Abstract
JAK2V617F is the most recurrent genetic mutation in Philadelphia-negative chronic Myeloproliferative Neoplasms (MPNs). Since the JAK2 locus is located on Chromosome 9, we hypothesized that Chromosome 9 copy number abnormalities may be a disease modifier in JAK2V617F-mutant MPN patients. In this study, we identified a subset of MPN patients with partial or complete Chromosome 9 trisomy (+9p patients), who differ from JAK2V617F-homozygous MPN patients as they carry three JAK2 alleles as well as three copies of all neighboring gene loci, including CD274, encoding immunosuppressive Programmed death-ligand 1 (PD-L1) protein. Investigation of the clonal hierarchy revealed that the JAK2V617F occurs first, followed by +9p. Functionally, CD34+ cells from +9p MPN patients demonstrated increased clonogenicity, generating a greater number of primitive colonies, due to high OCT4 and NANOG expression, with knock-down of these genes leading to a genotype-specific decrease in colony numbers. Moreover, our analysis revealed increased PD-L1 surface expression in malignant monocytes from +9p patients, while analysis of the T cell compartment unveiled elevated levels of exhausted cytotoxic T cells. Overall, here we identify a distinct novel subgroup of MPN patients, who feature a synergistic interplay between +9p and JAK2V617F that shapes immune escape characteristics and increased stemness in CD34+ cells.
Subject terms: Myeloproliferative disease, Cancer genomics, Cancer stem cells, Oncogenesis
Introduction
Philadelphia-negative Myeloproliferative Neoplasms (MPNs) encompass a range of clonal blood disorders originating from a single mutated hematopoietic stem cell, resulting in an overproduction of mature blood cells [1]. The classic MPN forms include polycythemia vera (PV), essential thrombocythemia (ET), and myelofibrosis (MF), which can either be primary or secondary to PV or ET [2]. While PV patients typically present with erythrocytosis [3], ET and MF patients show thrombocytosis [4, 5]. Strikingly, MF stands out as the most aggressive subtype, as it is also characterized by elevated white blood cell counts, development of bone marrow fibrosis, a higher risk of disease progression to secondary acute myeloid leukemia, and a median survival rate of five years from diagnosis [6–8].
Considerable efforts in the characterization of the molecular landscape of MPNs have led to the identification of driver mutations in JAK/STAT signaling pathway genes (JAK2 [9], CALR [10], or MPL [11]), with additional non-driver mutations being described more recently [12, 13]. A gain-of-function point mutation in JAK2, namely JAK2V617F, was discovered as being the most frequent lesion, affecting a significant proportion of MPN patients, particularly in PV and MF. Furthermore, the variant allele frequency (VAF) of JAK2V617F has emerged as a crucial factor in determining both severity and phenotypic characteristics of MPNs [14]. Additionally, cytogenetic abnormalities have frequently been observed, especially in MF, with about 30% of patients presenting with such lesions and aneuploidies of Chromosome 9, where the JAK2 gene locus is located, being among the most described karyotypic defects [15, 16]. Interestingly, somatic uniparental disomy (UPD) of Chromosome 9p has been reported as a common mechanism of copy-neutral loss of heterozygosity resulting in the acquisition of two mutated JAK2 alleles, thus leading to JAK2V617F homozygosity [17]. The JAK2 gene is situated on Chromosome 9p24.1, in close proximity to other gene loci like CD274, which encodes programmed death-ligand 1 (PD-L1) protein that serves as an immunosuppressive binding partner for PD-1 expressed on T cells. Strikingly, the PD-L1/PD-1 axis has been described to be exploited to evade immune surveillance by tumor cells in various myeloid malignancies [18], while recent studies report higher PD-L1 expression in granulocytes from MF patients harboring JAK2V617F compared to CALR-mutant patients or healthy individuals [17]. Notably, Chromosome 9 copy number gains have been described as more frequent in MF patients (3.8%), compared to ET and PV patients (0.58% and 2.25%) [19–21].
Despite the frequent occurrence of Chromosome 9 copy number gains observed in MPNs, their functional and clinical implications remain poorly understood, especially for JAK2V617F-mutated patients, where chromosomal amplifications may alter the mutational burden on a per-cell basis. These aberrations may in fact confer distinct biological features to MPN cells when compared to those with UPD, which carry only two copies of each gene located on Chromosome 9. To address this knowledge gap, we herein investigate the biological significance of Chromosome 9 copy number gains in MPNs in the context of JAK2V617F mutation. We show that somatic partial or complete trisomy of Chromosome 9 (+9p) involves the entire short arm and includes, among others, the JAK2 and CD274 loci. Furthermore, we demonstrate that +9p synergizes with JAK2V617F with functional implications, as hematopoietic stem/progenitor cells (HSPCs) retain more immature characteristics, while monocytes express more membrane PD-L1, leading to CD8 + T cell exhaustion.
Overall, this study sheds light on the biology of a previously uncharacterized subset of JAK2-mutated patients, with distinct karyotypic features.
Methods
Ethics statement
The study was conducted on a cohort of 18 healthy donors and 32 patients with a diagnosis of PV, ET, primary or secondary MF, under treatment in the Hematology department of Azienda Ospedaliera-Universitaria of Modena, Institutional MPN database of the Hematology Unit, University of Insubria (Varese, Italy), or Azienda Ospedaliero-Universitaria Careggi (Florence, Italy). PV, ET, and MF were diagnosed according to 2016 World Health Organization criteria [22], whereas the International Working Group for Myeloproliferative neoplasms Research and Treatment criteria were used for sMF diagnosis [23].
This study was conducted in accordance with the Declaration of Helsinki under the local Institutional Review Board’s approved protocol. All subjects involved in the study provided informed written consent.
Purification of cell populations from peripheral blood and culture conditions
Peripheral blood (PB) samples of MPN patients and healthy donors were collected and cell fractions were isolated and cultured as previously described [24, 25] and detailed in Supplementary Methods.
Next generation sequencing (NGS) and multiplex ligation-dependent probe amplification (MLPA) analyses
Targeted DNA sequencing on genomic DNA extracted from whole PB of MPN patients was performed as detailed in the Supplementary Methods [26]. 50 ng of DNA from whole PB, CD34+ cells, CD14+ cells, CD3+ cells, or granulocytes were further analysed by means of MLPA, using P474 MLPA kit (MRC-Holland, Amsterdam, Netherlands), following the manufacturer’s instructions. Fragment analysis was performed through the 3130xl Genetic Analyzer instrument (Applied Biosystems), and data were analysed using Coffalyser.net software (MRC-Holland), as previously described [27].
Methylcellulose and collagen clonogenic assays
For each analysed patient, 300 CD34+ cells were seeded in triplicate in 1 ml of semisolid methylcellulose-based medium (MethoCult GF H4434; StemCell Technologies Inc., Vancouver, Canada), as previously described [28]. Colonies were scored after 14 days of incubation.
MK colony forming units (CFU-MK) were assayed in collagen-based medium, by seeding 5000 CD34+ cells isolated from PB in each chamber of a double-chamber slide from a commercial MK assay detection kit (MegaCult-C; StemCell Technologies Inc.), as previously reported [29]. Colonies were scored after 11 days of incubation.
POU5F1 and NANOG silencing in CD34+ cells
Frozen human CD34+ cells were transfected with POU5F1 and NANOG small interfering RNAs (siRNA) by using the 4D-Nucleofector System (Lonza, Basel, Switzerland), with an optimized protocol from previously described methods (see Supplementary Methods). DNA and RNA from individual colonies isolated after 14 days of incubation underwent droplet digital PCR (ddPCR) analysis as detailed below.
DNA extraction
For each MPN patient, DNA was extracted from purified CD34+ cells, CD14+ cells, CD3+ cells, and granulocytes by means of DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the recommended protocol.
Single-cell derived colonies’ DNA was extracted using Arcturus PicoPure DNA Extraction Kit (Applied Biosystems, Waltham, Massachusetts, USA) according to manufacturer’s instructions.
RNA extraction and reverse transcription
Total RNA from Granulocytes, CD14+, or CD34+ cells was extracted using the miRNeasy Micro Kit (Qiagen, Hilden, Germany), as previously described [30]. Next, reverse transcription was performed starting from 100 ng of total RNA, using High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, Massachusetts, USA), as previously described [31].
For single-cell derived colonies, RNA was isolated and retro-transcribed by means of TaqMan™ Gene Expression Cells-to-CT™ Kit (ThermoFisher Scientific) following the manufacturer’s protocol.
Quantitative real-time PCR (qRT-PCR)
Quantitation of CD274, POU5F1, and NANOG gene expression in cDNA from Granulocytes, CD34+ cells, or CD14+ cells was carried out by means of qRT-PCR [32] as detailed in Supplementary Methods.
ddPCR assay
10 ng of each DNA sample were analysed through ddPCR to perform JAK2 mutation genotyping and copy number analysis (see Supplementary Methods). For coupled genomic-transcriptomic studies, isolated colonies were split in half to obtain DNA and RNA from each colony, as detailed above. JAK2V617F genotyping, JAK2 copy number analysis, CD274, POU5F1, and NANOG gene expression assessment were performed as detailed in Supplementary Methods [33].
Single cell DNA library preparation and sequencing
Frozen Peripheral Blood Mononuclear Cells (PBMCs) were thawed following 10× Genomics® “Sample Preparation Demonstrated Protocol” (10× Genomics, Pleasanton, CA, USA). PBMCs and CD34+ cells were mixed at a 1:1 ratio and were processed through Tapestri™ Platform (Mission Bio, San Francisco, CA, USA) as previously described [34]. Barcoded samples underwent targeted PCR amplification through a custom DNA panel (Supplementary Table 2). Tapestri workflow and data processing is detailed in the Supplementary Methods section.
Flow cytometry
CD14+ and CD3+ cells isolated from PB of patients were incubated with “FcR Blocking Reagent, human” (Cat. no. 130-059-901, Miltenyi Biotech, Cologne, Germany) and subsequently stained as detailed in Supplementary Methods. Stained cells were then analysed using BD FACSCanto II (BD Biosciences; San Jose, CA, USA), as previously described. Data were analysed by FlowJo (version 10.7.1) [35].
Immunofluorescence staining
Cytospins of CD14+ cells were fixed with 4% paraformaldehyde and stained with rabbit monoclonal anti-human PD-L1 antibody (Cell Signaling Technology, Inc., Danvers, Massachusetts, USA) and mouse anti-human CD14 FITC-conjugated antibody (Miltenyi Biotech) (see Supplementary Methods) [36, 37]
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 8.4.0 (GraphPad Software, San Diego, CA, USA) as detailed in the Supplementary Methods.
Results
Identification of chromosome 9p trisomy in a subset of MPN patients
We identified 12 JAK2V617F-mutant MPN patients with concurrent partial or complete Chromosome 9 gains through classical cytogenetics, NGS, or MLPA at the time of diagnosis (Figs. 1 and S1) (Table S1), with the minimal amplified region being the entire short arm of Chromosome 9. Therefore, these patients will be hereafter referred to as “+9p patients”. 20 additional JAK2V617F-mutant MPN patients were identified as Chromosome 9-diploidy controls.
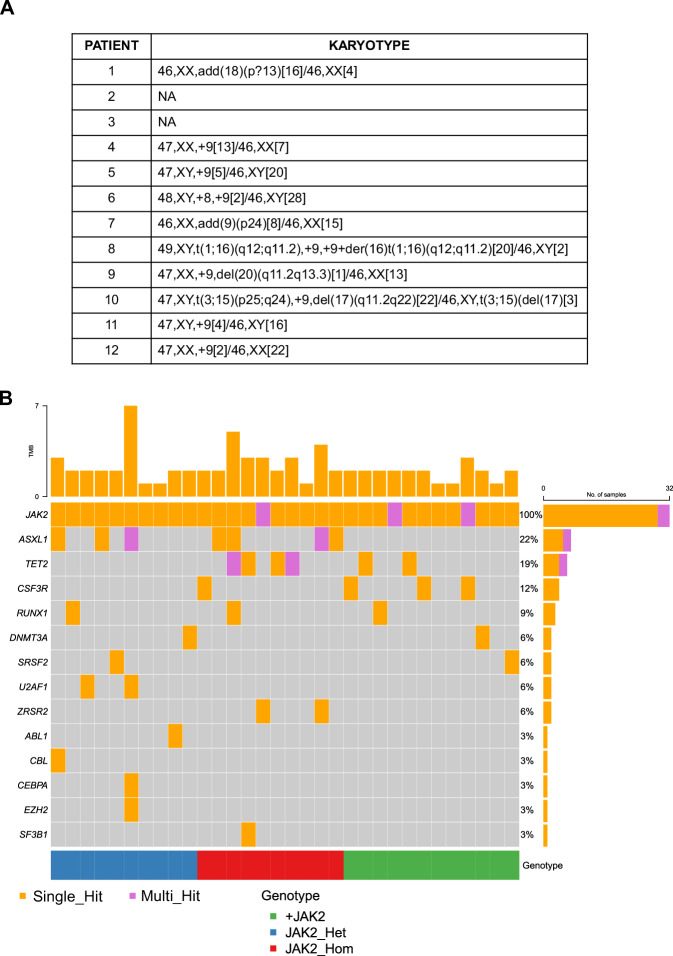
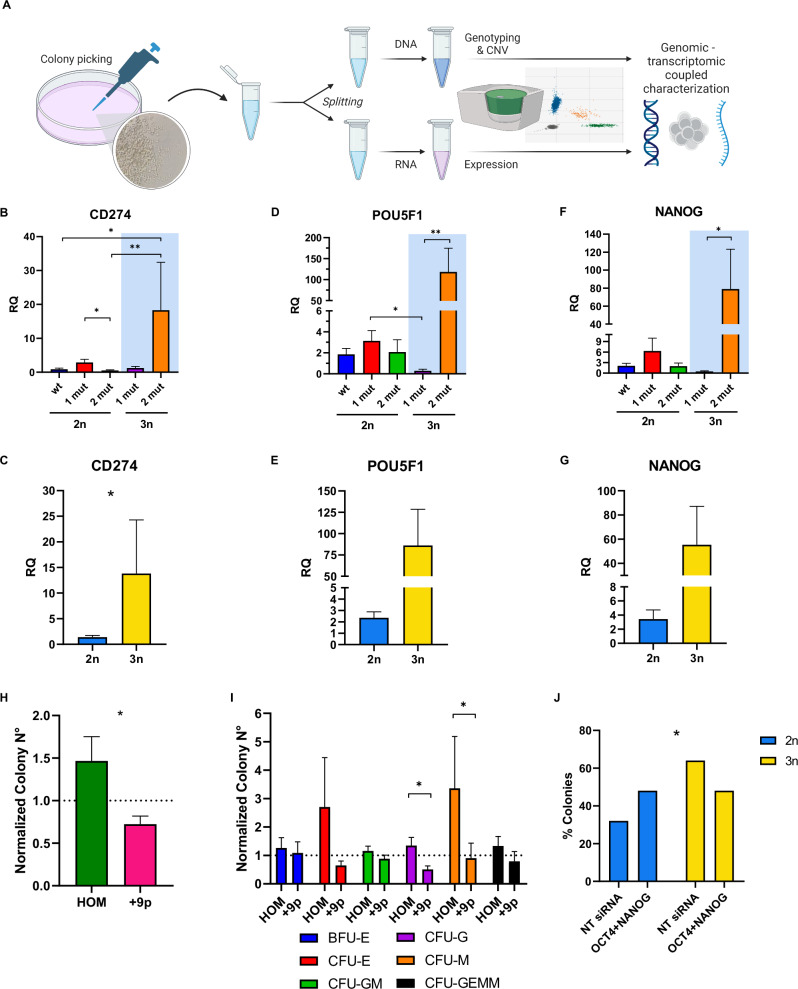
Fig. 1. Description of the genomic asset of the analyzed patients’ cohort.
A Karyotype of the +9p MPN patients (n = 12) at diagnosis. B Waterfall plot detailing the mutational profile of the entire cohort inferred from NGS data. Patients are split according to JAK2V617F allele frequency and/or JAK2 copy number (blue = normal copy number and heterozygous JAK2V617F, 20% < VAF < 50%, n = 10; red = normal copy number and homozygous JAK2V617F, VAF > 50%, n = 10; green = JAK2 mutation and trisomy, n = 12). For each patient, the mutational asset of a specific gene is reported as an orange box if the gene harbors a single variant or a purple box if multiple variants within the same gene are detected.
NGS analysis on DNA from whole PB allowed us to classify the Chromosome 9-diploidy control MPN patients in two additional sub-groups: those with normal copy numbers and JAK2 mutation in heterozygosity (20% < VAF < 50%; median 41.7%, range 22.9–55%; N = 10) or homozygosity (VAF > 50%; median 90.05%, range 71.2–99.6%; N = 10) (Fig. 1B). Correlation analysis revealed a negative association between +9p gain and the presence of co-occurring high-risk mutations or mutations in genes encoding epigenetic factors (Fig. S2).
CD274 expression is increased in +9p MPN cells
Next, we confirmed that Chromosome 9p was a somatic lesion by analyzing three malignant hematopoietic subpopulations (CD34+ cells, monocytes, granulocytes) and CD3 + T cells serving as a known germline control in MPN patients [38] through an MLPA assay on Chromosome 9p (Fig. 2A–D). Moreover, we observed the gain of an extra copy of the CD274 locus, which encodes the immune checkpoint molecule PD-L1 (Fig. 2A–D).
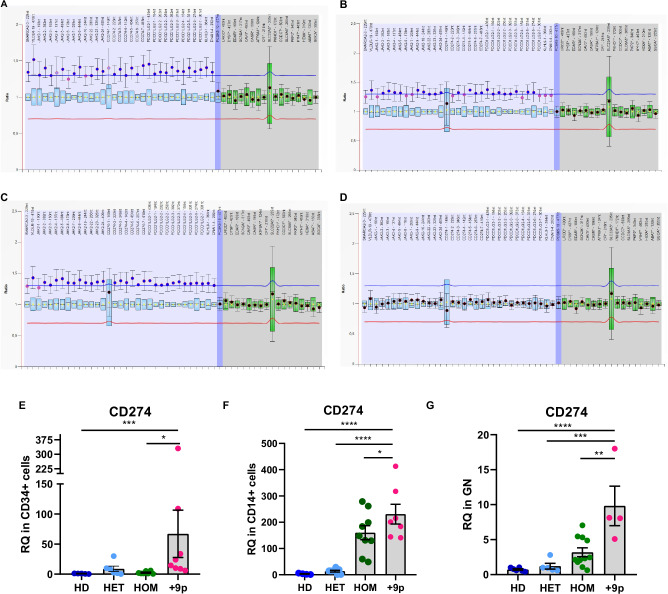
Fig. 2. Assessment of chromosome 9 trisomy and CD274 expression in hematopoietic cell subpopulations.
MLPA profiling of chromosome 9 copy number status in CD34+ cells (A), CD14+ cells (B), Granulocytes (C), or CD3+ cells (D) from patient #2. Labels on the horizontal axis list the genomic regions detected by MLPA probes, while on the vertical axis, the ratio of each probe signal between the patient and the reference samples is shown. The light blue region highlights the signal ratio of probe mapping on chromosome 9p, whereas the blue region represents the signal ratio of the probe mapping on chromosome 9q. The gray region highlights the signal ratio of reference probes. The horizontal lines represent the copy number thresholds beyond which an amplification (blue line) or a deletion (red line) is identified by Coffalyser.net software. Evaluation of CD274 expression at the transcriptional level in (E) CD34+ cells (n = 5 HD, n = 6 HET, n = 7 HOM, n = 8 + 9p), (F) CD14+ cells (n = 4 HD, n = 8 HET, n = 9 HOM, n = 7 + 9p) or (G) Granulocytes (n = 6 HD, n = 5 HET, n = 10 HOM, n = 5 + 9p) from JAK2-mutated MPN patients and healthy donors. Each barplot represents the relative quantity of CD274 mRNA quantified by means of qRT-PCR, whereas dots represent individual values. Results are presented as mean + SEM. Abbreviations: RQ = relative quantity; HD = healthy donors; HET = JAK2-mutated heterozygous patients; HOM = JAK2-mutated homozygous patients; +9p = JAK2-mutated patients with 9p trisomy. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
As previous research has demonstrated that JAK2 hyperactivation via p.V617F mutation can promote CD274 expression [18], we aimed to elucidate whether the JAK2V617F synergizes with Chromosome 9p trisomy in this context. To address this, we carried out gene expression analysis on CD34+ cells, CD14+ monocytes, and granulocytes isolated from +9p MPN patients compared with patients who do not present this chromosomal aberration. Our results show a significant upregulation of CD274 mRNA expression in +9p patients across all the analysed cell types, even when compared to JAK2V617F-homozygous MPN patients (Fig. 2E–G).
Chromosome 9p gain involves mutated JAK2 gene
Next, we aimed at investigating the genetic profile of +9p patients to assess whether the amplification of the JAK2 locus involved the mutated or the wild-type allele.
Given the inherent genetic heterogeneity within individual patients, we assessed JAK2 copy number and mutational status at single-cell resolution through ddPCR, using DNA extracted from single CD34+ cell-derived colonies cultured in methylcellulose-based medium.
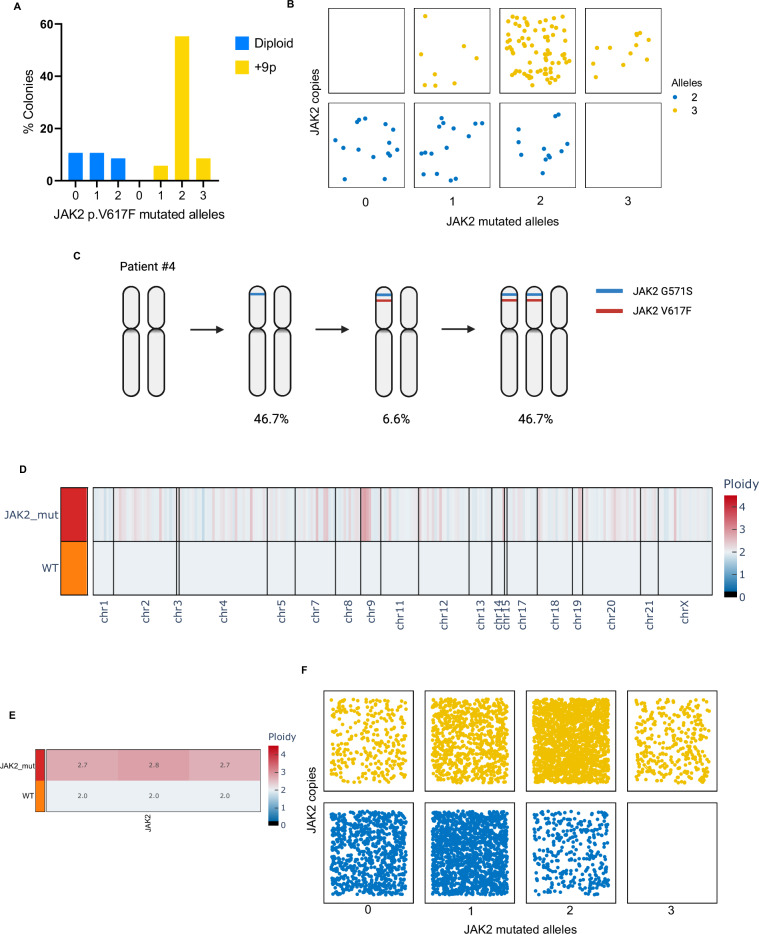
Our findings revealed that most colonies from patients with Chromosome 9p trisomy exhibited three copies of JAK2, with 2 out of 3 alleles harboring the JAK2 mutation (63.6% of colonies) (Fig. 3A). Given the relative abundance of each mutational status/copy number, our data suggest the chronological sequence of molecular events, with JAK2V617F occurring first, followed by Chromosome 9p gain carrying the JAK2-mutated allele, leading to the generation of cells with three JAK2 copies, two of which being mutated (Fig. 3B). Notably, one of the +9p patients carried both JAK2V617F and an additional JAK2G571S variant. In this case, most of the colonies exhibited 2 copies of JAK2 (53.3% of colonies), with JAK2G571S anteceding the acquisition of JAK2V617F. Both mutations were initially acquired in heterozygosity on the same allele, followed by chromosomal gain (46.2% of colonies) (Fig. 3C). Since these results may be affected by the capability of cells to grow in vitro, we opted to perform single-cell genomics on uncultured CD34+ cells and mononuclear cells from a selected +9p MPN patient by using Mission Bio TapestriTM platform. Here, we designed a custom DNA panel to be able to detect copy number alterations in chromosomes frequently affected by aneuploidies in MPN. This technique allowed us to independently corroborate our previous findings by screening a larger number of cells, confirming somatic Chromosome 9p trisomy in malignant cells (Fig. 3D, E). In addition, we found that the most abundant cell type had three JAK2 copies with two mutant alleles, followed by cells with two JAK2 copies with a single mutant allele, as already seen through ddPCR (Fig. 3F).
Fig. 3. Genetic profiling of + 9p patients by means of colony and single cell analysis.
A–C ddPCR analysis of the JAK2 mutational burden and copy number on DNA from single CD34+ -derived colonies from +9p patients (n = 139 colonies, with an average of 17 colonies/patient across 8 patients). A Barplot depicting the frequency of colonies split by the number of JAK2 mutated alleles and 9p ploidy. B Jitter plot displaying the distribution of JAK2 mutated alleles and copy number at the single-colony level. C Graphical representation of molecular events’ acquisition order of patient #4, as inferred from ddPCR analysis (n = 15 colonies). Percentages indicate the frequency of colonies carrying the above-depicted 9p ploidy and JAK2 genotype. D–F Single-cell genomic analysis of CD34+ cells and PBMCs from patient #1. The Copy Number Variation profiling of each clone, listed in the rows, at the amplicon level for the entire genome (D) or JAK2 locus (E), is reported in a heatmap. The shades of color depict the ratio between the median amplicon ploidy of a given clone and the one of the diploid reference, which was set to 2. The Wild-Type clone (WT) was set as diploid reference. F Jitter plot displaying the distribution of JAK2 mutated alleles and copy number at the single-cell level (n = 5617 cells). C Was created with Biorender.com.
Collectively, the application of both techniques facilitated the reconstruction of sequential events affecting Chromosome 9p in MPN patients, indicating that point mutations often serve as initial pathogenic events in clonal evolution. Subsequently, the JAK2-mutated allele undergoes amplification, leading to an increased burden of JAK2 mutations compared to JAK2-heterozygous diploid patients on a per-cell basis.
Chromosome 9p trisomy stimulates clonogenic potential of CD34+ cells through upregulation of POU5F1 and NANOG
Next, we sought to determine the impact of concurrent JAK2V617F and somatic Chromosome 9p trisomy on the clonogenic and differentiation potential of CD34+ cells in this subset of patients.
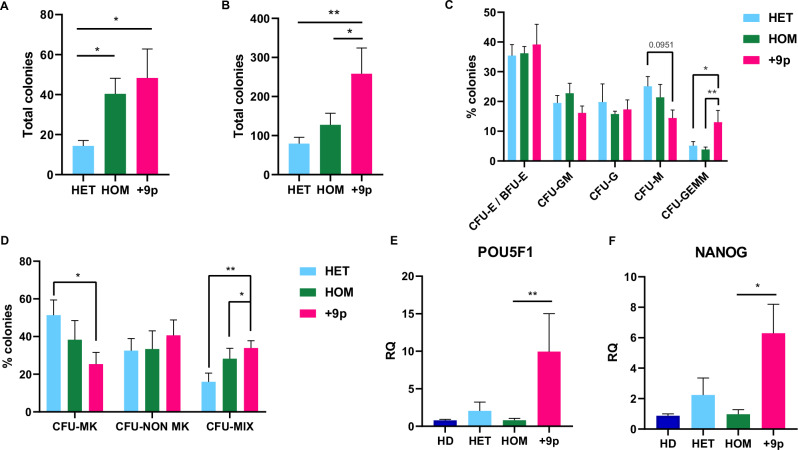
Both methylcellulose-based and collagen-based clonogenic assays revealed that CD34+ cells from +9p patients generated a significantly higher number of colonies compared to other MPN patients, indicating an increased clonogenic potential (Fig. 4A, B). Furthermore, we observed that CD34+ cells from +9p patients presented higher numbers of CFU-GEMM colonies in methylcellulose-based assay and “mixed colonies” in collagen-based assay, which both originated from more primitive progenitors (Fig. 4C, D).
Fig. 4. Clonogenic capacity of CD34+ cells in JAK2-mutated MF patients.
A Total number of colonies obtained from methylcellulose-based clonogenic assays (n = 7 HET, n = 7 HOM, n = 6 + 9p). B Total number of colonies obtained from collagen-based clonogenic assays (n = 7 HET, n = 7 HOM, n = 6 + 9p). C Percentage of the different Colony-Forming Units (CFUs) obtained from methylcellulose-based clonogenic assays. D Percentage of different Colony-Forming Units obtained from collagen-based clonogenic assays. Results are represented as mean + SEM. Evaluation of POU5F1 (E) and NANOG (F) expression at the transcriptional level in CD34+ cells from JAK2-mutated MPN patients and healthy donors (n = 5 HD, n = 6 HET, n = 7 HOM, n = 8 + 9p). Each barplot represents the relative quantity of the target mRNA quantified by means of qRT-PCR. Abbreviations: RQ = relative quantity; CFU-E/BFU-E = Colony-Forming Unit—Erythroid/Burst-Forming Unit—Erythroid; CFU-GM = Colony-Forming Unit—Granulocyte/Macrophage; CFU-G = Colony-Forming Unit—Granulocyte; CFU-M = Colony-Forming Unit—Macrophage; CFU-GEMM = Colony-Forming Unit—Granulocyte/Erythrocyte/Macrophage/Megakaryocyte; CFU-MK = Colony Forming Unit Megakaryocyte; CFU-NON MK = Colony Forming Unit Non-Megakaryocyte; CFU-MIX = Mixed CFU-Mk/Non CFU-Mk colonies; HD = healthy donors; HET = JAK2-mutated heterozygous patients; HOM = JAK2-mutated homozygous patients; +9p = JAK2-mutated patients with 9p trisomy. *p < 0.05; **p < 0.01.
As PD-L1 expression sustains the stemness of breast cancer cells via AKT signaling [39], we hypothesized that +9p cells were more sensitive to AKT inhibition. To test this, we treated CD34+ cells from JAK2V617F-homozygous and +9p patients with a potent AKT inhibitor, MK2206, and carried out clonogenic assays, which revealed decreased colony numbers for both patient subgroups, irrespective of the colony type (Fig. S3A, B). Given the heterogeneous clonal composition of +9p patients, we genotyped their colonies and found that triploid cell derived colonies were significantly more affected compared to diploid cell derived colonies (Fig. S3C).
Moreover, previous work has shown that the PD-L1/AKT axis may support stemness features through upregulation of OCT4 and NANOG [39]. Therefore, we assessed the gene expression levels of these genes in CD34+ cells from distinct MPN patient subsets, unveiling that high CD274 expression in CD34+ cells from +9p patients (Fig. 2E) is accompanied by upregulation of POU5F1 (encoding OCT4) and NANOG (Fig. 4E, F), suggesting that the more primitive features seen in +9p MPN CD34+ cells may be attributed to overexpression of these pro-stemness factors.
Since +9p MPN CD34+ cells are a mixture of different genotypes, we sought to unravel whether the increased expression of POU5F1 and NANOG may specifically be driven by high frequencies JAK2V617F-mutant cells with an extra copy of Chromosome 9p. To this end, we plated +9p CD34+ cells in semisolid medium and picked the resulting CD34+ cell derived colonies, which were simultaneously assessed for CD274, POU5F1, and NANOG RNA expression as well as JAK2 genetic status and Chromosome 9p copy number (Fig. 5A). Strikingly, we observed high CD274, POU5F1 and NANOG mRNA levels, almost exclusively expressed by cells carrying two mutant JAK2 copies out of three alleles (Fig. 5B–G). Furthermore, we knocked down POU5F1 and NANOG expression in CD34+ cells and found a decrease in colony numbers from +9p MPN patients compared to JAK2V617F- homozygous patients (Fig. 5H), which was mostly driven by a drop in myeloid colonies (Fig. 5I). Moreover, analysis of colony genotypes from +9p patients showed a higher sensitivity to POU5F1 and NANOG knock-down of +9p CD34+ cells compared to JAK2-homozygous cells within the same patient (Fig. 5J).
Fig. 5. Effect of 9p ploidy and JAK2 mutational status on PD-L1, OCT4, and NANOG expression and their role in CD34+ cells clonogenic potential.
A Experimental workflow of genomic/transcriptomic-coupled characterization of single-cell derived colonies by means of ddPCR. Evaluation of CD274 (B, C), POU5F1 (D, E), and NANOG (F, G) expression at the transcriptional level in single-cell derived colonies by ddPCR. In (B, D, and F) colonies (n = 54) were split according to the number of JAK2 mutated alleles and 9p ploidy status. The light blue area highlights +9p colonies. On the other hand, colonies were split according to 9p ploidy status in (C, E, and G). Results are represented as mean + SEM. H–J POU5F1 and NANOG silencing in CD34+ cells from HOM (n = 4) and +9p (n = 6) patients. H Ratio between the total number of colonies grown in methylcellulose-based clonogenic assays in OCT4siRNA+NANOGsiRNA and NTsiRNA samples, split between HOM and +9p patients. Results are represented as mean + SEM. I Ratio between the number of colonies grown in methylcellulose-based clonogenic assays in OCT4siRNA + NANOGsiRNA and NTsiRNA samples, split according to the different Colony-Forming Units (CFUs) in HOM and +9p patients. Results are represented as mean + SEM. J Barplot depicting the frequency of diploid and +9p colonies in +9p patients (n = 6) after POU5F1 and NANOG silencing or NTsiRNA nucleofection. RQ: relative quantity; 1 mut: one JAK2V617F-mutated allele; 2 mut: two JAK2V617F-mutated alleles; 2n: diploid colonies; 3n: +9p colonies; CFU-E/BFU-E: Colony-Forming Unit—Erythroid/Burst-Forming Unit—Erythroid; CFU-GM: Colony-Forming Unit—Granulocyte/Macrophage; CFU-G = Colony-Forming Unit—Granulocyte; CFU-M = Colony-Forming Unit—Macrophage; CFU-GEMM = Colony-Forming Unit—Granulocyte/Erythrocyte/Macrophage/Megakaryocyte; HOM: JAK2-mutated homozygous patients; +9p: JAK2-mutated patients with 9p trisomy; OCT4 + NANOG: POU5F1 siRNA + NANOG siRNA; NT siRNA: Non Targeting siRNA. *p < 0.05; **p < 0.01. A Was created with Biorender.com.
PD-L1 protein relocalizes on plasma membrane in monocytes and results in increased T cell exhaustion in + 9p MPN patients
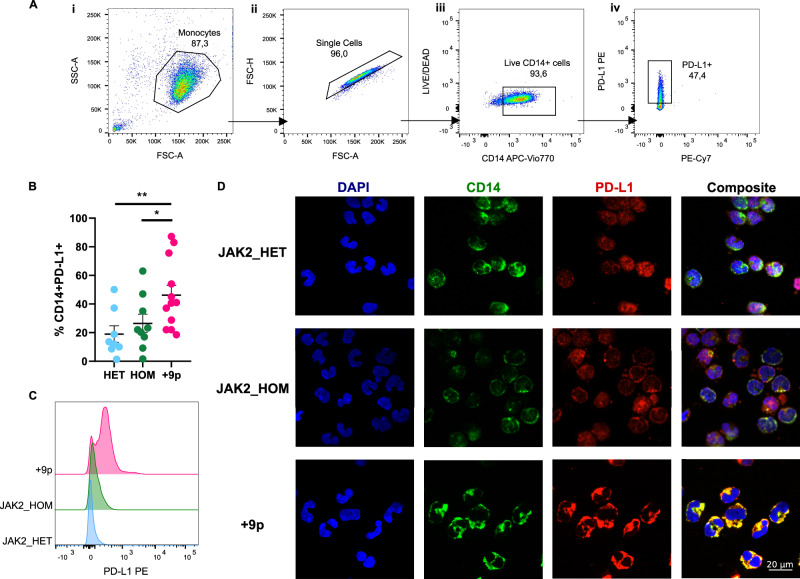
In order to evaluate alterations in PD-L1 expression at the protein level in monocytes in JAK2V617F-heterozygous, -homozygous, and +9p MPN patients, we carried out flow cytometry analysis, which revealed a statistically significant increase in the frequency of CD14 + PD-L1+ cells in the PB of +9p MPN patients (Fig. 6A, B), accompanied by increased PD-L1 surface expression levels (Fig. 6C).
Fig. 6. PD-L1 expression on monocytes of JAK2-mutated MF patients.
A Representative gating strategy used to quantify CD14 + PD-L1+ cells. (i) Side scatter area (SSC-A) vs forward scatter area (FSC-A) plot, showing the monocyte gate. (ii) Forward scatter height (FSC-H) vs forward scatter area (FSC-A) plot defining the single cell gate. (iii) LIVE/DEAD vs CD14 APC-Vio770 plot gating for live cells. (iv) PD-L1 PE vs PE-Cy7 plot depicting PD-L1+ cell gate. B Percentage of circulating CD14 + PD-L1+ cells in patients. Results are represented as means ± SEM. C Representative histograms for flow cytometry detection of PD-L1 staining in CD14+cells from HET (n = 8), HOM (n = 9) and +9p (n = 12) patients. D PD-L1 cellular localization in monocytes of JAK2-mutated MPN patients. The figure displays representative confocal microscopy images of immunofluorescence staining performed on monocytes coming from a JAK2_HET patient, a JAK2_HOM patient, and a + 9p patient. In every panel, the image on the far right is a merge of the other 3 images. Cells were labeled with anti-CD14 antibody (green fluorescence) and with anti-PD-L1 antibody (red fluorescence); nuclear counterstaining was performed with DAPI (blue fluorescence). HD: healthy donors; HET: JAK2-mutated heterozygous patients; HOM: JAK2-mutated homozygous patients; +9p: JAK2-mutated patients with 9p trisomy. *p < 0.05; **p < 0.01.
Additionally, we carried out immunofluorescence staining of PD-L1 on CD14+ monocytes, which highlighted differences in PD-L1 cellular localization among distinct patient subgroups. Specifically, our data indicate that PD-L1 is mostly cytoplasmic in JAK2V617F-heterozygous patients, while JAK2V617F-homozygous patients exhibited a concurrent PD-L1 cell membrane localization. Notably, PD-L1 expression was exclusively localized on the plasma membrane in monocytes from +9p patients (Fig. 6D).
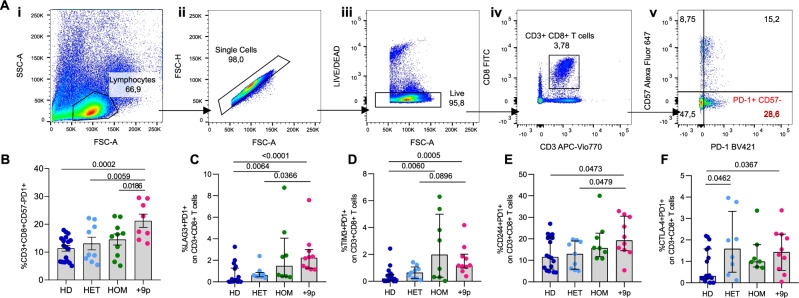
As elevated PD-L1 expression may contribute to compromised immune surveillance, we next investigated the potential activation of the PD-1/PD-L1 axis in +9p MPN patients by assessing T cell exhaustion levels. Flow cytometry analysis showed that +9p patients display a significantly higher frequency of CD3+/CD8+/CD57-/PD-1+ exhausted T cells compared to JAK2V617F-heterozygous and -homozygous MPN patients, as well as compared to healthy donors (HDs) (Fig. 7B). Flow cytometry analysis of other inhibitory receptors (CTLA-4, LAG-3 CD244, and TIM-3) revealed that T cells from +9p patients invariably showed higher expression of all the assessed surface proteins compared to HDs, unlike Chromosome 9-diploid JAK2V617F-mutant patients (Fig. 7C–F). Moreover, since the analysis of the JAK2V617F VAF of sorted cell fractions showed that a small fraction of T cells are mutant (Table S3), we assessed and failed to detect co-expression of PD-1/PD-L1 surface proteins within CD3+ T cells through flow cytometry (Fig. S4).
Fig. 7. T-cell exhaustion in MPN patients and healthy donors.
A Representative gating strategy used to quantify CD3 + CD8 + CD57-PD-1+ cells. (i) Side scatter area (SSC-A) vs forward scatter area (FSC-A) plot, showing the lymphocyte gate. (ii) Forward scatter height (FSC-H) vs forward scatter area (FSC-A) plot defining the single cell gate. (iii) LIVE/DEAD vs FSC-A plot gating for live cells. (iv) CD8-FITC vs CD3-APC Vio770 plot depicting CD3 + CD8+ cells. (v) CD57-Alexa Fluor 647 vs PD-1 BV421 plot, showing the percentage of PD-1 + CD57- cells. Flow-cytometric assessment of circulating exhausted CD3 + CD8 + CD57-PD1+ (B), CD3 + CD8 + PD1 + LAG3+ (C), CD3 + CD8 + PD1 + TIM3+ (D), CD3 + CD8 + PD1 + CD244+ (E), CD3 + CD8 + PD1 + CTLA4+ (F) T cells in MPN patients and healthy donors. Results are represented as means ± SEM. HD: healthy donors; HET: JAK2-mutated heterozygous patients; HOM: JAK2-mutated homozygous patients; +9p: JAK2-mutated patients with 9p trisomy.
Overall, these findings suggest that the increased PD-L1 expression on the cell membrane observed in +9p patients may overstimulate PD-1 on T cells, which may ultimately lead to the establishment of T cell exhaustion as an immune escape mechanism.
Discussion
MPNs are a group of blood disorders characterized by excessive production of mature blood cells, arising from mutated hematopoietic stem cells [1]. JAK2 mutations, particularly JAK2V617F, are the most common genetic alterations in MPNs, affecting ~50–60% of ET, MF, and 95% of PV patients [9]. Its VAF is a crucial disease modifier, with lower allele burden generally associated with higher platelet counts and higher mutant allele burden associated with erythrocytosis, thrombosis and higher risk of disease progression [40]. Interestingly, JAK2V617F homozygosity can occur during cell division due to aberrant recombination events, which lead to acquired UPD of Chromosome 9p [41].
As MPNs display different types of chromosomal aberrations, we report for the first time that Chromosome 9 copy number gains significantly impact the biological characteristics of JAK2V617F-mutant MPNs. Moreover, Chromosome 9 aneuploidies also affect other gene loci, which may contribute to the resulting phenotype. Therefore, further investigation is needed to disentangle the interplay between these two genetic events.
In this study, we included 12 JAK2-mutated MPN patients with (partial or complete) Chromosome 9 copy number trisomy as identified through cytogenetic or NGS analysis and compared them to JAK2V617F-heterozygous and -homozygous Chromosome 9-diploid patients. The CD274 gene locus, which encodes the immune checkpoint PD-L1, was also found duplicated, with its corresponding mRNA levels increased in different +9p cell types. ddPCR and single cell genomics analysis revealed that most cells in +9p patients carried three copies of Chromosome 9p, with two out of three JAK2 allele copies harbouring the mutation. Moreover, our results strongly suggest that the JAK2 mutation occurred as the first event in CD34+ cells in these patients, and the subsequent Chromosome 9p gain affected the JAK2-mutated allele. The reported order of mutational events is in agreement with a compelling body of evidence suggesting that JAK2 activation induces DNA damage, promotes homologous recombination, and ultimately triggers genomic instability [42, 43].
Interestingly, +9p MPN cells with two out of three JAK2-mutated alleles exhibited a competitive advantage over other cells with different genetic compositions within the same patient, including JAK2V617F-homozygous cells. Remarkably, +9p MPN CD34+ HSPCs were more clonogenic and formed more primitive colonies with high POU5F1 and NANOG expression levels. These genes encode OCT4 and NANOG, which are mostly embryonically-expressed transcription factors but have also been reported to be expressed in hematopoietic CD34+ cells [44–46]. Importantly, Chaurasia and colleagues report induction of their expression as correlated to increased numbers of hematopoietic stem cells in vitro [46]. Here, we show that +9p CD34+ cells are dependent on the PD-L1/AKT/NANOG-OCT4 axis as these cells result strikingly sensitive to a selective and potent AKT inhibitor as well as to NANOG and OCT4 knockdown. Additionally, flow cytometry analysis confirmed that +9p MPN monocytes have increased PD-L1 protein levels compared to other JAK2V617F-positive MPN patients, which may constitute an important mechanism of immune escape for malignant cells. Moreover, immunofluorescence analysis revealed distinct PD-L1 localization, with +9p MPN monocytes exhibiting the strongest surface PD-L1 staining. Several mechanisms regulate PD-L1 trafficking and stability onto the membrane, including post-translational modifications as glycosylation, ubiquitination, phosphorylation, and endosomal-mediated recycling [47]. Specifically, activation of the JAK/STAT pathway was reported as involved in PD-L1 relocalization in a STT3-dependent way through glycosylation, stabilization, and eventual trafficking to the cell membrane [48]. These findings are consistent with a progressive increase in surface localization from the cytoplasm in JAK2V617F-heterozygous, -homozygous, and +9p MPN patients. Nevertheless, the precise mechanism through which PD-L1 relocalizes on the surface warrants further future investigations [49].
As malignant +9p MPN patients express higher PD-L1, we hypothesized a heightened engagement with its cognate receptor PD-1, expressed on the surface of CD8+ T cells. Activation of the PD-1/PD-L1 axis is widely associated with decreased immune surveillance, which was confirmed by flow cytometry analysis with an increased exhaustion immunophenotype of CD8+ T cells.
Overall, the distinctive biological characteristics exhibited by +9p cells cannot be solely attributed to the presence of two mutated JAK2 alleles out of the three carried. Notably, +9p cells significantly differ from MPN cells carrying homozygous JAK2V617F. While a growing body of literature has previously described how JAK2 hyperactivation leads to increased PD-L1 expression, here we report that +9p cells with two mutant JAK2V617F alleles and three copies of JAK2 as well as of CD274 overall have significantly higher mRNA and protein expression levels of PD-L1. This finding holds true within +9p patients as well, where we could find small JAK2V617F-homozygous clones, indicating that the reported higher PD-L1 expression is a cell-intrinsic feature of +9p cells [17, 18]. Hence, this synergy sets +9p MPNs apart from previously described copy-neutral cytogenetic aberrations involving Chromosome 9, such as UPD [17], as our data show that +9p cells express higher levels of PD-L1 even compared to JAK2V617F-homozygous cells. Notably, among the 435 genes on Chromosome 9p, copy gain of the gene encoding the transcription factor MLLT3 may concur on the increased stemness features observed for +9p HSPCs, while genes encoding cytokines, including IL-33, may influence the myeloproliferative phenotype. In this work, we cannot exclude the possible role of other loci on Chromosome 9p, with further investigation being needed to shed light on the role of additional genes.
In conclusion, in this study, we present a distinct subset of JAK2V617F-positive patients with unique biological features. These findings highlight the hijacked interplay between JAK2 and the PD-L1/PD-1 axis in +9p MPNs, which contributes to stemness in CD34+ cells and immune escape characteristics in malignant myeloid cells. These observations pave the way for future investigations to further refine the clinical understanding and tailored management of +9p MPN patients, involving more targeted approaches that take into account the specific role of PD-L1 in this context.
Supplementary information
Acknowledgements
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) AIRC 5 per 1000 project #21267 and IG project #29077, Italian Ministry of University and Research (PRIN 2022 project # 2022F4WMR3 and PRIN PNRR 2022 project #P202259EM5). The research leading to these results has received funding from the European Union—NextGenerationEU through the Italian Ministry of University and Research under PNRR—M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to Rossella Manfredini and Sebastiano Rontauroli (CUP E93C22001860006, University of Modena and Reggio Emilia, Modena, Italy). The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them. ”Chiara Carretta was supported by an AIRC fellowship for Italy (project number 27959 2022).
Author contributions
RM designed the study and supervised the manuscript; CC, SP, and MBe performed in vitro experiments and analyzed data; FB, Mmal, and EP performed collagen and methylcellulose clonogenic assays; SR extracted DNA for NGS analysis; LT and EG performed CD3+ cells isolation and flow cytometry analysis; SR, AN, and CT performed POU5F1 and NANOG silencing experiments; SS and LC performed ddPCR analysis; EE performed confocal microscopy imaging; MMi and FP performed immunofluorescence staining; BM, MMaf, MMo, MBr, MMac, LP and ML provided patient samples; ETe, SM and NB performed NGS analysis; EB performed MLPA analysis; ETa, PG, MS, and FPa supervised the manuscript; RN and AMV co-supervised the experimental design and wrote the manuscript.
Data availability
Single-cell DNA sequencing data are available via NCBI Gene Expression Omnibus repository, accession number GSE251689.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
These authors contributed equally: Chiara Carretta, Sandra Parenti, Matteo Bertesi.
These authors jointly supervised this work: Ruggiero Norfo, Alessandro Maria Vannucchi, Rossella Manfredini.
Contributor Information
Rossella Manfredini, Email: rossella.manfredini@unimore.it.
on behalf of MYNERVA (Myeloid NEoplasms Research Venture AIRC):
Chiara Carretta, Sandra Parenti, Matteo Bertesi, Sebastiano Rontauroli, Lara Tavernari, Elena Genovese, Marica Malerba, Elisa Papa, Margherita Mirabile, Anita Neroni, Camilla Tombari, Elena Tenedini, Niccolò Bartalucci, Elisa Bianchi, Enrico Tagliafico, Paola Guglielmelli, Ruggiero Norfo, Alessandro Maria Vannucchi, and Rossella Manfredini
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02373-w.
References
- 1.Grabek J, Straube J, Bywater M, Lane SW. MPN: the molecular drivers of disease initiation, progression and transformation and their effect on treatment. Cells. 2020;9:1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, de Araujo IBO, Berti E, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36:1720–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinshelwood S, Bench AJ, Green AR. Pathogenesis of polycythaemia vera. Blood Rev. 1997;11:224–32. [DOI] [PubMed] [Google Scholar]
- 4.Barbui T, Barosi G, Grossi A, Gugliotta L, Liberato LN, Marchetti M, et al. Practice guidelines for the therapy of essential thrombocythemia. A statement from the Italian Society of Hematology, the Italian Society of Experimental Hematology and the Italian Group for Bone Marrow Transplantation. Haematologica. 2004;89:215–32. [PubMed] [Google Scholar]
- 5.Rumi E, Pietra D, Pascutto C, Guglielmelli P, Martínez-Trillos A, Casetti I, et al. Clinical effect of driver mutations of JAK2, CALR, or MPL in primary myelofibrosis. Blood. 2014;124:1062–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rumi E, Cazzola M. Diagnosis, risk stratification, and response evaluation in classical myeloproliferative neoplasms. Blood. 2017;129:680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tefferi A. Primary myelofibrosis: 2017 update on diagnosis, risk-stratification, and management. Am J Hematol. 2016;91:1262–71. [DOI] [PubMed] [Google Scholar]
- 8.Orvain C, Luque Paz D, Dobo I, Cottin L, Le Calvez G, Chauveau A, et al. Circulating Cd34+ cell count differentiates primary myelofibrosis from other Philadelphia-negative myeloproliferative neoplasms: a pragmatic study. Ann Hematol. 2016;95:1819–23. [DOI] [PubMed] [Google Scholar]
- 9.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–90. [DOI] [PubMed] [Google Scholar]
- 10.Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLOS Med. 2006;3:e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129:667–79. [DOI] [PubMed] [Google Scholar]
- 13.Rontauroli S, Carretta C, Parenti S, Bertesi M, Manfredini R. Novel molecular insights into leukemic evolution of myeloproliferative neoplasms: a single cell perspective. Int J Mol Sci. 2022;23:15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32:2601–13. [DOI] [PubMed] [Google Scholar]
- 15.Dunlap J, Kelemen K, Leeborg N, Braziel R, Olson S, Press R, et al. Association of JAK2 mutation status and cytogenetic abnormalities in myeloproliferative neoplasms and myelodysplastic/myeloproliferative neoplasms. Am J Clin Pathol. 2011;135:709–19. [DOI] [PubMed] [Google Scholar]
- 16.Molina O, Abad MA, Solé F, Menéndez P. Aneuploidy in cancer: lessons from acute lymphoblastic leukemia. Trends Cancer. 2021;7:37–47. [DOI] [PubMed] [Google Scholar]
- 17.Milosevic Feenstra JD, Jäger R, Schischlik F, Ivanov D, Eisenwort G, Rumi E, et al. PD-L1 overexpression correlates with JAK2-V617F mutational burden and is associated with 9p uniparental disomy in myeloproliferative neoplasms. Am J Hematol. 2022;97:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prestipino A, Emhardt AJ, Aumann K, O’Sullivan D, Gorantla SP, Duquesne S, et al. Oncogenic JAK2V617F causes PD-L1 expression, mediating immune escape in myeloproliferative neoplasms. Sci Transl Med. 2018;10:eaam7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuykendall A, Talati C, Al Ali NH, Padron E, Sallman D, Lancet JE, et al. Characterization of cytogenetic abnormalities in myelofibrosis and relationship to clinical outcome. Blood. 2016;128:1937. [Google Scholar]
- 20.Sever M, Kantarjian H, Pierce S, Jain N, Estrov Z, Cortes J, et al. Cytogenetic abnormalities in essential thrombocythemia at presentation and transformation. Int J Hematol. 2009;90:522–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sever M, Quintás-Cardama A, Pierce S, Zhou L, Kantarjian H, Verstovsek S. Significance of cytogenetic abnormalities in patients with polycythemia vera. Leuk Lymphoma. 2013;54:2667–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbui T, Thiele J, Gisslinger H, Kvasnicka HM, Vannucchi AM, Guglielmelli P, et al. The 2016 WHO classification and diagnostic criteria for myeloproliferative neoplasms: document summary and in-depth discussion. Blood Cancer J. 2018;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barosi G, Mesa RA, Thiele J, Cervantes F, Campbell PJ, Verstovsek S, et al. Proposed criteria for the diagnosis of post-polycythemia vera and post-essential thrombocythemia myelofibrosis: a consensus statement from the international working group for myelofibrosis research and treatment. Leukemia. 2008;22:437–8. [DOI] [PubMed] [Google Scholar]
- 24.Carretta C, Mallia S, Genovese E, Parenti S, Rontauroli S, Bianchi E, et al. Genomic analysis of hematopoietic stem cell at the single-cell level: optimization of cell fixation and Whole Genome Amplification (WGA) Protocol. Int J Mol Sci. 2020;21:7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rontauroli S, Castellano S, Guglielmelli P, Zini R, Bianchi E, Genovese E, et al. Gene expression profile correlates with molecular and clinical features in patients with myelofibrosis. Blood Adv. 2021;5:1452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parenti S, Rontauroli S, Carretta C, Mallia S, Genovese E, Chiereghin C, et al. Mutated clones driving leukemic transformation are already detectable at the single-cell level in CD34-positive cells in the chronic phase of primary myelofibrosis. Npj Precis Oncol. 2021;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parenti S, Rabacchi C, Marino M, Tenedini E, Artuso L, Castellano S, et al. Characterization of new ATM deletion associated with hereditary breast cancer. Genes. 2021;12:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salati S, Lisignoli G, Manferdini C, Pennucci V, Zini R, Bianchi E, et al. Co-culture of hematopoietic stem/progenitor cells with human osteblasts favours mono/macrophage differentiation at the expense of the erythroid lineage. PLOS ONE. 2013;8:e53496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zini R, Norfo R, Ferrari F, Bianchi E, Salati S, Pennucci V, et al. Valproic acid triggers erythro/megakaryocyte lineage decision through induction of GFI1B and MLLT3 expression. Exp Hematol. 2012;40:1043–54.e6. [DOI] [PubMed] [Google Scholar]
- 30.Fantini S, Rontauroli S, Sartini S, Mirabile M, Bianchi E, Badii F, et al. Increased plasma levels of lncRNAs LINC01268, GAS5 and MALAT1 correlate with negative prognostic factors in myelofibrosis. Cancers. 2021;13:4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertesi M, Fantini S, Alecci C, Lotti R, Martello A, Parenti S, et al. Promoter methylation leads to decreased ZFP36 expression and deregulated nlrp3 inflammasome activation in psoriatic fibroblasts. Front Med. 2021;7. https://www.frontiersin.org/articles/10.3389/fmed.2020.579383. [DOI] [PMC free article] [PubMed]
- 32.Salati S, Genovese E, Carretta C, Zini R, Bartalucci N, Prudente Z, et al. Calreticulin Ins5 and Del52 mutations impair unfolded protein and oxidative stress responses in K562 cells expressing CALR mutants. Sci Rep. 2019;9:10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sperduti S, Lazzaretti C, Paradiso E, Anzivino C, Villani MT, De Feo G, et al. Quantification of hormone membrane receptor FSHR, GPER and LHCGR transcripts in human primary granulosa lutein cells by real-time quantitative PCR and digital droplet PCR. Gene Rep. 2021;23:101194. [Google Scholar]
- 34.Calabresi L, Carretta C, Romagnoli S, Rotunno G, Parenti S, Bertesi M, et al. Clonal dynamics and copy number variants by single-cell analysis in leukemic evolution of myeloproliferative neoplasms. Am J Hematol. 2023;98:1520–31. [DOI] [PubMed] [Google Scholar]
- 35.Genovese E, Mirabile M, Rontauroli S, Sartini S, Fantini S, Tavernari L, et al. The response to oxidative damage correlates with driver mutations and clinical outcome in patients with myelofibrosis. Antioxidants. 2022;11:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salati S, Prudente Z, Genovese E, Pennucci V, Rontauroli S, Bartalucci N, et al. Calreticulin affects hematopoietic stem/progenitor cell fate by impacting erythroid and megakaryocytic differentiation. Stem Cells Dev. 2018;27:225–36. [DOI] [PubMed] [Google Scholar]
- 37.Enzo E, Secone Seconetti A, Forcato M, Tenedini E, Polito MP, Sala I, et al. Single-keratinocyte transcriptomic analyses identify different clonal types and proliferative potential mediated by FOXM1 in human epidermal stem cells. Nat Commun. 2021;12:2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenedini E, Bernardis I, Artusi V, Artuso L, Roncaglia E, Guglielmelli P, et al. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia. 2014;28:1052–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, et al. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141:1402–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moliterno AR, Kaizer H, Reeves BN. JAK2V617F allele burden in polycythemia vera: burden of proof. Blood. 2023;141:1934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106:2162–8. [DOI] [PubMed] [Google Scholar]
- 42.Karantanos T, Moliterno AR. The roles of JAK2 in DNA damage and repair in the myeloproliferative neoplasms: opportunities for targeted therapy. Blood Rev. 2018;32:426–32. [DOI] [PubMed] [Google Scholar]
- 43.Plo I, Nakatake M, Malivert L, de Villartay JP, Giraudier S, Villeval JL, et al. JAK2 stimulates homologous recombination and genetic instability: potential implication in the heterogeneity of myeloproliferative disorders. Blood. 2008;112:1402–12. [DOI] [PubMed] [Google Scholar]
- 44.Świstowska M, Gil-Kulik P, Czop M, Wieczorek K, Macheta A, Petniak A, et al. Comparison of SOX2 and POU5F1 gene expression in leukapheresis-derived CD34+ cells before and during cell culture. Int J Mol Sci. 2023;24:4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mintz PJ, Huang KW, Reebye V, Nteliopoulos G, Lai HS, Sætrom P, et al. Exploiting human CD34+ stem cell–conditioned medium for tissue repair. Mol Ther. 2014;22:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaurasia P, Gajzer DC, Schaniel C, D’Souza S, Hoffman R. Epigenetic reprogramming induces the expansion of cord blood stem cells. J Clin Investig. 2014;124:2378–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemma EY, Letian A, Altorki NK, McGraw TE. Regulation of PD-L1 trafficking from synthesis to degradation. Cancer Immunol Res. 2023;11:866–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Xiao Y, Yu H, Jin X, Fan S, Liu W. Mutual connected IL-6, EGFR and LIN28/Let7-related mechanisms modulate PD-L1 and IGF upregulation in HNSCC using immunotherapy. Front Oncol. 2023;13. https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2023.1140133/full. [DOI] [PMC free article] [PubMed]
- 49.Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell. 2019;76:359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell DNA sequencing data are available via NCBI Gene Expression Omnibus repository, accession number GSE251689.