Abstract
TRIM5 is a determinant of species-specific differences in susceptibility to infection by retroviruses bearing particular capsids. Human immunodeficiency virus type 1 (HIV-1) infection is blocked by the alpha isoform of macaque TRIM5 (TRIM5αrh) or by the product of the owl monkey TRIM5-cyclophilin A gene fusion (TRIMCyp). Human TRIM5α potently restricts specific strains of murine leukemia virus (N-MLV) but has only a modest effect on HIV-1. The amino termini of TRIM5 orthologues are highly conserved and possess a coiled-coil domain that promotes homomultimerization. Here we show that heterologous expression of TRIM5αrh or TRIMCyp in human cells interferes with the anti-N-MLV activity of endogenous human TRIM5α (TRIM5αhu). Deletion of the cyclophilin domain from TRIMCyp has no effect on heteromultimerization or colocalization with TRIM5αhu but prevents interference with anti-N-MLV activity. These data demonstrate that TRIM5 orthologues form heteromultimers and indicate that C-terminal extensions alter virus recognition by multimers of these proteins.
TRIM5 proteins inhibit the infectivity of a range of different retroviruses in a species-specific fashion (10). The capsid protein (CA) is the viral determinant for susceptibility to this restriction (3, 17). Rhesus macaque TRIM5α (TRIM5αrh) restricts human immunodeficiency virus type 1 (HIV-1) replication (18). Human TRIM5α (TRIM5αhu) restricts “N-tropic” strains of the murine leukemia virus (N-MLV) (8, 9, 13, 21). In owl monkey cells, HIV-1 is inhibited by TRIMCyp, the product of the TRIM5-cyclophilin (CypA) gene fusion (12, 15). The restriction activities of TRIM5αrh and TRIMCyp are conferred to nonrestrictive cells upon transduction of the respective cDNAs (15, 18). CypA modulates the restriction of HIV-1 in human and owl monkey cells in opposite ways: HIV-1 CA binding to the CypA domain of TRIMCyp (4, 5) is necessary for inhibition of HIV-1 in owl monkey cells, while “free” CypA appears to protect HIV-1 from restriction in human cells (20).
At the C-terminus of TRIM5α is a variable SPRY domain that determines the species specificity of restriction (8, 9, 21). In owl monkeys, the SPRY domain was replaced by CypA via L1-mediated retrotransposition (12, 15). TRIM5α and TRIMCyp both contain a tripartite motif, composed of RING finger, B-Box, and coiled-coil domains (14, 15), that exhibits E3 ubiquitin ligase activity (6, 18). The coiled-coil domain promotes the formation of TRIM5 homomultimers (14). Here we asked whether TRIM5α or TRIMCyp associates with TRIM5αhu and alters the antiviral activity of the human protein.
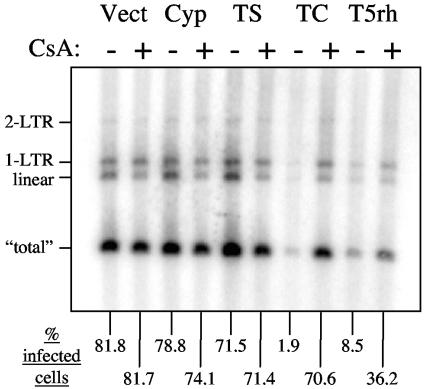
We transduced human rhabdomyosarcoma TE671 cells (19) with previously described LPCX vectors (15, 18) bearing cDNAs for TRIM5αrh, TRIMCyp, or owl monkey CypA. Cells were also transduced with a vector expressing TRIMStop, a truncated version of TRIMCyp lacking the CypA domain (15). Pools of transduced cells were selected in puromycin and then assessed for susceptibility to infection with HIV-1NL-GFP (2). For each transduced population we also tested the effect on HIV-1 infectivity of cyclosporine A (CsA), a drug that competes with HIV-1 CA for binding to CypA (11). We monitored the percentage of infected (GFP-expressing) cells by fluorescence-activated cell sorting (FACS) and the synthesis of viral cDNA in the infected cells using a Southern blot designed to detect full-length, linear viral cDNA and circular viral cDNAs that form in the nucleus (1, 2, 22).
As expected, both TRIM5αrh and TRIMCyp inhibited HIV-1 infection of TE671 cells and inhibited HIV-1 cDNA synthesis (Fig. 1). TRIMCyp inhibited HIV-1 replication roughly fivefold more efficiently than TRIM5αrh, consistent with the higher levels of HIV-1 restriction in owl monkey cells than in macaque cells (1, 15). CsA treatment rescued HIV-1 replication in TRIMCyp-expressing cells, as previously reported (15). CsA also enhanced HIV-1 infection of TE671-TRIM5αrh, indicating that CsA partially countered HIV-1 restriction when TRIM5αrh was expressed in human cells. This result was expected, as CsA also counteracts the restriction to HIV-1 in Old World monkey cells (1). At the high multiplicity of infection used here, CsA had little effect on HIV-1 infectivity; in experiments using a lower multiplicity of infection, CsA modestly decreased HIV-1 infection of control TE671 cells, as previously reported (data not shown and reference 20).
FIG. 1.
Inhibition of HIV-1 in human TE671 cells expressing owl monkey TRIMCyp or Rhesus macaque TRIM5α. LPCX-based retroviral vectors were used to transduce the indicated genes into human TE671 cells. Vect, Cyp, TS, TC and T5rh designate, respectively, Vector control, owl monkey CypA, TRIMStop, TRIMCyp, and TRIM5αrh. These cells were then infected for 16 h with HIV-1NL-GFP (pseudotyped vesicular stomatitis virus G protein) in the presence (+) or absence (-) of CsA (5 μM). One twentieth of the cells were maintained in culture for another day and used to determine the percentage of GFP-positive cells by FACS (bottom of the figure). Total DNA was extracted from the remainder of the cells 16 h after infection, and 5 μg of each DNA sample was analyzed by Southern blotting. The positions of the linear, 1-LTR, and 2-LTR HIV-1 cDNA species are indicated on the left. “Total” DNA refers to a band specific to all HIV-1 cDNA forms, including the integrated DNA.
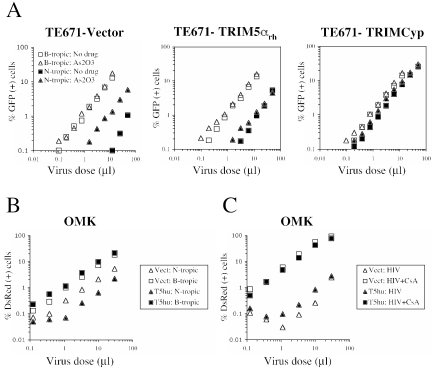
Next, we analyzed MLV replication in TE671-TRIM5αrh, TE671-TRIMCyp, and the control TE671-vector cells. We infected these cells with N- or B-tropic, GFP-expressing MLV vectors (2) that had identical titers in nonrestrictive Mus dunni tail fibroblasts and were normalized based on infection of these cells (19). In control cells, B-MLV was ∼200-fold more infectious than N-MLV (Fig. 2A). As2O3, a drug that counteracts restriction to N-MLV in TE671 cells (2, 9), specifically increased N-MLV infectivity by 10-fold or more (Fig. 2). In cells expressing TRIM5αrh, the N-MLV replication defect was reduced to about 20-fold (Fig. 2A); in cells expressing TRIMCyp, N-tropic restriction was fully abrogated (Fig. 2A). In either case, As2O3 no longer enhanced infectivity, as would be expected for cells lacking the antiviral activity targeting N-MLV.
FIG. 2.
TRIMCyp and TRIM5αrh abrogate the antiviral activity of endogenous TRIM5αhu. (A) TE671 cells expressing the indicated LPCX-derived constructs were infected with GFP-expressing B-MLV and N-MLV vectors at multiple doses. Cells were infected in the presence or absence of As2O3 (3 μM). Two days later, the percentage of infected cells was determined by FACS. (B) OMK cells were transduced with MIG (Vect) or with MIG-TRIM5αhu (T5hu) and challenged with DsRed-expressing N-MLV or B-MLV vectors. Two days later, the percentage of infected cells was determined by FACS. (C) OMK/MIG and OMK/MIG-T5hu cells were challenged with HIV-1-derived, DsRed-expressing CSRW vectors in the presence or absence of Cyclosporine A (2.5 μM).
We also performed the reciprocal experiment, transducing owl monkey OMK cells with TRIM5αhu and challenging them with MLV or HIV-1. TRIM5αhu had only a small (approximately twofold) restrictive effect on the replication of N-MLV and had no effect on B-MLV (Fig. 2B). Restriction of HIV-1 was similar in cells expressing TRIM5αhu to that in the control cells (Fig. 2C), and HIV-1 replication was rescued by CsA in both cell lines, consistent with previous reports (15, 20). Altogether, the data in Fig. 2 suggest that TRIMCyp is dominant over TRIM5αhu.
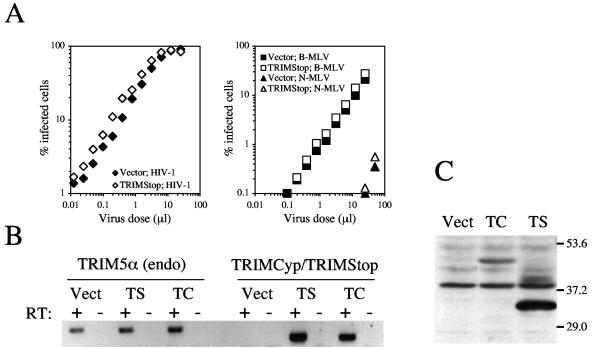
Since TRIMCyp (but not TRIM5αrh) completely suppressed the antiviral activity of TRIM5αhu, we further investigated the effects of this orthologue. TRIMStop, a truncated version of TRIMCyp lacking the CypA domain (15), did not affect the capacity of TE671 cells to restrict N-MLV replication (Fig. 3A). To determine if this was because TRIMStop was expressed at lower levels than TRIMCyp, we performed reverse transcription-PCR (RT-PCR) using conventional methods (15). TRIMStop mRNA was expressed at least as well as TRIMCyp mRNA (Fig. 3B); TRIM5αhu was expressed at similar levels in all three cell lines (Fig. 3B). By using Western blotting, we could not detect TRIM5 protein in the TE671 cell lines, but when 293T cells were transfected with the LPCX plasmids, TRIMStop was expressed at higher levels than TRIMCyp (Fig. 3C).
FIG. 3.
Effect of TRIMStop on TRIM5αhu antiviral activity. (A) TE671-Vector (Vector) cells and TE671-TRIMStop cells were infected with HIV-1NL-GFP (left panel) or with N- and B-MLV (right panel). Two days later, the percentage of infected (GFP-expressing) cells was determined by FACS. (B) Total RNA was prepared from TE671-Vector (Vect), TE671-TRIMCyp (TC), and TE671-TRIMStop (TS), and RT-PCR was used to detect endogenous TRIM5α or TRIMCyp/TRIMStop. RT was performed in the presence (+) or absence (-) of reverse transcriptase. (C) 293T cells were transfected with LPCX (Vect), LPCX-TRIMCyp (TC), or LPCX-TRIMStop (TS), and Western blotting was performed using an antibody specific to TRIM5.
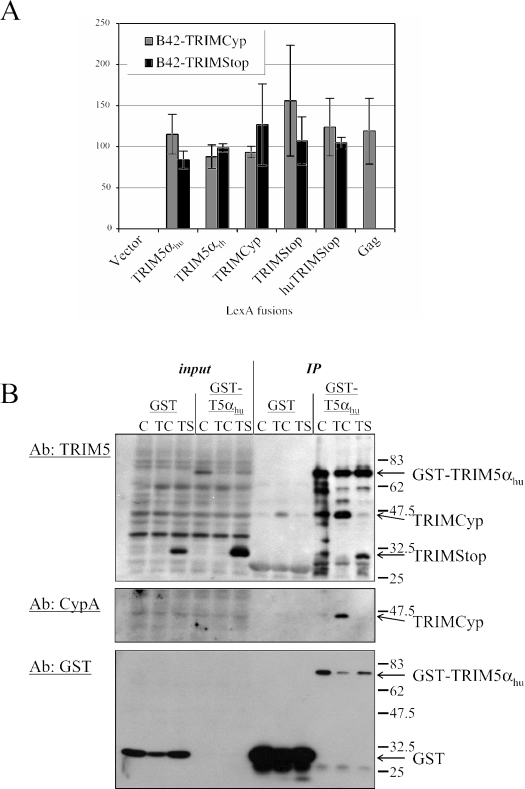
TRIM5 homomultimerization is promoted by the coiled-coil domain (14), which TRIMCyp also possesses. We hypothesized that the different TRIM5 orthologues heteromultimerize with each other and that TRIMStop did not interfere with TRIM5αhu activity because these particular proteins are incapable of interacting. We used the yeast two-hybrid system to analyze interactions between TRIMStop, TRIMCyp, TRIM5αhu, TRIM5αrh, HIV-1 Gag, and huTRIMStop, a version of TRIM5αhu which, like TRIMCyp, lacks the C-terminal SPRY domain. Fusions of these proteins with LexA and/or B42 were constructed, and the interactions between fusion proteins were analyzed using a previously described system (7) in which reporter gene β-galactosidase activity was assessed with a quantitative assay (16). TRIMCyp interacted equally well with TRIMCyp, TRIM5αrh, TRIM5αhu, TRIMStop, huTRIMStop, and HIV-1 Gag (Fig. 4A). Though TRIMStop was not able to interact with Gag, presumably because the CypA domain was deleted, this protein was as competent as TRIMCyp in interactions with each of the TRIM5 orthologues.
FIG. 4.
TRIMCyp and TRIMStop both bind TRIM5αhu. (A) Yeast two-hybrid system. TRIM5αhu, TRIM5αrh, TRIMCyp, TRIMStop, huTRIMStop, and HIV-1 Gag were fused to lexA. TRIMCyp and TRIMStop were expressed in fusion with the B42 activation domain. Pairs of fused LexA and B42 expression plasmids were transformed into Saccharomyces cerevisiae strain EGY48. For each transformant, β-galactosidase activity for three colonies is reported in Miller units with the standard deviation. (B) Binding in mammalian cells. TRIM5αhu was fused to GST. 293T cells were transfected with GST or GST-TRIM5αhu (GST-T5α) and cotransfected with LPCX (C), LPCX-TRIMCyp (TC), or LPCX-TRIMStop (TS). Thirty-six hours later, the cells were lysed in 50 mM Tris-Cl (pH 8.0), 150 mM NaCl, 1% NP40, 0.1% SDS, and GST was pulled down using glutathione-coated Sepharose beads (Pharmacia). One percent of the pre-pull-down lysate and 25% of the bound proteins were analyzed by Western blotting, using polyclonal antibodies directed against TRIM5, cyclophilin A, or GST.
To investigate TRIM5:TRIMCyp heterodimerization in mammalian cells, we expressed TRIM5αhu in fusion with glutathione-S-transferase (GST) and transfected 293T cells with either GST or GST-TRIM5αhu. These cells were cotransfected with the LPCX plasmid constructs described above that express TRIMCyp or TRIMStop. GST pull down on glutathione-Sepharose beads (Sigma) followed by Western blotting with anti-TRIM5 antibody showed that both TRIMCyp and TRIMStop associated with TRIM5αhu (Fig. 4B). The blot was also probed with an anti-cyclophilin A antibody, confirming the identity of TRIMCyp (Fig. 4B).
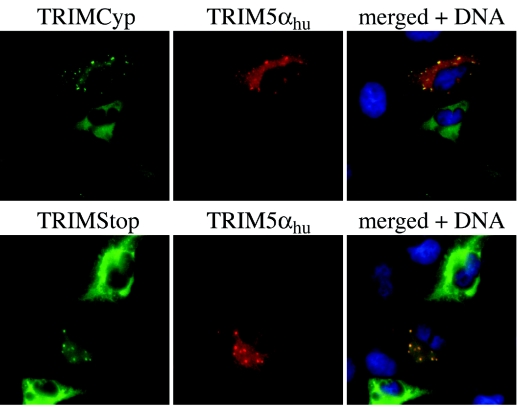
Finally, to examine the distribution of the tagged proteins in cells by immunofluorescence, we cloned TRIMCyp and TRIMStop into p3xFLAG.CMV (SIGMA), encoding N-terminal FLAG-tagged versions of the two proteins. These constructs were each transfected into TE671 cells and detected with anti-FLAG antibody. Both TRIMCyp and TRIMStop were diffusely distributed in the cytoplasm and concentrated around the nucleus in about half of the cells. TRIM5αhu, expressed in fusion with GST and detected with anti-GST antibody, was cytoplasmic as well and was partly localized to cytoplasmic bodies as previously reported (14). When TRIMCyp or TRIMStop were coexpressed in the same cells with TRIM5αhu, both TRIMCyp and TRIMStop showed partial localization to TRIM5αhu cytoplasmic bodies, suggesting that TRIMCyp and TRIMStop are both capable of heterodimerizing with TRIM5αhu in cells. Altogether, the results shown in Fig. 4 and 5 show that TRIM5 orthologues from different primate species heteromultimerize with each other and that failure of TRIMStop to block restriction activity of TRIM5αhu is not due to failure to multimerize.
FIG. 5.
Colocalization of TRIMCyp and TRIMStop with TRIM5αhu. TE671 cells were cotransfected with GST-TRIM5αhu and with 3× FLAG N-terminal-tagged versions of either TRIMCyp or TRIMStop. Thirty-six hours later, cells were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, and probed with antibodies against GST (rabbit polyclonal; Chemicon International) and FLAG (mouse monoclonal; Sigma). Fluorescent staining was done using Alexa488-conjugated goat anti-mouse and Alexa594-conjugated goat anti-rabbit antibodies and Hoechst33342 (all from Molecular Probes) to reveal DNA. Pictures were generated using a Nikon TE300 microscope with the Openlab 3.0 software.
In this work we show that TRIM5 proteins from different primate species interact with each other and can interfere with each other's function. The CypA domain of TRIMCyp is necessary for interference with endogenous TRIM5α (Fig. 3). CsA did not affect interference by TRIMCyp (data not shown), demonstrating that CypA peptidyl-isomerase activity was not relevant for the effect. A likely possibility is that when TRIM5αhu/TRIMCyp heterodimers are formed, the cyclophilin A domain of TRIMCyp interferes with TRIM5αhu's ability to recognize the N-MLV target. Perhaps TRIM5α and TRIMCyp function as multimers, with the TRIM5 C-terminus facing the viral target (consistent with the role of CypA in binding HIV-1 CA). Similar to the results reported here with different TRIM5 orthologues, the gamma isoform of macaque TRIM5 was found to inhibit TRIM5αrh anti-HIV restriction activity (18).
Acknowledgments
We thank Joseph Sodroski and Matthew Stremlau for reagents.
This work was funded by National Institutes of Health grant RO1 AI36199 and used core facilities of the Columbia-Rockefeller Center for AIDS Research. L.B. was supported by a fellowship (106524-35-RFHF) from the American Foundation for AIDS Reseach (AmFAR). D.M.S. was supported by the Columbia University College of Physicians and Surgeons Medical Scientist Training Program.
REFERENCES
- 1.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2004. Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J. Virol. 78:11739-11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthoux, L., G. J. Towers, C. Gurer, P. Salomoni, P. P. Pandolfi, and J. Luban. 2003. As2O3 enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J. Virol. 77:3167-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 4.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colgan, J., H. E. Yuan, E. K. Franke, and J. Luban. 1996. Binding of the human immunodeficiency virus type 1 Gag polyprotein to cyclophilin A is mediated by the central region of capsid and requires Gag dimerization. J. Virol. 70:4299-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freemont, P. S. 2000. RING for destruction? Curr. Biol. 10:R84-R87. [DOI] [PubMed] [Google Scholar]
- 7.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 8.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, K., and V. N. KewalRamani. 2004. In defense of the cell: TRIM5α interception of mammalian retroviruses. Proc. Natl. Acad. Sci. USA 101:10496-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 12.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 16.Stern, M., R. Jensen, and I. Herskowitz. 1984. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178:853-868. [DOI] [PubMed] [Google Scholar]
- 17.Stoye, J. P. 2002. An intracellular block to primate lentivirus replication. Proc. Natl. Acad. Sci. USA 99:11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 19.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 21.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]