Abstract
We isolated a human cDNA by expression cloning and characterized its gene product as a new human protein that enables entry and infection of herpes simplex virus (HSV). The gene, designated hfl-B5, encodes a type II cell surface membrane protein, B5, that is broadly expressed in human primary tissue and cell lines. It contains a high-scoring heptad repeat at the extracellular C terminus that is predicted to form an α-helix for coiled coils like those in cellular SNAREs or in some viral fusion proteins. A synthetic 30-mer peptide that has the same sequence as the heptad repeat α-helix blocks HSV infection of B5-expressing porcine cells and human HEp-2 cells. Transient expression of human B5 in HEp-2 cells results in increased polykarocyte formation even in the absence of viral proteins. The B5 protein fulfills all criteria as a receptor or coreceptor for HSV entry. Use by HSV of a human cellular receptor, such as B5, that contains putative membrane fusion domains provides an example where a pathogenic virus with broad tropism has usurped a widely expressed cellular protein to function in infection at events that lead to membrane fusion.
Herpes simplex virus type 1 (HSV-1) and HSV-2 are prevalent human pathogens that infect a broad range of animal cells. They establish lifelong latency in human neurons from which reactivation to lytic replication leads to recurrent herpes lesions. Entry into cells involves attachments of viral glycoproteins to multiple alternative cellular receptors (2, 10, 30). However, the molecular mechanisms of entry have not yet been defined.
The human proteins identified as receptors for HSV include heparan sulfate (HS) proteoglycans and several integral membrane proteins that are members of well-characterized families (30). Herpesvirus entry mediator HVEM (HveA) is a member of the tumor necrosis factor receptor family (20). Nectin 2 (HveB) and nectin 1 (HveC) or herpes immunoglobulin-like receptor are adhesion molecules in the immunoglobulin superfamily (7, 12, 35). d-Glucosaminyl 3-O-sulfotransferase (3-OS) modifies specific sites in cellular HS to generate binding sites for the essential HSV attachment glycoprotein D (gD) (27). Nectin 1, which was originally isolated as poliovirus receptor-related protein, allows entry of most HSV-1 and HSV-2 strains and is broadly expressed on a range of human tissues. HVEM and nectin 2 are more limited by either tissue distribution or strain specificity for transfer of HSV susceptibility. HVEM does not support entry of mutant virus HSV-1(Rid-1) that has a point mutation in gD (20). Nectin 2 is reported to allow entry of HSV-2 strains and HSV-1(Rid-1), but not most HSV-1 strains (35). Isolation of animal homologs for nectin 1 and HVEM raises the possibility that they may engage HSV during infection of animal cells.
With the exception of HS, HSV gD is a viral ligand for these receptors. Some regions of gD involved in receptor interactions have been defined (3, 8). How the cellular proteins interact with other viral proteins (each other or other cellular proteins) to carry out entry has not been determined. Recent reports confirm previous indications that gD can function during infection to attach virus to receptors, such as HVEM or nectin 1, and also at a different event that leads to conformational changes important for HSV fusion for penetration (6, 11, 36).
To identify and explore HSV receptors and the mechanisms for its pH-independent entry, we used an expression cloning approach with well-characterized replication-competent, but entry-defective porcine cells (23, 33). In this report, we describe a previously uncharacterized human gene whose product, currently designated B5, fits all criteria to establish it as a receptor for HSV entry. A unique structure, broad prevalence in human cells and tissues, and effects on virus entry suggest that it represents a new class of receptor or coreceptor for HSV infection.
MATERIALS AND METHODS
Cells and viruses.
Cell lines previously described are SK6-A7 cells (A7), a clonal nonpermissive porcine kidney cell line (23); HB1-9, an A7 cell line that constitutively expresses HVEM (25); and Neo, vector-only A7 cells. All porcine cells were maintained in Dulbecco modified Eagle medium (DMEM) with 5% fetal bovine serum (Gemini). Those cells transformed with drug selection markers were periodically passaged in 400 μg/ml G418 (Bethesda Research Laboratories [BRL]). Other cells were HEL, a fibroblast-like cell line from human embryonic lung; 293, adenovirus-transformed primary human embryonic kidney cells; A549, a human lung carcinoma; HEp-2, epidermoid carcinoma from human larynx; HeLa, human cervical epithelial carcinoma; SJNB 7, a human neuroblastoma cell line; and mouse L cells. Wild-type HSV-1 strains are F, KOS, and HFEM. HSV-2 strains are G and 333. Mutant viruses that encode lacZ, HSV-1(KOS)ICP4−lacZ+ (ICP4−/+), HSV-1(KOS)Rid1-lacZ+ (tkRid12) (a gift of P. Spear), and HSV-1(SC11)gH− lacZ+ (gH−/+) were propagated in supporting cell lines. The tkRid12, complemented gH−/+ and ICP4−/+ viruses enter cells to express lacZ under a constitutive cytomegalovirus promoter.
Virus infection.
Cells exposed to virus at 37°C for 70 min were treated briefly with citrate buffer (40 mM, pH 3.0) to inactivate extracellular virions, washed with phosphate-buffered saline (PBS), and incubated in DMEM at 37°C as previously described (33). To detect infection of reporter viruses, monolayers were washed three times with cold PBS and lysed for β-galactosidase (β-Gal) enzyme-linked immunosorbent assay (ELISA) performed according to the manufacturer's instructions (Roche). For quantifying blue foci by light microscopy, cells were fixed in 2% formaldehyde-0.2% glutaraldehyde for 5 min at 20 h postinfection and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Roche) and blue foci were counted. The substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) (3 mg/ml ONPG and 0.5% NP-40 in PBS) was used in colorimetric determination at optical density at 450 nm of β-Gal enzymatic activity. The titers of HSV progeny were determined in duplicate on Vero cells as described previously (33).
Screening of human cDNA library.
The human fetal lung cDNA library (catalog number A900; Invitrogen) contained 2 × 106 primary recombinants. Pools of cDNA plasmids were transiently transfected into A7 cells using Lipofectamine (Lifetech/BRL). After 24 h, cells were infected at 25 PFU/cell with ICP4−/+ or gH−/+ virus. Thirty-six hours later, monolayers were X-Gal stained to detect β-Gal in infected cells. Plasmid pools containing cDNAs that allowed highest infection above background were subdivided and tested in the same manner until individual cDNAs in bacterial colonies were identified that consistently transferred HSV susceptibility to A7 cells.
Nucleotide sequencing of cDNA clones and computer-assisted analyses.
DNA sequencing (Biomedical Research Core of the University of Michigan) of both strands of isolated cDNA clones used T7 or SP6 primer followed by nested internal primers. Lasergene software (DNASTAR) was used for computer-based analyses of nucleotide and predicted amino acid sequences. A BLAST program was used to search the GenBank database (NCBI).
Computer predictions.
Coiled-coil predictions were made with the COILS program (19, 21) on the basis of a prediction protocol proposed by Parry (22). The amino acid sequence is compared to a database of known parallel two-stranded coiled coils to derive a similarity score. Sequences of B5 and published sequences of other HSV receptors and HSV-1 glycoproteins were analyzed.
Construction of B5 expression vectors.
The hfl-B5 open reading frame (ORF) was amplified with primer pair 5′-GAATTCGTCCGCTGTGC (5′ primer) and 5′-AAAACTCAGGTATCAGAA (3′ primer) with added restriction enzyme digestion sites. The PCR product was digested and ligated into a pcDNA3.1-Zeo-CAT (Invitrogen) to generate plasmid pB5-CAT that has the chloramphenicol acetyltransferase (CAT) gene placed downstream of an untagged B5 sequence under the control of the same promoter. For C-terminal epitope-tagged B5 (pB5-myc), the B5 sequence was placed into pcDNA3.1-myc-his (Invitrogen) to place the ORF in frame with a short sequence encoding a C-terminal myc epitope (9 amino acids [aa]) and a polyhistidine epitope (6 aa). An N-terminal epitope-tagged B5 plasmid (pHis-B5) contained the B5 ORF downstream of a polyhistidine epitope and an Xpress epitope in pcDNA3.1-His (Invitrogen). For tetracycline induction by doxycycline (DOX), hfl-B5 was placed into the BamHI site of pTRE (Clontech) to produce a vector with a hygromycin resistance gene.
Transient and stable expression of B5 protein.
A7 porcine cells were transiently transfected with vector only or with the indicated purified plasmid containing hfl-B5. At 24 h posttransfection, cells were harvested and analyzed for B5 expression with antitag antibody. For untagged B5, we monitored expression of the B5 ORF indirectly by CAT activity of transfected cells.
B5 polyclonal cell lines (B5-1 or B5-2) were made by cotransfecting pB5-CAT and pSV2Neo into A7 cells and double selection with zeocin (0.6 mg/ml) (Invitrogen) and G418 (400 μg/ml) (BRL). Multiple cell colonies selected in one flask after 14 days were trypsinized and grown as B5-1 or B5-2 polyclonal monolayers. Epitope-tagged clonal porcine cell lines were CB5 that expressed myc-tagged B5 (pB5myc) and NB5 that expressed N-tagged B5 (pHisB5). CB5-1 and NB5-1 polyclonal cell lines were made by transfection of pB5myc or pHisB5 and G418 selection for 7 days followed by harvesting all the cell colonies from one dish. Neo, Zeo, and Seo cell lines were A7 cells that contain vectors without inserts. HB1-9/Zeo is an HB1-9 cell line transfected with vector encoding zeocin resistance. For all cell lines, the presence of the transfected plasmid was confirmed by PCR. mRNA was detected by reverse transcription-PCR (RT-PCR), and tagged protein was detected by Western blot analyses of cell lysates with anti-myc (9E-10) or anti-Xpress antibody (Invitrogen).
Over 30 stable clonal porcine cells lines, such as 10-1G1, were made using A7 cells and untagged B5 (pDNA-B5) pcDNA3.1 (Invitrogen) (G. DeLassus and A. O. Fuller, unpublished). After selection in G418 (400 μg/ml) (BRL), individual cell colonies were isolated and propagated as cell lines.
Inducible B5 expression.
Transfected A7 cells were subjected to two cycles of drug selection. Cell colonies from transfection of pTet-ON plasmid (neomycin resistant) were propagated and screened by transient transfection with reporter plasmid pTRE-luc, which expresses luciferase under the control of the TRE promoter when 1 μg/ml of doxycycline (DOX) is in the medium. Two cell lines with a relatively high level of induction and minimum background without induction were used in a second transfection with pTRE-B5. Double selected (neomycin and hygromycin) clones were screened for B5 mRNA by RT-PCR. The presence of functional B5 upon DOX induction was confirmed by RT-PCR detection of mRNA and fluorescence-activated cell sorting (FACS) detection of HSV binding.
Immunostaining and FACS to detect epitope-tagged B5 protein.
Parallel wells containing about 5 × 105 cells or cells on coverslips were transiently transfected with pB5-myc using Lipofectamine (Lifetech/BRL). Forty-eight hours later, cells were left alone or permeabilized and fixed in methanol-acetone at −20°C for 10 min or fixed by treating the cells with 2% paraformaldehyde in PBS at room temperature for 20 min. Cells were incubated with anti-myc 9E-10 antibody and goat anti-mouse secondary antibody (1:100) (HyClone). For immunostaining, cells at about 100% confluency were fixed with 2% paraformaldehyde-0.2% glutaraldehyde and stained with anti-myc (1:1,000) followed by anti-mouse secondary antibody and diaminobenzidine substrate (Lifetech).
For FACS of transiently expressed CB5, A7 monolayers were transfected with pB5myc or vector only. At 48 h posttransfection, cells were gently removed from the monolayer, and unfixed cells were exposed to anti-myc 9E10 antibody and secondary fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibody (Sigma) and analyzed as described below. For FACS of NB5, HB1-9 or Neo stable cell lines, 1 × 106 cells harvested by gentle trypsinization were allowed to recover in growth medium without serum for 20 min at room temperature. Cells were gently pelleted, washed in cold PBS, and either left alone or fixed and permeabilized in cold methanol-acetone for 10 min. To cells on ice, we added anti-Xpress antibody (Invitrogen) (1:500) followed by a secondary FITC-conjugated antibody. Controls lacked the primary anti-Xpress antibody. Cells were washed with cold PBS between incubations and analyzed immediately with a FACScan (Becton Dickinson).
FACS of virus binding.
Virus binding was determined as described previously (23) using PBS wash with or without heparin (100 μg/ml). Cell-bound virus was determined with a FACScan (Becton-Dickinson), a cocktail of monoclonal antibodies to HSV glycoproteins and FITC-conjugated anti-mouse secondary antibody.
Northern blotting and RT-PCR to detect mRNA.
Total RNA was extracted from cell lines using Trizol reagent (BRL) and electrophoresed in a gel containing 1% formaldehyde before transfer to a nylon membrane (MSI). A nonradioactive probe was generated by PCR incorporation of digoxigenin (DIG)-11-dUTP (Roche) with a primer pair that amplifies a fragment of 500 bp from the 3′ end of the B5 ORF. Membranes were prehybridized, washed, and incubated with alkaline phosphatase-conjugated anti-DIG-11-dUTP antibody according to the manufacturer's instructions (Roche). After serial washes, the nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) substrate was added to visualize hybridized bands.
RT-PCR was performed with a Titan One Tube kit (Roche). Oligo(dT) primer was used to initiate reverse transcription. A B5-specific upper primer (GAATTCGTCCGCTGTGC) amplified hfl-B5. β-Actin primers served as a control for RNA. The PCR products were analyzed by electrophoresis on 1% agarose gels and representative DNA bands eluted and sequenced using an automated sequencer ABI377 (Perkin-Elmer).
Immunoprecipitation and Western blots of biotinylated proteins.
Flasks (75 cm2) of confluent NB5 or HB1-9 cells were trypsinized gently to detach cells and allowed to recover for 20 min at 37°C without serum. Cells were washed twice in PBS before biotin labeling (1). Briefly, pelleted cells were resuspended in sulfosuccinimidyl-6-(biotinamido)hexanoate (Sigma) in PBS and rotated at room temperature for 30 min. After PBS washes, cells were lysed in detergent buffer (50 mM Tris, 100 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, 1 mM phenylmethylsulfonyl fluoride) for 30 min at 4°C before centrifugation at 2,400 × g for 5 min.
After preclearing by incubation for 3 h at 4°C with protein A beads, lysate supernatants were divided into three parallel samples and 100-μl samples incubated overnight at 4°C with preimmune serum or with anti-HVEM or anti-Xpress antibodies. After 50 μl of protein A beads were added for 3 hours at 4°C, protein A-bound precipitants were processed for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Electrophoresed proteins were transferred to a nitrocellulose membrane (Bio-Rad) where biotinylated proteins were detected in Western blots with streptavidin-conjugated peroxidase (1:500) and visualized with enhanced chemiluminescence (Amersham). The same membrane was exposed to anti-Xpress monoclonal antibody (1:1,000) followed by alkaline phosphatase-conjugated anti-mouse secondary antibody at 1:1,000 and an NBT/BCIP substrate mix (Lifetech).
Inhibition by synthetic peptides.
Synthetic peptides were a B5 peptide corresponding to aa 344 to 374 (KQQWQQLYDTLNAWKQNLNKVKNSLLSLSD) that covered the C-terminal heptad repeat, a gL 28-mer peptide corresponding to a predicted α-helix region from aa 114 to 141 (ANTQETETRLALYKEIRQALDSRKQAAS), and a randomly chosen hydrophilic HVEM peptide corresponding to aa 41 to 60 (CKEDEYPVGSECCPKCSPG). They were synthesized and purified by the University of Michigan protein core facility. Peptides were acetylated at the N terminus, amidated at the C terminus, and dissolved in PBS containing 1% bovine serum albumin. For inhibition of binding or entry, cells were incubated with the indicated concentrations of B5 peptide for 1 hour at 37°C. Cells were washed twice with cold PBS supplemented with 1% bovine serum albumin and exposed to 10 PFU/cell for 1 hour at 4°C. After a peptide dose curve was performed, 42 mM solutions of the indicated peptide were used in inhibition experiments. In an alternative procedure, viruses were incubated with 42 mM of peptide for 1 hour at 4°C before infecting cells that had not been exposed to peptide.
The effects of peptides on virus binding were determined by FACS as described above and previously (23). Peptide was added either to cells and washed out, or peptide was preincubated with virus and this material was added to cells. After infection, parallel cell samples were washed with heparin (100 μg/ml) (Sigma) or PBS followed by anti-HSV antibodies in FACS as described above.
For inhibition of HSV infection detected by ELISA, confluent cell monolayers in six-well plates were incubated with peptides (42 mM/1 × 106 cells) at 37°C for 1 hour. Unbound peptide was removed by washing with PBS. Alternatively, gH−/+ virus was pretreated for 1 hour with peptide. Peptide-treated or untreated virus at 10 PFU/cell was exposed to cells at 34°C for 90 min, followed by a 40 mM citrate buffer (pH 3.0). Monolayers were overlaid with DMEM, incubated at 37°C for 8 h, and analyzed for the level of β-Gal protein by ELISA (Roche) or by counting blue foci after X-Gal staining.
Transient transfection of B5 into human cells.
HEp-2 cells seeded to 30 to 40% confluency in six-well dishes or cells on coverslips in these dishes were transfected with either pcDNA3 or pB5-myc (5 μg) and incubated at 37°C. For peptide inhibition, at 3 hours posttransfection, 30-mer peptide in PBS was added to the medium to a final concentration of 42 mM. Forty-eight hours posttransfection, cells were washed and fixed with 0.25% glutaraldehyde on ice for 10 min. Anti-myc (9E-10) antibody was added (1:300) for 1 h at room temperature followed by FITC-conjugated anti-mouse secondary antibody (Sigma) (1:100) for 30 min. Cells were visualized by UV light microscopy. A fusion event was defined as a polykaryocyte with three or more nuclei stained with 4′,6′-diamidino-2-phenylindole (DAPI). The total numbers of polykaryons with three or more nuclei were counted, and the number of nuclei per polykaryon was scored directly from fluorescence microscopy of coverslips or from photographs of six randomly chosen fields.
RESULTS
Isolation and sequence predictions of hfl-B5.
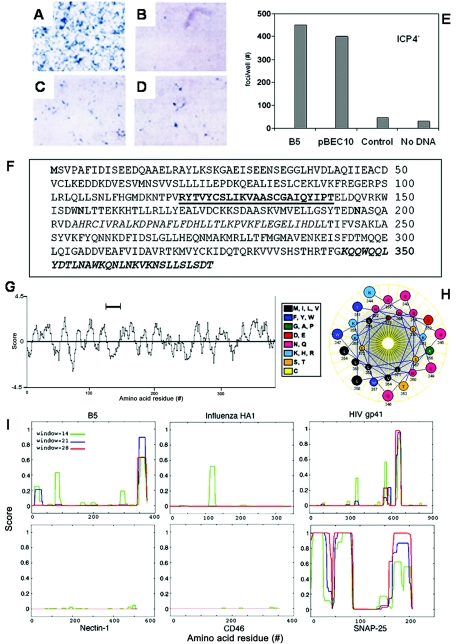
We screened a human fetal lung cDNA library for sequences that transfer susceptibility for HSV infection to entry-defective porcine kidney SK6-A7 (A7) cells (23, 32, 33). The susceptibility of transiently transfected A7 porcine cells was determined by blue foci detected as individual cells or clusters of cells infected by either reporter HSV-1(KOS)ICP4 (ICP4−/+) or HSV-1(SCgHZ) (gH−/+) that encode lacZ (Fig. 1A to E). Of several novel uncharacterized human genes that produced a protein product from in vitro translation (data not shown), we first examined the nucleotide sequence of one transfer-positive clone, designated hfl-B5 for human fetal lung B5 (Fig. 1E).
FIG.1.
Transfer of HSV susceptibility to porcine cells and computer predictions from the hfl-B5 sequence. A7 cells exposed to lacZ+ reporter virus HSV-1(KOS) ICP4−/+ after transient transfection with pools of individual human cDNAs were X-Gal stained to detect blue cell foci. (A) HEp-2 cells infected with 3 PFU/cell for 8 h. (B-D) A7 cells infected at 48 h posttransfection with 50 PFU/cell stained at 24 h postinfection. (B) Vector only or (C and D) two different pools of human cDNAs were used to transfect the cells. (E) Number of blue foci in a representative experiment after transfection with individual plasmids. pBEC10 encodes HVEM. Control is vector only. Relative susceptibility transfer for the plasmids was similar in multiple experiments where the overall transfection efficiencies varied. (F-I) Computer predictions from the hfl-B5 cDNA sequence (AY769947, NCBI). (F) Predicted amino acid sequence from the hfl-B5 nucleotide sequence. A putative membrane-spanning region is underlined, and amino acids that compose a C-terminal heptad repeat motif are shown in bold type. (G) A Kyte-Doolittle hydropathy plot generated from the B5 amino acid sequence. The predicted transmembrane domain is indicated. (H) A helix-wheel diagram of the predicted coiled-coil domain in the carboxy terminus shows leucine and valine alignment to one side of the helix. (I) COILS prediction plots for the indicated proteins. The y axis shows the COILS prediction score, and the amino acid residue position is shown on the x axis. Green lines represent the likelihood of a given region forming a two-helix bundle, blue lines a three-helix bundle and red lines a four-helix bundle.
Computer analyses of hfl-B5.
Analyses of the nucleotide sequence of hfl-B5, National Center for Biotechnology Information (NCBI) GenBank accession number AY769947, predicts expression of an approximately 42,500-Da type II membrane protein that contains 374 aa residues (Fig. 1F). According to the human genome sequence (34), hfl-B5 cDNA is identical to the uncharacterized gene XM-034431 located on human chromosome 11. It includes a strong Kozak's consensus start signal and a polyadenylation sequence, but no cleavable signal peptide sequence. Computer predictions of hydropathy revealed possible membrane-spanning domains at approximately aa 122 to 144 and aa 232 to 253 and two potential N-glycosylation sites at aa 155 and aa 196 (Fig. 1F and G). Infection of SF9 cells with a baculovirus vector that encodes a truncated B5 protein beginning at aa 142 produced a glutathione S-transferase (GST)-B5 fusion protein that is secreted into the medium. This indicated that the hydrophobic span from aa 232 to 253 was not sufficient for anchoring in SF9 cell membranes. In contrast, the full-length B5-GST fusion protein that contains aa 122 to 144 remains cell associated when expressed from baculovirus infection of SF9 cells (data not shown).
A search for other motifs revealed the presence of a heptad repeat in the C terminus from aa 344 to 374. This region was predicted to form an α-helix for coiled-coil structure (Fig. 1H to I) (19, 22). The heptad repeat in B5 was similar to that in human immunodeficiency virus (HIV) gp41 and scored higher in tetramer and trimer formation than influenza virus hemagglutinin (HA) (Fig. 1I). Comparisons were made to cellular proteins nectin 1 as an HSV receptor, CD46 as a common cell surface protein, and SNAP-25 as a cellular SNARE (Fig. 1I). The B5 C-terminal motif fits predicted coiled-coil structure where apolar residues align to one region of a helical wheel (Fig. 1H).
Search of the NCBI databases revealed that the amino acid sequence predicted from hfl-B5 had 99% identity with a Homo sapiens cDNA with accession number NM006360 referred to as dendritic cell protein. No further characterizations of the putative dendritic cell protein or its nucleotide sequence were found in the literature.
Cell surface location and orientation of B5.
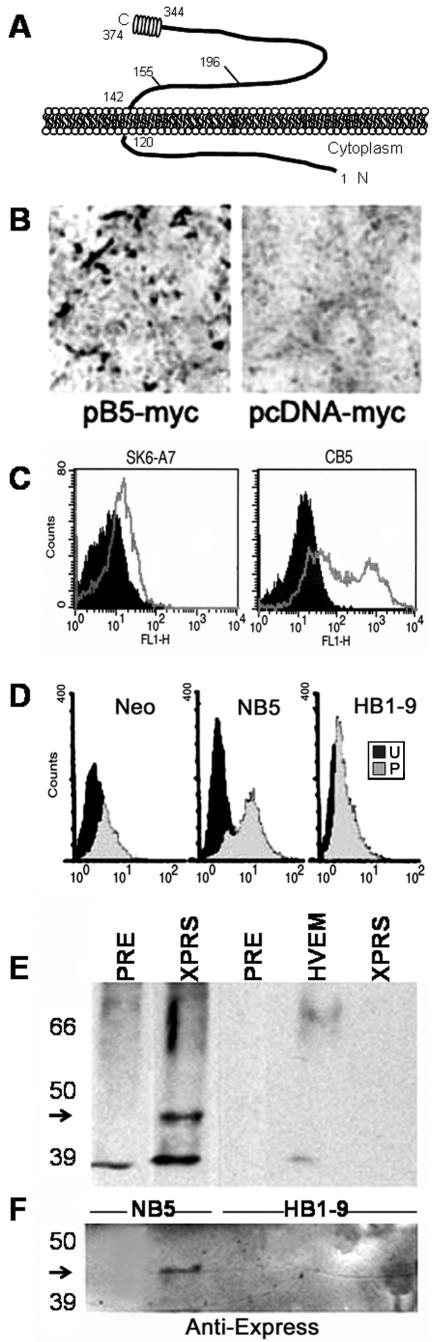
We expressed the hfl-B5 ORF, transiently or stably, in porcine A7 cells to explore the computer predictions (Fig. 2). Cell surface location and orientation of epitope-tagged proteins were determined by several approaches using vectors with C-terminal (pB5myc) or N-terminal (pHisB5) epitope tags (Fig. 2A). Both by light microscopy of immunostained cells and FACS anti-myc antibody detected C-terminally tagged B5 in nonpermeabilized A7 cells that were transiently transfected with pB5myc (Fig. 2B and C).
FIG.2.
Cell surface expression of B5 in porcine cells. (A) Cartoon of the predicted structure of B5 as an integral membrane protein with a predicted C-terminal α-helix. The C terminus and N terminus are labeled. (B) Cells transiently transfected with pB5-myc or vector only (pcDNA-myc) fixed at 48 h posttransfection and stained with anti-myc antibody, a secondary, peroxidase-conjugated antibody and diaminobenzidine substrate. (C) FACS of SK6-A7 cells transiently transfected with pcDNA vector (left) or pB5-myc (right) and stained with anti-myc (9E10) and a secondary, FITC-conjugated antibody (gray lines). Black peaks are cells in the absence of primary anti-myc antibody. (D) FACS of the indicated stable porcine cell line exposed to anti-Xpress and a secondary, FITC-conjugated antibody. Cells were permeabilized (P) with methanol-acetone or not permeabilized (unpermeabilized [U]) before staining. Neo is a vector-only-transformed cell line. NB5 is a stable cell line made with an N-terminal Xpress epitope-tagged B5. HB1-9 is a stable untagged HVEM-expressing porcine cell line. (E and F) NB5 and HB1-9 cells surface labeled with biotin and lysed as described in Materials and Methods. Lysates normalized for protein concentration were immunoprecipitated with preimmune serum (Pre) or with anti-HVEM polyclonal serum (HVEM), or anti-Xpress monoclonal antibody (XPRS). Proteins visualized in Western blots hybridized with streptavidin-conjugated peroxidase and enhanced chemiluminescence substrate (Amersham) (E) or anti-Xpress monoclonal antibody and alkaline phosphatase-conjugated anti-mouse secondary antibody and NBT/BCIP (F). The positions of molecular mass markers (in kilodaltons) are shown to the left of the gels. The arrow points to B5 at about 43 kDa.
We made a stable clonal porcine cell line, NB5, that constitutively produced N-terminal Xpress-tagged B5 (pHisB5). Anti-Xpress antibody was positive by FACS only when NB5 cells were permeabilized (Fig. 2D). There was no signal above background for nonpermeabilized NB5 cells, or in any condition for HB1-9 cells that constitutively express HVEM (25), or vector-only (Neo) cells (Fig. 2D). There was no signal when anti-Xpress antibody was omitted (data not shown).
These results show the following: (i) epitope-tagged B5 proteins were transported to the cell surface, (ii) the myc epitope at the C terminus of B5 was exposed extracellularly on intact cells (Fig. 2B and C), and (iii) the Xpress epitope at the N terminus was intracellular and accessible to anti-Xpress antibody only in permeabilized cells (Fig. 2D). As predicted, hfl-B5 encodes a type II cell surface integral membrane protein (Fig. 2A).
In a different approach to confirm cell surface presence, we biotinylated surface proteins for streptavidin detection in immunoprecipitated cell lysates. Lysates of porcine cells that produce Xpress-tagged B5 (NB5) or untagged HVEM (HB1-9) were immunoprecipitated. Separated proteins were blotted with streptavidin-conjugated peroxidase to visualize biotinylated proteins (Fig. 2E) or with anti-Xpress antibody to detect tagged B5 (Fig. 2F). For NB5, but not HB1-9 cells, a prominent biotinylated 43-kDa band precipitated only with anti-Xpress at the size expected of B5 monomers. This was confirmed as B5 in Western blots with anti-Xpress (Fig. 2F). As a control for labeling of cell surface proteins, a biotinylated protein of approximately 65 kDa (20) was immunoprecipitated from HB1-9 cell lysates by anti-HVEM (Fig. 2E). We conclude that B5 is a cell surface type II protein with an extracellular C terminus accessible for interactions with virus (Fig. 2A).
Tissue distribution of hfl-B5 mRNA expression.
Distribution of hfl-B5 mRNA expression was examined in commonly used tissue culture cell lines and in primary human tissues. RNA of approximately 1,300 nucleotides was detected by Northern blot analyses (Fig. 3A) and RT-PCR amplification (data not shown) of total RNA from a range of human cell lines, mouse L cells, and a CB5 stable transformed porcine A7 cell line. It was not detected in a vector-only porcine cell line (Seo) or in Vero cells. Nucleotide sequencing of representative RT-PCR products confirmed amplified products as hfl-B5. hfl-B5 transcripts were detected in preparations of human fetal lung, brain, liver, and muscle and adult lung tissue (Fig. 3B). These results indicate broad expression.
FIG. 3.
Detection of B5 mRNA. Northern blot analyses to detect the presence of hfl-B5 mRNA. (A) Total RNA from the indicated cell line or (B) commercially available polyadenylated RNA from primary human fetal tissues or adult lung tissue probed with a DIG-11-dUTP-labeled hfl-B5 or β-actin-specific sequence. In panel A, the porcine cells are Seo vector only or CB5 that expresses myc-tagged B5. CB5 mRNA runs slightly larger due to the added epitope tags. The presence and quality of mRNA from all the cell lines were monitored by RT-PCR amplification of β-actin (not shown). In panel B, probes of the same primary tissue blots are shown with a B5-specific sequence (top) or a β-actin sequence (bottom).
Susceptibilities of porcine cell lines that express B5.
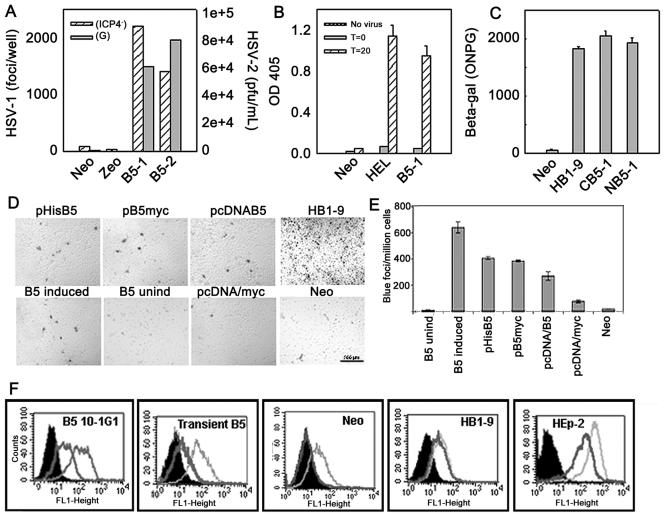
Transfer of susceptibility to HSV by the hfl-B5 gene product was examined by expressing B5 in porcine A7 cells under different conditions (Fig. 4). These cells contain heparan sulfate but lack receptors that allow stable attachment and entry of HSV (23, 32, 33). Polyclonal cell lines with untagged B5 (B5-1 and B5-2) were exposed to reporter HSV-1(lacZ+) viruses and infection determined by counting blue cell foci, ELISA of β-Gal protein, or spectrophotometer detection of enzymatic activity of β-Gal (Fig. 4A to E). HSV-2(G) infection was determined by progeny yields. Early passage B5-1 and human HEL cells showed similar levels of infection by a β-Gal reporter virus at 20 h postinfection (Fig. 4B).
FIG. 4.
Transfer of susceptibility to HSV infection by expression of B5. (A-C) Polyclonal porcine cell lines express untagged B5 (B5-1 and B5-2) or epitope-tagged B5 (NB5-1 and CB5-1). Clonal cell lines express HVEM (HB1-9) or vector only (Neo). HEL is a highly susceptible human cell line. HSV-1 strains ICP4−/+ and gH−/+ constitutively express lacZ. (A) Cell monolayers were exposed to 25 PFU/cell of ICP4−/+ or 3 PFU/cell of HSV-2(G) at 37°C for 90 min followed by citrate buffer inactivation of extracellular virus. Infection was determined by X-Gal staining of infected blue cell foci (ICP4−/+) or by progeny yields (G). (B) Infection at 25 PFU/cell to detect β-Gal protein by ELISA of lysates harvested at the time of infection (T0) or 20 h postinfection (T20). OD 405, optical density at 405 nm. (C) Cells infected with 10 PFU/cell of gH−/+ for 8 h were harvested for ONPG detection of β-Gal enzymatic activity. (D and E) Infection of porcine cells that transiently express B5. The indicated plasmids were transfected into porcine A7 cells and 48 h later infected with gH−/+ at 10 PFU/cell for 90 min followed by citrate buffer wash. After another 8 h, monolayers were X-Gal stained for infected blue foci. Plasmids are tagged B5 (NB5 or pcDNA3/His-B5; CB5 or pcDNA3/myc-His-B5), untagged B5 (pcDNA/B5), and vector only (pcDNA/myc). For inducible expression, a Tet-ON porcine cell line was transfected with pTRE-B5 and incubated for 24 h before induction with 1 μg/ml DOX or were left uninduced (unind). Forty-eight hours after induction, cells were infected and blue foci determined. Controls are the susceptible HB1-9 cell line infected at 3 PFU for 8 h and vector-only (Neo) porcine cells. (D) Photographs at a magnification of ×40 as viewed with an inverted microscope. (E) The average number of blue foci counted in four randomly chosen fields from transiently transfected cell monolayers. (F) HSV binding to cells. Profiles of virus binding to cells after exposure to 10 PFU/cell of purified HSV-1(F) at 4°C for 60 min. 10-1G1 cells constitutively express untagged B5. Transient B5 is A7 cells transfected with pcDNA/B5 for 48 h. Cells are as described above. Parallel cell samples were washed with either PBS to determine total bound virus (light gray line) or with buffer containing 100 μg/ml of heparin for heparin-resistant stable binding (thick black line) or were mock infected (black area). Virus was detected by FACS with a monoclonal antibody cocktail to HSV glycoproteins and FITC-conjugated anti-mouse secondary antibody.
All HSV-1 and HSV-2 strains tested could infect B5-1 cells. A derivative of HSV-1(Rid1) mutant virus, Rid1(tk12), that encodes lacZ produced yields of 1,780 ± 80 foci/well from B5-1 cells, but only background blue foci in HB1-9 porcine cells (<10) in triplicate wells. Rid-1 virus has a single point mutation in gD such that it cannot use HVEM as the only HSV entry receptor in CHO cells (20) or porcine cells (25). These results show that although B5 and HVEM allow HSV entry into porcine cells, they function differently as receptors for HSV.
We examined the susceptibilities of stable clonal porcine cells that expressed different levels of epitope-tagged B5 (CB5 or NB5), untagged B5 (10-1G1), or inducible B5 from a tetracycline-controlled promoter (B5 Tet-ON). Stable cell lines, such as 10-1G1, produced no viral reporter gene product or progeny (data not shown), even though they allowed HSV binding (Fig. 4F). This occurred for 10-1G1 and each of over 20 cell lines that stably express untagged B5 (DeLassus and Fuller, unpublished). When the same B5 plasmids used to make stable cell lines were transiently expressed in A7 porcine cells, each mediated entry detected as infected-cell foci (Fig. 4D and E) or β-Gal in ELISA (data not shown). Transiently transfected cells with DOX induction of a tetracycline-controlled hfl-B5 (B5-Tet-ON) were infected, but not uninduced transfected cells (Fig. 4D and E).
To further explore the effects of B5 expression, we used epitope-tagged hfl-B5 to make polyclonal cells similar to B5-1. CB5-1 and NB5-1 cells were infected at levels similar to that for HB1-9 cells (Fig. 4C). These results and those from transient transfections show that lack of infection by HSV of clonal B5 porcine cell lines is not related to the presence of C- or N-terminal epitope tags, but to some feature of clonal selection and stable expression over time of hfl-B5 in porcine cells. The B5 protein mediated HSV entry as the only human protein in polyclonal porcine cells or when transiently expressed (Fig. 1E and 4).
Stable virus binding of HSV to B5 expressed in porcine cells.
We examined HSV binding to cells to explore virus interaction with B5. Porcine kidney (SK-6) cells lack a protein that allows HSV to bind in a manner that resists heparin elution (23, 32, 33). HSV binding was compared in porcine cells that express B5, HVEM, or vector only (Fig. 4F).
Like stable virus binding to human HEp-2 cells and HB1-9 porcine cells that express only human HVEM, HSV stably bound to clonal 10-1G1 cells and to cells transiently transfected with B5 (Fig. 4F). There was no heparin-resistant stable binding to vector-only Neo porcine cells. Binding was detected for DOX-induced B5-Tet-ON cells, but not for uninduced cells (data not shown). That HSV can stably bind to all the porcine cells that express B5 indicates that lack of infection of clonal B5 cell lines is likely due to an effect other than stable binding.
Inhibition of HSV entry or stable binding that is mediated by B5.
Blocking of entry by a specific inhibitor is one of four criteria to establish a protein as a viral entry receptor. Despite several approaches, antibody that specifically recognizes human B5 has not yet been produced. The coiled coil predicted in the B5 sequence C-terminal heptad repeat at residues 344 to 374 (Fig. 1I) was comparable to those in fusion proteins of influenza virus HA and HIV gp41. For both of these viral proteins, some peptides that mimic the heptad repeat motifs specifically interfere with virus infection or membrane fusion (16, 28). In a similar approach, we examined effects on HSV interactions with cells of a 30-aa (30-mer) synthetic peptide that has the same sequence as the C-terminal coiled coil predicted for B5.
The 30-mer B5 peptide specifically inhibited HSV infection of B5-1 cells when preincubated with virus or when added to cells before or at the same time as virus (Fig. 5A and B). It did not block HSV infection when added after virus had been given time to penetrate (Fig. 5C). This indicated that the peptide affects an early event of HSV binding or events during penetration. From a dose-dependent analysis (data not shown), we found that 42 mM of the 30-mer peptide blocked most virus stable binding to clonal B5 10-1G1 cells (Fig. 5D). Compared to control peptides, it reduced binding and infection significantly for B5-expressing porcine cells. There was no substantial effect on HB1-9 porcine cells where HSV attachment and entry occur through human HVEM (Fig. 5B and E). Specific inhibition of HSV entry by the 30-mer B5 peptide fulfills blocking as one of the criteria to designate B5 as an HSV entry receptor.
FIG. 5.
Inhibition of HSV entry by synthetic peptides. A7 and Neo cells are parental or vector-only porcine cells. B5-1 is a polyclonal cell line that expresses untagged B5. HB1-9 constitutively expresses HVEM. (A-C) Cells were preincubated with equimolar amounts of peptide for 30 min before exposure to gH−/+ at 25 PFU/cell in the presence of peptide for 70 min at 37°C. At 24 h postinfection, β-Gal was detected by either blue foci (A) or ELISA of B-Gal protein (B and C). Peptides (described in Materials and Methods) are a B5 30-mer, an HVEM 26-mer, a gL 28-mer, or a control peptide, UOM-59, composed of randomly chosen 26 amino acids (B and C). In panel B, 100% infectivity shows the infection without peptide measured by ELISA for the amount of β-Gal protein in the indicated cells. In panel C, B5 peptide was added after citrate buffer wash (B5 peptide post C.T.). (D and E) Effects of a 30-mer B5 peptide on HSV stable binding detected by FACS. The filled histograms in panels D and E are a no-virus control. (D) 10-1G1 clonal B5 cells were incubated without peptide or with 42 mM of 30-mer B5 peptide before adding to virus. Stably bound virus is indicated by the thick light gray line. (E) Effects of 42 mM of the 30-mer B5 peptide on HSV stable binding to susceptible HB1-9, Neo porcine cells, or human HEp-2 cells. Thick white lines show stable virus binding in the absence of peptide. The thick gray lines show stable binding in the presence of peptide. (F) Virus preincubated with 42 mM of the indicated peptides or no peptide before adding to the indicated cells. O.D. 405 nm, optical density at 405 nm; B5pep, B5 peptide.
B5 involvement for HSV infection of human cells.
Human B5 can function for entry and infection of both HSV serotypes as the only human protein present in porcine cells (Fig. 1, 4, and 5A to C). The hfl-B5 mRNA is found in a range of human cells, including HEp-2 cells that are well characterized for susceptibility to HSV infection (Fig. 3). To determine the relevance of B5 to HSV infection of human cells as the natural host for this virus, we examined the effects of B5 on HEp-2 cells using the 30-mer peptide and transient overexpression of B5. Although HEp-2 cells likely contain multiple receptors that bind HSV to initiate the events of entry, the 30-mer B5 peptide blocked most of the HSV entry (Fig. 5F). Neither a control peptide consisting of a randomly chosen highly antigenic hydrophilic region of HVEM nor a 28-mer peptide that mimics an α-helix region predicted in gL (P. Perez-Romero and A. O. Fuller, unpublished) blocked HSV infection of HEp-2 cells (Fig. 5B and F). None of the peptides blocked HSV entry into HB19 porcine cells that express only human HVEM. These results establish that B5 has some interaction with HSV during entry into human cells.
Effects of B5 overexpression in human HEp-2 cells.
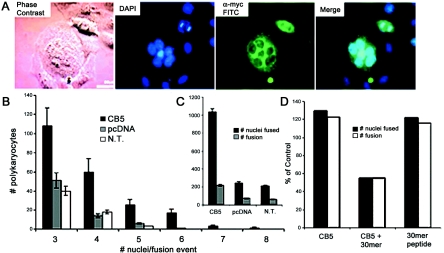
To further explore the effects of expression of the hfl-B5 gene product, we transfected HEp-2 cells with pB5myc. Immunostaining to follow epitope-tagged B5 and DAPI staining to identify DNA in cell nuclei indicates that B5-expressing HEp-2 cells fused to form into multinucleated polykaryons (Fig. 6A). There were a higher number of polykaryons and a higher average number of cell nuclei in each polykaryon for cells transfected with B5 compared to vector only or to nontransfected HEp-2 monolayers (Fig. 6B and C). Since no fusion was observed with transient (Fig. 4D) or stable (data not shown) expression of B5 in porcine cells, the fusion phenotype likely involves B5 interactions with or modification by some factor found in the human HEp-2 background. That HEp-2 polykaryons formed in the absence of HSV glycoproteins raised the possibility that B5 might affect human cell fusion through the predicted C-terminal α-helix region.
FIG. 6.
Effects in human cells of synthetic peptides and of overexpression of hfl-B5. (A) HEp-2 cells transiently transfected with C-terminal tagged B5 or vector were stained at 20 h with anti-myc and FITC-conjugated antibody (α-myc FITC) and DAPI for cellular DNA. The lower bar represents 60 μM. (B) The average number of nuclei in each polykaryocyte for those in four dishes of transfected cells. N.T., no transfection. (C) The total number of cell nuclei found in polykaryons (# nuclei fused) and the total number of fused polykaryons (# fusion) are defined as FITC- and DAPI-stained cells foci with three or more nuclei. (D) HEp-2 cells grown on coverslips were transiently transfected as described above. Three hours posttransfection, monolayers were incubated in PBS with or without 42 mM of the 30-mer B5 peptide. Forty-five hours later, the cells were washed, fixed, stained, and scored as described above. Data are expressed as percentages of the totals counted, where 100% is set as the level of polykaryons in vector-transfected cells without peptide.
As an initial test of a function in polykaryon formation for the putative C-terminal coiled coil, we examined the effects of the 30-mer B5 peptide on fusion from overexpressing CB5 in HEp-2 cells. In the presence of transiently overexpressed CB5, the 30-mer peptide reduced the number of polykaryocytes below that of the vector-only control (Fig. 6D). When only B5 peptide was added to HEp-2 cells, fusion increased to levels similar to those found with overexpression of CB5. Interestingly, either presence of excess B5 protein from transient expression or availability in the medium of the 30-mer peptide that mimics the C-terminal 30 residues enhanced the number of polykaryocytes formed in HEp-2 monolayers. In contrast, B5 overexpression and presence of the 30-mer peptide together reduced polykaryon formation. Although there is no obvious fusion peptide, these result indicate that the C terminus of B5 can affect membrane fusion of human HEp-2 cells. Because B5-transfected HEp-2 cells fuse, effects on HSV infection could not be reliably determined. However, these results suggested that B5 may function in HSV entry at a postattachment step that leads to molecular changes for membrane fusion.
DISCUSSION
We isolated a previously uncharacterized human gene designated hfl-B5 and show that it encodes a type II cell surface membrane protein that can serve as a receptor for entry of HSV. Features that establish it as an HSV entry receptor follow. (i) It is a cell surface integral membrane protein. (ii) It is broadly expressed in cells naturally susceptible to HSV. (iii) It allows stable binding of virus and entry into entry-defective porcine cells when expressed transiently or in polyclonal B5 porcine cells. (iv) Binding and infection of porcine and human cells were specifically blocked with a synthetic peptide identical to the B5 C terminus. Moreover, expression in human HEp-2 cells, even in the absence of viral proteins, altered the level of cell membrane fusion. Among currently known HSV receptors, B5 is unique structurally and functionally. These results establish that the hfl-B5 gene product can function as a new class of receptor, or possibly as a coreceptor, for entry of HSV.
B5 as the product of an uncharacterized novel human gene.
The hfl-B5 cDNA has the same sequence as the predicted gene XM_034431 located on human chromosome 11 (34). A cDNA deposited into the GenBank database (NM_006360), designated dendritic cell protein or GA17, has 99% identity with B5 and also is located on chromosome 11. In another entry (NM_068217), a Caenorhabditis elegans nucleotide sequence encodes a predicted ORF with 97% identity to hfl-B5 (4). Both putative proteins are predicted to contain a PINT motif at a location that overlaps the C-terminal heptad repeat of B5 (Fig. 1F). PINT motifs are thought to allow protein interactions among large intracellular multimeric complexes, such as components of proteosomes or signalosomes (14). Computer predictions and the results of several experimental approaches indicate that the hfl-B5 sequence encodes a type II integral membrane protein that has an extracellular C-terminal heptad repeat (Fig. 2). There are no published reports to characterize the predicted dendritic cell protein GA17 or the product of the C. elegans ORF. Lack of further information in the databases and in the literature suggests that these sequences have not been studied for protein production, structure, or function.
If, and how, B5 and these genes or their putative proteins are related remains to be determined. We speculate that hfl-B5 may be a highly conserved eukaryotic gene on the basis of the possible presence of a C. elegans homolog, expression of hfl-B5 in all tested human tissues and in mouse L cells (Fig. 3), and the difficulty of producing antibody in rabbits, mice, or chickens that specifically recognize human B5.
B5 differs in function and structure from known HSV receptors.
Several features of the hfl-B5 gene product make it unique among characterized HSV entry receptors. HSV infection occurs when porcine cells express B5 transiently or as a polyclonal mixture of B5-transformed cells. However, unlike clonal porcine cells that express HVEM (25) or nectin 1 (Perez-Romero and Fuller, unpublished), stable clonal B5 cell lines bind virus but do not allow entry. Some feature of the B5 clonal cell lines interferes with an event of penetration, uncoating, nucleocapsid transport, or initiation of viral gene expression. All of the B5 vectors transiently expressed in porcine cells allow HSV infection (Fig. 4D). Thus, something about the microenvironment of clonal B5 porcine cells affects HSV entry at events during or after stable attachment. Interestingly, resistance to HSV infection at an event during entry has been reported from overexpression of viral glycoprotein gD (30) and recently for gH (26). We report here resistance to HSV infection that results from stable clonal overexpression of a cellular receptor protein.
Expression of human B5 in porcine cells is not cytotoxic. Clonal cell lines, such as CB5, NB5, 10-1G1, and others, can be propagated and shown to express the B5 mRNA and epitope-tagged protein (DeLassus and Fuller, unpublished). Protein purified from these cells or the truncated protein from infection with a B5 baculovirus vector forms poorly soluble, high-molecular-weight oligomers that are confirmed as B5 in Western blots (DeLassus and Fuller, unpublished). Oligomerization is a feature characteristic of cellular SNARE proteins that contain coiled coils (16, 28, 31). Members of this family of proteins are not closely related by amino acid sequence but function at docking and membrane fusion for vesicle transport in protein trafficking (16). B5 is similar in some features to plasma membrane SNAREs but differs in others. The extracellular location of the putative coiled coil for B5 is unlike the intracellular oriented coiled coils in plasma membrane SNAREs. It also does not contain the hydrophobic 28 aa residues with Asn or Arg at position 13 as found in most SNAREs (16). Although there is no obvious fusion peptide sequence, fusion of human cells that overexpress only human B5 (Fig. 6) raises the possibility of a natural function in human cells in some manner to influence membrane fusion. Interestingly, engineering of flipped SNAREs suggests that sequences within SNARE proteins are sufficient to mediate membrane fusion (15). We speculate that for human cells, B5 may function naturally in fusion as a SNARE-like protein.
A B5 synthetic peptide blocks HSV entry.
The C-terminal heptad repeat of B5 scores at a high probability of forming coiled coils (Fig. 1H and I) as reported for many viral glycoproteins involved in virus-cell fusion (28). Some synthetic peptides to coiled coils in viral proteins block membrane fusion mediated by these glycoproteins. For influenza virus HA or HIV gp41, peptides to defined α-helix regions interfere with fusion between the virus envelope and a cell membrane (5, 13). A synthetic peptide that mimics the predicted coiled coil of B5 specifically inhibits HSV stable binding and entry into B5 polyclonal or transiently transfected porcine cells (Fig. 4 and 5). It blocks HSV-1 infection of human HEp-2 cells, but not HB1-9 cells that have only human HVEM (Fig. 5F). The presence of only the C-terminal peptide influences cell fusion of HEp-2 cells (Fig. 6D).
Point mutations predicted to break the α-helix in the C terminus of B5 result in proteins that do not support HSV entry (24). This indicates an important function of the C-terminal region. While biophysical confirmation of the presence of the coiled coil is required, these results from a genetic approach support the presence of a coiled coil. Peptides to this region will be useful probes to explore molecular mechanisms of how HSV induces membrane fusion. As with HIV and other viruses, the site at which an inhibitory peptide engages the virus could be a vulnerable target site to develop potent antiviral agents (5, 13, 17).
The B5 receptor may function in HSV entry at membrane fusion.
On the basis of analogies with viral entry or cell fusion proteins (5, 13, 16, 17, 28, 31), we hypothesize that the predicted B5 C-terminal coiled coil may engage those found in an essential HSV glycoprotein or possibly in an as-yet-unknown accessory cellular protein. Computer-based analyses of HSV glycoprotein sequences show that only gH and gL contain heptad repeats with predicted high scoring α-helices (24). gH and gL are essential for HSV entry and cell fusion (2, 10, 29). Although not yet experimentally shown to interact with B5, they fit the putative coiled coil containing criteria as possible ligands for B5. HSV entry involves stable binding through gD to one of its receptors (e.g., nectin 1 and HVEM). This could bring about rearrangements that allow α-helices in B5 and its ligand(s) to engage (Fig. 7). While aspects of this process remain to be proven, several reports provide results that support such events in HSV entry. Functions of gD in binding and fusion have been reported (2, 9, 11, 23, 29, 30). A physical interaction of gD, HVEM, and gH can be detected in HSV-infected human or porcine cells (25). Multiple receptor options for HSV binding using gD could trigger interactions with a broadly available cellular protein, such as B5 or others, to lead to protein and lipid rearrangement. Such a scenario would be a convenient means for HSV to initiate and regulate membrane fusion at neutral pH. Further support comes from fusion of human cells that overexpress B5 (Fig. 6), structural and functional features of B5, gH, and gL, and the requirement of gH/gL (2, 10, 29, 30) for HSV entry or virus-induced membrane fusion.
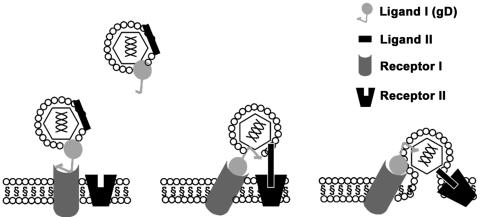
FIG. 7.
Model of minimum required receptors for HSV entry. A cartoon model to simplify use of multiple receptors into two minimum types of required receptor interactions to mediate HSV entry. The working model proposes that at least two types of viral glycoprotein and cellular receptor interactions are required to trigger pH-independent virus-cell membrane fusion. Ligands I and II on the virion are in an entry-incompetent state until ligand I (gD) encounters any one of its cell receptors (HVEM, nectin 1, nectin 2, or 3-OS-HS). It then undergoes conformational changes to an entry-active form. These changes would induce rearrangements in viral ligand II to an entry-competent form that can interact with host cell receptor II (B5 and possibly others). Interactions between ligand II and receptor II drive changes in the attachment-fusion complex to eventually trigger localized fusion of the viral envelope and host membrane. Similar to SNARE interactions or interactions in class I viral fusion proteins that contain α-helices, the putative coiled coils in B5 (a type of receptor II) and partner coils in its viral ligand(s) could mediate the interactions required to trigger membrane fusion at entry.
Relevance of B5 to HSV infection of human cells.
HSV can infect entry-defective porcine cells using the B5 receptor (Fig. 1, 4, and 5). Blocking by the 30-mer B5 peptide of HSV entry into porcine and HEp-2 cells establishes its relevance to HSV infection of human cells. How the peptide interferes with functions when B5 is the only human HSV receptor in porcine cells may differ from its interactions with human cells that have a number of cellular receptors for HSV. Of the characterized HSV receptors, direct evidence for relevance to virus infection of human cells has been shown only for nectin 1 (2) and for B5 (this report). Interestingly, B5, nectin 1, and d-glucosaminyl 3-O-sulfotransferase are broadly expressed receptors used by HSV.
The growing list of HSV entry receptors indicates that this prevalent human viral pathogen can engage multiple alternative receptors to successfully infect host cells (2, 10, 30). Figure 7 shows a model for minimum receptor requirements for HSV entry. It incorporates a new class of receptor, such as B5, with what is known about HSV receptors.
We propose that at a minimum two types of receptor-ligand interactions are needed. These are (i) a receptor for stable binding of gD to a human attachment receptor or animal homolog and (ii) a receptor, such as B5 or other proteins to be identified, for fusion involving a viral ligand that contains partner coiled coils such as predicted for gH/gL. HSV may have evolved to obtain these two functions by combinations of a repertoire of receptors that can bind to gD or participate in membrane fusion. A strategy of engaging multiple receptors that supply the two minimum required interactions of viral proteins and cell receptors is consistent with features of HSV entry and spread. It agrees with saturation blocking that indicates that HSV-1, HSV-2, and pseudorabies virus can engage common and also unique receptors (18, 35).
To our knowledge, this is the first indication that a virus has usurped what may be a cellular protein that contains membrane fusion motifs in coiled-coil structure. Utilization of a widely expressed cellular protein that contains membrane fusion motifs fits with tropism and lifelong infection of HSV and with multiple attachments at neutral pH that trigger fusion events (9, 30). As a new class of HSV receptor, B5, and possibly other proteins to be identified, may provide the missing link in the known functions of HSV glycoproteins and their interactions with receptor for entry or membrane fusion. Functional regions of this receptor or of its viral ligands provide a new potential target for development of novel therapeutics to intervene at entry as a critical event in HSV infection.
ADDENDUM IN PROOF
A recent article (T. Gianni, P. L. Martelli, R. Casadio, and G. Campadelli-Fiume, J. Virol. 79:2931-2940, 2005) identifies a region in HSV gH that contains an α-helix that may function in membrane fusion. This may represent one candidate region to interact with the α-helix in B5 as predicted with gH/gL as a potential B5 receptor ligand.
Acknowledgments
We thank M. Raghavan and K. Spindler for helpful discussions and D. Miller and D. Friedman for manuscript critique.
A. Perez and P. Perez-Romero were supported by UM Rackham Fellowships, G. DeLassus and S. Lopez by an NIH-funded Training Program in Genetics, Q.-X. Li by a Herpes Foundation Award, and S. Sutter by an NIH-funded Cellular and Molecular Biology Training Program. The research was supported by grants from NIAID and from the University of Michigan Technology Development Office to A.O.F.
REFERENCES
- 1.Alexeyev, M. F. 1995. Improved nonradioactive cell surface labelling technique for immunoprecipitation. BioTechniques 18:58-60. [PubMed] [Google Scholar]
- 2.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 3.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 4.C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012-2018. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 6.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA 101:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocchi, F., M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume. 1998. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc. Natl. Acad. Sci. USA 95:15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA. J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller, A. O., and W.-C. Lee. 1992. Herpes simplex virus entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J. Virol. 66:5002-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller, A. O., and P. Perez-Romero. 2002. Mechanisms of DNA virus infection: entry and early events. Front. Biosci. 7:D390-D406. [DOI] [PubMed] [Google Scholar]
- 11.Fuller, A. O., and P. G. Spear. 1987. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus type 1 prevent virion-cell fusion at the cell surface. Proc. Natl. Acad. Sci. USA 84:5454-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 13.Gruenke, J. A., R. T. Armstrong, W. W. Newcomb, J. C. Brown, and J. M. White. 2002. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J. Virol. 76:4456-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann, K., and P. Bucher. 1998. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem. Sci. 23:204-205. [DOI] [PubMed] [Google Scholar]
- 15.Hu, C., M. Ahmed, T. J. Melia, T. H. Sollner, T. Mayer, and J. E. Rothman. 2003. Fusion of cells by flipped SNAREs. Science 300:1745-1749. [DOI] [PubMed] [Google Scholar]
- 16.Jahn, R., and T. C. Sudhof. 1999. Membrane fusion and exocytosis. Annu. Rev. Biochem. 68:863-911. [DOI] [PubMed] [Google Scholar]
- 17.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 18.Lee, W.-C., and A. O. Fuller. 1993. Herpes simplex virus type 1 and pseudorabies bind to a common saturable receptor on Vero cells that is not heparan sulfate. J. Virol. 67:5088-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 20.Montgomery, R. I., M. S. Warner, B. Lum, and P. G. Spear. 1996. Herpes simplex virus 1 entry into cells mediated by a novel member of the TNF/NGR receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 21.Pallen, M. J., G. Dougan, and G. Frankel. 1997. Coiled-coil domains in proteins secreted by type III secretion systems. Mol. Microbiol. 25:423-425. [DOI] [PubMed] [Google Scholar]
- 22.Parry, D. A. 1982. Coiled coils in α-helix-containing proteins: analysis of the residue types within the heptad repeat and the use of the data in the prediction of coiled coils in other proteins. Biosci. Rep. 2:1017-1024. [DOI] [PubMed] [Google Scholar]
- 23.Perez, A., and A. O. Fuller. 1998. Stable attachment for HSV penetration into human cells requires gD in the virion and cell receptors that are missing for entry defective porcine cells. Virus Res. 58:21-34. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Romero, P., and A. O. Fuller. 2005. The C terminus of the B5 receptor for herpes simplex virus contains a functional region important for infection. J. Virol. 79:7431-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Romero, P., A. Perez, A. Capul, R. Montgomery, and A. O. Fuller. 2005. Herpes simplex virus entry mediator associates in cells in a complex with viral proteins gD and at least gH. J. Virol. 79:4540-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanlan, P. M., V. Tiwari, S. Bommireddy, and D. Shukla. 2003. Cellular expression of gH confers resistance to herpes simplex virus type-1 entry. Virology 312:14-24. [DOI] [PubMed] [Google Scholar]
- 27.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 28.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 29.Spear, P. G. 2001. A first step toward understanding membrane fusion induced by herpes simplex virus. Mol. Cell 8:2-4. [DOI] [PubMed] [Google Scholar]
- 30.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Subramaniam, V. N., F. Peter, R. Philp, S. H. Wong, and W. Hong. 1996. GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science 272:1161-1163. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian, G., R. A. LeBlanc, R. C. Wardley, and A. O. Fuller. 1995. Defective entry of herpes simplex virus type 1 and type 2 into porcine cells and lack of infection in infant pigs indicate species tropism. J. Gen. Virol. 76:2375-2379. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian, G., D. S. McClain, A. Perez, and A. O. Fuller. 1994. Swine testis cells contain functional heparan sulfate but are defective in entry of herpes simplex virus. J. Virol. 68:5667-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 35.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2 and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, G., E. Avitabile, G. Campadelli-Fiume, and B. Roizman. 2003. The domains of glycoprotein D required to block apoptosis induced by herpes simplex virus 1 are largely distinct from those involved in cell-cell fusion and binding to nectin1. J. Virol. 77:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]