Abstract
Interferon regulatory factors (IRFs) are a family of transcription factors involved in the regulation of the interferons (IFNs) and other genes that may have an essential role in antiviral defense in the central nervous system, although this is currently not well defined. Therefore, we examined the regulation of IRF gene expression in the brain during viral infection. Several IRF genes (IRF-2, -3, -5, -7, and -9) were expressed at low levels in the brain of uninfected mice. Following intracranial infection with lymphocytic choriomeningitis virus (LCMV), expression of the IRF-7 and IRF-9 genes increased significantly by day 2. IRF-7 and IRF-9 gene expression in the brain was widespread at sites of LCMV infection, with the highest levels in infiltrating mononuclear cells, microglia/macrophages, and neurons. IRF-7 and IRF-9 gene expression was increased in LCMV-infected brain from IFN-γ knockout (KO) but not IFN-α/βR KO animals. In the brain, spleen, and liver or cultured glial and spleen cells, IRF-7 but not IRF-9 gene expression increased with delayed kinetics in the absence of STAT1 but not STAT2 following LCMV infection or IFN-α treatment, respectively. The stimulation of IRF-7 gene expression by IFN-α in glial cell culture was prevented by cycloheximide. Thus, (i) many of the IRF genes were expressed constitutively in the mouse brain; (ii) the IRF-7 and IRF-9 genes were upregulated during viral infection, a process dependent on IFN-α/β but not IFN-γ; and (iii) IRF-7 but not IRF-9 gene expression can be stimulated in a STAT1-independent but STAT2-dependent fashion via unidentified indirect pathways coupled to the activation of the IFN-α/β receptor.
Alpha interferon (IFN-α) and IFN-β belong to a family of cytokines that have a function in innate defense against viral infection by directly inhibiting viral replication (17, 29), inducing apoptosis of infected cells, mobilizing macrophages and NK cells, and stimulating chemokine and cytokine secretion (8, 18). In addition, the IFNs regulate the adaptive arm of the immune response that spearheads antiviral defenses such as CD4+ and CD8+ T-cell responses and antibody production (12, 13). Thus, IFNs play a vital role in limiting the dissemination and promoting the clearance of pathogens. However, IFN overproduction is also associated with the development and progression of certain autoimmune disorders such as type I diabetes (32) and other diseases such as the neurological disorder Aicardes-Goutieres syndrome (20). As such, interferon production is normally tightly regulated in a process that involves the interferon regulatory factors (IRFs).
IRFs are a family of transcription factors with diverse functions that include host defense, cell cycle regulation, apoptosis, oncogenesis, and immune cell development and homeostasis (for reviews, see references 9, 21, and 33). Currently, there are 10 members of the mammalian IRF family (IRFs 1 to 10), all of which contain a conserved DNA binding domain. The DNA binding domain is located at the amino termini of IRFs and consists of a five-tryptophan repeat that binds to a specific GAAA genomic sequence that is similar to the IFN-stimulated response element (ISRE). The IRFs become activated via phosphorylation at their carboxyl termini, after which they translocate from the cytoplasm to the nucleus to affect transcription of ISRE-containing genes. The various IRFs differ in cellular localization, structural properties, and activation-induced stimuli, thus conferring each IRF with unique functions. For example, IRF-2 has an inhibitory domain that represses the transcriptional activation of other IRFs such as IRF-1. IRF-5 is localized primarily to lymphoid tissue except for the thymus, while IRF-3 and IRF-7 are both constitutively expressed, but only IRF-7 expression is regulated. Interestingly, a well-characterized collaboration exists between IRF-3 and IRF-7 that serves to amplify the antiviral response. Specifically, during viral infection, IRF-3 has an early role in inducing transcription of IFN-α4 and IFN-β that can then induce IRF-7 expression. The newly synthesized IRF-7 then promotes transcription of different IFN subunits, thereby creating a positive-feedback mechanism that augments the host antiviral response. The IRFs are therefore vital components of the host defense against viral and bacterial infections in the periphery, and previous studies have made significant contributions toward our understanding of the biology of these transcription factors.
The central nervous system (CNS) is susceptible to viral infection, and mounting an appropriate antiviral response to clear infectious agents has to be balanced against the potential for immune-mediated neuronal injury and loss that would be catastrophic for the host. Currently, it is unknown the extent to which, or even whether, IRFs participate in antiviral responses in the CNS. Therefore, as a first step towards clarifying some of these issues, here we investigated the expression and regulation of the IRF genes in the mouse brain following intracranial (i.c.) infection with the Armstrong (ARM) strain of lymphocytic choriomeningitis virus (LCMV).
MATERIALS AND METHODS
Animals.
Transgenic mice that were null for gamma interferon (GKO) (strain C57BL/6 × 129/Sv; obtained from Timothy Stewart, Genentech Inc., San Francisco, CA), IFN-α/β receptor (IFNAR KO) (C57BL/6 backcrossed from 129/Sv/Ev × C57BL/6; kindly provided by Luca Guidotti, The Scripps Research Institute, La Jolla, CA), signal transducers and activators of transcription (STAT)-1 (STAT1 KO) (strain C57BL/6 × 129/Sv/Ev; obtained from Robert Schreiber, Washington University School of Medicine, St. Louis, MO), STAT-2 (STAT2 KO) (strain C57BL/6 × 129/Sv; provided by Chris Schindler, Columbia University, New York, NY), and wild-type (WT) (strain C57BL/6 × 129/Sv/Ev) mice were used in this study. The genotypes of the mice were maintained by interbreeding and verified by PCR analysis of tail DNA. Handling of mice and experimental procedures were conducted in accordance with the National Institutes of Health Guidelines for Animal Care and Use under an approved permit from The Scripps Research Institute Animal Ethics Committee.
LCMV infection.
Adult mice for each genotype, aged around 3 months, were injected intracranially into the frontal cortex with 250 PFU of the Armstrong 53b strain of LCMV as described previously (15). At 2, 4, and 6 days following infection, the animals were euthanized and their brains and other organs were removed for RNase protection assay (RPA), histological analysis, and in situ hybridization.
Mixed glial cell and splenocyte culture.
Glial cell cultures (astrocytes and microglia) were derived from the brain of 4-day-old WT, STAT1 KO, and STAT2 KO mice. Briefly, the cerebral cortices from three mice of each genotype were placed in modified Eagle's medium (Invitrogen, Carlsbad, CA) with penicillin-streptomycin-l-glutamine (Invitrogen), and the meninges were removed. The cortices were then placed in 1 ml of complete Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and penicillin-streptomycin-l-glutamine (all from Invitrogen), minced, vortexed at high speed for 1 min, and passed though an 18.5-gauge needle. The ensuing mixture was filtered successively through sterile 80-μm and 11-μm filters (Millipore, Bedford, MA) using a 25-mm Swinnex syringe filter holder (Millipore). The filtered cells were then diluted up to 1 ml with complete DMEM, plated into three 75-cm2 tissue culture flasks containing 10 ml of complete DMEM, and placed in a 5% aerated CO2 incubator kept at 37°C.
Erythrocyte-free spleen cell suspensions were prepared from 2- to 3-month-old wild-type, STAT1 KO, or STAT2 KO mice. The splenocytes (5 × 106 viable cells) were resuspended in RPMI 1640 medium (Invitrogen) containing 10% bovine serum albumin and used immediately for IFN-α stimulation as described below.
Murine recombinant IFN-α (PBL Biomedical Laboratories, New Brunswick, NJ) was added (final concentration, 1,000 U/ml) to the flasks of confluent glial cells or splenocytes, and the cultures were placed in a 5% aerated CO2 incubator at 37°C for 2, 6, or 12 h. Thereafter, total RNA was extracted using TRIZOL reagent (Life Technologies, Gaithersburg, MD).
RNase protection assay.
Poly(A)+ RNA was isolated from snap-frozen hemibrain by an oligo(dT) cellulose (Ambion, Austin, TX) method (7). RPAs were performed, and RNA levels were quantified from autoradiographs by densitometry using NIH Image software (version 1.31) as described previously (31). A set of RPA probes for the different IRF mRNAs was constructed as described previously (26). Briefly, target sequences were synthesized by reverse transcription-PCR, cloned into the pGEM-4Z vector (Promega, Madison, WI), and verified by sequence analysis using methods described previously (31). The targeted sequences were IRF-1, nucleotides (nt) 211 to 326 (GenBank accession number M21065); IRF-2, nt 431 to 561 (GenBank accession number J03168); IRF-3, nt 1219 to 1399 (GenBank accession number U75839); IRF-5, nt 61 to 286 (GenBank accession number AF028725); IRF-6, nt 301 to 551 (GenBank accession number U73029); IRF-7, nt 481 to 766 (GenBank accession number U73037); and IRF-9, nt 481 to 631 (GenBank accession number U51992). The construction and characterization of the other multiprobe set for the IFNs were described previously (3).
Dual in situ hybridization and immunohistochemistry.
LCMV-infected or control, uninfected brains were removed and fixed overnight in ice-cold 4% paraformaldehyde in phosphate-buffered saline (pH 7.4). Paraffin-embedded sections (8 μm) were incubated with 33P-labeled cRNA probes transcribed from linearized IRF-7, IRF-9, or LCMV-nucleoprotein (NP) RPA plasmids and processed for in situ hybridization as described previously (4, 15). Sections were then processed for immunohistochemistry to detect CD3+ T cells (rabbit anti-human CD3 antibody; DAKO Cytomation, California Inc., Carpinteria, CA), microglia (biotinylated lectin from Lycopersicon esculentum; Sigma, St. Louis, MO), neurons (mouse anti-human neurofilament antibody; Sternberger-Monoclonal Inc., Lutherville, MD), and astrocytes (rabbit anti-cow glial fibrillary acidic protein antibody; DAKO Cytomation). All antibodies were used at a final concentration of 5 μg/ml. Bound antibody was detected using Vectastain ABC kits (Vector Laboratories, Burlingame, CA), and diaminobenzidine/H2O2 reagent (Vector Laboratories) was used as the immunoperoxidase substrate.
RESULTS
IRFs are expressed in the uninfected and LCMV-infected murine brain.
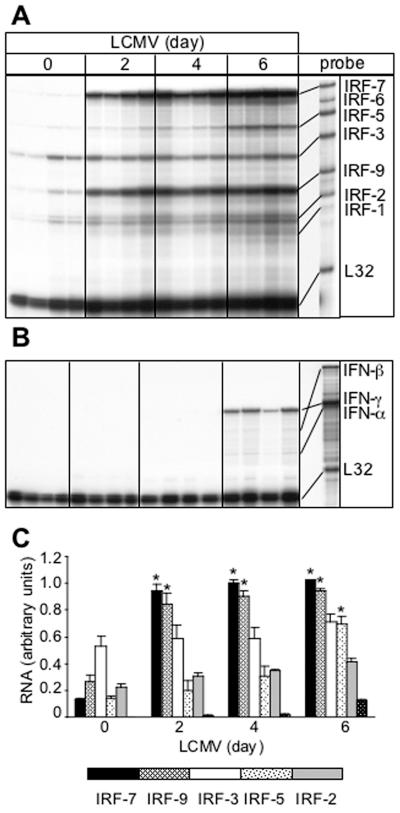
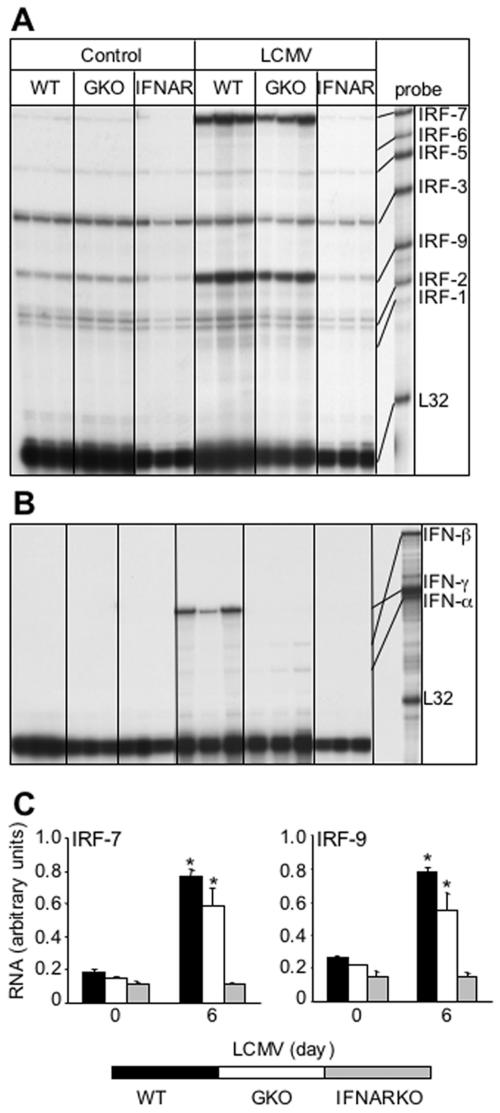
In brains from normal, uninfected mice, a number of IRF RNA transcripts were found to be constitutively present by RPA analysis. These included IRFs 2, 3, 5, 7, and 9, with IRF-3 showing the highest levels, while IRF-1 and IRF-6 RNA transcripts were not detectable (Fig. 1A and C). Following intracranial infection with LCMV, WT mice developed a classic lymphocytic choriomeningitis (LCM) phenotype (Table 1). Thus, by 4 to 5 days postinfection, these animals exhibited a huddled, hunched posture along with ruffled fur, ataxia, and progressive immobility. A few animals underwent seizure-induced death before the experimental endpoint at day 6 postinfection. Following intracranial infection with LCMV, a significant increase in the cerebral levels of the IRF-7 and IRF-9 mRNA transcripts was seen at day 2 postinfection and was maintained through day 6 postinfection (Fig. 1A and C). A significant increase in IRF-5 mRNA transcript was also observed at day 6 postinfection. IRF-2 and IRF-3 mRNA levels remained unaltered in the presence of LCMV, while no IRF-6 mRNA transcript was detected by RPA in the control or infected brains (Fig. 1A).
FIG. 1.
IRF (A) and IFN (B) mRNA levels in the brain following LCMV infection. In this representative experiment, C57BL/6 × 129/Sv/Ev WT mice were injected i.c. with saline or 250 PFU LCMV (ARM); poly(A)+ RNA was isolated from brains at 0, 2, 4, and 6 days after infection, and 2 μg was analyzed by RPA as outlined in Materials and Methods. (A) IRF-2, -3, -5, -7, and -9 mRNA transcripts were constitutively present in normal, uninfected brain (day 0). The IRF-1, -2, and -3 mRNA transcripts were unchanged following LCMV infection, whereas IRF-7 and IRF-9 mRNA transcripts were significantly (P < 0.05) upregulated from day 2 to day 6 postinfection. An increase in IRF-5 mRNA was also seen at day 6 following LCMV infection. (B) High IFN-γ but low IFN-α and IFN-β mRNA levels were detectable at day 6 postinfection only. (C) Quantification of IRF gene expression. Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31). Statistical analysis was performed using the Student's t test.
TABLE 1.
Phenotypes of wild-type and mutant mice infected intracranially with LCMV
| Genotype | Phenotype |
|---|---|
| WT | Classic LCM; hunchbacked huddled posture, piloerection, progressive immobility, convulsive seizures, and death from 6-8 days postinfection |
| GKO | Classic LCM; hunchbacked huddled posture, piloerection, progressive immobility, convulsive seizures, and death from 6-8 days postinfection |
| IFNAR KO | No physical or behavioral abnormalities; splenomegaly |
| STAT1 KO | Hunched posture, ruffled fur, unsteady gait, no convulsive seizures, moribund, with death from 7-14 days postinfection |
| STAT2 KO | No overt physical or behavioral abnormalities; splenomegaly |
Since the IRFs are known to be involved in the regulation of the IFN genes and these cytokines, in turn, can regulate IRF gene expression, we examined for the cerebral expression of IFN-β and IFN-α1 and IFN-γ. Similar to uninfected brain, no detectable level of any IFN-α/β mRNA was seen at day 2 or day 4 following LCMV infection, but very low levels of IFN-α and IFN-β mRNAs were detectable at day 6 postinfection (Fig. 1B). While IFN-γ mRNA was also undetectable in brains from uninfected mice and from mice at early stages of LCMV infection, mRNA transcripts for this cytokine were detectable at day 6 postinfection (Fig. 1B).
IRF-7 and IRF-9 RNA transcripts were localized to immune cells, neurons, and glial cells in LCMV-infected brain.
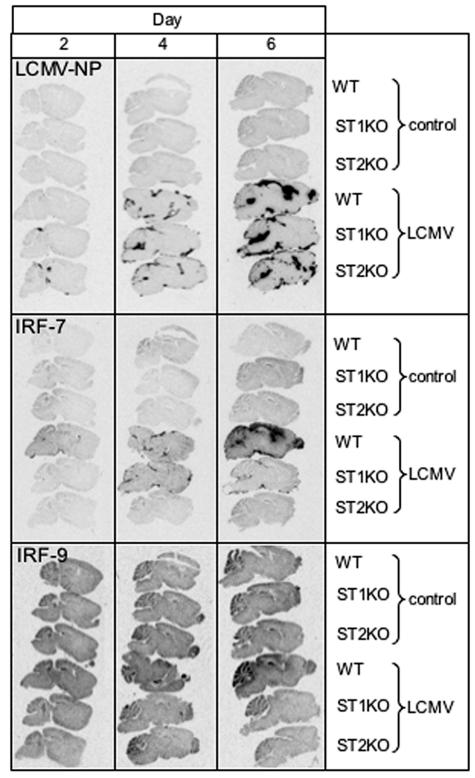
To determine the gross and cellular anatomical localization of IRF genes in the brain, in situ hybridization was performed in control and infected brains of WT animals. No detectable IRF-7 RNA hybridization above background levels was observed in uninfected brain, while diffuse hybridization of the IRF-9 probe was evident in adjacent sections (Fig. 2). Following LCMV infection, there was a progressive increase in IRF-7 RNA from day 2 to day 6 in WT brains (Fig. 2). At day 2 postinfection, some hybridization was visible in the choroid plexus, while strong hybridization for the IRF-7 cRNA probe was additionally observed in the meninges and ventricular surfaces at day 4 and day 6 postinfection. For IRF-9, increased RNA hybridization was localized to the meninges, hippocampus, and cerebellum from LCMV-infected brain at day 6 (Fig. 2). RNA hybridization corresponding to IRF-5 was detectable at low levels at day 6 postinfection only (not shown). In comparison with LCMV-NP, the IRF-7 and IRF-9 RNA hybridizations mapped to the principal sites of the virus infection in the brain at each time point examined (Fig. 2).
FIG. 2.
Localization of LCMV-NP, IRF-7, and IRF-9 mRNA in the brain. WT, STAT1 KO, and STAT2 KO mice were injected i.c. with saline or 250 PFU LCMV (ARM), and brains were removed at day 0, 2, 4, and 6 for in situ hybridization. Paraffin-embedded sagittal sections (8 μm) were hybridized with 33P-labeled cRNA probes transcribed from linearized IRF-7, IRF-9, or LCMV-NP RPA plasmids and exposed to Kodak MR film for 5 days as outlined in Materials and Methods. No hybridization of the LCMV-NP, IRF-7, or IRF-9 cRNA probes was evident in uninfected (control) brain. Following LCMV infection, LCMV-NP hybridization was evident from day 2 to day 6 postinfection in the meninges and ventricles of WT, STAT1 KO, and STAT2 KO brain sections. A similar pattern of hybridization was observed for IRF-7 RNA in infected WT brain, while STAT1 KO and STAT2 KO brain sections showed lower and no hybridization, respectively. For IRF-9, overall, there was higher constitutive RNA hybridization, but increased levels were seen in LCMV-infected WT brains at day 6 only in cortical and cerebellar regions as well as meninges.
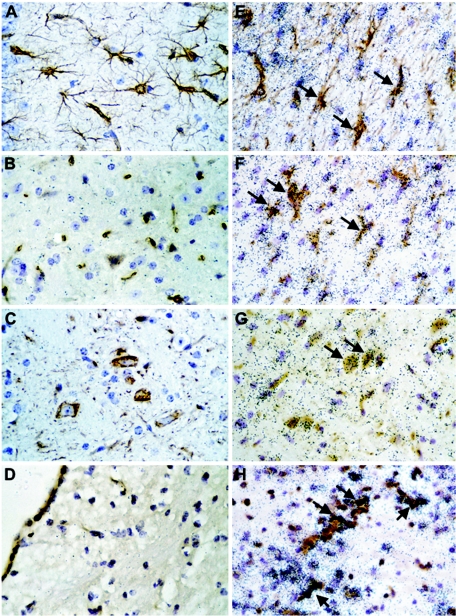
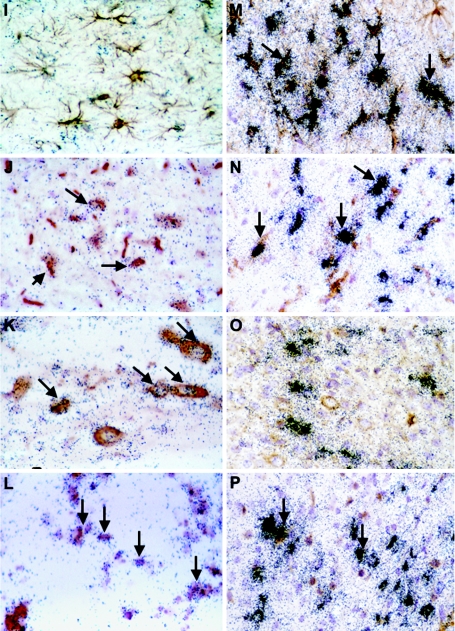
To determine which cells in the brain of infected WT mice were the sources of the IRF-7 and IRF-9 RNA, dual-label in situ hybridization and immunohistochemistry were performed. Compared with uninfected controls (Fig. 3A to D), in LCMV-infected WT mice, IRF-7 mRNA was predominantly localized to astrocytes (Fig. 3E, arrows), microglia (Fig. 3F, arrows), some neurons (Fig. 3G, arrows), and CD3+ T cells (Fig. 3H, arrows), while IRF-9 RNA was found to be mainly in neurons (Fig. 3K, arrows) and CD3+ T cells (Fig. 3L, arrows). In general, there was a correlation between IRF-7 and IRF-9 mRNA levels and the presence of LCMV-NP RNA. The viral RNA was detected in astrocytes (Fig. 3M, arrows), microglia (Fig. 3N, arrows), some neurons (Fig. 3O), and CD3+ T cells (Fig. 3P, arrows). In contrast with IRF-7 and IRF-9, the IRF-5 mRNA was localized exclusively to infiltrating mononuclear cells (not shown). Thus, the findings indicated that in addition to peripheral immune leukocytes, the IRF-7 and IRF-9 genes were also expressed by glial and neuronal cells in the brain during LCMV infection.
FIG. 3.
Cellular localization of IRF-7, IRF-9, and LCMV-NP mRNA in brain. WT mice were injected i.c. with saline or 250 PFU LCMV (ARM), and brains were removed at day 6 for in situ hybridization and immunohistochemistry. Eight-micron-thick paraffin-embedded sections were hybridized with 33P-labeled cRNA probes transcribed from linearized IRF-7, IRF-9, or LCMV-NP RPA plasmids and then processed for immunohistochemistry for cell-specific markers to identify CD3+ T cells, microglia, neurons, and astrocytes as outlined in Materials and Methods. Slides were coated with photographic emulsion, developed after 2 weeks, and visualized using bright-field microscopy. Photomicrographs of control (A to D) and day 6 LCMV-infected (E to P) brain sections hybridized for IRF-7 (E to H), IRF-9 (I to L), and LCMV-NP (M-P) mRNA and stained for astrocytes (A, E, I, M), microglia (B, F, J, N), neurons (C, G, K, O), and CD3+ T cells (D, H, L, P) are shown. IRF-7 hybridization was localized to astrocytes (E; arrows), microglia (F; arrows), neurons (G; arrows), and CD3+ T cells (H; arrows), while mainly microglia (I; arrows), neurons (K; arrows), and CD3+ T cells (L; arrows) expressed IRF-9 mRNA. Strong LCMV-NP hybridization was seen in astrocytes (M; arrows), microglia (N; arrows), some neurons (O), and CD3+ T cells (P; arrows).
Increased expression of the IRF-7 and IRF-9 genes in the brain during LCMV infection is dependent on IFN-α/β but not IFN-γ.
To determine whether IFN-α/β or IFN-γ regulated IRF expression in the CNS, mice that were null for either the IFN-α/β receptor (IFNAR KO) or IFN-γ (GKO) were infected with LCMV, and the IRF RNA levels were then determined by RPA. Consistent with previous reports (11, 25), following intracranial infection with LCMV, IFNAR KO mice failed to develop LCM, while GKO mice developed LCM indistinguishable from that of WT controls (Table 1). IFNAR KO animals did not show any overt physical signs of illness even up to 2 months postinfection. The only observable abnormality in these mice was splenomegaly.
Analysis of IRF gene expression revealed similar patterns and levels of IRF mRNAs in brains from uninfected wild-type, IFNAR KO, and GKO mice (Fig. 4). Compared with the IRF-2, IRF-3, and IRF-5 mRNA transcript levels, which remained largely unchanged, the IRF-7 and IRF-9 mRNA levels in the brain following LCMV infection were increased significantly at day 2 (not shown) and were maintained through day 6 postinfection in GKO mice, paralleling changes seen in these mRNA transcripts in similarly infected wild-type mice (Fig. 4A and C). However, in brain from IFNAR KO mice, there was no detectable change in the levels of either the IRF-7 or IRF-9 mRNAs at any time point following infection with LCMV (Fig. 4A and C). Therefore, following intracranial LCMV infection, IFN-γ is not required for development of LCM or the upregulation in IRF-7 and IRF-9 gene expression, while IFN-α/β is essential for both the development of LCM and the upregulation in IRF-7 and IRF-9 gene expression. These findings also suggested that the upregulation of IRF-7 and IRF-9 gene expression following LCMV infection is not due to the virus per se.
FIG. 4.
Comparison of IRF and IFN mRNA levels in the brain from WT, IFN-γ (GKO) KO, and IFN-α/β (IFNAR) KO mice. Mice were injected i.c. with saline (Control) or 250 PFU LCMV (ARM), and at day 6, poly(A)+ RNA was isolated from the brain and analyzed by RPA as outlined in Materials and Methods. (A) IRF-7 and IRF-9 mRNA levels were significantly (P < 0.05) upregulated in WT and GKO mice in response to the virus but remained unchanged in IFNAR KO animals. (B) High IFN-γ mRNA levels were observed only in WT animals, while very low levels of IFN-α and IFN-β RNA transcripts were present in WT and GKO brains. (C) Quantification of IRF gene expression. Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31).
With regard to IFN gene expression, IFN-γ mRNA was detectable only in WT mice at day 6 (Fig. 4B) but not at day 2 or day 4 postinfection (not shown). IFN-γ RNA was not detectable in brain from infected IFNAR KO mice or, as expected, in brain from GKO infected mice (Fig. 4B). Very low levels of IFN-α and IFN-β mRNA were detected in WT and GKO brains at day 6 postinfection only; however, no detectable IFN-α or IFN-β mRNA expression was seen in the brains of IFNAR KO mice at any time point postinfection (Fig. 4B).
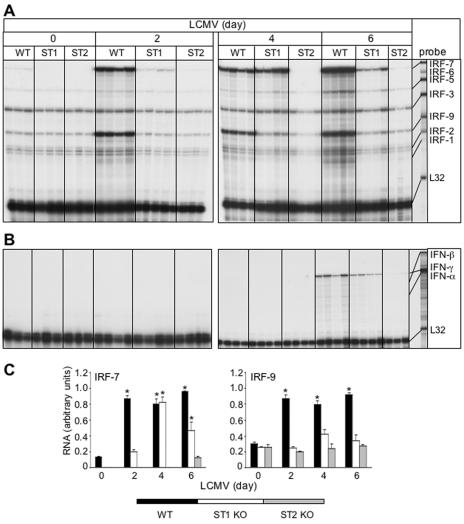
The IRF-7 and IRF-9 genes have a differential requirement for STAT1 following LCMV infection.
To determine whether the IRF-7 and IRF-9 genes were regulated through the classical interferon stimulated gene factor 3 (ISGF3)/ISRE IFN-α/β pathway that requires STAT1 and STAT2, we next investigated IRF-7 and IRF-9 gene expression in LCMV-infected mice that were deficient in either STAT1 or STAT2. These two groups of gene-disrupted mice differed in their phenotypic response to the LCMV (Table 1). STAT2 KO mice, like the IFNAR KO animals, showed no obvious signs of illness except for splenomegaly. By contrast, the STAT1 KO animals developed physical signs of illness that included a hunched posture, ruffled fur, immobility, and progressive weight loss, and they died between day 7 and day 14 postinfection. However, unlike typical LCM in WT and GKO mice, the LCMV-infected STAT1 KO animals did not have convulsive seizures.
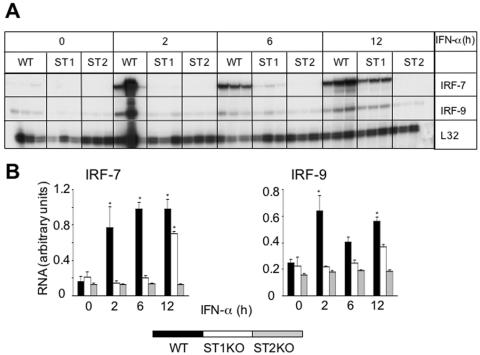
IRF gene expression was then determined in the brain of the STAT knockout mice. The brains from uninfected STAT1 KO and STAT2 KO mice displayed constitutive expression of IRF-2, -3, -5, and -9 mRNAs (Fig. 5A) but no detectable RNA transcripts for IRFs 1, 6, or 7 (Fig. 5A). Following LCMV infection, IRF-7 but not IRF-9 mRNA levels were upregulated in STAT1 KO mice (Fig. 5A and C). In the LCMV-infected STAT1 KO brain, increased IRF-7 gene expression was detectable at day 2 and reached maximum levels at day 4 postinfection and decreased at day 6 (Fig. 5A and C). By comparison, in WT animals, increased IRF-7 mRNA expression was maximal at day 2 postinfection and remained elevated to day 6 postinfection. Neither the level of IRF-7 mRNA nor the IRF-9 mRNA showed any significant alteration in brain from LCMV-infected STAT2 KO mice (Fig. 5A and C). Thus, these findings highlighted a differential requirement for STAT1 in the regulation of the IRF-7 and IRF-9 genes in the brain following LCMV infection.
FIG. 5.
Comparison of IRF and IFN mRNA levels in brains from WT, STAT1 KO (ST1), and STAT2 KO (ST2) mice. Mice were injected i.c. with saline or 250 PFU LCMV (ARM), and at 0, 2, 4, and 6 days, poly(A)+ RNA was isolated from the brains and analyzed by RPA as outlined in Materials and Methods. (A) IRF-7 and IRF-9 mRNA levels were significantly (P < 0.05) upregulated from day 2 to day 6 postinfection in WT brains. In STAT1 KO brain, a small increase in IRF-7 mRNA was detectable at day 2 postinfection and reached maximum levels at day 4 before decreasing at day 6 postinfection. By contrast, IRF-9 mRNA transcript levels remained unregulated throughout. LCMV did not modulate transcription of the IRF genes at any time point in STAT2 KO mice. (B) High levels of IFN-γ mRNA were seen only in infected WT and STAT1 KO brains at day 6 with a barely detectable level in STAT2 KO brain. IFN-α and IFN-β RNA levels were seen at low levels only in WT and STAT1 KO brains at day 6 postinfection. (C) Quantification of IRF gene expression. Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31).
We next examined the expression of the IFN genes in the different STAT KO mice following LCMV infection. The pattern of IFN-γ gene expression was similar for wild-type and STAT1 KO mice, with high levels evident in the brain at day 6 postinfection only (Fig. 5B). By contrast, this cytokine RNA was present at only barely detectable levels in the STAT2 KO brains. In the case of IFN-α and IFN-β, RNA transcripts for these cytokines were detectable at low levels at day 6 in the LCMV-infected WT and STAT1 KO mice but were absent in STAT2 KO mice (Fig. 5B).
To determine whether the IRF-7 and IRF-9 genes were differentially distributed in STAT1 KO and STAT2 KO brains compared with WT animals, gross anatomical localization of these genes was determined by in situ hybridization (Fig. 2). Similar to WT animals, no IRF-7 or IRF-9 RNA hybridization was detectable in uninfected STAT1 KO and STAT2 KO brains. In STAT1 KO brain following LCMV infection, no significant hybridization of the IRF-7 cRNA probe was detectable at day 2, while increased hybridization was evident mainly in the meninges and choroid plexus at day 4 before decreasing at day 6 and overlapped with sites of LCMV-NP RNA. In terms of cellular localization at day 4 postinfection, high IRF-7 hybridization was present in meningeal and ependymal cells as well as the choroid plexus cells and vascular endothelium. In adjacent regions, there were also numerous parenchymal cells positive for IRF-7 hybridization that included astrocytes and microglia. For the IRF-9 gene, in STAT1 KO mice, no significant mRNA hybridization was detectable above control levels at any time point postinfection. Similarly, no detectable change in hybridization for both IRF-7 and IRF-9 RNA transcripts was evident in brain sections from LCMV-infected STAT2 KO mice. Thus, LCMV-infected STAT1 KO and STAT2 KO mice differed in their hybridization levels and localization of the IRF-7 and IRF-9 mRNA. However, both of these genotypes had a similar temporal and spatial localization pattern of hybridization for the LCMV-NP gene (Fig. 2). Like WT mice, a progressive increase in hybridization and distribution of the LCMV-NP RNA was evident from day 2 to day 6 in infected STAT1 KO and STAT2 KO brains, with the choroid plexus, meninges, and ventricle surfaces being the principal areas of expression (Fig. 2).
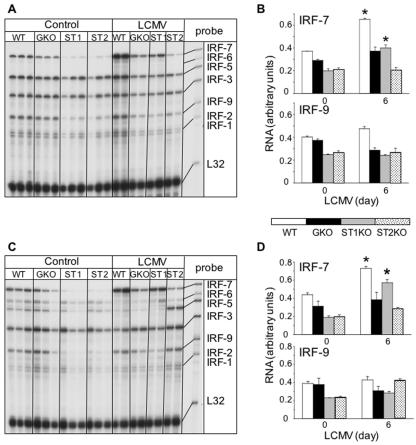
To clarify whether the changes in IRF gene expression in the brain were also found in the periphery, we examined the spleen and liver of mice infected with LCMV (Fig. 6). Tissue- and genotype-specific differences in the expression of individual IRF genes were apparent between the spleen and liver from uninfected mice. For example, high levels of IRF-5 mRNA were present in spleen (Fig. 6A) compared with those in the liver (Fig. 6C), and while IRF-6 mRNA was not detectable in spleen, this transcript was present in the liver. Compared with the WT animals, the levels of IRF-7 and IRF-9 mRNAs were lower in spleen and liver from uninfected STAT1 KO and STAT2 KO mice. At day 6 following LCMV infection, IRF-7 but not IRF-9 mRNA levels were upregulated significantly in both spleen and liver from STAT1 KO mice but not STAT2 KO mice (Fig. 6A to D). Thus, these findings indicated that, like the brain, there is a differential requirement for STAT1 in the regulation of the IRF-7 and IRF-9 genes in the spleen and liver following LCMV infection
FIG. 6.
IRF mRNA levels in spleen (A, B) and liver (C, D) of uninfected (Control) and LCMV-infected WT, IFN-γ (GKO), STAT1 KO (ST1), and STAT2 KO (ST2) mice. Mice were injected i.c. with saline or 250 PFU LCMV (ARM), poly(A)+ RNA was isolated from the spleen and liver 6 days after injection, and 2 μg was analyzed by RPA as outlined in Materials and Methods. (A, C) Autoradiographs showing that IRF-2, -3, -5, -7, and -9 mRNA transcripts were constitutively present in normal, uninfected (Control) spleen and liver and additionally showing IRF-6 in livers of WT and GKO mice. In STAT1 KO and STAT2 KO animals, IRF-2, -3, -5, -7, and -9 mRNA transcripts were also constitutively expressed in spleen and liver, but the levels of IRF-7 and IRF-9 mRNAs were lower than those of the wild type. (B, D) Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31). In both spleen (B) and liver (D), the level of IRF-7 mRNA increased significantly (*P < 05) after LCMV infection in wild-type and STAT1 KO mice compared with that of uninfected controls.
The IRF-7 and IRF-9 genes are differentially regulated in STAT1 KO and STAT2 KO glial cells and splenocytes following IFN-α stimulation.
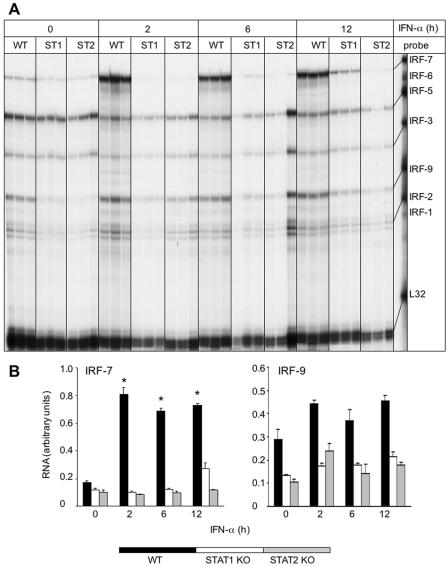
To further explore the regulation of IRF-7 and IRF-9 gene expression and identify potential mediators that influence the expression of these genes in the CNS, we employed mixed glial cell cultures that were isolated from the brain of neonatal WT, STAT1 KO, and STAT2 KO mice. These cells were stimulated with recombinant murine IFN-α for 2, 6, and 12 h, and the IRF-7 and IRF-9 mRNA levels were determined by RPA analysis. Unstimulated control glial cells from all three genotypes contained detectable levels of the IRF-2, IRF-3, IRF-5, IRF-7, and IRF-9 mRNAs, with WT cells displaying a slightly higher baseline expression (Fig. 7A and B). Similar to LCMV-infected mice, in response to IFN-α stimulation, IRF-7 RNA levels were significantly and maximally increased by 2 h in WT glial cells. In STAT1 KO glial cells, IRF-7 RNA levels were also significantly increased but only after a considerable delay, at 12 h post-IFN-α-stimulation. In contrast to WT and STAT1 KO cells, IRF-7 RNA levels remained unaltered in STAT2 KO glial cells following treatment with IFN-α. Furthermore, in contrast with IRF-7 and with the exception of a modest increase in WT glial cells, IRF-9 RNA levels were not significantly altered in the glial cells from all three genotypes (Fig. 7A and B).
FIG. 7.
Effect of IFN-α on IRF mRNA levels in mixed glial cell cultures. (A) Glial cells were prepared from the cerebral cortex of WT, STAT1 KO (ST1), or STAT2 KO (ST2) neonatal mice, and confluent cultures were incubated with 1,000 U/ml IFN-α for 2, 6, and 12 h as described in Materials and Methods. Total RNA was isolated with TRIZOL reagent, and 10 μg RNA was analyzed by RPA as outlined in Materials and Methods. (A) In glia treated with IFN-α, IRF-7 mRNA transcript showed a significant (P < 0.05) increase by 2 h in WT and at 12 h in STAT1 KO cells but was unchanged in STAT2 KO cells. For IRF-9 mRNA transcript, no significant increase was evident in glial cells following IFN-α stimulation. (B) Quantification of IRF-7 and IRF-9 gene expression. Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31).
Results which were essentially similar to those for the glial cells were observed for IRF-7 and IRF-9 gene expression in splenocytes after exposure to IFN-α (Fig. 8A and B). Thus, in response to IFN-α stimulation, IRF-7 RNA levels were significantly and maximally increased by 2 h in WT cells. In STAT1 KO splenocytes, IRF-7 RNA levels increased significantly after a considerable delay, at 12 h post-IFN-α-stimulation. In contrast to WT and STAT1 KO cells, IRF-7 RNA levels remained unaltered in STAT2 KO splenocytes following treatment with IFN-α. However, IRF-9 RNA levels were not significantly altered by IFN-α treatment in the splenocytes from any of the genotypes.
FIG. 8.
Effect of IFN-α on IRF mRNA levels in splenocytes. (A) Splenocytes were prepared from WT, STAT1 KO (ST1), or STAT2 KO (ST2) adult mice and incubated with 1,000 U/ml IFN-α for 2, 6, and 12 h as described in Materials and Methods. Total RNA was isolated with TRIZOL reagent, and 10 μg RNA was analyzed by RPA as outlined in Materials and Methods. (A) In splenocytes treated with IFN-α, IRF-7 mRNA transcript showed a significant (P < 0.05) increase by 2 h in WT and at 12 h in STAT1 KO cells but was unchanged in STAT2 KO cells. For IRF-9 mRNA transcript, no significant increase was evident in splenocytes following IFN-α stimulation. (B) Quantification of IRF-7 and IRF-9 gene expression. Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31).
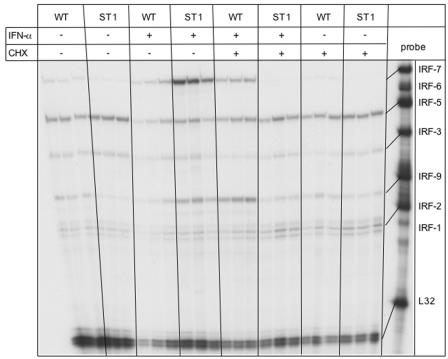
To determine whether production of an intermediate factor(s) may be required for the IFN-α-mediated upregulation of IRF-7 mRNA in STAT1 KO glial cells, WT and STAT1 KO glial cultures were incubated with IFN-α plus cycloheximide (CHX) for 12 h. CHX blocked the ability of IFN-α to upregulate IRF-7 RNA levels in STAT1 KO glial cells and reduced the level of IFN-α-upregulated IRF-7 mRNA in WT cells (Fig. 9). No detectable change in IRF-7 mRNA levels was seen in cells treated with CHX alone.
FIG. 9.
Effect of CHX on IFN-α-stimulated IRF7 mRNA levels in mixed glial cell cultures. Glial cell cultures were prepared and treated for 12 h with IFN-α as described in the legend of Fig. 6 and in Materials and Methods. Additional cultures were cotreated with CHX (20 μg/ml) as described in Materials and Methods. Total RNA was isolated with TRIZOL reagent, and 10 μg RNA was analyzed by RPA as outlined in Materials and Methods. IRF-7 mRNA transcript was increased in WT and STAT1 KO cells following IFN-α treatment, but this was blocked in STAT1 KO but not WT cells in the presence of CHX.
In summary, IRF-7 but not IRF-9 gene expression increased with delayed kinetics following IFN-α stimulation in STAT1 KO glial cells and splenocytes compared with that of the WT, while expression of this gene remained unresponsive to IFN-α stimulation in the absence of STAT2. Furthermore, upregulation of IRF-7 mRNA levels in glial cells lacking STAT1 could be blocked by the protein synthesis inhibitor cycloheximide.
IFN-α induction of IL-1α, IL-1β, and TNF gene expression in STAT1 KO glial cells.
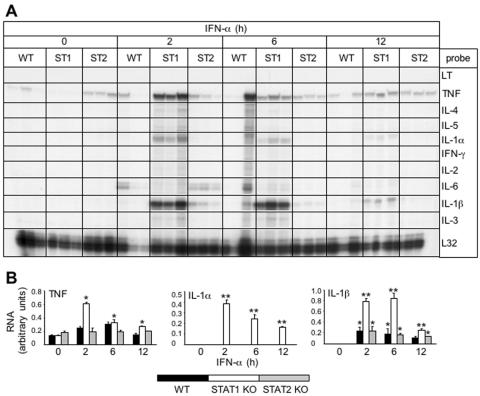
The temporal delay in IRF-7 gene expression in IFN-α-stimulated STAT1 KO glial cells and its inhibition by cycloheximide suggested the likely involvement of an indirect pathway leading to the activation of this gene following IFN-α stimulation. Cytokines such as tumor necrosis factor (TNF) have been shown to induce IRF-7 expression in mononuclear cells through an NF-κB regulatory site in the IRF-7 gene promoter (23). To determine whether cytokines might be induced in a STAT1-independent fashion by exposure to IFN-α, we tested for the expression of a number of proinflammatory cytokine genes in glial cell cultures (Fig. 10A and B). The expression of three cytokine genes, interleukin-1α (IL-1α), IL-1β, and TNF, was increased significantly in response to IFN-α treatment. In STAT1 KO glia, following exposure to IFN-α, a significant increase in the level of TNF RNA was seen at 2 h which remained elevated at 6 and 12 h. Similar to TNF, significantly increased levels of IL-1α and IL-1β RNA were also evident by 2 h following IFN-α treatment of the STAT1 KO glia. With the exception of a small increase in the levels of IL-1β and IL-6 RNA in STAT2 KO glia, IFN-α treatment did not significantly alter the level of RNA for any of the cytokines examined in these or WT glial cells. The results showed that depletion of STAT1 in glial cells resulted in a marked increase in IFN-α-stimulated TNF and IL-1 gene expression at a time that preceded IRF-7 gene upregulation (Fig. 7A and B).
FIG. 10.
Cytokine mRNA levels in mixed glial cell cultures. Glial cell cultures were prepared and treated for various times with IFN-α as described in the legend of Fig. 6 and in Materials and Methods. Total RNA was isolated with TRIZOL reagent, and 10 μg RNA was analyzed by RPA as outlined in Materials and Methods. (A) IL-1α, IL-1β, and TNF mRNA transcript levels increased significantly (*P < 0.05; **P < 0.005) from 2 h to 12 h in IFN-α-stimulated STAT1 KO cells. No detectable IL-1α mRNA transcripts were seen in WT and STAT2 KO glia, while a small but significant increase in IL-1β mRNA was detectable in these two genotypes after stimulation with IFN-α. TNF mRNA was present constitutively in WT and STAT2 KO glia and remained unchanged following IFN-α treatment. (B) Quantification of TNF, IL-1α, and IL-1β gene expression. Densitometric analysis of each lane was performed on scanned autoradiographs using NIH Image software (version 1.31).
DISCUSSION
The IRF transcription factors are vital components of host defense where they regulate the production of IFNs and are mediators important for mobilizing the innate and adaptive immune responses against invading pathogens (9, 21, 33). Our investigation of the temporal and spatial regulation of IRF gene expression in the murine CNS during LCMV infection revealed fundamental differences in the expression patterns of the different IRF genes. Thus, while IRF-2, -3, -5, -7, and -9 genes were constitutively expressed at different levels in the uninfected murine CNS, only the expression of the IRF-5, IRF-7, and IRF-9 genes was altered following LCMV infection. The expression of these latter three genes, which were all significantly upregulated, also displayed differences in their temporal and spatial expression. Thus, while IRF-7 and IRF-9 were upregulated early and expressed by both infiltrating immune cells and cells intrinsic to the brain, that of IRF-5 occurred late in infection and was restricted to the immune cell compartment. In all, these findings provide novel insights into the expression patterns and regulation of the IRF gene family following a viral infection in vivo and point to likely differing roles of the IRFs in antiviral defense in the brain.
A number of IRF genes (IRFs 2, 3, 5, 7, and 9) were expressed constitutively in the CNS. Since few immune cells circulate within the normal brain, it is likely that resident glia and neuronal cells were the source of these IRFs. However, due to the low levels of expression, it was not possible to localize precisely which cells expressed these IRF genes. These findings are on the whole consistent with previous reports showing constitutive expression of IRF-2 (33), IRF-3 (5), and IRF-9 (14) genes by a wide variety of cell types, while IRF-5 (10) and IRF-7 (6) genes are expressed predominantly by hematopoietic cells. However, after LCMV infection, the expression of the IRF-7 and IRF-9 genes was increased significantly and localized to the neural cells, astrocytes, microglia, and neurons as well as infiltrating CD3+ T cells. Therefore, in addition to peripheral cells, many of the major cells intrinsic to the CNS can at least express the IRF-7 and IRF-9 transcription factor genes. This is also supported by the in vitro data showing that IRF-7 and IRF-9 gene expression occurred in mixed glial cell cultures that consisted of astrocytes and microglia. These cultures also constitutively expressed IRF-2, -3, and -5 mRNA that was not regulated by IFN-α. The constitutive expression of these IRFs in CNS cells may comprise a “ready-to-battle” antiviral mechanism in the brain similar to the well-defined interaction between IRF-3 and IRF-7 demonstrated in peripheral cells during viral infection (9, 21, 33). In those cells, constitutively expressed IRF-3 is directly phosphorylated and thus activated by viruses to mediate the gene induction of various IFN species and IRF-7 that create a potent antiviral response mechanism. Thus, it is likely that an IRF-3-IRF-7 interaction might also exist in the CNS cells. In support of this, of all the IRF genes studied, we noted that the IRF-3 gene was expressed at the highest levels in the brain and remained unregulated following intracranial LCMV infection, while IRF-7 mRNA increased significantly following viral infection. Such a “ready-to-battle” mechanism could be very important in providing an early innate antiviral response capability in the CNS since mobilization of the adaptive antiviral immune response to LCMV occurs with a considerable delay compared with the periphery.
Previous studies using lymphocytes, B cells, monocytes, fibroblasts, or epithelial cells have shown that many IRF genes including IRF-1, IRF-2, IRF-5, IRF-7, and IRF-9 are induced by IFN-α/β and viral infection, whereas IRF-8 is IFN-γ inducible (9, 21, 33). Consistent with their critical dependence on IFN-α/β, we demonstrated here that the increased expression of IRF-7 and IRF-9 in the CNS following LCMV infection was completely abrogated in the absence of IFN-α/β signaling but not by the absence of IFN-γ signaling. This finding indicated that during LCMV infection of the brain, (i) the regulation of these IRF genes does not involve direct stimulation by the virus and (ii) the alternate regulation of the IRF-9 gene by IFN-γ through an interaction between the transcription factor C/EBO-β (CCAAT/enhancer-binding protein beta) and GATE (IFN-γ activated transcriptional element) located within the IRF-9 promoter region (34) does not play a role in the modulation of this IRF in the brain.
Despite the clear dependence on IFN-α/β signaling for the stimulation of IRF-7 and IRF-9 gene expression, low cerebral expression of the IFN-α/β genes was detectable only late in the course of LCMV infection, well after the early increases detected in the IRF gene expression. It is therefore possible that the levels of IFN-α/β required to stimulate IRF-7 and IRF-9 gene expression in the brain early after LCMV infection fall below the detection limits of our RPA. It also cannot be ruled out that IFN-α/β derived from peripheral production and entering the brain might also contribute to the stimulation of IRF gene expression. More robust expression of the IFN-α/β genes has been noted to occur in the periphery after LCMV infection, with high levels of IFN-α detectable in the plasma 1 to 3 days postinfection (27). Previous studies have detected increased expression of the IFN-α/β genes in the CNS as LCMV infection progresses in mice (27), and astrocytes and microglia have been shown to produce IFN-α/β in vitro, suggesting that these cells might provide a local source of IFN production to defend the brain against viral insult (1, 22). The importance of locally produced IFN-α/β in antiviral defense is highlighted by transgenic mice with astrocyte production of IFN-α which are resistant to infection by LCMV (2) as well as other viruses including herpes simplex virus type 1 (16) and Borna disease virus (30).
Interestingly, and despite their dependence on IFN-α/β receptor signaling, the regulation of the IRF-7 and IRF-9 genes had differential requirements for the molecular components of the IFN-α/β signaling pathway. No upregulation of IRF-9 mRNA levels occurred in either STAT1 KO or STAT2 KO infected mice, indicating that IRF-9 transcription was dependent on both STAT1 and STAT2 and that this IRF gene was regulated via the canonical IFN-α/β receptor-activated ISGF3/ISRE-mediated signaling pathway in the CNS. By contrast, the stimulation of IRF-7 gene expression in the CNS, although delayed, occurred in part independently of STAT1 but not STAT2. Although it might be argued that the increased IRF-7 expression in this setting is simply due to the infiltration and accumulation of IRF-7 mRNA-positive mononuclear cells in the brain, our data do not support this notion. First, microscopic analysis clearly showed that the majority of IRF-7 mRNA-expressing cells were intrinsic to the brain. Second, if the kinetics of IFN-γ gene expression are examined, it was found that IFN-γ mRNA was maximal in both WT and STAT1 KO brains at day 6 but not detectable at day 4 postinfection, at which time IRF-7 gene expression was maximal. IFN-γ is a product and therefore a specific marker of activated T cells. The absence of detectable IFN-γ at day 4 postinfection suggests the absence of a significant infiltrating mononuclear cell population that could account for IRF-7 gene expression in the LCMV-infected STAT1 KO brain. Third, other IRFs expressed by hematopoietic cells, such as IRF-5 and IRF-9, were not similarly increased in the LCMV-infected STAT1 KO brain. Finally, parallel findings in IFN-α-treated glial cells confirmed the delayed STAT1-independent, STAT2-dependent stimulation of IRF-7 gene expression that was prevented by cycloheximide. This latter finding indicated that IFN-α-stimulated IRF-7 gene expression in the absence of STAT1 required the synthesis of an intermediary protein factor(s) and would thus account for the delayed kinetics of this response. The fact that IRF-7 gene expression remained increased in IFN-α-stimulated WT cells treated with cycloheximide indicated that expression of this IRF can also be induced directly by IFN-α when the IFN-α/β receptor-activated ISGF3/ISRE-mediated signaling pathway remains intact. Although it had been thought that IFN-α/β stimulation of IRF-7 gene expression was STAT1 dependent, this was based on relatively short-term studies in cultured STAT1 KO fibroblasts infected with Newcastle disease virus (24). As far as we are aware, the effect of IFN-α on the expression of the IRF-7 gene in STAT1 KO cells has not been previously studied. Thus, our findings here clearly show that both in CNS glial cells as well as in splenocytes, stimulation of the IRF-7, but not the IRF-9, gene can be mediated by IFN-α/β through two distinct pathways which are either STAT1 dependent or STAT1 independent.
Regardless of which IFN-α/β receptor-activated pathway is involved, we showed that there is an obligatory requirement for STAT2 in IFN-α-stimulated IRF-7 gene expression. One consequence of this is that in addition to the classical IFN-α/β receptor-activated STAT1/STAT2/IRF-9 signaling pathway, STAT2 also must be involved in an alternative signaling pathway that does not depend upon STAT1. Interestingly, Huang and colleagues demonstrated that one of the components of the SW1/SNF complex, Brahma-related gene (BRG)-1, can associate with STAT2 in the nucleus and stimulate certain IFN-α-regulated genes, such as ISG-12, which contain ISRE binding sites (19). In recent studies, we have observed significant STAT2-dependent, STAT1-independent upregulation of the ISG-12 gene in the brain following LCMV infection (S. S. Ousman and I. L. Campbell, unpublished observation), suggesting that a STAT2/BRG-1 signaling complex might play a role in IFN-α signaling in the absence of STAT1 that ultimately leads to increased IRF-7 gene expression. This potential mechanism for the STAT1-independent actions of IFN-α in the CNS is the focus of ongoing studies by us.
If STAT2/BRG-1 signaling is involved in IFN-α/β-stimulated STAT1-independent signaling, it is unlikely that this pathway regulates the IRF-7 gene directly. Such a mechanism would be inconsistent with both the delayed nature of this response and its requirement for protein synthesis in glial cells. Therefore, we suggest that the STAT2/BRG-1 or perhaps some other STAT2-dependent, STAT1-independent transcriptional factor that is activated by the IFNAR induces the production of an intermediary factor(s) that then stimulates the expression of the IRF-7 gene. Other than IFN-α, a number of factors including TNF and tetradecanoyl phorbol acetate or topoisomerase inhibitors are known to stimulate IRF-7 gene expression via an NF-κB binding site in the promoter or changes in chromatin structure, respectively (23). Interestingly, expression of both the TNF and IL-1 genes was increased significantly by IFN-α treatment of glial cells derived from STAT1 KO but not STAT2 KO mice, and this occurred prior to the increased expression of the IRF-7 gene, which suggested the possible involvement of either of these proinflammatory cytokines as the intermediary factors in IFN-α-stimulated STAT1-independent regulation of IRF-7. However, in preliminary studies, neither neutralization of these specific cytokines in the presence of IFN-α nor their direct application to the glial cells altered IRF-7 gene expression (Ousman and Campbell, unpublished). At present, the putative intermediary factors involved in the STAT1-independent, IFN-α-stimulated IRF-7 gene expression remain to be identified.
A number of conclusions can be drawn from our study. First, many of the IRF genes are expressed constitutively in the mouse in an organ-specific fashion, with the IRF-7, IRF-9, and IRF-5 genes alone being highly upregulated locally following a viral infection. Second, this upregulation of both the IRF-7 and IRF-9 genes in the brain is dependent on IFN-α/β but not IFN-γ. Third, a fundamental difference exists in the mechanism of regulation of the IRF-7 gene compared to the IRF-9 gene, with the IRF-7 gene but not the IRF-9 gene being stimulated in a STAT1-independent fashion via unidentified indirect pathways coupled to the activation of the IFN-α/β receptor. It remains to be determined what the exact roles of IRFs are in the CNS during pathogen infection and whether these transcription factors may participate in other neurological disorders. The development of mice with targeted disruption of specific IRF genes (28) should aid in addressing these outstanding issues.
Acknowledgments
This work was supported by NIH grants (MH62231, MH62261, and NS36979) to I.L.C. and postdoctoral fellowships to S.S.O. from the National Multiple Sclerosis Society (NMSS) and the Multiple Sclerosis Society of Canada (MSSC).
We also thank Heather Weinkauf and Mary Kies-Wagner for managing the mouse colonies.
This is manuscript number 16767-NP from The Scripps Research Institute.
REFERENCES
- 1.Akiyama, H., K. Ikeda, M. Katoh, E. G. McGeer, and P. L. McGeer. 1994. Expression of MRP14, 27E10, interferon-α and leukocyte common antigen by reactive microglia in postmortem human brain tissue. J. Neuroimmunol. 50:195-201. [DOI] [PubMed] [Google Scholar]
- 2.Akwa, Y., D. E. Hassett, M. L. Eloranta, K. Sandberg, E. Masliah, H. Powell, J. L. Whitton, F. E. Bloom, and I. L. Campbell. 1998. Transgenic expression of IFN-α in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J. Immunol. 161:5016-5026. [PubMed] [Google Scholar]
- 3.Asensio, V. C., C. Kincaid, and I. L. Campbell. 1999. Chemokines and the inflammatory response to viral infection in the central nervous system with a focus on lymphocytic choriomeningitis virus. J. Neurovirol. 5:65-75. [DOI] [PubMed] [Google Scholar]
- 4.Asensio, V. C., S. Lassmann, A. Pagenstecher, S. C. Steffensen, S. J. Henriksen, and I. L. Campbell. 1999. C10 is a novel chemokine expressed in experimental inflammatory demyelinating disorders that promotes the recruitment of macrophages to the central nervous system. Am. J. Pathol. 154:1181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Au, W.-C., P. A. Moore, W. Lowther, Y. T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of the interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Au, W.-C., P. A. Moores, D. W. LaFleur, B. Tombal, and P. M. Pitha. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 273:29210-29217. [DOI] [PubMed] [Google Scholar]
- 7.Badley, J. E., G. A. Bishop, T. St. John, and J. A. Frelinger. 1988. A simple, rapid method for the purification of poly A+ RNA. BioTechniques 6:114-116. [PubMed] [Google Scholar]
- 8.Barber, G. N. 2000. The interferons and cell death: guardians of the cell or accomplices of apoptosis? Semin. Cancer Biol. 10:103-111. [DOI] [PubMed] [Google Scholar]
- 9.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 10.Barnes, B. J., P. A. Moore, and P. M. Pitha. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon α genes. J. Biol. Chem. 276:23382-23390. [DOI] [PubMed] [Google Scholar]
- 11.Bartholdy, C. J., J. P. Christensen, D. Wodarz, and A. R. Thomsen. 2000. Persistent virus infection despite chronic T-lymphocyte activation in gamma-interferon-deficient mice infected with lymphocytic choriomeningitis virus. J. Virol. 74:10304-10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belardelli, F. 1995. Role of interferons and other cytokines in the regulation of the immune response. APMIS 103:161-179. [DOI] [PubMed] [Google Scholar]
- 13.Biron, C. A. 2001. Interferons alpha and beta as immune regulators—a new look. Immunity 14:661-664. [DOI] [PubMed] [Google Scholar]
- 14.Bluyssen, H. A., R. Muzaffar, R. J. Vlieststra, A. C. J. Van Der Made, S. Leung, G. R. Stark, I. M. Kerr, J. Trapman, and D. E. Levy. 1995. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc. Natl. Acad. Sci. USA 92:5645-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell, I. L., M. V. Hobbs, P. Kemper, and M. B. A. Oldstone. 1994. Cerebral expression of multiple cytokine genes in mice with lymphocytic choriomeningitis. J. Immunol. 152:716-723. [PubMed] [Google Scholar]
- 16.Carr, D. J., L. A. Veress, S. Noisakran, and I. L. Campbell. 1998. Astrocyte-targeted expression of IFN-alpha1 protects mice from acute ocular herpes simplex virus type 1 infection. J. Immunol. 161:4859-4865. [PubMed] [Google Scholar]
- 17.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 18.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 19.Huang, M., F. Qian, Y. Hu, C. Ang, Z. Li, and Z. Wen. 2002. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-α-inducible genes. Nat. Cell Biol. 4:774-781. [DOI] [PubMed] [Google Scholar]
- 20.Lebon, P., J. F. Meritet, A. Krivine, and F. Rozenberg. 2002. Interferon and Aicardi-Goutieres syndrome. Eur. J. Paediatr. Neurol. 6:A47-A53. [DOI] [PubMed] [Google Scholar]
- 21.Levy, D. E., I. Marie, and A. Prakash. 2003. Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr. Opin. Immunol. 15:52-58. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman, A. P., P. M. Pitha, H. S. Shin, and M. L. Shin. 1989. Production of tumor necrosis factor and other cytokines by astrocytes stimulated with lipopolysaccharide or a neurotropic virus. Proc. Natl. Acad. Sci. USA 86:6348-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, R., P. A. Moore, and P. M. Pitha. 2002. Stimulation of IRF-7 gene expression by tumor necrosis factor α. Requirement for the NFκB transcription factor and gene accessibility. J. Biol. Chem. 277:16592-16598. [DOI] [PubMed] [Google Scholar]
- 24.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou, R., S. Zhou, L. Huang, and D. Moskophidis. 2001. Critical role for alpha/beta and gamma interferons in persistence of lymphocytic choriomeningitis virus by clonal expansion. J. Virol. 75:8407-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ousman, S. S., and I. L. Campbell. Analysis of murine interferon regulatory factor (IRF) gene expression by multiprobe RNase protection assay. Methods Mol. Med., in press. [DOI] [PubMed]
- 27.Sandberg, K., M.-L. Eloranta, and I. L. Campbell. 1994. Expression of alpha/beta interferons (IFN-α/β) and their relationship to IFN-α/β-induced genes in lymphocytic choriomeningitis. J. Virol. 68:7358-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato, M., T. Taniguchi, and N. Tanaka. 2001. The interferon system and interferon regulatory factor transcription factors—studies from gene knockout mice. Cytokine Growth Factor Rev. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 29.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 30.Staeheli, P., M. Sentandreu, A. Pagenstecher, and J. Hausmann. 2001. Alpha/beta interferon promotes transcription and inhibits replication of Borna disease virus in persistently infected cells. J. Virol. 75:8216-8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stalder, A., A. Pagenstecher, C. Kincaid, and I. L. Campbell. 1999. Analysis of gene expression by multiprobe RNase protection assay, p. 53-66. In J. Harry and H. A. Tilson (ed.), Neurodegeneration methods and protocols. Human Press, Totowa, N.J. [DOI] [PubMed]
- 32.Stewart, T. A. 2003. Neutralizing interferon alpha as a therapeutic approach to autoimmune disease. Cytokine Growth Factor Rev. 14:139-154. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 34.Weihua, X., J. Hu, S. K. Roy, S. B. Mannino, and D. V. Kalvakolanu. 2000. Interleukin-6 modulates interferon-regulated gene expression by inducing ISGF3 gamma gene using CCAAT/enhancer binding protein-beta (C/EBP-beta). Biochim. Biophys. Acta 1492:163-171. [DOI] [PubMed] [Google Scholar]