Abstract
The pathogenesis of severe acute respiratory syndrome (SARS) remains unclear. Macrophages are key sentinel cells in the respiratory system, and it is therefore relevant to compare the responses of human macrophages to infections with the SARS coronavirus (SARS-CoV) and other respiratory viruses. Primary human monocyte-derived macrophages were infected with SARS-CoV in vitro. Virus replication was monitored by measuring the levels of positive- and negative-strand RNA, by immunofluorescence detection of the SARS-CoV nucleoprotein, and by titration of the infectious virus. The gene expression profiles of macrophages infected with SARS-CoV, human coronavirus 229E, and influenza A (H1N1) virus were compared by using microarrays and real-time quantitative reverse transcriptase PCR. Secreted cytokines were measured with an enzyme-linked immunosorbent assay. SARS-CoV initiated viral gene transcription and protein synthesis in macrophages, but replication was abortive and no infectious virus was produced. In contrast to the case with human coronavirus 229E and influenza A virus, there was little or no induction of beta interferon (IFN-β) in SARS-CoV-infected macrophages. Furthermore, SARS-CoV induced the expression of chemokines such as CXCL10/IFN-γ-inducible protein 10 and CCL2/monocyte chemotactic protein 1. The poor induction of IFN-β, a key component of innate immunity, and the ability of the virus to induce chemokines could explain aspects of the pathogenesis of SARS.
A novel coronavirus was identified as the causative agent of severe acute respiratory syndrome (SARS) (20) and is believed to be of zoonotic origin (10). The previously known human coronaviruses 229E (HCoV-229E) and OC43 have only been linked with the common cold. However, several animal coronaviruses result in severe animal diseases affecting the respiratory or gastrointestinal tract or cause disseminated infections.
Compared to common respiratory viral infections, SARS is unusually severe, with an overall case fatality rate of about 10%. The infection is not localized to the respiratory tract, and the causative virus is also detected in the gastrointestinal and urinary systems (21). Contrary to what is seen with other common respiratory viral infections, such as infections with HCoV-229E (2) and influenza A virus (13, 14), the viral load in the upper respiratory tract in patients with SARS progressively increases, peaking on about day 10 after the onset of symptoms (21). This suggests that the innate arm of the immune system may be unable to adequately control SARS-CoV infection. The appearance of antibodies in the second week of the illness appears to coincide with a falling viral load in the upper respiratory tract.
Macrophages are key cells for host defense and are abundant within all tissues of the body, including the respiratory system. They are potent producers of cytokines that are crucial components of innate immunity and potential mediators of immunopathology (9). Genetic resistance to strains of the coronavirus mouse hepatitis virus is associated with the ability of the virus to replicate in macrophages (1, 31). On the other hand, feline infectious peritonitis is a disease caused by a coronavirus in which prior immunity or passive antibodies increase the severity of the disease (33). In this disease, macrophages are the main target cells for virus replication, and antiviral antibodies enhance the replication of the virus in macrophage cultures in vitro (12). This has led to concern about whether antibody-mediated enhancement of disease may be relevant to the pathogenesis of SARS.
It is therefore relevant to study the interaction between SARS-CoV and macrophages. In this study, we investigate the response of human macrophages to infection with SARS-CoV and compare it with the responses to HCoV-229E and influenza A virus.
MATERIALS AND METHODS
Viruses.
SARS-CoV (strain HK39849) was propagated in FRhK-4 cells (20). All procedures involving the use of live SARS-CoV were performed in a biosafety level 3 facility. The influenza A/Hong Kong/54/98 (H1N1) virus was propagated in MDCK cells (5). HCoV-229E was propagated in MRC-5 cells, and both the virus and cells were obtained from the American Type Culture Collection (Manassas, Va.). Amicon Ultra-15 centrifugal filter units (Millipore Corporation, Bedford, Mass.) were used for the differential filtration of SARS-CoV cultures.
Cells.
Primary human monocyte-derived macrophages were prepared as previously described and used after 14 days of differentiation in vitro in 5% autologous serum (5). Two days prior to the experiment, the cell culture medium was changed to macrophage serum-free medium (Invitrogen, Carlsbad, Calif.).
Infection of cells.
Differentiated macrophages from monocytes were seeded at 2 × 105 cells per well in 24-well tissue culture plates and were infected at a multiplicity of infection (MOI) of 1 to 2. After 60 min of virus adsorption at 37°C, the virus inoculums were removed, and the cells were washed and incubated in macrophage serum-free medium supplemented with 0.6 mg/liter penicillin and 60 mg/liter streptomycin. Samples of culture supernatants were collected and stored at −70°C for virus titration or cytokine analysis.
Other cells were seeded at 2 × 105 cells per well in 24-well tissue culture plates and were infected at an MOI of 1 to 2. After 60 min of virus adsorption at 37°C, the virus inoculums were removed, and infected cells were washed with warmed culture medium and incubated with the original maintenance medium.
Infectious viral titers in the supernatants were determined by titration at half-log10 dilutions on FRhK-4 cell monolayers in quadruplicate. Cells were seeded in 96-well plates and used at about 90% confluence. The 50% tissue culture infective dose was determined by the method of Reed and Muench (24).
RNA extraction.
Total cellular RNAs were extracted with an RNeasy RNA Mini kit (QIAGEN, Hilden, Germany), with DNase digestion, according to the manufacturer's instructions. Extracted RNAs were stored at −70°C until use.
Microarray analysis.
Extracted RNAs were examined for human genome-wide gene expression with a Human Genome U133A GeneChip probe array (Affymetrix, Inc., Santa Clara, Calif.) by the use of oligonucleotide probe sets interrogating approximately 21,000 transcripts. Quality control, GeneChip hybridization, and raw data analysis were performed according to the manufacturer's instructions (Genome Centre, University of Hong Kong). Total RNAs from macrophages isolated from three experiments from separate donors were extracted and pooled. Double-stranded cDNAs were synthesized by means of the SuperScript Choice system (Invitrogen) and a GeneChipT7-Oligo(dT) promoter primer kit (Affymetrix, Inc.). The cDNAs were subjected to in vitro transcription in the presence of biotin-labeled ribonucleotides by means of a BioArray High-Yield RNA transcript labeling kit (Affymetrix, Inc). The biotin-labeled cRNAs were chemically fragmented and hybridized to the GeneChip probe array. Using the EukGE-WS2 fluidics protocol, we stained the probe array with a streptavidin R-phycoerythrin conjugate (Molecular Probes, Eugene, Oreg.). The image was scanned by a GeneChip scanner (Affymetrix, Inc.) at an excitation wavelength of 488 nm. The amount of light emitted at 570 nm was proportional to the bound target for each probe set on the probe array. The data generated were analyzed with Microarray Suite Expression Analysis software, version 5.1 (Affymetrix, Inc.).
Real-time quantitative RT-PCR for cytokine and viral gene expression.
Superscript II reverse transcriptase (RT; Invitrogen) and an oligo(dT) primer (Invitrogen) were used to convert mRNAs to cDNAs according to the manufacturer's instructions. The levels of cytokine mRNA and viral RNA were measured by real-time quantitative PCR using a LightCycler (Roche, Mannheim, Germany).
Each 20-μl reaction mixture was composed of 5 μl of diluted cDNA added to a master mix consisting of 2 μl of DNA Master SYBR green, 4 mM MgCl2, and a 0.5 μM concentration of each primer. The primer sequences were as follows: for beta interferon (IFN-β), forward (3′-GCC GCA TTG ACC ATC T-5′) and reverse (3′-CAC AGT GAC TGT ACT CCT-5′); for CXCL10/IFN-γ-inducible protein 10 (IP-10), forward (3′-CTG ACT CTA AGT GGC ATT-5′) and reverse (3′-TGA TGG CCT TCG ATT CTG-5′); and for CCL2/monocyte chemotactic protein 1 (MCP-1), forward (3′-CAT TGT GGC CAA GGA GAT CTG-5′) and reverse (3′-CTT CGG AGT TTG GGT TTG CTT-5′). Fluorescence readings were taken at the end of each extension cycle. After a denaturation cycle at 95°C for 10 min, the temperature cycling conditions for IFN-β and CCL2/MCP-1 were 40 cycles consisting of denaturation at 95°C for 10 s, annealing at 65°C for 5 s, and extension at 72°C for 11 s, with acquisition at 72°C. For CXCL10/IP-10, the conditions were 40 cycles consisting of denaturation at 95°C for 10 s, annealing at 60°C for 5 s, and extension at 72°C for 9 s, with extension at 72°C. After a melting curve analysis from 60°C to 95°C with fluorescence readings taken at 0.1°C/s, a final cooling step was carried out to reduce the rotor temperature to 40°C. The primers and conditions for the SARS-CoV Orf1b and nucleoprotein genes have been described previously (22, 23). Positive and negative controls were included in each run, and the levels of cytokine mRNA were normalized to the β-actin level.
Quantitation of cytokines.
Cytokines produced from macrophages were measured by specific enzyme-linked immunosorbent assays (ELISA) (R&D Systems, Minneapolis, Minn.) according to the manufacturer's instructions. Culture supernatants were UV irradiated for 20 min to inactivate infectious viruses prior to assay in a biosafety level 3 facility. Previous experiments have confirmed that cytokine levels are not affected by the dose of UV radiation used (5).
RESULTS
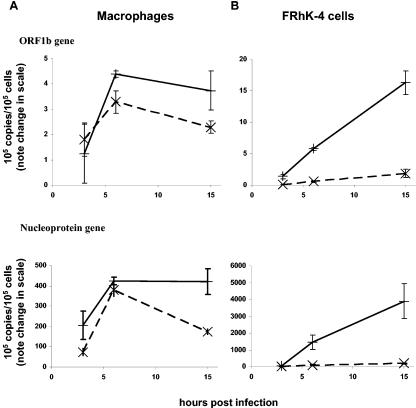
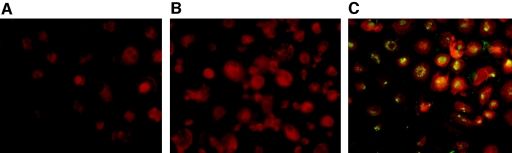
In SARS-CoV-infected macrophages, there was an increase in the copy numbers of both the positive and negative RNA strands of the SARS-CoV ORF-1b and nucleocapsid genes (Fig. 1A) over the first few hours postinfection. Viral RNA levels in macrophages peaked at modest levels at about 6 h postinfection, but they continued to increase in FRhK-4 cells, reaching much higher absolute levels (Fig. 1A and B). In SARS-CoV-permissive cells (e.g., FRhK-4 cells), there was a marked excess of positive-strand RNA for both Orf1b and the nucleoprotein (Fig. 1B), as expected (26). In contrast, comparable amounts of positive- and negative-sense RNAs for both Orf1b and the nucleoprotein were detected in macrophages, suggesting that macrophages do not support efficient viral replication. SARS-CoV nucleoprotein expression in SARS-CoV-infected macrophages was demonstrated by use of a mouse monoclonal antibody (4D11), and >90% of macrophages showed nucleoprotein expression when infected at an MOI of 1 to 2 (Fig. 2). However, no infectious virus was detected in the supernatant of virus-infected macrophages for up to 7 days postinfection, indicating that virus infection of these cells was abortive (Table 1). Increasing the MOI infecting macrophages to 20 did not further increase viral RNA levels or lead to productive virus replication. In contrast, virus-infected FRhK-4 cells produced infectious virus titers up to 105 50% tissue culture infective doses/ml (data not shown), peaking at about 2 to 3 days postinfection.
FIG. 1.
Nonproductive replication of SARS-CoV in human macrophages. Differentiated primary human monocyte-derived macrophages (A) and FRhK-4 cells (B) were seeded in 24-well plates (2 × 105 cells per well) on glass coverslips. Cells were infected at an MOI of 1 to 2, and RNAs were extracted at 3, 6, and 15 h postinfection. The levels of positive (solid lines) and negative (dotted lines) RNA strands of the SARS-CoV Orf1b and nucleoprotein genes were determined by real-time quantitative RT-PCR. The data show means of duplicate cultures from the same donor and are representative of three independent experiments with similar results.
FIG. 2.
Human macrophages were mock treated (A) or infected with SARS-CoV (B and C) and fixed with methanol for 15 min at 15 h postinfection. A mouse monoclonal antibody (4D11) specific for the SARS-CoV nucleoprotein (K. H. Chan, unpublished results) was tested on SARS-CoV-infected and uninfected macrophages (A and C). A mouse monoclonal antibody against influenza A virus hemagglutinin was used as a control (B). All three cell smears were stained with a secondary fluorescein isothiocyanate-conjugated anti-mouse antibody (Zymed Laboratories, San Francisco, Calif.), and Evans blue was used as a counterstain.
TABLE 1.
Infectious virus yields from macrophages infected with SARS-CoV, HCoV-229E, or influenza A (H1N1) virusa
| Virus | Yield (log10 TCID50/ml) at indicated time postinfection
|
||||
|---|---|---|---|---|---|
| 3 h | 1 day | 2 days | 3 days | 7 days | |
| SARS-CoV | 1 | 1 | <1 | <1 | <1 |
| HCoV-229E | 1 | <1 | 3 | <1 | <1 |
| Influenza A (H1N1) virus | 2 | 3 | 5 | 6 | <1 |
Differentiated human monocyte-derived macrophages were infected with SARS-CoV, HCoV-229E, and influenza A/HK/54/98 (H1N1) virus, and culture supernatants were titrated on monolayers of FRhK-4 (SARS-CoV), MRC-5 (HCoV-229E), and MDCK (H1N1) cells. The data shown are averages from three independent experiments corrected to 1 significant figure.
There was no increase in positive- or negative-strand viral RNA or viral antigen expression, as detected by immunofluorescence, in macrophages or FRhK-4 cells infected with UV-inactivated SARS-CoV (data not shown).
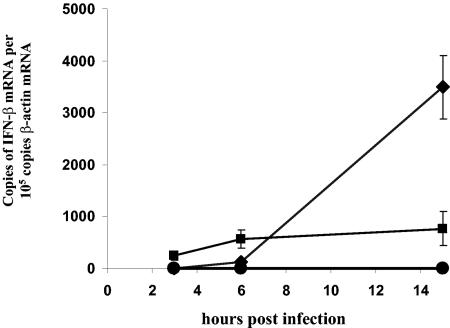
In a microarray analysis of the cellular gene expression of SARS-CoV-infected human macrophages, it was striking that in contrast to influenza A virus, SARS-CoV failed to induce significant IFN-α/β gene expression (Table 2). These findings were confirmed by independent experiments in which the IFN-α and -β mRNA levels were determined by real-time quantitative RT-PCR (Fig. 3) and the amounts of secreted protein were assayed by ELISA (data not shown). Parallel experiments with influenza A virus and HCoV-299E showed that, unlike SARS-CoV, these viruses were strong inducers of IFN-β. It is intriguing that despite the absence of IFN-α/β induction in SARS-CoV-infected cells, many of the IFN-stimulated gene (ISG) products were up-regulated (Table 2).
TABLE 2.
Microarray analysis of IFN-related genesa
| Gene product | Log2 fold change compared to mock-treated cells
|
|
|---|---|---|
| SARS-CoV | Influenza A virus | |
| Beta interferon | NC | 7.1 |
| Alpha interferon 1, 2, 4, 5, 8, 10, and 21b | NC | NC |
| Omega interferon | NC | NC |
| Interferon-stimulated gene, 20 kDa | 6.8 | NC |
| Interferon-induced protein with tetratricopeptide repeats 1 | 4.9 | 3.5 |
| Alpha interferon-inducible protein (clone IFI-15K) | 3.8 | 2.5 |
| Interferon-inducible guanylate binding protein 1, 67 kDa | 3.7 | 2.6 |
| Interferon-induced protein with tetratricopeptide repeats 4 | 3.7 | 2.6 |
| Interferon-induced transmembrane protein 1 (9-27) | 3.4 | NC |
| Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | 3 | 1.1 |
| Interferon-induced protein 44 | 2.7 | NC |
| Interferon-induced protein 35 | 1.9 | NC |
| Retinoic acid- and interferon-inducible protein (58 kDa) | 1.9 | 3.3 |
| Alpha interferon-inducible protein 27 | 1.7 | NC |
| Interferon-induced transmembrane protein 3 (1-8U) | 1.7 | NC |
| Protein kinase, interferon-inducible double-stranded RNA dependent | 1.6 | NC |
| Alpha interferon-inducible protein (clone IFI-6-16) | 1.5 | NC |
| Interferon-induced transmembrane protein 2 (1-8D) | 1.2 | NC |
| Gamma interferon-inducible protein 16 | 1.1 | NC |
| Interferon-stimulated transcription factor 3, gamma 48 kDa | 1.1 | NC |
| Interferon-inducible guanylate binding protein 2 | 1 | NC |
Macrophages were infected at an MOI between 1 and 2, and RNAs were extracted at 3 h postinfection. RNAs were pooled from three different donors, and gene expression was analyzed with an Affymetrix U133A Genechip microarray. Many of the IFN-stimulated genes, but not IFN- α/β, were up-regulated in SARS-CoV-infected macrophages and are shown relative to influenza A (H1N1) virus infection. A significant up-regulation in gene expression is indicated by an increase of twofold or more and was accompanied by a positive signal intensity call by the Microarray Suite Expression Analysis software, version 5 (Affymetrix Inc.). Only genes that have at least twofold change compared to mock-treated cells are shown. NC, no change compared with mock-treated macrophages.
Alpha interferon 1, 2, 4, 5, 8, 10, and 21 were tested individually, but none of their genes were up-regulated.
FIG. 3.
No induction of IFN-β gene expression in SARS-CoV-infected macrophages. Levels of IFN-β mRNA were determined by real-time quantitative RT-PCR. (A) Macrophages were infected with SARS-CoV (•), HCoV-229E (♦), and influenza A (H1N1) virus (▪) at an MOI of 1 to 2, and RNAs were extracted at 3, 6, and 15 h postinfection. SARS-CoV-infected macrophages did not induce IFN-β at any of the three time points, in contrast with infections with influenza A (H1N1) virus and HCoV-299E.
RT-PCRs for the interferon-like cytokines interleukin-28 (IL-28) and IL-29 (not included in the Genechip microarray) demonstrated that there was no up-regulation of either of these genes by SARS-CoV. In contrast, both HCoV-229E and influenza A virus induced the gene expression of both of these cytokines (data not shown).
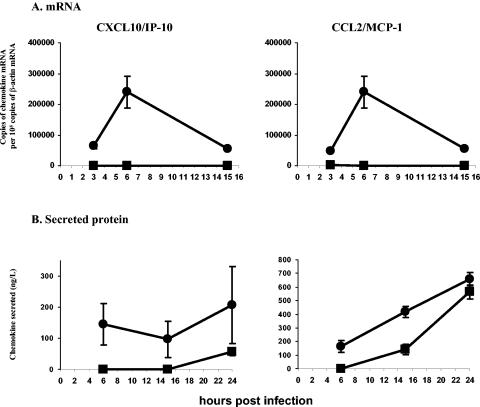
Real-time quantitative RT-PCR analysis was used to confirm the microarray data, which showed an early induction of several chemokines, such as CXCL10/IP-10 and CCL2/MCP-1, in SARS-CoV-infected macrophages (Table 3 and Fig. 4A). ELISAs for CXCL10/IP-10 and CCL2/MCP-1 in macrophage culture supernatants confirmed that SARS-CoV induced CXCL10/IP-10 and CCL2/MCP-1 secretion in macrophages in the first few hours after infection (Fig. 4B). UV irradiation of SARS-CoV partially abrogated the induction of these chemokines (by 40 to 60%), despite a complete loss of SARS-CoV gene expression (data not shown), suggesting that viral replication was only partly necessary for the induction of these chemokine genes.
TABLE 3.
Microarray analysis of chemokinesa
| Gene product | Fold change compared to mock-treated cells
|
|
|---|---|---|
| SARS-CoV | Influenza A virus | |
| Chemokine (C-X-C motif) ligand 11/MIG | 7.9 | 7.7 |
| Chemokine (C-C motif) ligand 8/MCP-2 | 6.5 | 5.5 |
| Chemokine (C-C motif) ligand 20/MIP-3α | 6.5 | 3.9 |
| Chemokine (C-X-C motif) ligand 10/IP-10 | 5.7 | 5.2 |
| Chemokine (C-C motif) ligand 5/RANTES | 3 | 2.1 |
| Chemokine (C-X-C motif) ligand 2/MCP-1 | 1.9 | 2.1 |
| Chemokine (C-C motif) ligand 7/MCP-3 | 1.7 | NC |
| Chemokine (C-C motif) ligand 3/MIP-1α | 1.2 | 2.2 |
Macrophages were infected at an MOI between 1 and 2, and RNAs were extracted at 3 h postinfection. RNAs were pooled from three different donors, and gene expression was analyzed with an Affymetrix U133A Genechip microarray. Many of the chemokine genes were up-regulated in SARS-CoV-infected macrophages and are shown relative to influenza A (H1N1) virus-infected macrophages. A significant up-regulation in gene expression is indicated by an increase of twofold or more and was accompanied by a positive signal intensity call by the Microarray Suite Expression Analysis software, version 5 (Affymetrix Inc.). Only genes that have at least twofold change compared to mock-treated cells are shown. NC, no change compared with mock-treated macrophages.
FIG. 4.
Levels of CXCL10/IP-10 and CCL2/MCP-1 were elevated in SARS-CoV-infected macrophages. Macrophages were infected with SARS-CoV at an MOI of 1 to 2. (A) RNAs were extracted at 3, 6, and 15 h postinfection, and the levels of mRNA for CXCL10/IP-10 and CCL2/MCP-1 were determined by real-time quantitative RT-PCR. (B) Aliquots of the culture supernatant were taken at 6, 15, and 24 h postinfection, and the levels of secreted CXCL10/IP-10 and CCL2/MCP-1 were determined by specific ELISAs. The data shown are means (± standard deviations) of duplicate cultures from the same donor and are representative of three independent experiments with similar results. SARS-CoV infection (•) of macrophages induced higher levels of gene expression and secretion of CXCL10/IP-10 and CCL2/MCP-1 than did mock infection (▪).
In order to confirm that cytokines carried over in the SARS-CoV inoculum were not responsible for the induction of these chemokine genes, we tested the effect of differential filtration on the virus inoculum. After filtering the virus inoculum through a 0.2-μm filter to remove large debris, we passed it through filters with a 30-kDa cutoff. Titration of the filtrate on FRhK-4 cells and real-time quantitative RT-PCR with viral RNA indicated that most of the SARS-CoV was retained by the 30-kDa-cutoff filter (Table 4). In human macrophages, the retained material eluted from the filter induced the gene expression of CXCL10/IP-10 and CCL2/MCP-1 to similar levels as those induced by the initial virus inoculum. An assay of the filtrate was negative for SARS-CoV and did not up-regulate CXCL10/IP-10 and CCL2/MCP-1 gene expression in human macrophages.
TABLE 4.
Differential filtration of SARS-CoV inoculum to confirm that the induction of chemokines was not attributable to cytokines from the FRhK-4 culturea
| Exptl condition | SARS-CoV RNA level | Chemokine mRNA level
|
|
|---|---|---|---|
| CXCL10/IP-10 | CCL2/MCP-1 | ||
| Mock | Not detectable | 80 (81) | 700 (230) |
| Filtered SARS-CoV (0.2 μm) | 690 (7.1) | 9,100 (560) | 11,000 (1,300) |
| SARS-CoV filtered with 30-kDa cutoff | |||
| Filter eluate (>30 kDa) | 1,200 (4.2) | 15,000 (4,000) | 5,000 (2,600) |
| Filtrate (<30 kDa) | Not detectable | 20 (9.2) | 860 (150) |
The ability of the fractions to stimulate chemokine gene expression in macrophages was determined. The values shown are mean levels of mRNA (no. of copies/105 copies of β-actin mRNA) and standard deviations (in parentheses) for duplicate cultures.
DISCUSSION
SARS-CoV infections of macrophages lead to the initiation of viral gene transcription and viral protein synthesis. No infectious virus was produced, and hence SARS-CoV infections of macrophages appeared to be abortive (Table 1). The copy number of Orf1b or nucleoprotein RNA was found to increase with time, reaching a plateau at 6 h postinfection, with the ratio of positive- to negative-strand RNA being about 1. The viral nucleoprotein was expressed in >90% of infected macrophages (Fig. 1 and 2). In contrast, the amounts of positive-sense RNA for Orf1b and the nucleoprotein progressively increased in infected FRhK-4 cells (MOI, 1 to 2). These results confirmed that virus gene transcription and translation were initiated in infected macrophages and that the block in productive virus replication occurred subsequently.
These findings suggest that double-stranded RNAs are generated in SARS-CoV-infected macrophages. Double-stranded RNA is a potent inducer of IFN-β, triggering cell signaling pathways such as those mediated through RNA-dependent protein kinases and IFN regulatory factor 3 (IRF-3) (32). These would be expected to lead to the induction of IFN-β, even in the absence of productive virus replication. Thus, the lack of IFN-β induction in SARS-CoV-infected macrophages is interesting. Viral proteins from other viruses, such as the nonstructural 1 protein of influenza virus, play a major role in suppressing the IFN-α/β response by the host (8). Whether SARS-CoV encodes similar proteins to counteract the host's antiviral response has yet to be determined. SARS-CoV also failed to induce IL-28 and -29, two other recently discovered interferon-like cytokines with antiviral activities (27). In contrast, both HCoV-229E and influenza A virus induced IFN-β as well as IL-28 and IL-29 in macrophages.
IFN-α/β is considered one of the body's key first-line antiviral defenses. IFN-β exerts its effects on neighboring uninfected cells by binding to cell surface receptors, leading to the induction of IFN-α/β and ISGs, and is an important aspect of host defense (7, 28). ISGs are integral components in the development of the cellular antiviral state, and it appears paradoxical that several ISGs are activated in SARS-CoV-infected macrophages in the absence of IFN-α/β induction. Recently, evidence was reported showing that the transcription of ISGs could be activated without the need for prior de novo synthesis of IFNs and could bypass the JAK-STAT pathway (16). The mechanism of ISG induction in SARS-CoV-infected macrophages will require further investigation. However, the direct induction of ISGs in SARS-CoV-infected macrophages does not replace the protective role of IFN-α/β in vivo, as interferons act on neighboring cells and protect them from infection.
Macrophages are key sentinel cells of the immune system and are known to be a major source of cytokines, including IFN-α/β, in response to viral infections (17). In addition to its direct antiviral effect, IFN-α/β has a host of other effects on the immune response and plays a central role in the host defense against infections (18). Therefore, impairment of the IFN-α/β response in macrophages can lead to a significant deficit of innate immunity. We have not been able to assess whether a similar defect in the IFN response is operative in pulmonary epithelial cells (e.g., type 2 pneumocytes), which appear to be the target cell for the virus in the lungs (19). Irrespective of whether or not respiratory epithelial cells exhibit a similar lack of IFN-α/β response in vivo, the lack of this response in the macrophage is likely relevant to pathogenesis. It may be noted that in preliminary studies, IFN-β mRNA levels in nasopharyngeal epithelial cells from patients with SARS were significantly lower than those observed for patients with influenza (unpublished data).
A striking observation regarding patients with SARS is that the viral load in the upper respiratory tract progressively increases, peaking on about day 10 after the onset of clinical disease, and its subsequent decline correlates with the appearance of an adaptive immune response (4, 21). This contrasts with findings for other respiratory viral infections such as the common cold and influenza, in which the viral load peaks soon after the onset of clinical symptoms (2, 13, 14). We hypothesize that the lack of IFN-α/β in macrophages may allow viral replication to proceed unchecked, leading to extensive infection within the respiratory tract by the end of the first week of illness. When the adaptive immune response appears during the second week of illness, it is faced with the task of clearing a widespread infection within the lung, thereby aggravating the disease pathology. A comparable situation was seen with patients with severe combined immune deficiency with persistent viral respiratory infections who received allogeneic bone marrow transplants. The respiratory pathology was greatly enhanced around the time of marrow engraftment, often leading to a fatal outcome (29). Hepatitis B virus infection of the liver offers a similar analogy, in which the cellular immune responses destroying infected hepatocytes cause acute hepatitis.
Chemokines such as CXCL10/IP-10 and CCL2/MCP-1 were shown to be up-regulated in macrophages by SARS-CoV. Since the UV-inactivated virus also had a similar, albeit reduced, effect, the stimulation of chemokines is at least partially independent of viral replication. Since cytokines are molecules smaller than 30 kDa and since filtration of the inoculum to exclude molecules of >30 kDa removed the ability of the virus to stimulate chemokines (Table 3), we concluded that the stimulation of chemokines is not simply an effect induced by cytokines carried over in the virus inoculum. These data, of course, do not rule out a role for larger molecules that might stimulate chemokines. Investigations are currently under way to identify the component of SARS-CoV which is responsible for this effect.
CXCL10/IP-10 and CCL2/MCP-1 are chemotactic for monocytes/macrophages, which are the predominant inflammatory cell type found in the lungs of patients with SARS (19). We (unpublished data) and others (34) have found significantly elevated blood levels of CXCL10/IP-10 and CCL2/MCP-1 in patients with SARS and have observed that both chemokines are significantly elevated during the early stage of the illness. The chemokines CCL3/macrophage inflammatory protein 1α, CCL7/MCP-3, and CCL8/MCP-2 were induced by SARS-CoV in the microarray analysis, and they have similar biological effects as CCL2/MCP-1 (30). Therefore, the members of the MCP and macrophage inflammatory protein families can synergistically induce a cycle of monocyte/macrophage recruitment and, potentially, monocyte/macrophage-induced immunopathology. In addition, both CXCL-10/IP-10 (25) and CCL2/MCP-1 (3) have also been shown to suppress hematopoietic progenitor cell growth, which may contribute to lymphopenia, a prominent manifestation in SARS.
In conclusion, our results indicate that unlike HCoV-229E or influenza A virus, SARS-CoV is unable to elicit strong induction of an IFN-α/β response in human macrophages. Together with the ability of SARS-CoV to trigger the release of certain chemokines from macrophages, these effects may explain some aspects of the pathogenesis of SARS. However, our studies have focused on one key component of the innate immune response, viz, the macrophage. Further investigations of this hypothesis with relevant animal models of SARS may be considered. It is known that SARS-CoV is highly susceptible to the antiviral effects of IFN-β in vitro (6). Furthermore, studies with primates have shown that IFN therapy is beneficial for both prophylaxis and early therapy (11), and a recent clinical study indicated that the use of IFN in combination with steroids was possibly associated with a clinical benefit (15). Thus, our results may provide a biological basis for the observed therapeutic benefit with IFNs and for treating SARS patients by early administration of IFNs.
ADDENDUM
It is relevant to report the findings of two recent papers that appeared before this paper went to press. One paper showed a lack of induction of beta interferon in SARS-CoV-infected 293 cells. Since SARS-CoV induced the nuclear translocation of IRF-3 in these cells, it was proposed that the lack of an interferon response is due to a block in the subsequent dimerization and hyperphosphorylation of IRF-3 (27a). In the second paper, a comparison of gene expression profiles of peripheral blood mononuclear cells obtained from patients with SARS and influenza revealed that unlike influenza, SARS failed to elicit alpha and beta interferon responses (24a). The latter findings, which were conducted ex vivo, parallel our observations with macrophages in vitro.
Acknowledgments
We are grateful for the help of Carol L. K. Chan and William Mak of the Genome Center, The University of Hong Kong, with performing the Affymetrix Genechip hybridization and analysis of the raw data. We thank Thomas Y. O. Chan, Charmaine H. K. Wong, Renee W. Y. Chan, and Bonnie W. Y. Wong for their technical assistance.
This research was supported by the National Institutes of Allergy and Infectious Diseases (Public Health research grant AI95357) and by grants HKU7542/03M from the Research Grants Council of Hong Kong and RFCID grant 03040722, the government of Hong Kong SAR.
REFERENCES
- 1.Bang, F. B., and A. Warwick. 1960. Mouse macrophages as host cells for the mouse hepatitis virus and the genetic basis of their susceptibility. Proc. Natl. Acad. Sci. USA 46:1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradburne, A. F., M. L. Bynoe, and D. A. Tyrrell. 1967. Effects of a “new” human respiratory virus in volunteers. Br. Med. J. 23:767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broxmeyer, H. E., B. Sherry, S. Cooper, L. Lu, R. Maze, M. P. Beckmann, A. Cerami, and P. Ralph. 1993. Comparative analysis of the human macrophage inflammatory protein family of cytokines (chemokines) on proliferation of human myeloid progenitor cells. Interacting effects involving suppression, synergistic suppression, and blocking of suppression. J. Immunol. 150:3448-3458. [PubMed] [Google Scholar]
- 4.Chan, K. H., L. L. Poon, V. C. Cheng, Y. Guan, I. F. Hung, J. Kong, L. Y. Yam, W. H. Seto, and K. Y. Yuen. 2004. Detection of SARS coronavirus in patients with suspected SARS. Emerg. Infect. Dis. 10:294-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, C. Y., L. L. Poon, A. S. Lau, W. Luk, Y. L. Lau, K. F. Shortridge, S. Gordon, Y. Guan, and J. S. Peiris. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360:1831-1837. [DOI] [PubMed] [Google Scholar]
- 6.Cinatl, J., B. Morgenstern, G. Bauer, P. Chandra, H. Rabenau, and H. W. Doerr. 2003. Treatment of SARS with human interferons. Lancet 362:293-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deonarain, R., A. Alcami, M. Alexiou, M. J. Dallman, D. R. Gewert, and A. C. Porter. 2000. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J. Virol. 74:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gendelman, H. E., and P. S. Morahan. 1992. Macrophages in viral infections, p. 168-172. In C. E. Lewis and J. O'D. McGee (ed.), The macrophage. Oxford University Press, New York, N.Y.
- 10.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 11.Haagemans, B. L., T. Kuiken, B. E. Martina, R. A. M. Fouchier, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, T. de Jong, S. Itamura, K. H. Chan, M. Tashiro, and A. D. Osterhaus. 2004. Pegylated IFN-a protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10:290-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohdatsu, T., M. Yamada, R. Tominaga, K. Makino, K. Kida, and H. Koyama. 1998. Antibody-dependent enhancement of feline infectious peritonitis virus infection in feline alveolar macrophages and human monocyte cell line U937 by serum of cats experimentally or naturally infected with feline coronavirus. J. Vet. Med. Sci. 60:49-55. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser, L., M. S. Briones, and F. G. Hayden. 1999. Performance of virus isolation and Directigen Flu A to detect influenza A virus in experimental human infection. J. Clin. Virol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser, L., R. S. Fritz, S. E. Straus, L. Gubareva, and F. G. Hayden. 2001. Symptom pathogenesis during acute influenza: interleukin-6 and other cytokine responses. J. Med. Virol. 64:262-268. [DOI] [PubMed] [Google Scholar]
- 15.Loutfy, M. R., L. M. Blatt, K. A. Siminovitch, S. Ward, B. Wolff, H. Lho, D. H. Pham, H. Deif, E. A. LaMere, M. Chang, K. C. Kain, G. A. Farcas, P. Ferguson, M. Latchford, G. Levy, J. W. Dennis, E. K. Lai, and E. N. Fish. 2003. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290:3222-3228. [DOI] [PubMed] [Google Scholar]
- 16.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct IFN-alpha genes by positive feedback through IFN regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogensen, S. C., and J. L. Virelizier. 1987. The IFN-macrophage alliance. Interferon 8:55-84. [PubMed] [Google Scholar]
- 18.Muller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 19.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen: SARS study group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen: HKU/UCH SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon, L. L., K. H. Chan, O. K. Wong, T. K. Cheung, I. Ng, B. Zheng, W. H. Seto, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2004. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 50:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon, L. L., O. K. Wong, K. H. Chan, W. Luk, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2003. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin. Chem. 49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 24a.Reghunathan, R., M. Jayapal, L. Y. Hsu, H. H. Chng, D. Tai, B. P. Leung, and A. J. Melendez. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarris, A. H., H. E. Broxmeyer, U. Wirthmueller, N. Karasavvas, S. Cooper, L. Lu, J. Krueger, and J. V. Ravetch. 1993. Human interferon-inducible protein 10: expression and purification of recombinant protein demonstrate inhibition of early human hematopoietic progenitors. J. Exp. Med. 178:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawicki, S. G., and D. L. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 27a.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. J. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, C. E., H. K. Osman, A. J. Turner, T. J. Flood, M. Abinun, B. Crooks, and A. J. Cant. 1998. Parainfluenza virus and respiratory syncytial virus infection in infants undergoing bone marrow transplantation for severe combined immunodeficiency. Commun. Dis. Public Health 1:202-203. [PubMed] [Google Scholar]
- 30.Van Damme, J., P. Proost, J. P. Lenaerts, and G. Opdenakker. 1992. Structural and functional identification of two human, tumor-derived monocyte chemotactic proteins (MCP-2 and MCP-3) belonging to the chemokine family. J. Exp. Med. 176:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virelizier, J. L., and A. C. Allison. 1976. Correlation of persistent mouse hepatitis virus (MHV-3) infection with its effect on mouse macrophage cultures. Arch. Virol. 50:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 33.Weiss, R. C., and F. W. Scott. 1981. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 4:175-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, C. K., C. W. Lam, A. K. Wu, W. K. Ip, N. L. Lee, I. H. Chan, L. C. Lit, D. S. Hui, M. H. Chan, S. S. Chung, and J. J. Sung. 2004. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 136:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]