Abstract
In addition to hepatocytes, hepatitis C virus (HCV) infects immune cells, including macrophages. However, little is known concerning the impact of HCV infection on cellular functions of these immune effector cells. Lipopolysaccharide (LPS) activates IκB kinase (IKK) signalsome and NF-κB, which leads to the expression of cyclooxygenase-2 (COX-2), which catalyzes production of prostaglandins, potent effectors on inflammation and possibly hepatitis. Here, we examined whether expression of HCV core interferes with IKK signalsome activity and COX-2 expression in activated macrophages. In reporter assays, HCV core inhibited NF-κB activation in RAW 264.7 and MH-S murine macrophage cell lines treated with bacterial LPS. HCV core inhibited IKK signalsome and IKKβ kinase activities induced by tumor necrosis factor alpha in HeLa cells and coexpressed IKKγ in 293 cells, respectively. HCV core was coprecipitated with IΚΚβ and prevented nuclear translocation of IKKβ. NF-κB activation by either LPS or overexpression of IKKβ was sufficient to induce robust expression of COX-2, which was markedly suppressed by ectopic expression of HCV core. Together, these data indicate that HCV core suppresses IKK signalsome activity, which blunts COX-2 expression in macrophages. Additional studies are necessary to determine whether interrupted COX-2 expression by HCV core contributes to HCV pathogenesis.
Hepatitis C virus (HCV), a flavivirus, causes hepatitis, cirrhosis, and hepatocellular carcinoma (18). Currently, almost 3% of the world population is infected by HCV, and these numbers seem to be increasing (3). One of the most remarkable features of HCV infection is that more than 85% of acutely infected patients become chronically infected (4). Although CD4+ and CD8+ T-cell responses are crucial for controlling HCV infection in acute HCV patients, these T-cell responses are significantly impaired in chronic HCV patients (16). Thus, this suggests that HCV evades host immune responses. While hepatocytes are a major target of HCV infection, recent studies showed that HCV can replicate in immune cells such as B and T lymphocytes and monocytes that express HCV receptors, such as CD81 and low-density lipoprotein receptor (1, 2, 52). Thus, it is possible that HCV infects immune effector cells, which contributes to evasion of host immune surveillance.
The HCV core protein, a nucleocapsid protein, binds to the cytoplasmic domain of tumor necrosis factor receptor (TNFR) and lymphotoxin-beta receptor to regulate apoptosis (16, 17, 45, 81). This viral protein is also involved in oncogenesis, as evidenced by the development of hepatocellular carcinoma in transgenic mice expressing HCV core in the liver (47). In addition, HCV core has been shown to affect diverse cellular and viral gene expressions (53, 55, 60) and, depending on subtypes, to activate or suppress NF-κB function that is involved in both innate and adaptive immunity (27, 44, 61, 79).
NF-κB is a homo- or heterodimer of five proteins: c-Rel, RelA (p65), RelB, p50, and p52. Under unstimulated conditions, NF-κB resides in the cytoplasm by forming complexes with inhibitory κB (IκB) (27). Proinflammatory stimuli, such as TNF-α and lipopolysaccharide (LPS), induce signal cascades through their cognate receptors, TNFR and Toll-like receptor 4 (TLR4), to activate IκB kinase (IKK) signalsome and subsequently NF-κB that is required for innate and adaptive immune responses to pathogens (13, 38). IKK signalsome is composed of at least two kinases, IKKα and -β, and a regulatory factor, IKKγ (26, 36). Among these proteins, IΚΚβ is known as a major kinase and IΚΚγ is required for the full activation of IΚΚβ upon proinflammatory stimulation (41-43, 56, 57, 59, 66, 78). Treatment of cells with TNF-α results in IKK signalsome recruitment to the type 1 TNF-α receptor (TNFR1), which induces phosphorylation at key residues in the activation loop of the kinase domain of IKKβ to be an active kinase (21). Activated IKK signalsome phosphorylates IκB, which subsequently undergoes ubiquitination and degradation, liberating NF-κB to translocate into the nucleus and activate target genes (37).
NF-κB influences gene expression of cyclooxygenase-2 (COX-2) along with other transcription factors, including CREB and C/EBP-β (15, 20, 33, 34, 48, 50, 63, 67, 72, 75). Accordingly, proinflammatory stimuli induce the expression of COX-2 to catalyze the production of prostaglandins, which promote inflammation through a variety of mechanisms.
Our hypothesis is that HCV evades innate immunity by directly infecting immune cells and altering gene expression of key inflammatory molecules. Here we tested whether the HCV core protein affects NF-κB activity and COX-2 production in macrophages. We demonstrate that the HCV core protein interacts with IKKβ, suppresses its kinase activity, and interferes with nuclear translocation of IKKβ, which are correlated with inhibition of NF-κB activity. Furthermore, we show that the HCV core protein suppresses NF-κB-dependent COX-2 expression. These data indicate that the HCV core protein interrupts diverse IKKβ functions, which results in blunted COX-2 expression.
MATERIALS AND METHODS
Cell lines.
Murine macrophage cell line RAW 264.7, murine alveolar macrophage cell line MH-S, and HEK 293 and HeLa cells were all obtained from the American Type Culture Collection (Rockville, MD). Cell lines were maintained according to the recommendations of the American Type Culture Collection.
Plasmids.
The coding sequence of HCV core (Hutchinson strain, genotype 1a) was inserted into the polylinker site of pCI:neo (Promega). The resultant construct, named pCI-HCV core, was engineered to express the 192-amino-acid-long HCV core protein. To generate a construct that encodes FLAG-tagged HCV core, pCI-HCV core was digested with restriction enzymes NheI and EcoRI (New England BioLabs) and inserted with a linker containing the FLAG sequence. The coding sequence of HCV core and E1 (Hutchinson strain, genotype 1a) was amplified by Pfu polymerase (Stratagene). The forward primer binds to the first 20 nucleotides of the 5′ end of the HCV core sequence and contains NheI restriction site, Kozak, and FLAG sequences (5′-GCGGCTAGCCACCATGGACTACAAAGACGATGACGATAAAGGGATGAGCACGAATCCTAAACC-3′; underlined is the core sequence). The reverse primer binds to 18 nucleotides of the 3′ end of the HCV E1 sequence and contains HindIII (5′-GCAAGCTTGCCGGCAAATAGCAGCAG-3′). The PCR product was digested with restriction enzymes NheI and HindIII (New England BioLabs) and inserted into the corresponding sites of pcDNA3(-)-Myc/His (Invitrogen). Candidate clones encoding HCV core and E1 were analyzed by sequencing (Vanderbilt DNA Sequencing Core Facility). Expression of the FLAG-tagged HCV core and Myc-tagged protein was determined by Western blotting with M2 antibody (Sigma) and anti-Myc antibody (Santa Cruz Biotechnology). Plasmids encoding a FLAG-tagged IKKα and IKKβ were kindly provided by R. Carter (Vanderbilt University) and F. Mercurio (Signal Research Division, Celgene Corp., New Jersey), respectively, and a plasmid expressing T7-tagged NEMO/IΚΚγ was a gift from E. S. Alnemri (Thomas Jefferson University). A plasmid encoding superrepressor IκBαS32,36A was a gift from K. Pennington (Vanderbilt University). The luciferase reporter plasmid NF-κB-Luc was purchased from Stratagene (La Jolla, CA).
Plasmid transfection, LPS treatment, and luciferase assay.
Cells were transfected with Superfect (QIAGEN), as specified by the manufacturer. Each transfection was normalized with appropriate empty vector plasmids. At 48 h posttransfection, transfected cells were harvested for the experiment. For the LPS treatment, after incubation for 14 to 16 h in a 37°C, CO2 incubator, transfected cells were further maintained without serum overnight and subsequently treated with LPS (1 μg/ml in phosphate-buffered saline; Sigma) for 4 h to induce COX-2 expression. For the luciferase assay, cells were cotransfected with luciferase reporter plasmids NF-κB-Luc and tk-Renilla Luc. Luciferase activity was measured by a dual-luciferase assay system (Promega) according to the protocol recommended by the manufacturer. NF-κB-mediated luciferase activity was normalized with Renilla luciferase activity. The luciferase assay was done in triplicate.
Viruses and infection.
Adenoviruses encoding IKKβ, superrepressor IkBαS32,36A, and β-galactosidase (β-Gal) were described previously (58). Viruses were propagated and virus titer was determined by standard protocols. RAW 264.7 cells were infected with adenoviruses at multiplicity of infection of 0.2 for 1 h. After inocula were removed, cells were placed in fresh medium and incubated overnight at 37°C in a CO2 incubator. At 24 h postinfection, cells were harvested for the experiment.
Immunoprecipitation and Western blotting.
Total cell lysate was prepared with RIPA (radioimmunoprecipitation assay) cell lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 1% sodium orthovanadate, 1% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) supplemented with phenylmethylsulfonyl fluoride (1 mM), aprotonin (2 μg/ml), leupeptin (2 μg/ml), and soybean trypsin inhibitor (37.5 μg/ml). Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (Bio-Rad). Specific bands were revealed using enhanced chemiluminescence (ECL plus; Amersham). For immunoprecipitation analysis, 1 to 2 μg of appropriate antibodies was added to precleared cell lysate that was normalized by protein content and incubated overnight at 4°C. Immune complexes were captured with 30 μl of protein A-Sepharose (Zymed) for 30 min at 4°C and washed five times with RIPA buffer. Antibodies used in this study were as follows: polyclonal anti-murine COX-2 (Cayman Chemical); monoclonal anti-FLAG M2 antibody (Sigma); polyclonal anti-IKKα/β and anti-IKKγ (Santa Cruz Biotechnology); monoclonal anti-IKKα and -β (PharMingen); monoclonal anti-HCV core antibody (Affinity Bioreagents, Inc.).
IKK assay.
Transfected HeLa cells were treated with 10 ng/ml of TNF-α (R&D Systems Inc.) for 15 min before lysis. Cytoplasmic extracts from HeLa or HEK 293 cells were prepared as described previously (49). To immunoprecipitate IKK signalsome, ELB buffer (49) was added to the prepared cytoplasmic extract. Kinase activity was measured in a reaction buffer containing 10 mM HEPES (pH 7.4), 5 mM MgCl2, 1 mM MnCl2, ATP (10 μM), [γ-32P]ATP (5 μCi), and recombinant glutathione S-transferase (GST)-IκBα, GST protein fused to amino acids 1 to 54 of IκBα, at 30°C for 30 min. Radiolabeled products were separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and revealed by autoradiography.
Preparation of cytoplasmic and nuclear fractions.
Cytoplasmic and nuclear fractions were prepared as described elsewhere (9). In brief, cells were washed three times with ice-cold phosphate-buffered saline and added to hypotonic buffer (10 mM HEPES [pH 7.9], 5 mM KCl, 1.5 mM MgCl2, 1 mM NaF, and 1 mM Na3VO4) supplemented with 1 mM dithiothreitol, phenylmethylsulfonyl fluoride (1 mM), aprotonin (2 μg/ml), leupeptin (2 μg/ml), and soybean trypsin inhibitor (37.5 μg/ml). After lysis for 15 min on ice, the cytoplasmic fraction was prepared by centrifugation at 3,000 rpm for 5 min. The cytoplasmic fraction was cleared by centrifugation at 13,000 rpm for 15 min before further analysis. To prepare the nuclear fraction, the cell pellet generated after initial centrifugation was washed with the hypotonic buffer three times and suspended in high-salt buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM EDTA [pH 8.0], 420 mM NaCl, 25% glycerol [vol/vol], 50 mM β-glycerophosphate, 1 mM NaF, and 1 mM Na3VO4) supplemented with 1 mM dithiothreitol, phenylmethylsulfonyl fluoride (1 mM), aprotonin (2 μg/ml), leupeptin (2 μg/ml), and soybean trypsin inhibitor (37.5 μg/ml). The suspended cell pellet was incubated for 30 min on ice with occasional vortexing, and the nuclear fraction was collected after centrifugation at 13,000 rpm for 10 min. Protein content from each fraction was quantified by the Bradford assay (Bio-Rad) as specified by the manufacturer to ensure equal loading.
Statistical analysis.
For comparison among groups, paired or unpaired t tests and one-way analysis of variance tests were used (with the assistance of InStat; Graphpad Software, Inc., San Diego, CA). P values of <0.05 are considered significant.
RESULTS
HCV core suppresses NF-κB activity in LPS-treated macrophages.
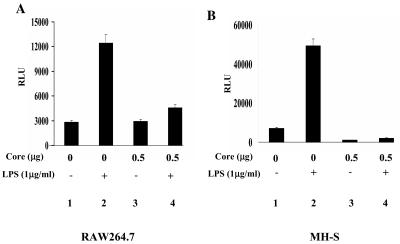
HCV core differentially modulates NF-κB activity, depending on the subtype of HCV (54). To examine whether the HCV core used in this study (genotype 1a) interferes with NF-κB activity in LPS-treated macrophages, RAW 264.7, a murine macrophage cell line, and MH-S, a murine alveolar macrophage cell line, were transfected with a plasmid encoding HCV core along with the NF-κB-luciferase reporter construct in the presence or absence of LPS. The results in Fig. 1 showed that HCV core, by itself, did not activate NF-κB but rather suppressed NF-κB activity elicited by LPS treatment of RAW 264.7 (Fig. 1A) and MH-S (Fig. 1B) cells. A similar experiment using TNF-α produced similar results (data not shown). These results demonstrate that HCV core suppresses NF-κB activity in macrophages.
FIG. 1.
HCV core inhibits NF-κB activated by LPS. RAW 264.7 (A) and MH-S (B) cells were transfected with 0.5 μg of NF-κB-Luc and 0.2 μg of tk-Renilla-Luc along with the indicated amounts of the HCV core-expressing plasmid. The total amount of transfected plasmid was adjusted with an empty vector plasmid. NF-κB activity elicited by LPS was measured after treating cells with LPS for 4 h (lanes 1 and 2). To examine an effect of the HCV core on NF-κB-mediated transcriptional activity, the plasmid encoding HCV core was transfected without or with treatment with LPS (1 μg/ml) for 4 h (lanes 3 and 4).
HCV core inhibits IKK signalsome kinase activity.
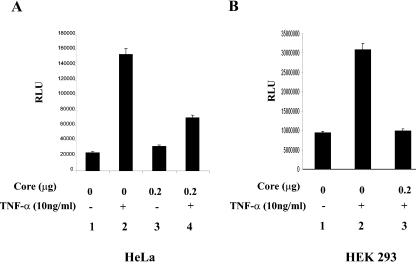
To investigate the mechanism by which HCV core inhibits NF-κB activity, we tested whether HCV core interferes with IKK kinase activity. For these studies, we employed HeLa and 293 cells because of the ability to achieve high transfection efficiencies in these cell lines. First, to test whether these cell lines were able to recapitulate the results shown in Fig. 1, we performed a similar experiment. As shown in Fig. 2A, when HeLa cells were treated with TNF-α, NF-κB-mediated transcriptional activity was increased (lanes 1 and 2). However, the presence of HCV core suppressed NF-κB activity elicited by TNF-α (lanes 2 and 4). Similarly, HCV core suppressed NF-κB activated by TNF-α in HEK 293 cells (Fig. 2B, lanes 2 and 3). These results suggest that HCV core also represses NF-κB activity in these cell lines.
FIG. 2.
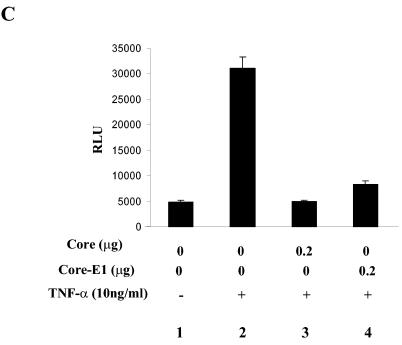
HCV core inhibits NF-κB activated by TNF-α in other cell types. HeLa (A) and HEK 293 (B) cells were transfected with 0.2 μg of the NF-κB-Luc and 0.1 μg of the tk-Renilla-Luc reporter construct, along with the indicated amount of the HCV core-expressing plasmid. The total amount of transfected plasmid was adjusted with an empty vector plasmid. NF-κB-mediated transcriptional activity was measured after treating cells with TNF-α (10 ng/ml) for 6 h. (C) A possible effect of HCV E1 on HCV core was examined. HeLa cells were transfected with two reporter constructs, indicated above, along with either the plasmid encoding HCV core or a plasmid encoding HCV core and E1, and treated with TNF-α as described above. NF-κB-mediated transcription activity was measured by assaying luciferase activity. The result shown here is a representative of three independent experiments.
We tested whether coexpression of HCV core and E1 alters the effect of HCV core on NF-κB activity. HeLa cells were transfected with a plasmid encoding HCV core and E1 along with the NF-κB reporter construct and treated with TNF-α. As shown in Fig. 2C, HCV core was able to suppress NF-κB activity elicited by TNF-α in the presence of HCV E1 (lane 4). A similar result was obtained in HEK 293 cells (data not shown). These results indicate that suppression of NF-κB activity by HCV core is not significantly affected by E1.
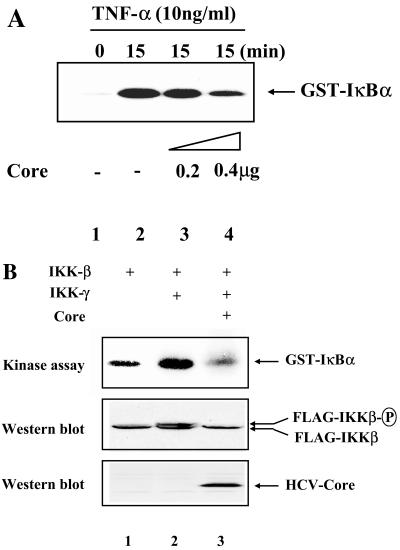
Next, we tested whether HCV core interferes with IKK signalsome kinase activity. HeLa cells were transfected with increasing amounts of a plasmid encoding HCV core and treated with TNF-α (10 ng/ml) for 15 min to induce IKK signalsome activity. The IKK signalsome was immunoprecipitated with anti-IKKβ antibody, extensively washed, and incubated with GST-IκBα and [γ-32P]ATP for kinase assays. As shown in Fig. 3A, HCV core reduced IKK kinase activity elicited by TNF-α treatment in a dose-dependent fashion.
FIG. 3.
The HCV core protein inhibits IKK kinase activity. (A) HeLa cells were transfected with either an empty vector (lanes 1 and 2) or increasing amounts of the HCV core-expressing plasmid (lanes 3 and 4). The total amount of transfected plasmid was normalized as indicated in Materials and Methods. At 48 h posttransfection, the cells were treated with TNF-α (10 ng/ml) for 15 min and the cytoplasmic fraction was prepared for immunoprecipitation of IKK with anti-IKKβ antibody. Immune complex, after being extensively washed, was incubated with GST-IκBα and [γ-32P]ATP to measure IKK kinase activity. (B) HEK 293 cells were transfected with 0.1 μg of the FLAG-tagged IKKβ-expressing plasmid along with 0.5 μg of a plasmid encoding IKKγ (lanes 2 and 3) and 0.5 μg of the HCV core-expressing plasmid (lane 3). At 48 h posttransfection, total cell lysate was prepared for immunoprecipitation with M2 antibody to capture IKKβ. Immune complexes were incubated with GST-IκBα and [γ-32P]ATP to measure IKKβ kinase activity (top panel). Expression levels of IKKβ and the HCV core were determined by Western blotting with M2 antibody and anti-HCV core antibody, respectively (middle and bottom panels).
Since IKKβ is known as a major kinase in inflammatory gene expression and overexpression of IKKβ results in kinase activity that is markedly increased by IKKγ (24), we examined whether HCV core inhibits IKKβ kinase activity in the absence of an activating stimulus (Fig. 3B). HEK 293 cells were transfected with plasmids expressing FLAG-tagged IΚΚβ and T7-tagged IKKγ in the presence or absence of HCV core. For IKKβ kinase assays, IΚΚβ was precipitated with M2 antibody. As shown in Fig. 3B, IKKβ kinase activity was markedly enhanced by IKKγ (lane 2 compared to lane 1); however, this was strongly suppressed by HCV core (lane 3). Western blot analysis also revealed that the phosphorylated form of IKKβ, an indicator of kinase activation, was induced by overexpression of IKKγ. The phosphorylation of IKKβ was blocked by ectopic expression of the HCV core protein (middle panel, lanes 2 and 3). These results indicate that HCV core specifically suppresses IKKβ kinase activity.
HCV core physically interacts with IΚΚβ.
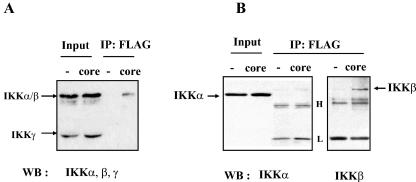
To understand how the HCV core protein suppresses IKKβ kinase activity, we examined the physical interaction between the two proteins by immunoprecipitation assay. HeLa cells were transfected with either an empty vector or a plasmid encoding FLAG-tagged HCV core. After cell lysis, the cytoplasmic fraction was incubated with M2 antibody to capture FLAG-tagged HCV core, and the immune complex was analyzed by SDS-PAGE and Western blotting. First, to detect any subunits of IKK signalsome interacting with the HCV core protein, the membrane was incubated with a mixture of anti-IKKα, -IKKβ, and -IKKγ antibodies. As shown in Fig. 4A, the HCV core protein coprecipitated with IKKα and/or IKKβ but not with IKKγ. To identify the precipitated band, coimmunoprecipitates of the HCV core protein were further analyzed by Western blotting with IKKα and IKKβ antibodies. The result in Fig. 4B indicated that IKKβ but not IKKα coimmunoprecipitated with the HCV core protein.
FIG. 4.
The HCV core protein interacts with IKKβ. HeLa cells were transfected with either 2 μg of an empty vector plasmid or 1 μg of a plasmid encoding FLAG-tagged HCV core that was normalized with the empty vector plasmid to 2 μg. At 48 h posttransfection, cell lysate was prepared for immunoprecipitation of FLAG-tagged HCV core with M2 antibody. Immune complex captured by protein A-Sepharose was separated by SDS-PAGE and analyzed by Western blotting. (A) HCV core protein immunoprecipitated with IKK. Blot was incubated with anti-IKKα, -β, and -γ antibodies (rabbit polyclonal antibodies) to reveal subunits of IKK that interact with the HCV core protein. One-tenth of the total cell lysate used for immunoprecipitation was loaded as a positive control for IKKα, -β, and -γ. (B) The HCV core protein interacts with IKKβ. Total cell lysate was prepared from transfected cells and incubated with M2 antibody. One-tenth of the total cell lysate used for immunoprecipitation was loaded as a positive control for IKKα. Precipitated immune complex was analyzed by Western blotting after SDS-PAGE separation. The blot was incubated with either anti-IKKα antibody or anti-IKKβ (mouse monoclonal) antibody to reveal IKKα and IKKβ, respectively.
The HCV core interferes with nuclear localization of IKKβ.
A recent discovery that IKKα and -β are located not only in the cytoplasm but also in the nucleus suggests additional functions of IKK signalsome in regulation of gene expression (12). Indeed, IKKα is recruited to the NF-κB-binding DNA element via physical interaction with CREB-binding protein, which enhances target gene expression (7, 76). IΚΚγ is also directly involved in transcription regulation by inhibiting the interaction between IKKα and CREB-binding protein (71). Therefore, we investigated the possibility that the HCV core protein affects IΚΚβ activity by interfering with nuclear-cytoplasmic partitioning of IKKβ.
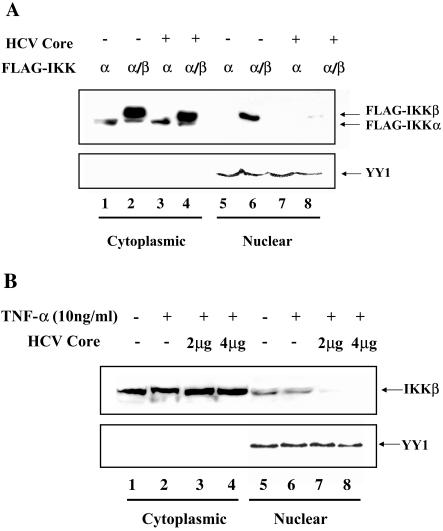
To test whether HCV core selectively interferes with IKKβ partitioning within a cell, HEK 293 cells were transfected with plasmids encoding FLAG-tagged IKKα and IKKβ along with the plasmid encoding HCV core. To determine partitioning of IKKα and IKKβ, cytoplasmic and nuclear fractions were prepared and analyzed by Western blotting with Μ2 antibody to reveal IKKα and -β. As shown in Fig. 5A, while the majority of IKKα resides in the cytoplasm, IKKβ was detected in both cytoplasmic and nuclear fractions (lanes 1, 2, 5, and 6). However, the presence of the HCV core protein decreased localization of IKKβ in the nuclear fraction (lanes 6 and 8), while not significantly affecting the amount of cytoplasmic IKKβ. To ensure equal loading of nuclear proteins, the membrane was stripped and probed with anti-YY1 antibody (8, 29, 46).
FIG. 5.
The HCV core protein interferes with localization of IKKβ to the nucleus. (A) The HCV core protein interferes with nuclear localization of transfected IKKβ. HEK 293 cells were transfected with 0.5 μg of the plasmid encoding FLAG-tagged IKKα and IKKβ with (lanes 3 and 4 and lanes 7 and 8) or without (lanes 1 and 2 and lanes 5 and 6) 2 μg of the plasmid encoding HCV core. The total amount of transfected plasmid was adjusted to 3 μg with an empty host plasmid. Cytoplasmic and nuclear fractions were prepared as described in Materials and Methods. Ten micrograms of cytoplasmic proteins and 50 μg of nuclear proteins were analyzed by SDS-PAGE and Western blotting with M2 antibody to reveal the location of IKKα and -β. To ensure equal loading of nuclear proteins, nuclear protein YY1 was Western blotted after stripping membrane. (B) The HCV core protein interferes with nuclear localization of endogenous IKKβ. HEK 293 cells were transfected with either 4 μg of an empty host plasmid (lanes 1 and 2 and lanes 5 and 6) or a different dose of the plasmid encoding HCV core (lanes 3 and 4 and 7 and 8). The total amount of transfected plasmid was adjusted to 4 μg with an empty host plasmid. At 48 h posttransfection, transfected cells were treated with TNF-α (10 ng/ml) for 30 min before preparing cytoplasmic and nuclear fractions. Fifty micrograms of each fraction was analyzed by Western blotting to reveal endogenous IKKβ. To ensure equal loading of nuclear protein, the membrane was stripped and reblotted to reveal nuclear protein YY1.
To further confirm this observation, a similar experiment was conducted to determine the distribution of endogenous IKKβ (Fig. 5B). HEK 293 cells were transfected with increasing amounts of the HCV core-encoding plasmid. Transfected cells were treated with 10 μg/ml of TNF-α to examine a possible effect on distribution of endogenous IKKβ. Western blotting analysis in Fig. 5B showed that distribution of IKKβ was not significantly affected by TNF-α treatment (compare lanes 1 and 2 and lanes 5 and 6), as indicated in another study (7). However, the HCV core protein reduced nuclear localization of IKKβ (lanes 6, 7, and 8). To ensure equal loading of nuclear protein, the membrane was treated as for Fig. 5A to reveal nuclear protein YY1. Taken together, these results indicate that the HCV core protein blocks IKKβ shuttling to the nucleus.
NF-κB activation elicited by IKKβ is sufficient to induce COX-2.
Involvement of NF-κB in COX-2 expression is well documented in a variety of cell types (15, 20, 77).
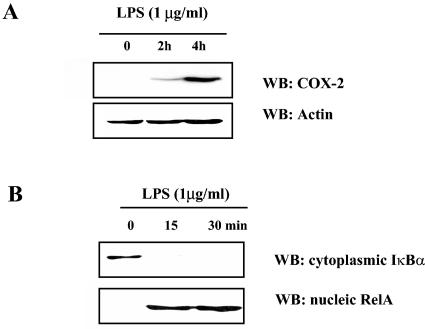
To define the relationship between NF-κB activation and COX-2 expression in RAW 264.7 cells, we treated the cells with endotoxin and measured COX-2 expression and NF-κB activation by Western blot analysis. As indicated in Fig. 6A, COX-2 protein production increased in response to treatment with LPS, and this was associated with degradation of cytoplasmic IκBα and the appearance of immunoreactive RelA in the nucleus (Fig. 6B). These data show that the macrophage cell line treated with LPS results in both activation of NF-κB and COX-2 protein production.
FIG. 6.
Casual relationship between NF-κB activation and COX-2 induction. (A) Murine macrophage cell line RAW 264.7 was treated with 1 μg/ml of LPS for the indicated times, and 20 μg of total cell lysate was analyzed for COX-2 expression. To ensure equal loading, membrane was stripped and Western blotted for actin. (B) After LPS treatment of RAW 264.7 cells for the indicated times, Western blotting was performed to reveal IκBα in the cytoplasm and RelA in the nucleus.
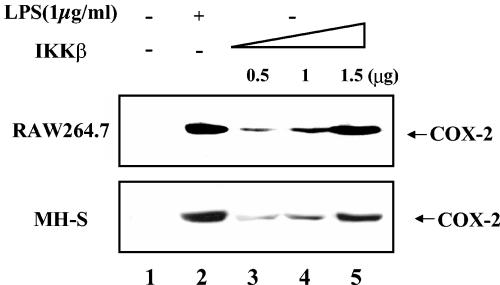
To assess the role of NF-κB in COX-2 expression in macrophages without ambiguity associated with cross talk by inflammatory signals, NF-κB was activated in a highly specific manner through ectopic expression of IKKβ (26), and resultant COX-2 expression was analyzed by Western blotting in two separate murine macrophage cell lines, RAW 264.7 and MH-S. As shown in Fig. 7, ectopic expression of IKKβ increases COX-2 protein production in both macrophage cell lines in a dose-dependent fashion. These data indicate that NF-κB activation by ectopic expression of IKKβ is sufficient to induce COX-2 expression.
FIG. 7.
NF-κB activation confers COX-2 expression. RAW 264.7 and MH-S cells were transiently transfected with either 1.5 μg of an empty vector plasmid (lanes 1 and 2) or an increasing amount of a plasmid that encodes FLAG-tagged IKKβ (lanes 3 to 5). The total amount of transfected DNA was normalized as described in Materials and Methods. Transfected cells were further incubated without serum overnight and treated with or without LPS (1 μg/ml) for 4 h. Expression of COX-2 was measured by Western blotting with 20 μg of total cell lysate.
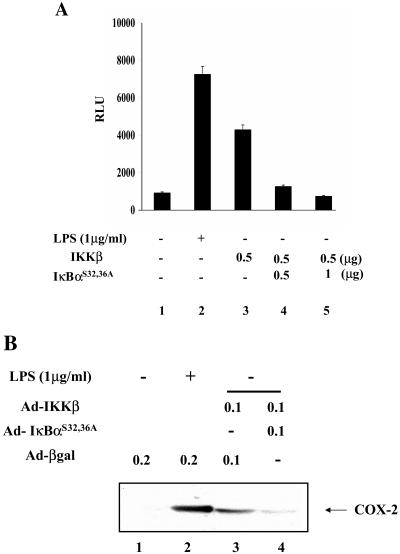
To assess the specificity of this response, we examined whether superrepressor IkBαS32,36A suppresses COX-2 expression induced by IKKβ. First, RAW 264.7 cells were transfected with an NF-κB luciferase reporter construct along with the IKKβ-expressing plasmid in the absence or presence of the superrepressor-expressing plasmid. As shown in Fig. 8A, transfection of the superrepressor IκBαS32,36A markedly inhibits NF-κB-mediated luciferase activity induced by IKKβ. Next, to test whether COX-2 protein production induced by IKKβ is suppressed by IκBαS32,36A, RAW 264.7 cells were infected with replication-deficient adenoviruses encoding kinase-active IKKβ, IκBαS32,36A, or β-Gal. Western blot analysis in Fig. 8B shows that infection with IKKβ-expressing adenovirus induced COX-2 expression, while infection with β-Gal-expressing adenovirus failed to do so. Coinfection with two adenoviruses encoding IKKβ and IκBαS32,36A blunted the production of COX-2. Taken together, these data demonstrate that selective NF-κB activation results in COX-2 expression in macrophage cell lines.
FIG. 8.
COX-2 induction by NF-κB is suppressed by the superrepressor IκBαS32,36A. (A) IκBαS32,36A inhibits NF-κB activated by IKKβ. RAW 264.7 cells were transfected with 0.2 μg of a tk-Renilla luciferase construct (tk-Renilla-Luc) and 0.5 μg of a luciferase reporter construct containing NF-κB-binding sites (NF-κB-Luc) along with an empty vector plasmid (lanes 1 and 2) or the indicated amounts of the IKKβ-expressing plasmid (lanes 3 to 5). To examine the inhibitory effect of IκBαS32,36A on NF-κB activated by IKKβ, an increasing amount of a plasmid encoding IκBαS32,36A was cotransfected (lanes 4 and 5). At 48 h posttransfection, the cell extract was prepared and enzymatic activity was measured. The result shown here is a representative of three independent experiments. (B) COX-2 expression elicited by activated NF-κB is inhibited by IκBαS32,36A. RAW 264.7 cells were infected with adenovirus either encoding β-Gal (lanes 1 and 2) or IKKβ (lanes 3 and 4) along with IκBαS32,36A (lane 4). Each infection was normalized with adenovirus encoding β-Gal to a multiplicity of infection of 0.2. At 24 h postinfection, the total cell lysate was prepared and 20 μg of total protein was used for Western blotting of COX-2. Infected cells were serum starved overnight before LPS treatment for 4 h (lane 2). COX-2 was measured by Western blotting with 20 μg of total cell lysate.
HCV core suppresses COX-2 expression induced by LPS.
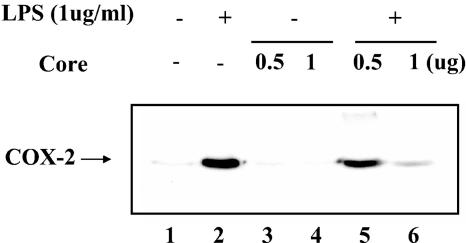
Finally, we examined whether the inhibitory effect of HCV core on NF-κB activity leads to down regulation of COX-2 expression. RAW 264.7 cells were transfected with either an empty host plasmid or a plasmid expressing the HCV core protein. Subsequently, transfected cells were treated with LPS to induce COX-2. As indicated in Fig. 9, HCV core protein, by itself, did not induce COX-2 expression (lanes 3 and 4). In contrast, HCV core protein strongly suppressed COX-2 expression induced by LPS treatment (lanes 5 and 6).
FIG. 9.
The HCV core protein suppresses COX-2 expression. RAW 264.7 cells were transfected with either empty vector (lanes 1 and 2) or the HCV core-expressing plasmid (lanes 3 to 6). Transfected cells were treated with LPS (1 μg/ml) for 4 h to induce COX-2 (lanes 2, 5, and 6). Expressed COX-2 was measured by Western blotting with 20 μg of total protein.
DISCUSSION
Primary HCV infection evades protective immunity and establishes a chronic infection (4, 16). Chronic HCV-infected patients have reduced Th1-type CD4+ T-cell activity and increased Th2-type CD4+ T-cell activity (16, 23). Some chronic hepatitis C patients treated with alpha interferon (IFN-α) (14) and ribavirin show enhanced Th1 CD4+ T-cell effector function with improvement of the medical condition (32, 65, 69). An HCV murine model also showed the compromised production of Th1 cytokines, such as interleukin-2 (IL-2) and IFN-γ, by HCV core (39). Similarly, there is a decreased Th1 cytokine production in transgenic mice that express HCV core (62). These data, together, suggest that HCV evades the host immune response through a mechanism that is mediated by HCV core. Recent experimental evidence shows that HCV infection occurs with a broad cellular tropism encompassing not only hepatocyte cells but also immune cells, such as B and T lymphocytes and monocytes (1). However, the impact of HCV on infected immune cells is not understood. In this study, we addressed this issue by examining an inhibitory effect of HCV core on NF-κB, a key transcription factor in innate and adaptive immunity. Our data showed that, through inhibition of IKK kinase activity, HCV core hinders NF-κB activity, which results in blunted expression of an important inflammatory protein, COX-2, in macrophages.
Along with constitutively expressed COX-1, COX-2 metabolizes arachidonic acid to prostaglandin H2, which is further metabolized to generate various prostanoids (24). Prostanoids comprise three groups: prostaglandins, such as D2 (PGD2), E2 (PGE2), and F2 (PGF2); prostacylin, such as PGI2; and thromboxane A2. In general, prostanoids are considered to promote inflammation by inducing vasodilation and increasing vascular permeability and cellular migration to the site of inflammation (22, 73, 74). Notably, prostanoids are also important modulators of adaptive immunity (28, 30); however, effects of prostanoids on immunity are complex, and the net result depends on the surrounding milieu. For example, PGE2 promotes dendritic cell maturation and migration to lymphoid organs in peripheral tissue, while it suppresses T-cell activation by dendritic cells in lymphoid organs (28). In the case of PGD2, while enhancing the Th2-type immune response, PGD2 and its derivative cyclopentenone prostaglandins are known to have anti-inflammatory activities by inhibiting NF-κB, inducing apoptosis of macrophages and neutrophils in inflammatory lesions, and activating anti-inflammatory transcription factor PPAR-γ (64). Other prostanoids, such as PGI2 and thromboxane A2, also increase T-cell functions, and PGI2 enhances natural killer (NK) cell functions (19, 35, 68, 70). Nevertheless, these results clearly illustrate that prostanoids are implicated in innate and adaptive immunity.
It is not well documented that there is a relationship between HCV infection and COX-2 expression that might influence HCV pathogenesis. As yet, combined treatment with IFN-α and ribavirin is the popular measure to control hepatitis C, albeit with limited success. A report that IFN-α increased PGE2 production suggested that elevated PGE2 might be attributed to unresponsiveness to IFN-α treatment, given the immunosuppressive nature of PGE2 (6). Accordingly, IFN-α signaling can be enhanced by indomethacin, one of the nonsteroidal anti-inflammatory drugs (NSAIDs) that blocks prostanoids production by inhibiting COX-1 and COX-2 activities (25). However, combined treatment of HCV patients with NSAIDs and IFN-α has shown limited success and is less effective than treatment with ribavirin (5). This result might be due to complex effects of prostanoids on host immunity. Interestingly, recent studies have shown that arachidonic acid is able to suppress HCV genomic replication and has a beneficial effect on HCV-infected patients (40, 51). Although it is not understood how arachidonic acid affects HCV, it is conceivable that impaired arachidonic acid metabolism might be favorable for HCV survival.
Our data implicate HCV in COX-2 expression. Although additional studies are apparently required to understand the biological significance of deregulated COX-2 expression in HCV-infected cells, possible roles of COX-2 in HCV pathogenesis can be hypothesized. It is possible that HCV avoids unwanted stimulation of adaptive immune responses against itself by down regulating COX-2 expression in HCV-infected antigen-presenting cells, such as macrophages and dendritic cells. Another possibility is that, by down regulating COX-2 expression, HCV-infected immune effector cells avoid lysis by NK cells and/or apoptosis caused by cyclopentenone 15d-PGJ2, which is derived from PGD2 during inflammatory responses. Lastly, HCV avoids evoking antiviral activities of prostanoids by suppressing COX-2 expression. Although a precise mechanism is not known, PGA and PGJ series prostaglandins have been reported to have antiviral activities against various DNA and RNA viruses (64). Recently, PGI2 was reported to have antiviral activity against respiratory syncytial virus in a respiratory syncytial virus-induced disease mouse model (31). Similarly, in a vaccinia virus-induced disease mouse model, PGI2-treated mice showed enhanced survival, whereas NSAIDs-treated mice succumbed to virus-induced mortality (80). Therefore, impaired COX-2 expression could be attributable to host immune evasion by HCV, favoring persistent HCV infection.
COX-2 expression is regulated by diverse transcription factors that include NF-κB, CREB, and C/EBP-β. However, the relative importance of these pathways is not clear and, more importantly, the impact of NF-κB on COX-2 expression in macrophages is controversial. Our results show that COX-2 is induced by selective activation of NF-κB alone and is suppressed by expression of superrepressor IκBα, suggesting that NF-κB activation is sufficient for COX-2 expression.
Our data in Fig. 8 show that COX-2 induction by LPS is substantially inhibited by HCV core. Given the complexity of cox-2 transcriptional regulation and the diverse transcriptional modulating function of HCV core, the possibility that HCV core also influences function of other transcription factors that are involved in COX-2 expression cannot be excluded. It is also possible that the significant decrease of COX-2 is due to a direct interruption of TNF-α or other cytokine production, via NF-κB suppression by HCV core, which otherwise contributes to COX-2 expression in an autocrine fashion, as LPS treatment induces TNF-α and other cytokines that, along with TLR4 signaling, enhance COX-2 expression (11). Finally, it is conceivable that HCV core could alter the half-life or increase the turnover rate of COX-2 protein, although transcription of the cox-2 gene is closely related to the level of COX-2 protein expression (M. D. Bryer, personal communication), and the half-life of COX-2 mRNA is significantly prolonged by LPS treatment (10).
NF-κB is either activated or suppressed by HCV core, depending on subtype. For example, while the core protein of HCV type 1b activates NF-κB activity, that of HCV type 1a (Hutchinson strain) suppresses NF-κB activity (54). A mutagenesis study showed that a single amino acid substitution in the core protein of HCV type 1a converting either from Lys at 9 (K9) to Arg or from Asn at 11 (N11) to Thr is sufficient to relieve its inhibitory function (54). However, it is not understood how these amino acid residues affect modulation of NF-κB activity.
Previously, it was proposed that activation of NF-κB by type 1b HCV core is related to IKKβ, although a direct biochemical interaction between the two proteins and direct functional interference by the HCV core protein were not presented (79). In the present study, we report evidence that type 1a HCV core protein biochemically interacts with IKKβ and inhibits IKK kinase activity (Fig. 6A and 7). Although we do not fully understand the mechanism that, unlike type 1a HCV core, type 1b HCV core activates IKK signalsome and NF-κB activity, our data raise the possibility that a capability of HCV core to physically interact with IKKβ might determine differential regulation of NF-κB activity. It is possible that the type 1a HCV core protein sequesters IKKβ and prevents formation of a fully functional IKK signalsome, which impairs NF-κB signaling. Alternatively, it is possible that the HCV core protein interferes with stoichiometric interactions between IKKβ and, subsequently, other subunits of IKK signalsome. More importantly, given that a single amino acid change at either K9 or N11 relieves inhibitory functions of the HCV core protein type 1a, it is conceivable that microenvironmental changes such as electric charge or physical hindrance affect interactions between IKKβ and its substrates.
Another observation in our study that might lead to understanding of the mechanism by which HCV core suppresses IKK activity is the disruption of cytoplasmic-nuclear partitioning of IKKβ. As shown in Fig. 4, either transfected or endogenous IKK-β is located in both cytoplasmic and nuclear fractions; however, the HCV core protein selectively disturbs the nuclear localization of IKK-β. In similar experiments to measure cellular distributions of IKKα and IKKγ, HCV core did not affect the distributions of these two subunits of the IKK signalsome (data not shown). The functional significance of nuclear IKKβ and the biological significance of blocked translocation of IKKβ by the HCV core protein are areas of intense investigation.
In summary, we studied the inhibitory function of HCV core on NF-κB activity in macrophages. Our data indicate that inhibition of NF-κB activity by the HCV core protein might be related to its physical interaction with and interrupted nuclear localization of IKKβ. Furthermore, our data show that COX-2 expression is induced by NF-κB activation and suppressed by the HCV core protein. These results suggest the possibility that HCV-infected macrophages have impaired immune responses at least in part due to impaired COX-2 expression.
Acknowledgments
This work was supported by the Department of Veterans Affairs and National Institutes of Health grants AI057591 (Y.S.H.), HL075557 (J.W.C.), and HL061419 (T.S.B.) and program project HL066196.
We thank Tamara Lasakow for editorial assistance with the manuscript.
REFERENCES
- 1.Afonso, A. M., J. Jiang, F. Penin, C. Tareau, D. Samuel, M. A. Petit, H. Bismuth, E. Dussaix, and C. Feray. 1999. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J. Virol. 73:9213-9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed] [Google Scholar]
- 4.Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, R. E. Sampliner, E. L. Meeks, M. J. Beach, et al. 1992. The natural history of community-acquired hepatitis C in the United States. N. Engl. J. Med. 327:1899-1905. [DOI] [PubMed] [Google Scholar]
- 5.Andreone, P., A. Gramenzi, C. Cursaro, M. Biselli, S. Lorenzini, E. Loggi, F. Felline, S. Fiorino, L. Di Giammarino, F. Porzio, S. Galli, and M. Bernardi. 2003. Interferon-alpha combined with ketoprofen as treatment of naive patients with chronic hepatitis C: a randomized controlled trial. J. Viral Hepat. 10:306-309. [DOI] [PubMed] [Google Scholar]
- 6.Andreone, P., C. Cursaro, and G. Gasbarrini. 1993. Interferon-alpha increases prostaglandin E2 production by cultured liver biopsy in patients with chronic viral hepatitis: can non-steroidal anti-inflammatory drugs improve the therapeutic response to interferon? J. Hepatol. 19:228-231. [DOI] [PubMed] [Google Scholar]
- 7.Anest, V., J. L. Hanson, P. C. Cogswell, K. A. Steinbrecher, B. D. Strahl, and A. S. Baldwin. 2003. A nucleosomal function for IκB kinase-αin NF-κB-dependent gene expression. Nature 423:659-663. [DOI] [PubMed] [Google Scholar]
- 8.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 9.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Short protocols in molecular biology, 4th ed., p. A1-33. John Wiley & Sons, New York, N.Y.
- 10.Barrios-Rodiles, M., G. Tiraloche, and K. Chadee. 1999. Lipopolysaccharide modulates cyclooxygenase-2 transcriptionally and posttranscriptionally in human macrophages independently from endogenous IL-1 beta and TNF-alpha. J. Immunol. 163:963-969. [PubMed] [Google Scholar]
- 11.Beutler, B., and E. T. Rietschel. 2003. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3:169-176. [DOI] [PubMed] [Google Scholar]
- 12.Birbach, A., P. Gold, B. R. Binder, E. Hofer, R. de Martin, and J. A. Schmid. 2002. Signaling molecules of the NF-kappa B pathway shuttle constitutively between cytoplasm and nucleus. J. Biol. Chem. 277:10842-10851. [DOI] [PubMed] [Google Scholar]
- 13.Caamano, J., and C. A. Hunter. 2002. NF-κB family of transcription factors: central regulators of innate and adaptive immune functions. Clin. Microbiol. Rev. 15:414-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cacciarelli, T. V., O. M. Martinez, R. G. Gish, J. C. Villanueva, and S. M. Krams. 1996. Immunoregulatory cytokines in chronic hepatitis C virus infection: pre- and posttreatment with interferon alpha. Hepatology 24:6-9. [DOI] [PubMed] [Google Scholar]
- 15.Caivano, M., B. Gorgoni, P. Cohen, and V. Poli. 2001. The induction of cyclooxzygenase-2 mRNA in macrophages is biphasic and requires both CCAAT enhancer-binding protein β (C/EBPβ) and C/EBPδ transcription factors. J. Biol. Chem. 276:48693-48701. [DOI] [PubMed] [Google Scholar]
- 16.Cerny, A., and F. V. Chisari. 1999. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 30:595-601. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C. M., L. R. You, L. H. Hwang, and Y. H. Lee. 1997. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-beta receptor modulates the signal pathway of the lymphotoxin-beta receptor. J. Virol. 71:9417-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 19.Costantini, V., A. Giampietri, M. Allegrucci, G. Agnelli, G. G. Nenci, and M. C. Fioretti. 1991. Mechanisms of the antimetastatic activity of stable prostacyclin analogues: modulation of host immunocompetence. Adv. Prostaglandin Thromboxane Leukoc. Res. 21B:917-920. [PubMed] [Google Scholar]
- 20.D'Acquisto, F., T. Iuvone, L. Rombola, L. Sautebin, M. Di Rosa, and R. Carnuccio. 1997. Involvement of NF-κB in the regulation of cyclo-oxygenase-2 protein expression in LPS-stimulated J774 macrophages. FEBS Lett. 418:175-178. [DOI] [PubMed] [Google Scholar]
- 21.Devin, A., A. Cook, Y. Lin, Y. Rodriguez, M. Kelliher, and Z. Liu. 2000. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity 12:419-429. [DOI] [PubMed] [Google Scholar]
- 22.Dubois, R. N., S. B. Abramson, L. Crofford, R. A. Gupta, L. S. Simon, L. B. Van De Putte, and P. E. Lipsky. 1998. Cyclooxygenase in biology and disease. FASEB J. 12:1063-1073. [PubMed] [Google Scholar]
- 23.Eckels, D. D., H. Wang, T. H. Bian, N. Tabatabai, and J. C. Gill. 2000. Immunobiology of hepatitis C virus (HCV) infection: the role of CD4 T cells in HCV infection. Immunol. Rev. 174:90-97. [DOI] [PubMed] [Google Scholar]
- 24.FitzGerald, G. A. 2003. COX-2 and beyond: approaches to prostaglandin inhibition in human disease. Nat. Rev. Drug Discov. 2:879-890. [DOI] [PubMed] [Google Scholar]
- 25.Giambartolomei, S., M. Artini, C. Almerighi, S. M. Moavero, M. Levrero, and C. Balsano. 1999. Nonsteroidal anti-inflammatory drug metabolism potentiates interferon alpha signaling by increasing STAT1 phosphorylation. Hepatology 30:510-516. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109:S81-S96. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 28.Gualde, N., and H. Harizi. 2004. Prostanoids and their receptors that modulate dendritic cell-mediated immunity. Immunol. Cell Biol. 82:353-360. [DOI] [PubMed] [Google Scholar]
- 29.Guo, B., P. R. Odgren, A. J. van Wijnen, T. J. Last, J. Nickerson, S. Penman, L. B. Lian, J. L. Stein, and G. S. Stein. 1995. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc. Natl. Acad. Sci. USA 92:10526-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris, S. G., J. Padilla, L. Koumas, D. Ray, and R. P. Phipps. 2002. Prostaglandins as modulators of immunity. Trends Immunol. 23:144-150. [DOI] [PubMed] [Google Scholar]
- 31.Hashimoto, K., B. S. Graham, M. W. Geraci, G. A. FitzGerald, K. Egan, W. Zhou, K. Goleniewska, J. F. O'Neal, J. D. Morrow, R. K. Durbin, P. F. Wright, R. D. Collins, T. Suzutani, and R. S. Peebles, Jr. 2004. Signaling through the prostaglandin I2 receptor IP protects against respiratory syncytial virus-induced illness. J. Virol. 78:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hultgren, C., D. R. Milich, O. Weiland, and M. Sallberg. 1998. The antiviral compound ribavirin modulates the T helper (Th) 1/Th2 subset balance in hepatitis B and C virus-specific immune responses. J. Gen. Virol. 79:2381-2391. [DOI] [PubMed] [Google Scholar]
- 33.Hwang, D., B. C. Jang, G. Yu, and M. Boudreau. 1997. Expression of mitogen-inducible cyclo-oxygenase induced by lipopolysaccharide: mediation through both mitogen-activated protein kinase and NF-κB signaling pathways in macrophages. Biochem. Pharmacol. 54:87-96. [DOI] [PubMed] [Google Scholar]
- 34.Inoue, H., T. Nanayama, S. Hara, C. Yokoyama, and T. Tanabe. 1994. The cyclic AMP response element plays an essential role in the expression of the human prostaglandin-endoperoxide synthase 2 gene in differentiated U937 monocytic cells. FEBS Lett. 350:51-54. [DOI] [PubMed] [Google Scholar]
- 35.Jaffar, Z., K. S. Wan, and K. Roberts. 2002. A key role for prostaglandin I2 in limiting lung mucosal Th2, but not Th1, responses to inhaled allergen. J. Immunol. 169:5997-6004. [DOI] [PubMed] [Google Scholar]
- 36.Karin, M., and M. Delhase. 2000. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin. Immunol. 12:85-98. [DOI] [PubMed] [Google Scholar]
- 37.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 38.Kontgen, F., R. J. Grumont, A. Strasser, D. Metcalf, R. Li, D. Tarlinton, and S. Gerondakis. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965-1977. [DOI] [PubMed] [Google Scholar]
- 39.Large, M. K., D. J. Kittlesen, and Y. S. Hahn. 1999. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J. Immunol. 162:931-938. [PubMed] [Google Scholar]
- 40.Leu, G. Z., T. Y. Lin, and J. T. Hsu. 2004. Anti-HCV activities of selective polyunsaturated fatty acids. Biochem. Biophys. Res. Commun. 318:275-280. [DOI] [PubMed] [Google Scholar]
- 41.Li, Q., D. Van Antwerp, F. Mercurio, K. F. Lee, and I. M. Verma. 1999. Severe liver degeneration in mice lacking the IκΒ kinase 2 gene. Science 284:321-325. [DOI] [PubMed] [Google Scholar]
- 42.Li, Z. W., W. Chu, Y. Hu, M. Delhase, T. Deerinck, M. Ellisman, R. Johnson, and M. Karin. 1999. The IKK beta subunit of IκΒ kinase (IKK) is essential for NF-κB activation and prevention of apoptosis. J. Exp. Med. 189:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makris, C., V. L. Godfrey, G. Krahn-Senftleben, T. Takahashi, J. L. Roberts, T. Schwarz, L. Feng, R. S. Johnson, and M. Karin. 2000. Female mice heterozygous for IKKγ/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol. Cell 5:969-979. [DOI] [PubMed] [Google Scholar]
- 44.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-κB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto, M., T. Y. Hsieh, N. Zhu, T. VanArsdale, S. B. Hwang, K. S. Jeng, A. E. Gorbalenya, S. Y. Lo, J. H. Ou, C. F. Ware, and M. M. Lai. 1997. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J. Virol. 71:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNeil, S., B. Guo, J. L. Stein, J. B. Lian, S. Bushmeyer, E. Seto, M. L. Atchison, S. Penman, A. J. V. Wijnen, and G. S. Stein. 1998. Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J. Cell. Biochem. 65:500-510. [PubMed] [Google Scholar]
- 47.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 48.Morris, J. K., and J. S. Richards. 1996. An E-box region within the prostaglandin endoperoxide synthase-2 (PGS-2) promoter is required for transcription in rat ovarian granulosa cells. J. Biol. Chem. 271:16633-16643. [DOI] [PubMed] [Google Scholar]
- 49.Muller, M. M., E. Schreiber, W. Schaffner, and P. Matthias. 1989. Rapid test for in vivo stability and DNA binding of mutated octamer binding proteins with “mini-extracts” prepared from transfected cells. Nucleic Acids Res. 17:6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogasawara, A., T. Arakawa, T. Kaneda, T. Takuma, T. Sato, H. Kaneko, M. Kumegawa, and Y. Hakeda. 2001. Fluid shear stress-induced cyclooxygenase-2 expression is mediated by C/EBP beta, cAMP-response element-binding protein, and AP-1 in osteoblastic MC3T3-E1 cells. J. Biol. Chem. 276:7048-7054. [DOI] [PubMed] [Google Scholar]
- 51.Okita, M., K, Tomioka, Y. Ota, T. Sasagawa, T. Osawa, N. Sakai, M. Kawaguchi, and T. Itoshima. 2003. Arachidonic acid in mononuclear cells and its clinical significance in HCV cirrhotic patients. Nutrition 19:727-732. [DOI] [PubMed] [Google Scholar]
- 52.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 53.Ray, R. B., L. M. Lagging, K. Meyer, R. Steele, and R. Ray. 1995. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 37:209-220. [DOI] [PubMed] [Google Scholar]
- 54.Ray, R. B., R. Steele, A. Basu, K. Meyer, M. Majumder, A. K. Ghosh, and R. Ray. 2002. Distinct functional role of hepatitis C virus core protein on NF-κB regulation is linked to genomic variation. Virus Res. 87:21-29. [DOI] [PubMed] [Google Scholar]
- 55.Ray, R. B., R. Steele, K. Meyer, and R. Ray. 1998. Hepatitis C virus core protein represses p21 waf1/Cip1/Sid1 promoter activity. Gene 208:331-336. [DOI] [PubMed] [Google Scholar]
- 56.Rothwarf, D. M., E. Zandi, G. Natoli, and M. Karin. 1998. IKKγ is an essential regulatory subunit of the IκB kinase complex. Nature 395:297-300. [DOI] [PubMed] [Google Scholar]
- 57.Rudolph, D., W. C. Yeh, A. Wakeham, B. Rudolph, D. Nallainathan, J. Potter, A. J. Elia, and T. W. Mak. 2000. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ-deficient mice. Genes Dev. 14:854-862. [PMC free article] [PubMed] [Google Scholar]
- 58.Sadikot, R. T., W. Han, M. B. Everhart, O. Zoia, R. S. Peebles, E. D. Jansen, F. E. Yull, J. W. Christman, and T. S. Blackwell. 2003. Selective I kappa B kinase expression in airway epithelium generates neutrophilic lung inflammation. J. Immunol. 170:1091-1098. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt-Supprian, M., W. Bloch, G. Courtois, K. Addicks, A. Israel, K. Rajewsky, and M. Pasparakis. 2000. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol. Cell 5:981-992. [DOI] [PubMed] [Google Scholar]
- 60.Shih, C. M., C. M. Chen, S. Y. Chen, and Y. H. Lee. 1995. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J. Virol. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shrivastava, A., S. K. Manna, R. Ray, and B. B. Aggarwal. 1998. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J. Virol. 72:9722-9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soguero, C., M. Joo, K. A. Chianese-Bullock, D. T. Nguyen, K. Tung, and Y. S. Hahn. 2002. Hepatitis C virus core protein leads to immune suppression and liver damage in a transgenic murine model. J. Virol. 76:9345-9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sorli, C. H., H. J. Zhang, M. B. Armstrong, R. V. Rajotte, J. Maclouf, and R. P. Robertson. 1998. Basal expression of cyclooxygenase-2 and nuclear factor-interleukin 6 are dominant and coordinately regulated by interleukin 1 in the pancreatic islet. Proc. Natl. Acad. Sci. USA 95:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Straus, D. S., and C. K. Glass. 2001. Cyclopentenone prostaglandins: new insights on biological activities and cellular targets. Med. Res. Rev. 21:185-210. [DOI] [PubMed] [Google Scholar]
- 65.Tam, R. C., B. Pai, J. Bard, C., Lim, D. R. Averett, U. T. Phan, and T. Milovanovic. 1999. Ribavirin polarizes human T cell responses towards a type 1 cytokine profile. J. Hepatol. 30:376-382. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka, M., M. E. Fuentes, K. Yamaguchi, M. H. Durnin, S. A. Dalrymple, K. L. Hardy, and D. V. Goeddel. 1999. Embryonic lethality, liver degeneration, and impaired NF-κB activation in IKKβ-deficient mice. Immunity 10:421-429. [DOI] [PubMed] [Google Scholar]
- 67.Thomas, B., F. Berenbaum, L. Humbert, H. Bian, G. Bereziat, L. Crofford, and J. L. Olivier. 2000. Critical role of C/EBPδ and C/EBPβ factors in the stimulation of the cyclooxygenase-2 gene transcription by interleukin-1β in articular chondrocytes. Eur. J. Biochem. 267:6798-6809. [DOI] [PubMed] [Google Scholar]
- 68.Thomas, D. W., P. N. Rocha, C. Nataraj, L. A. Robinson, R. F. Spurney, B. H. Koller, and T. M. Coffman. 2003. Proinflammatory actions of thromboxane receptors to enhance cellular immune responses. J. Immunol. 171:6389-6395. [DOI] [PubMed] [Google Scholar]
- 69.Thomas, H. C., M. E. Torok, D. M. Forton, and S. D. Taylor-Robinson. 1999. Possible mechanisms of action and reasons for failure of antiviral therapy in chronic hepatitis C. J. Hepatol. 31:152-159. [DOI] [PubMed] [Google Scholar]
- 70.Ushikubi, F., Y. Aiba, K. Nakamura, T. Namba, M. Hirata, O. Mazda, Y. Katsura, and S. Narumiya. 1993. Thromboxane A2 receptor is highly expressed in mouse immature thymocytes and mediates DNA fragmentation and apoptosis. J. Exp. Med. 178:1825-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verma, U. N., Y. Yamamoto, S. Prajapati, and R. B. Gaynor. 2004. Nuclear role of IKKγ/NEMO in NF-kappa B-dependent gene expression. J. Biol. Chem. 279:3509-3515. [DOI] [PubMed] [Google Scholar]
- 72.Wadleigh, D. J., S. T. Reddy, E. Kopp, S. Ghosh, and H. R. Herschman. 2000. Transcriptional activation of the cyclooxygenase-2 gene in endotoxin-treated RAW 264.7 macrophages. J. Biol. Chem. 275:6259-6266. [DOI] [PubMed] [Google Scholar]
- 73.Williams, J. A., and E. Shacter. 1997. Regulation of macrophage cytokine production by prostaglandin E2. Distinct roles of cyclooxygenase-1 and -2. J. Biol. Chem. 272:25693-25699. [DOI] [PubMed] [Google Scholar]
- 74.Williams, T. J., and J. Morley. 1973. Prostaglandins as potentiators of increased vascular permeability in inflammation. Nature 246:215-217. [DOI] [PubMed] [Google Scholar]
- 75.Xie, W., B. S. Fletcher, R. D. Andersen, and H. R. Herschman. 1994. v-src induction of the TIS10/PGS2 prostaglandin synthase gene is mediated by an ATF/CRE transcription response element. Mol. Cell. Biol. 14:6531-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 423:655-659. [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto, K., T. Arakawa, N. Ueda, and S. Yamamoto. 1995. Transcriptional roles of nuclear factor kappa B and nuclear factor-interleukin-6 in the tumor necrosis factor alpha-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J. Biol. Chem. 270:31315-31320. [DOI] [PubMed] [Google Scholar]
- 78.Yamaoka, S., G. Courtois, C. Bessia, S. T. Whiteside, R. Weil, F. Agou, H. E. Kirk, R. J. Kay, and A. Israel. 1998. Complementation cloning of NEMO, a component of the IκΒ kinase complex essential for NF-κB activation. Cell 93:1231-1240. [DOI] [PubMed] [Google Scholar]
- 79.Yoshida, H., N. Kato, Y. Shiratori, M. Otsuka, S. Maeda, J. Kato, and M. Omata. 2001. Hepatitis C virus core protein activates nuclear factor kappa B-dependent signaling through tumor necrosis factor receptor-associated factor. J. Biol. Chem. 276:16399-16405. [DOI] [PubMed] [Google Scholar]
- 80.Zavagno, G., B. Jaffe, and M. Esteban. 1987. Role of prostaglandins and non-steroid anti-inflammatory drugs in the pathogenicity of vaccinia virus. J. Gen. Virol. 68:593-600. [DOI] [PubMed] [Google Scholar]
- 81.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]