficiently recognized with all antibodies used. However, while the polyclonal antibody could detected truncated CD46 forms containing the CCP1 (pI/2) and CCP1 and CCP2 (pI+II/4) domains (Fig. 6A), it could not stain cells expressing the CCP2 domain only (pII/4). Staining of transfected cells with CCP1-specific Tra2.10 or J4-48 monoclonal antibodies revealed that the CCP1 domain is efficiently recognized by these two monoclonal antibodies independently of whether the CCP1 domain is expressed alone (pI/4) or in combination with CCP2 (pI+II/4) (Fig. 6B and C). In contrast, while staining of transfected cells with CCP2-specific monoclonal antibody M75 allowed for detection of the CD46 form containing both CCP1 and CCP2 domains (comparable to wild-type CD46 levels [Fig. 6D]), this antibody failed to recognize the CCP2 domain when it was expressed alone (pII/4) (Fig. 6D). Taken together, these data demonstrate that the CCP2 domain of CD46 changes its conformation when expressed as an individual domain. The change of conformation results in poor recognition of this domain by both polyclonal and monoclonal antibodies; however, when it is joined with the CCP1 domain, the conformation of the CCP2 domain is similar to that present in the wild-type full-length CD46 molecule (Fig. 6D). The antibody staining experiments demonstrate that the CCP1 domain maintains its wild-type conformation independently whether it is expressed as an individual domain (pI/4) or in combination with CCP2 (pI+II/4). Since the CCP1 domain, when it was expressed alone, was unable to support Ad5/35GFP infection (Fig. 5B), our data indicate that an Ad binding site is localized within conformationally sensitive areas of the CCP2 domain.
FIG. 6.
Detection of conformational changes in N-terminal CCP1 and CCP2 domains by antibody staining. CHO cells were transfected with plasmids expressing full-length or truncated forms of CD46 as described for Fig. 5A, and 24 hours later, cells were stained with indicated antibodies (Ab). Staining of primary antibodies was detected by Alexaflour-488-conjugated antibody and flow cytometry. All stainings were done in triplicate in two independent experiments. Error bars indicate standard deviations.
Amino acid substitutions at positions 130 to 135 or 152 to 156 of the CCP2 domain abrogate Ad35 receptor function of CD46.
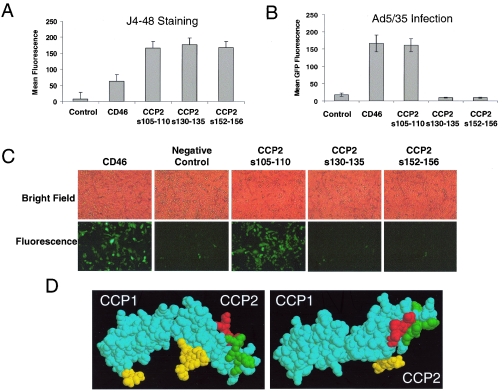
In an attempt to localize the binding domain of CD46 more precisely, we used a set of plasmids expressing full-length CD46 with mutations within the CCP2 domain. These mutants were created by replacing stretches of CCP2 with corresponding regions of the highly related complement control protein decay acceleration factor (CD55) (reviewed in reference 24). Plasmids expressing three mutant CD46 proteins with substitutions of amino acids (aa) 105 to 110 (CD46105-110, replacing PLNGQA with VPNAQP), 130 to 135 (CD46130-135, replacing GYYLIG with SFVKIP), or 152 to 156 (CD46152-156, replacing GKPPI with DIEEF), as well as a wild-type control CD46 molecule, were transiently expressed in CHO cells. Forty-eight hours later, cells either were assessed for CD46 expression by using CCP1-specific J4-48 monoclonal antibody staining or were infected with Ad5/35GFP. These analyses revealed that at 48 h posttransfection, all mutated CD46 proteins were expressed at similar levels (Fig. 7A). However, while cells expressing CD46105-110 showed levels of viral transduction similar to those of the wild-type CD46, cells expressing CD46130-135, and CD46152-156 failed to support Ad5/35GFP infection (Fig. 7B and C). It is interesting that when these two regions of CCP2 were highlighted on the crystal structure for the first two N-terminal CD46 domains, they localized to a planar face on the same outer surface of the molecule (Fig. 7D). In contrast, amino acids 105 to 110 are localized in an area on the opposite plane of CCP2, and substitution of these amino acids had no effect on the level of Ad5/35GFP infection.
FIG. 7.
Substitutions of amino acid residues 130 to 135 and 152 to 156 in the CCP2 domain completely abolish adenovirus receptor function of CD46. (A) CHO cells were transfected with plasmids expressing either wild-type CD46 (CD46) or full-length CD46 mutants possessing four amino acid substitutions of the indicated amino acids within the CCP2 domain. The levels of CD46 protein expression were determined by J4-48 antibody staining and flow cytometry 48 h posttransfection. (B) Mean GFP fluorescence of CHO cells transfected with plasmids described for panel A and infected with Ad5/35GFP vector. Error bars indicate standard deviations; n = 4. (C) Bright-field and fluorescence photographs of transfected CHO cells 24 h after Ad5/35GFP infection shown in panel B. (D) Crystal structure representation of domains CCP1 and CCP2 of CD46, with residues 96 to 101 shown in green, residues 118 to 122 shown in red, and N-linked glycosylations shown in yellow.
To further delineate amino acid residues that are critical for Ad5/35 binding to CD46 on the single-amino-acid level, we used a set of CD46 mutants in which each amino acid in regions 130 to 135 and 152 to 156 was subsequently changed to an alanine. To determine whether these CD46 mutants allow for viral infection, we transfected CHO cells with plasmids encoding these mutants, and 48 h later transfected cells were either stained with anti-CD46 antibody or infected with Ad5/35GFP. Staining of cells with polyclonal anti-CD46 antibody confirmed similar levels of mutant CD46 expression following transfection of plasmid DNAs. However, analysis of virus infection efficiency revealed that no single-amino-acid substitution was sufficient to abolish Ad5/35 binding (data not shown).
The fact that substitution of amino acids 130 to 135 or 152 to 156 within the CCP2 domain of the full-length CD46 (which contains intact CCP3 and -4) completely abolished its receptor function excludes a major role of CCP3 and CCP4 in Ad binding. However, since CCP3 and CCP4 deletion mutants were not tested in our study, a minor contribution of these domains cannot be excluded.
The CD46 CCP2 domain is required to support cell infection with adenovirus possessing human serotype Ad11-derived fibers.
Earlier studies suggested that CD46 can serve as a functional receptor for all group B Ad serotypes, including Ad3, Ad11, Ad16, Ad21, Ad35, and Ad50 (9, 40, 48, 55). To analyze whether the CD46 CCP2 domain is also critical for binding of group B Ad serotypes (other than Ad35), we transfected CHO cells with plasmids expressing mutated full-length CD46 protein possessing the amino acid substitutions within the CCP2 domain described above. Twenty-four hours after transfection, one set of cells was stained with either CCP1-specific J4-48 antibody to compare the levels of mutated CD46 expression or M75 antibody to assess the conformation of the CCP2 domain. The other set of transfected cells was infected with either Ad5/35GFP or Ad5/11GFP vector. Ad5/11GFP vector possesses an Ad5 capsid; however, the fiber shaft and knob domains of Ad5 fiber were replaced with the corresponding domains of a fiber derived from Ad11 (49). The two vectors possess identical GFP gene expression cassettes, and therefore the efficiency of Ad35 or Ad11 fiber-dependent infection can be compared by flow cytometry for GFP expression.
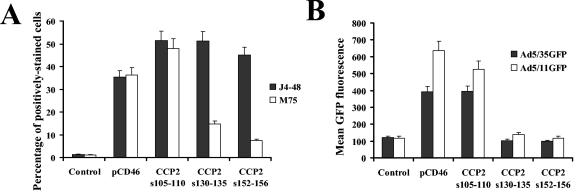
Staining of transfected cells with J4-48 antibody demonstrated that all mutated forms of CD46 as well as the wild-type CD46 molecule were expressed on cells at comparable levels (Fig. 8A). However, staining with CCP2-specific M75 antibody demonstrated that the CCP2 domain is efficiently recognized by this antibody only in the wild-type CD46 and the CD46105-110 mutant and not in the CD46130-135 and CD46152-156 mutants (Fig. 8A). This suggests that the overall conformation of CCP2 was affected by introduction of these mutations and/or that these areas of CCP2 overlap with epitopes which are recognized by the M75 antibody. Ad5/35 and Ad5/11 transduction studies demonstrated that CD46105-110-expressing CHO cells can be efficiently infected with both vectors, whereas the CD46130-135 and CD46152-156 mutants were unable to support Ad vector transduction. These data indicate that Ad vectors possessing Ad35 or Ad11 fibers bind to two specific areas in CCP2 (aa 130 to 135 and aa 152 to 156) and that an intact CCP2 domain is required for CD46 to serve as a functional receptor for Ad5/35 and Ad5/11.
FIG. 8.
Infection of cells expressing CCP2-mutated CD46 with vectors possessing Ad35 or Ad11 fibers. (A) Staining of CHO cells transfected with plasmids expressing mutated and wild-type CD46 with CCP1-specific J4-48 or CCP2-specific M75 antibodies 24 hours after transfection. Staining of primary antibodies was developed with secondary Alexa-488-conjugated antibody, and the percentage of positively stained cells was measured by flow cytometry. (B) Susceptibility of CHO cells transfected with plasmids expressing CCP2-mutated CD46 to infection with viruses possessing either Ad35 or Ad11 fibers. Twenty-four hours after transfection, cells were infected with the indicated vectors at an MOI of 10 PFU/cell. Twenty four hours later, cells were analyzed by flow cytometry for the expression of virus-encoded GFP transgene. Error bars indicate standard deviations; n = 4.
DISCUSSION
Understanding the precise mode of virus interaction with its cellular receptor is fundamental for unraveling the mechanisms underlying virus pathogenesis and for the development of novel antiviral therapeutics. Several groups have recently identified CD46 as a cellular receptor for adenovirus serotypes from group B as well as serotype 37 from group D (9, 40, 48, 55). The goal of this study was to map the Ad binding site(s) within CD46. Because the crystal structure of the full-length CD46 is not available, most of the published studies on the interaction between CD46 and its natural ligands have been done using antibody mapping and CD46 mutants (3, 25). Utilizing similar approaches, we demonstrate here that a major Ad35 and Ad11 fiber binding site is localized within the CCP2 domain of CD46. Several lines of evidence support this conclusion. (i) CCP1- and CCP2-interacting measles virus H protein efficiently competes with Ad35 fiber knob domain for binding to CD46-expressing CHO-C2 cells (Fig. 1). (ii) CCP2 domain-specific antibody M75 blocks Ad35 fiber-mediated infection of CD46-expressing CHO-C2 cells (Fig. 3). (iii) Truncated CD46 containing CCP1 and CCP2 domains (when expressed together) supports virus infection on otherwise resistant CHO cells (Fig. 5B). (iv) Amino acid substitutions at positions 130 to 135 or 152 to 156 within the CCP2 domain abrogate the adenovirus receptor function of the full-length CD46 protein (Fig. 7 and 8).
Although we showed a crucial role of CCP2 in Ad binding, our data also indicate that the presence of an intact CCP1 domain is ultimately required for CCP2 to correctly present Ad35 binding sites. Detailed analyses of measles virus and C4b binding epitopes using mutated CD46 and CCP domain-specific antibodies showed that while CD46 is a highly flexible molecule (21, 34), its conformation can change significantly after introduction of mutations (8, 15, 16, 34). Utilizing CCP1-specific monoclonal and polyclonal antibodies, we found that the CCP1 domain maintains its conformation independently of whether it is expressed alone or in combination with the CCP2 domain. Because if expressed alone the CCP1 domain could not support infection of CHO cells with Ad35 fiber-possessing vectors, we concluded that the Ad binding site is not localized within CCP1. Unexpectedly, the CCP2 domain alone was also unable to support Ad infection (plasmid pII/4, Fig. 5). Studies with CCP2-specific antibodies revealed that the CCP2 domain cannot be recognized when it is expressed as an individual domain. However, when CCP2 is expressed together with CCP1 (plasmid pI+II/4, Fig. 5B), CCP2 antibody recognition and Ad5/35 infection were restored. These data suggest that CCP1 is required to maintain CCP2 in a conformation that allows for Ad binding.
We showed that preincubation of CD46-expressing CHO-C2 cells with the CCP2-specific M75 antibody efficiently blocked Ad5/35GFP infection (Fig. 3). Furthermore, introduction of amino acid substitutions within CCP2 at positions 130 to 135, replacing GYYLIG with SFVKIP, or at positions 152 to 156, replacing GKPPI with DIEEF, completely abrogated both the CCP2 recognition by the M75 antibody, and the Ad receptor function of CD46. We therefore concluded that CCP2 areas encompassing amino acids 130 to 135 and 152 to 156 are critical Ad binding sites. These areas of CCP2 are conserved between humans and primates (15). (In this context, it is notable that, in contrast to the case for humans, CD46 is expressed on monkey erythrocytes and that an Ad35 fiber knob-possessing vector causes efficient hemagglutination of monkey but not human erythrocytes [45].) In the crystal structure of the two N-terminal domains of CD46, areas 130 to 135 and 152 to 156 localize to the same plane of the molecule, whereas the other CCP2 area that was analyzed (aa 105 to 110) is localized at the opposite plane of CD46. Because substitution of aa 105 to 110 had no effect on CCP2 recognition by the M75 antibody, and because the CD46105-110 mutant did not affect infection with Ad35 and Ad11 fiber-possessing vectors, it is unlikely that aa 105 to 110 represent a virus recognition site. Although we found that amino acid substitutions at positions 130 to 135 or 152 to 156 were sufficient to abrogate the Ad receptor function, this does not exclude the possibility that other areas in CD46 are involved in Ad binding. Furthermore, our finding that single amino acid substitutions within these two CCP2 domains have no significant effect on Ad-CD46 interaction indicates that the Ad binding site is formed by a complex structure or conformation.
Upon binding to CAR and following internalization into the cell, Ad5 fiber-containing particles rapidly escape from the endosomal compartment (11, 12, 43, 44). For endosome escape to efficiently occur, the fiber protein must be released from the virion (11). We showed, however, that an Ad5 vector (Ad5/35L) that possesses Ad35-derived fiber knob domains and binds to CD46 was unable to rapidly escape from endosomes (43). Since the only difference between Ad5 and Ad5/35L is the nature of the fiber knob domain and consequently the cellular attachment receptor (CD46 versus CAR), we concluded that Ad35 fiber knob binding to CD46 prevents rapid fiber release and endosome escape. Ad fibers possess trimeric knob domains, and this structure is critical for virus infection. While Ad5-CAR interaction occurs via binding of individual fiber knob domains to CAR monomers (20, 37), binding of the Ad12 fiber knob to CAR involves amino acid residues located within two adjacent knob domain monomers (2). We speculate that, similar to the case for Ad12, two Ad35 knob monomers interact with different CD46 areas and that this interaction produces a rigid virus-receptor complex and affects endosome escape. This hypothesis is supported by recent findings that the trimeric Ad fiber is a highly flexible molecule (53, 54), implying that rotation or shift of the fiber knob trimer along its vertical axis is required for its release from the virion body. While this is possible in Ad5-CAR interaction (because each single Ad5 knob domain monomer binds to a CAR monomer), the binding of Ad35 fiber knob monomers to multiple (different) areas within CD46 might inhibit the fiber knob motility and prevent fiber release from the virion body, thus trapping the virus within the endosomal compartment until the CD46-fiber knob complex is proteolytically processed. Clearly, without a crystal structure of the Ad35 knob-CD46 complex, these thoughts merely remain speculations.
Our data also indicate that the topology of Ad35 receptor recognition appears to be different from those of a number of other viruses. For human immunodeficiency virus, poliovirus, rhinovirus, CAR-binding adenovirus serotypes, and other viruses, the attachment site is localized predominantly at the tip of the most distal receptor domain. Measles virus, Epstein-Barr virus, and echovirus require interaction with two receptor domains for efficient infection (3, 7, 30). We demonstrate that Ad35 and Ad11 fiber knob binding domains are localized within the second N-terminal domain of the receptor. The functional significance of interaction with this area remains to be determined.
Several groups have recently identified CD46 as a cellular receptor for adenovirus serotypes from group B as well as serotype 37 from group D. The group B adenoviruses consist of 9 serotypes (Ad3, -7, -11, -14, -16, -21, -34, -35, and -50) that are highly diverse in their fiber knob domains (50). Previously, Segerman et al. used an antibody to CD46 directed against CCP3/4 to block binding of a group B adenovirus, Ad11 (40). In this study we demonstrate that an Ad5/Ad11 chimeric virus requires CCP2 for infection and that CCP3 and CCP4 are unlikely to be important for this interaction. It is noteworthy that Wu et al. found that Ad37, a group D adenovirus, interacts with CD46 and that this interaction can be blocked by an antibody directed against CCP1 (55). This indicates that Ad37 uses a different binding site than Ad35 and Ad11 fiber-containing Ads and agrees with previous observations that the cellular tropisms of these serotypes differ greatly; Ad35 and Ad11 (Slobitsky strain) primarily infect the urogenital tract (47), while Ad37 infects the conjunctiva (51). Recently it was also suggested that the involvement of different CD46 isoforms or coreceptors might account for this differential tropism (6). Our previous finding that Ad35 can bind to all four CD46 isoforms (9) argues somewhat against this idea.
The identification of the Ad35 binding domain within CD46 is important for a better understanding of virus-cell interaction and the identification of the domains within the Ad fiber knob that are involved in CD46 binding. The data obtained in this study might help in the design of antiviral therapeutics against adenovirus serotypes that use CD46.
Acknowledgments
We thank Roberto Cattaneo, Yasuko Mori, and Eric Hsu for providing valuable materials.
This work was supported by NIH grants CA80192, DK47754, and HL-00-008 (to A.L.) and AI060807 (to D.M.S.) and grants from the Cystic Fibrosis Foundation and the Doris Duke Charitable Foundation.
REFERENCES
- 1.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 2.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz, C. J., D. Koller, P. Devaux, C. Mumenthaler, J. Schneider-Schaulies, W. Braun, D. Gerlier, and R. Cattaneo. 1997. Mapping of the primary binding site of measles virus to its receptor CD46. J. Biol. Chem. 272:22072-22079. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, C. J., U. Schneider, P. Devaux, D. Gerlier, and R. Cattaneo. 1996. Cell entry by measles virus: long hybrid receptors uncouple binding from membrane fusion. J. Virol. 70:3716-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casasnovas, J. M., M. Larvie, and T. Stehle. 1999. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 18:2911-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattaneo, R. 2004. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens' magnet. J. Virol. 78:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarkson, N. A., R. Kaufman, D. M. Lublin, T. Ward, P. A. Pipkin, P. D. Minor, D. J. Evans, and J. W. Almond. 1995. Characterization of the echovirus 7 receptor: domains of CD55 critical for virus binding. J. Virol. 69:5497-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devaux, P., C. J. Buchholz, U. Schneider, C. Escoffier, R. Cattaneo, and D. Gerlier. 1997. CD46 short consensus repeats III and IV enhance measles virus binding but impair soluble hemagglutinin binding. J. Virol. 71:4157-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 10.Giannakis, E., T. S. Jokiranta, R. J. Ormsby, T. G. Duthy, D. A. Male, D. Christiansen, V. A. Fischetti, C. Bagley, B. E. Loveland, and D. L. Gordon. 2002. Identification of the streptococcal M protein binding site on membrane cofactor protein (CD46). J. Immunol. 168:4585-4592. [DOI] [PubMed] [Google Scholar]
- 11.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 12.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 13.Greenstone, H. L., F. Santoro, P. Lusso, and E. A. Berger. 2002. Human herpesvirus 6 and measles virus employ distinct CD46 domains for receptor function. J. Biol. Chem. 277:39112-39118. [DOI] [PubMed] [Google Scholar]
- 14.Hara, T., A. Kojima, H. Fukuda, T. Masaoka, Y. Fukumori, M. Matsumoto, and T. Seya. 1992. Levels of complement regulatory proteins, CD35 (CR1), CD46 (MCP) and CD55 (DAF) in human haematological malignancies. Br. J. Haematol. 82:368-373. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, E. C., R. E. Dorig, F. Sarangi, A. Marcil, C. Iorio, and C. D. Richardson. 1997. Artificial mutations and natural variations in the CD46 molecules from human and monkey cells define regions important for measles virus binding. J. Virol. 71:6144-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, E. C., S. Sabatinos, F. J. Hoedemaeker, D. R. Rose, and C. D. Richardson. 1999. Use of site-specific mutagenesis and monoclonal antibodies to map regions of CD46 that interact with measles virus H protein. Virology 258:314-326. [DOI] [PubMed] [Google Scholar]
- 17.Kallstrom, H., D. Blackmer Gill, B. Albiger, M. K. Liszewski, J. P. Atkinson, and A. B. Jonsson. 2001. Attachment of Neisseria gonorrhoeae to the cellular pilus receptor CD46: identification of domains important for bacterial adherence. Cell Microbiol. 3:133-143. [DOI] [PubMed] [Google Scholar]
- 18.Kallstrom, H., M. K. Liszewski, J. P. Atkinson, and A. B. Jonsson. 1997. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25:639-647. [DOI] [PubMed] [Google Scholar]
- 19.Kinugasa, N., T. Higashi, K. Nouso, H. Nakatsukasa, Y. Kobayashi, M. Ishizaki, N. Toshikuni, K. Yoshida, S. Uematsu, and T. Tsuji. 1999. Expression of membrane cofactor protein (MCP, CD46) in human liver diseases. Br. J. Cancer 80:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol. 74:2804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirkitadze, M. D., and P. N. Barlow. 2001. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol. Rev. 180:146-161. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., V. R. Melanson, A. M. Mirza, and R. M. Iorio. 2005. Decreased dependence on receptor recognition for the fusion promotion activity of L289A-mutated Newcastle disease virus fusion protein correlates with a monoclonal antibody-detected conformational change. J. Virol. 79:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liszewski, M. K., and J. P. Atkinson. 1996. Membrane cofactor protein (MCP; CD46). Isoforms differ in protection against the classical pathway of complement. J. Immunol. 156:4415-4421. [PubMed] [Google Scholar]
- 24.Liszewski, M. K., T. C. Farries, D. M. Lublin, I. A. Rooney, and J. P. Atkinson. 1996. Control of the complement system. Adv. Immunol. 61:201-283. [DOI] [PubMed] [Google Scholar]
- 25.Liszewski, M. K., M. Leung, W. Cui, V. B. Subramanian, J. Parkinson, P. N. Barlow, M. Manchester, and J. P. Atkinson. 2000. Dissecting sites important for complement regulatory activity in membrane cofactor protein (MCP; CD46). J. Biol. Chem. 275:37692-37701. [DOI] [PubMed] [Google Scholar]
- 26.Maisner, A., J. Alvarez, M. K. Liszewski, D. J. Atkinson, J. P. Atkinson, and G. Herrler. 1996. The N-glycan of the SCR 2 region is essential for membrane cofactor protein (CD46) to function as a measles virus receptor. J. Virol. 70:4973-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manchester, M., J. E. Gairin, J. B. Patterson, J. Alvarez, M. K. Liszewski, D. S. Eto, J. P. Atkinson, and M. B. Oldstone. 1997. Measles virus recognizes its receptor, CD46, via two distinct binding domains within SCR1-2. Virology 233:174-184. [DOI] [PubMed] [Google Scholar]
- 28.Manchester, M., A. Valsamakis, R. Kaufman, M. K. Liszewski, J. Alvarez, J. P. Atkinson, D. M. Lublin, and M. B. Oldstone. 1995. Measles virus and C3 binding sites are distinct on membrane cofactor protein (CD46). Proc. Natl. Acad. Sci. USA 92:2303-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer, K., T. Krey, V. Moennig, H. J. Thiel, and T. Rumenapf. 2004. CD46 is a cellular receptor for bovine viral diarrhea virus. J. Virol. 78:1792-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, M. D., M. J. Cannon, A. Sewall, M. Finlayson, M. Okimoto, and G. R. Nemerow. 1991. Inhibition of Epstein-Barr virus infection in vitro and in vivo by soluble CR2 (CD21) containing two short consensus repeats. J. Virol. 65:3559-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori, Y., X. Yang, P. Akkapaiboon, T. Okuno, and K. Yamanishi. 2003. Human herpesvirus 6 variant A glycoprotein H-glycoprotein L-glycoprotein Q complex associates with human CD46. J. Virol. 77:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray, K. P., S. Mathure, R. Kaul, S. Khan, L. F. Carson, L. B. Twiggs, M. G. Martens, and A. Kaul. 2000. Expression of complement regulatory proteins—CD 35, CD 46, CD 55, and CD 59—in benign and malignant endometrial tissue. Gynecol. Oncol. 76:176-182. [DOI] [PubMed] [Google Scholar]
- 33.Okada, N., M. K. Liszewski, J. P. Atkinson, and M. Caparon. 1995. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc. Natl. Acad. Sci. USA 92:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Leary, J. M., K. Bromek, G. M. Black, S. Uhrinova, C. Schmitz, X. Wang, M. Krych, J. P. Atkinson, D. Uhrin, and P. N. Barlow. 2004. Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces. Protein Sci. 13:1238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipson, L., K. Lonberg-Holm, and U. Pettersson. 1968. Virus-receptor interaction in an adenovirus system. J. Virol. 2:1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell, D. F., S. M. Russell, N. J. Deacon, M. A. Brown, D. J. Hooker, and I. F. McKenzie. 1991. Alternatively spliced RNAs encode several isoforms of CD46 (MCP), a regulator of complement activation. Immunogenetics 33:335-344. [DOI] [PubMed] [Google Scholar]
- 37.Roelvink, P. W., G. Mi Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 38.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 39.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 40.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seya, T., A. Hirano, M. Matsumoto, M. Nomura, and S. Ueda. 1999. Human membrane cofactor protein (MCP, CD46): multiple isoforms and functions. Int. J. Biochem. Cell Biol. 31:1255-1260. [DOI] [PubMed] [Google Scholar]
- 42.Seya, T., J. R. Turner, and J. P. Atkinson. 1986. Purification and characterization of a membrane protein (gp45-70) that is a cofactor for cleavage of C3b and C4b. J. Exp. Med. 163:837-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shayakhmetov, D. M., Z. Y. Li, V. Ternovoi, A. Gaggar, H. Gharwan, and A. Lieber. 2003. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J. Virol. 77:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shenk, T. 2001. Adenoviridae, p. 2265-2301. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 47.Shields, A. F., R. C. Hackman, K. H. Fife, L. Corey, and J. D. Meyers. 1985. Adenovirus infections in patients undergoing bone-marrow transplantation. N. Engl. J. Med. 312:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Sirena, D., B. Lilienfeld, M. Eisenhut, S. Kalin, K. Boucke, R. R. Beerli, L. Vogt, C. Ruedl, M. F. Bachmann, U. F. Greber, and S. Hemmi. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 78:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stecher, H., D. M. Shayakhmetov, G. Stamatoyannopoulos, and A. Lieber. 2001. A capsid-modified adenovirus vector devoid of all viral genes: assessment of transduction and toxicity in human hematopoietic cells. Mol. Ther. 4:36-44. [DOI] [PubMed] [Google Scholar]
- 50.Stone, D., A. Furthmann, V. Sandig, and A. Lieber. 2003. The complete nucleotide sequence, genome organization, and origin of human adenovirus type 11. Virology 309:152-165. [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi, R., Y. Nomura, M. Kojima, E. Uchio, N. Kobayashi, and M. Matumoto. 1990. A nosocomial outbreak of epidemic keratoconjunctivitis due to adenovirus type 37. Microbiol. Immunol. 34:749-754. [DOI] [PubMed] [Google Scholar]
- 52.Thorsteinsson, L., G. M. O'Dowd, P. M. Harrington, and P. M. Johnson. 1998. The complement regulatory proteins CD46 and CD59, but not CD55, are highly expressed by glandular epithelium of human breast and colorectal tumour tissues. APMIS 106:869-878. [DOI] [PubMed] [Google Scholar]
- 53.Wu, E., and G. R. Nemerow. 2004. Virus yoga: the role of flexibility in virus host cell recognition. Trends Microbiol. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 54.Wu, E., L. Pache, D. J. Von Seggern, T. M. Mullen, Y. Mikyas, P. L. Stewart, and G. R. Nemerow. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J. Virol. 77:7225-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, E., S. A. Trauger, L. Pache, T. M. Mullen, D. J. von Seggern, G. Siuzdak, and G. R. Nemerow. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J. Virol. 78:3897-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]