Abstract
Conventional influenza vaccines can prevent infection, but their efficacy depends on the degree of antigenic “match” between the strains used for vaccine preparation and those circulating in the population. A universal influenza vaccine based on invariant regions of the virus, able to provide broadly cross-reactive protection, without requiring continuous manufacturing update, would solve a major medical need. Since the temporal and geographical dominance of the influenza virus type and/or subtype (A/H3, A/H1, or B) cannot yet be predicted, a universal vaccine, like the vaccines currently in use, should include both type A and type B influenza virus components. However, while encouraging preclinical data are available for influenza A virus, no candidate universal vaccine is available for influenza B virus. We show here that a peptide conjugate vaccine, based on the highly conserved maturational cleavage site of the HA0 precursor of the influenza B virus hemagglutinin, can elicit a protective immune response against lethal challenge with viruses belonging to either one of the representative, non-antigenically cross-reactive influenza B virus lineages. We demonstrate that protection by the HA0 vaccine is mediated by antibodies, probably through effector mechanisms, and that a major part of the protective response targets the most conserved region of HA0, the P1 residue of the scissile bond and the fusion peptide domain. In addition, we present preliminary evidence that the approach can be extended to influenza A virus, although the equivalent HA0 conjugate is not as efficacious as for influenza B virus.
Infection by influenza virus is responsible for 20,000 to 40,000 deaths and over 100,000 hospitalizations each year in the United States alone (50, 57). Globally, about 20% of children and 5% of adults worldwide develop symptomatic influenza each year (39). There are two influenza viruses of public health concern, A and B. Influenza A virus replicates in a wide range of avian and mammalian hosts. Subtypes are defined based on the immunological specificity of the hemagglutinin (HA) and neuraminidase (NA) envelope proteins (15). To date, three subtypes of influenza A virus have established stable lineages in humans, H1N1, H2N2, and H3N2 (15, 39, 41), only two of which, H1N1 and H3N2, have been circulating exclusively since 1968.
The influenza B virus, which is found almost exclusively in humans, has only one recognized subtype (39). However, two genetically distinct lineages are cocirculating in humans, represented by the B/Yamagata/16/88 and B/Victoria/2/87 viruses (9, 19, 46, 48). The two lineages are antigenically distinct, such that little or no postinfection cross-neutralizing antibody response is observed (45). Although the spectrum of disease caused by influenza B virus is generally milder than that by influenza A virus (15, 39), severe illness requiring hospitalization is still frequently observed (34).
Influenza A and B viruses continuously fluctuate in prevalence, with type and subtype dominance being different each year (9). The influenza B virus in particular has been the dominant one for 6 years between 1976 and 2001, accounting for >70% of laboratory-confirmed infections during those influenza seasons, and contributed ≥40% of infections for 3 more years (4). Because of the unpredictable type/subtype prevalence, the inactivated influenza vaccines currently in use must contain an influenza A virus H1N1, an influenza A virus H3N2, and an influenza B virus strain (41). These conventional vaccines represent an effective measure to prevent infection (20), but their efficacy depends primarily on the degree of antigenic “match” between the strains used for vaccine preparation and those circulating in the population. Since HA and NA readily undergo point mutations to evade the immune system (antigenic drift) (39, 41), the vaccine formulations need to be evaluated on a yearly basis and accordingly, vaccination must be performed annually. For influenza B virus, the emergence of new variants (36), coupled with the cocirculation of the different viral lineages (30, 46), makes the annual World Health Organization designation of the type B vaccine strain particularly problematic (48).
Against this background, the development of a universal influenza vaccine, effective against all circulating strains of both influenza A and B viruses and not requiring continuous manufacturing update, would meet a major medical need (59). Several laboratories have described important progress toward this goal for influenza A, but comparatively little attention has been dedicated to a universal influenza B vaccine. One reason is that the leading approach for the influenza A virus vaccine is based on the highly conserved, 24-amino-acid extracellular domain of the M2 protein (8, 10, 18, 33, 38), which has no equivalent in influenza B virus (20). Of the two influenza B virus candidate M2-like proteins, NB has been shown to be dispensable for viral replication in vitro (13), while BM2 has a very short extracellular ectodomain, with only five to six amino acids external to the membrane (32, 42). In addition, the antiviral drug amantadine, which blocks the acidification function of M2, is active against influenza A virus but not influenza B virus (40).
We describe here the design and validation in a preclinical animal model of a universal subunit vaccine for influenza B virus, based on the highly conserved maturational cleavage site of the precursor of the viral hemagglutinin HA0.
In addition, we present preliminary evidence that the same approach can be extended to influenza A virus, although the equivalent HA0 conjugate is less efficacious.
MATERIALS AND METHODS
Peptides and conjugates.
Synthetic peptides were prepared by standard solid-phase synthesis (2). The outer membrane protein complex (OMPC) of Neisseria meningitidis was purified by Merck & Co. Inc (14), and OMPC conjugates of peptides A/H3/HA0 and B/HA0 were prepared as previously described (8, 29, 31). Peptide loading for the A/H3/HA0 and the B/HA0 conjugates was 3,236 and 6,500 mol peptide/mol OMPC, respectively (49).
Viruses, cells, and other reagents.
Mouse-adapted A/Puerto Rico/8/34 (PR8; H1N1), A/Hong Kong/68 × PR8 reassortant (HKxPR8; H3N1), B/Ann Arbor/4/55, B/Hong Kong/330/2001, and B/Yamanashi/166/1998 viruses were produced as previously described (8) (unpublished data). The adjuvant QS21 was from Antigenics (Framingham, MA).
Animal studies and experimental protocols.
BALB/c mice (Charles River Laboratories, Wilmington, MA) and BALB/c mice lacking the γ subunit of the Fc receptor (Taconic, Germantown, NY) were maintained in accordance with institutional guidelines. Vaccination and challenge protocols and nasal and lung wash collection were all as previously described (8). Briefly, animals (10 per group) were vaccinated intramuscularly with a given vaccine on alum adjuvant, with or without addition of the adjuvant QS21, and boosted twice at 2-week intervals. Mice were challenged intranasally with 20 μl of virus, corresponding to the 90% lethal dose (LD90) (10.24 hemagglutinin units), and body weight and the survival rate were recorded daily. For passive protection studies, serum (200 μl) from mice vaccinated with B/HA0-OMPC or with unconjugated OMPC, or monoclonal antibody (MAb) ascites (2 ml, diluted 1:5) of the three protective MAbs was transferred to naïve BALB/c or FcR(−) mice via intraperitoneal injection. Mice were challenged 4 h following serum transfer (unpublished data).
Fluorescence polarization competitive immunoassay.
For the fluorescence polarization competitive immunoassay (FP-IA) assay, the B/HA0 peptide was covalently labeled with Alexa Fluor 488. A 25 μM solution in phosphate-buffered saline of the peptide, containing an extra cysteine at the N terminus, was reacted with Alexa Fluor 488 C5 maleimide (20:1 [wt/wt] fluorophore/peptide ratio), for 2 h at room temperature. The labeled peptide was purified by reversed-phase high-pressure liquid chromatography (HPLC). The concentration of the fluorescent-labeled peptide was determined using a BCA protein assay (Pierce, Rockford, IL) following the supplier's procedure.
When the labeled peptide, the MAb, and an unlabeled alanine mutant peptide were mixed in the assay, labeled and unlabeled peptide competed for antibody binding, and this competition was detected by changes in fluorescence polarization. Quantitation was made by running a dilution series for each peptide, using the same labeled control peptide for each. Each peptide generated its own binding isotherm and yielded a 50% inhibitory concentration (IC50) value. The IC50s of MAb D9E and O8F12 for the B/HA0 were 86.2 nM and 52.5 nM, respectively. The IC50 for the same MAbs of a set of alanine-substituted B/HA0 peptides was used as a measure of the relative contribution of each residue to binding.
RESULTS
Vaccine design.
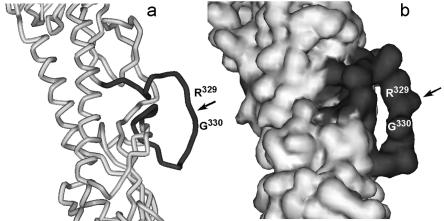
The mature influenza virus HA is composed of two subunits, HA1 and HA2, which are cleaved from the precursor, HA0 (5, 51). The sensitivity of HA to host proteases is mainly determined by the composition of the proteolytic site of the HA0 precursor, whose crystallographic structure was solved for the influenza A virus (5, 11, 55). The cleavage site forms an extended, highly exposed surface loop (Fig. 1). Upon cleavage, the carboxy terminus of HA1 and the amino terminus of HA2 separate. The newly formed N-terminal region of HA2 hosts the fusion peptide, which mediates fusion of the viral and cellular membranes. Therefore, cleavage of HA0 into HA1-HA2 activates virus infectivity (22, 25) and is crucial to pathogenicity (21, 53). In all human viruses, HA0 maturation occurs extracellularly, being mediated by proteases secreted by the cells in which the virus is replicating (47, 60).
FIG. 1.
(a) Alpha-carbon trace and (b) solvent-accessible surface of the cleavage site region (dark grey) of the HA0 precursor of the HA from the A/Hong Kong/68 influenza virus. Coordinates from reference 5.
The HA0 sequence is thus subject to a dual functional constraint, i.e., to remain a suitable substrate for host-encoded proteases and to maintain a functional fusion domain. Accordingly, a search in the influenza virus sequence database (28) shows that the HA0 cleavage site is highly conserved in all the available sequences of influenza B viruses, belonging to either one of the two cocirculating lineages. In addition, HA0 is also conserved within each subtype of influenza A virus. The HA2 side, which corresponds to the fusion peptide domain, is even conserved across the two influenza virus types (Table 1). This sequence stability extends over a remarkably long period of time (>60 years of known records) (28). Of interest, the cleavage site sequence of the pandemic 1918 A/H1N1 strain was found to be identical to that of the current H1N1 consensus (44).
TABLE 1.
Consensus sequence of the solvent-exposed region of the influenza A and B virus maturational cleavage sites
| Virus/subtype | Strain | Sequencea |
|---|---|---|
| A/H3/HA0 | Consensus | NVPEKQTR↓GIFGAIAGFIE |
| A/H1/HA0 | Consensus | NIPSIQSR↓GLFGAIAGFIE |
| B/HA0 | Consensusb | PAKLLKER↓GFFGAIAGFLE |
The position of cleavage between HA1 and HA2 is indicated by the arrow.
The consensus is the same for both the Victoria and Yamagata lineages.
Based on the crystal structure of HA0 from A/Hong Kong/68 (5), and assuming a similar structure for the influenza B virus, the B/HA0 peptide was designed to include all the cleavage site residues which are exposed to solvent (Fig. 1). The specific peptide sequence was then simply the consensus sequence in the chosen region (Table 1).
Since it is well known that low-molecular-weight synthetic peptides are rarely immunogenic, even with an adjuvant (1), we conjugated the peptides to a carrier protein. We selected the outer membrane protein complex (OMPC) of Neisseria meningitidis, a carrier widely used in human vaccines (8, 23, 31, 58), which we have recently employed for a candidate influenza A virus M2 vaccine (8).
Immunogenicity and protection of the HA0-based influenza B vaccine.
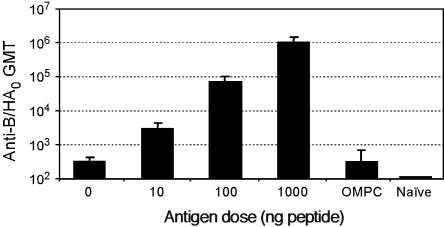
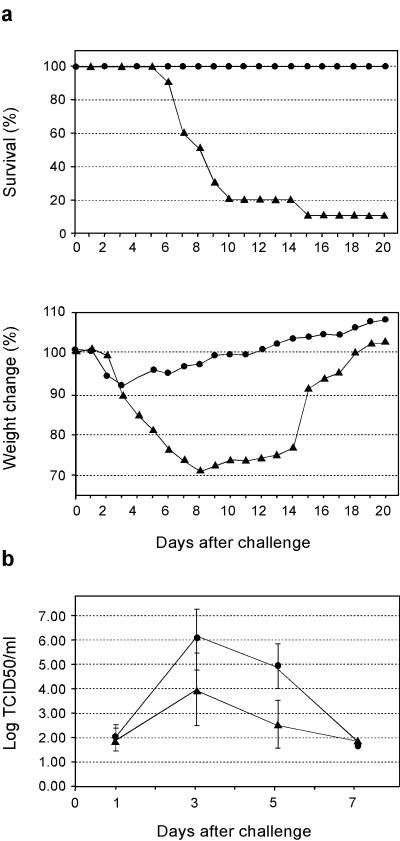
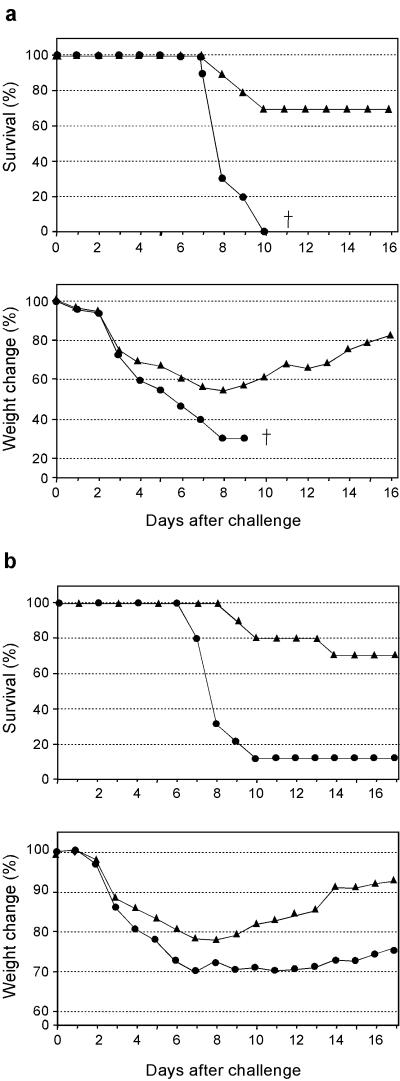
Mice were vaccinated with different doses of the B/HA0-OMPC conjugate, in the presence of the adjuvant QS21 (7, 26), which we had found particularly effective for the M2-conjugate vaccine (8). The vaccine was highly immunogenic, in a dose-dependent manner, with the highest dose producing a peptide-specific enzyme-linked immunosorbent assay (ELISA) geometric mean titer (GMT) of 1,080,941 (Fig. 2). We then tested the protection afforded by the conjugate by challenging vaccinated mice with LD90 of mouse-adapted influenza B/Ann Arbor/4/55 virus. As expected, only 10% of the animals in the control group survived, while 100% of the vaccinated mice were protected (Fig. 3a, top panel). In addition, significant protection from weight loss was also observed (Fig. 3a, bottom panel). Weight change is a sensitive parameter associated with the morbidity induced by the virus. Mice receiving the B/HA0 vaccine had only 10% maximum weight loss, compared to the 30% weight loss in control mice. Comparison with the results obtained in the same animal model with M2-based influenza A vaccines (8, 38) shows that the protection observed with B/HA0 is much more robust (see also below).
FIG. 2.
Induction of B/HA0 specific antibody responses in mice. Female BALB/c mice, 10 per group, were vaccinated intramuscularly with the indicated dose of B/HA0 conjugate formulated on alum and boosted once with the same antigen 4 weeks later. Blood samples were collected at 2 weeks after the second vaccination and tested for B/HA0 peptide-specific antibody titers by ELISA.
FIG. 3.
(a) B/HA0 vaccine protects against lethal viral challenge. BALB/c mice, 10 per group, were vaccinated with 1 μg of the B/HA0 conjugate formulated in alum and challenged with LD90 of B/Ann Arbor/4/55. After the challenge animals were monitored for survival (top) and weight change (bottom) for a total of 20 days. Solid circles, B/HA0 vaccinated, Solid triangles, control (OMPC vaccinated). The difference between the curves is statistically significant (P < 0.0001) (b) B/HA0 vaccine protects against respiratory viral replication in the lungs. BALB/c mice (32/group) were vaccinated with the B/HA0 conjugate (1 μg) formulated in alum plus 20 μg QS21 and challenged with 0.2 LD50 of B/Ann Arbor/4/55. The 50% tissue culture infectious dose (TCID50) of lung washes are represented as the mean of eight mice at each time point. Solid triangles, B/HA0 vaccinated; solid circles, OMPC vaccinated. The difference of overall lung viral shedding between the vaccine and control groups is statistically significant (P < 0.01), and the differences at day 3 and day 5 are also statistically significant (P < 0.05 and P < 0.01, respectively).
HA0-based influenza B vaccine reduces viral replication in the lungs.
In the same experiment, we collected nasal and lung washes from a separately vaccinated group (8, 37). On the basis of the viral shedding kinetics, we found that the B/HA0 vaccine reduced viral replication in the lungs for the entire viral infection course (Fig. 3b), while having no effect on nasal viral shedding. This is similar to influenza A virus M2 vaccines (8, 38).
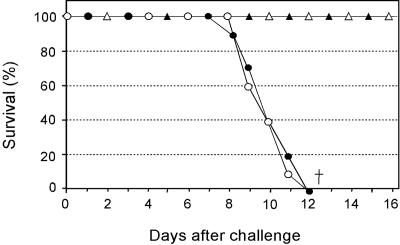
HA0-based influenza B vaccine is broadly protective.
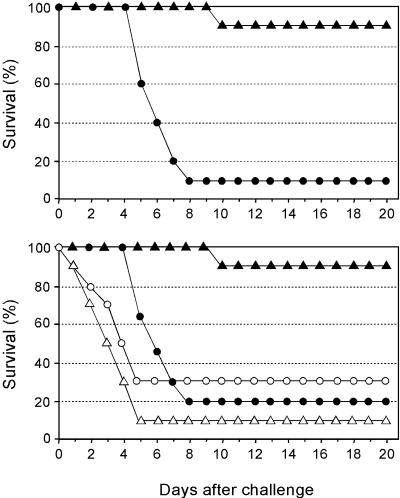
As previously discussed, the commercially available vaccines are manufactured with only one influenza B virus strain, belonging to either one of the two antigenically distinct, cocirculating lineages B/Victoria/2/87 and B/Yamagata/16/88 (19, 41, 45, 46, 48), despite the fact that these lineages are not immunologically cross-reactive (45). We tested the B/HA0 vaccine against LD90 challenge by mouse-adapted viruses corresponding to the currently recommended strains for the two lineages, B/Hong Kong/330/2001 and B/Yamanashi/166/1998. As expected from sequence conservation of the cleavage site, complete protection was observed in both cases (Fig. 4).
FIG. 4.
B/HA0 vaccine protects against both lineages of influenza B. The percentage of mice (10 animals/group) surviving challenge is graphed versus days postchallenge. Solid circles, OMPC vaccinated, challenged with B/Hong Kong/330/2001. Solid triangles, B/HA0 vaccinated (1 μg, formulated on alum), challenged with B/Hong Kong/330/2001. Open circles, OMPC vaccinated, challenged with B/Yamanashi/166/1998. Open triangles, B/HA0 vaccinated (1 μg, formulated on Alum), challenged with B/Yamanashi/166/1998. The dagger indicates that no animal in the group was alive at this time point. All vaccines were given together with 20 μg of QS21.
Protection by the HA0-based vaccine is mediated by antibodies.
To establish that the observed protection was due to antibodies targeting the cleavage site sequence, we performed two experiments. First, serum from B/HA0-immunized animals was transferred by intraperitoneal injection to naïve animals, which were challenged with LD90 of B/Ann Arbor/4/55 virus: 90% of the mice receiving immune serum were protected, compared to 10% in the control group (Fig. 5a).
FIG. 5.
Mechanism of protection. (a) Passive transfer of B/HA0 serum protects mice. The percentage of mice (10 animals/group) surviving challenge with LD90 of B/Ann Arbor/4/55 is graphed versus days postchallenge. Solid triangles, B/HA0 sera; solid circles, OMPC sera. (b) Protection by B/HA0 vaccine depends on the FcγR effector pathway. The effect of vaccination with B/HA0 against lethal challenge is compared in FcR(−) and normal FcR(+) BALB/c mice (10 animals/group). Solid triangles, BALB/c mice vaccinated with B/HA0. Open triangles, FcR(−) mice vaccinated with B/HA0. Solid circles, BALB/c mice vaccinated with OMPC; Open circles, FcR(−) mice vaccinated with OMPC. Vaccines (1 μg, formulated on Alum), were given together with 20 μg of QS21.
Second, we tested the protective efficacy of three anti-B/HA0 MAbs (unpublished data). Naïve mice were administered MAb ascites by intraperitoneal injection and challenged as indicated above; 70 to 100% of the mice receiving the MAbs were protected, compared to 10% in the control group (unpublished data). The study was repeated with two purified MAbs (D9E and O8F12), with identical results (unpublished data).
Major part of the protective antibody response targets the fusion peptide.
We then mapped the epitope of the three protective MAbs with a series of progressively N- and C-terminally truncated peptides. Remarkably, the same pattern was observed for all three MAbs: while truncation of several residues of the HA1 moiety was tolerated, truncation of even one C-terminal residue of the fusion peptide region completely ablated binding (unpublished data). To investigate the protective epitope in more detail, we prepared a panel of alanine-scanning mutants of the B/HA0 peptide and used a competitive fluorescence polarization immunoassay to establish the key binding determinants of the two most potent MAbs, O8F12 and D9E (Table 2). Both MAbs bind across the cleavage site, with only three critical residues: the arginine at the P1 position of the scissile bond (Arg8) and two phenylalanines in the fusion peptide moiety (Phe11 and Phe17). The two antibodies therefore bind the same residues, although differences in the binding affinities to each of the three critical amino acids were observed.
TABLE 2.
Relative binding affinity of MAbs D9E and O8F12 for single-point alanine mutants of the B/HA0 epitope
| Sequencea | Relative IC50b
|
|
|---|---|---|
| MAb D9E | MAb O8F12 | |
| PAKLLKER↓GFFGAIAGFLE | 1.0 | 1.0 |
| AAKLLKER↓GFFGAIAGFLE | 0.8 | 1.1 |
| PAALLKER↓GFFGAIAGFLE | 1.1 | 1.0 |
| PAKALKER↓GFFGAIAGFLE | 1.0 | 1.3 |
| PAKLAKER↓GFFGAIAGFLE | 1.0 | 0.9 |
| PAKLLAER↓GFFGAIAGFLE | 1.1 | 2.3 |
| PAKLLKAR↓GFFGAIAGFLE | 1.1 | 0.9 |
| PAKLLKEA↓GFFGAIAGFLE | 1,331.3 | 45.0 |
| PAKLLKER↓GAFGAIAGFLE | 1.5 | 1.2 |
| PAKLLKER↓GFAGAIAGFLE | 19,334.3 | 25,826.6 |
| PAKLLKER↓GFFGAAAGFLE | 0.9 | 1.9 |
| PAKLLKER↓GFFGAIAGALE | 14,635.1 | 288,386.0 |
| PAKLLKER↓GFFGAIAGFAE | 2.0 | 2.0 |
| PAKLLKER↓GFFGAIAGFLA | 1.1 | 1.0 |
All peptides have the additional N-terminal sequence biotin-6-aminohexanoic acid-Glu-Gly and an additional Glu residue at the C terminus.
IC50 = MAb concentration for 50% inhibition of binding of the reference peptide. Relative IC50 = IC50(mutant)/IC50(wild type).
The above data strongly suggest that in the B/HA0-OMPC conjugate, the most conserved part of the cleavage site, which corresponds to the scissile bond and the fusion peptide domain, is both the immunodominant and the key protective epitope.
Protection by HA0-based vaccine is Fc receptor mediated.
To further investigate the mechanism of protection by the B/HA0 vaccine, we immunized mice genetically deficient in the γ subunit of Fc receptor (FcR). These mice completely lack expression of FcγRI and FcγRIII and display severely impaired antibody-mediated immune responses, including loss of natural killer (NK) cell-mediated antibody-dependent cellular cytotoxicity, macrophage phagocytosis, and mast cell degranulation in response to FcR cross-linking (56). When the FcR(−)-vaccinated mice were challenged with LD90 of B/Ann Arbor/4/55 virus, their survival rate was comparable to that of nonimmunized controls (10 to 30%), while, as expected, 90% of the normal FcR(+) mice receiving the vaccine were protected (Fig. 5b). It therefore appears that the FcγR effector pathway is an important mechanism of protection of the humoral response against the HA0 vaccine.
Immunogenicity and protection of an HA0-based influenza A vaccine.
Having established the efficacy of an HA0-based vaccine for influenza B, we examined if the same approach could be extended to influenza A. In the past, partial (20 to 40%) protection from lethal viral challenge had been reported with a peptide spanning the HA0 sequence of influenza A subtype H1 (16, 35). In these studies, animals vaccinated with the free peptide developed GMT of 1 × 104 to 3 × 104. We investigated if a more immunogenic HA0-OMPC conjugate could provide greater efficacy. In a first experiment, we tested the protection afforded by a subtype H3 HA0 vaccine in a mouse challenge model (Table 1). BALB/c mice vaccinated with the A/H3/HA0-OMPC conjugate in the presence of QS21 developed very high titers against the HA0 peptide, with a GMT of 4.5 × 106. The higher antipeptide titers translated into increased protection: when the mice were challenged with LD90 of influenza A/H3 virus, the majority (70%) of the vaccinated mice survived, compared to none in the control group (Fig. 6a, top). Unlike what was seen for influenza B, vaccinated animals still suffered significant weight loss, albeit less severe than that of the controls (Fig. 6a, bottom). This indicates that the HA0 vaccine, while effective in reducing mortality, is not as efficacious in preventing morbidity, in analogy to M2-based vaccines (8).
FIG. 6.
HA0-based vaccine against influenza A. (a) A/H3/HA0 vaccine protects against challenge with the homologous subtype of influenza A. Mice (10 animals/group) were vaccinated with the A/H3/HA0 conjugate (1 μg) formulated with 20 μg of QS21, and challenged with LD90 of influenza A/Hong Kong/68 × PR8 reassortant (H3N1). Animals were monitored for survival (top) and weight change (bottom). Solid triangles, A/H3/HA0 vaccinated, solid circles, OMPC vaccinated.The dagger indicates that no animal in the group was alive at this time point. (b) A/H3/HA0 vaccine protects against challenge with the heterologous subtype of influenza A. Mice (10 animals/group) were vaccinated with the A/H3/HA0 conjugate (3 μg), and challenged with LD90 of influenza A/Puerto Rico/8/34 (H1N1). Solid triangles, A/H3/HA0 vaccinated; solid circles, OMPC vaccinated. The difference between the curves is statistically significant (P < 0.001).
We then examined the possibility that the HA0 vaccine could provide cross-protection against heterosubtypic viral challenge, a possibility suggested by the high degree of sequence homology between the H3 and H1 subtype consensus sequences (Table 1). Mice vaccinated with the A/H3/HA0 vaccine (reaching a GMT of 6.5 × 106) were indeed protected against LD90 challenge with the A/H1 subtype virus (Fig. 6b).
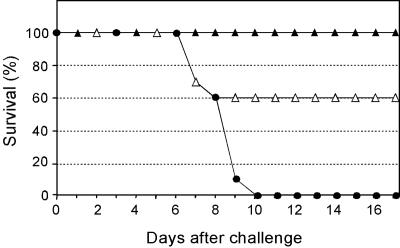
Sequence conservation of the fusion peptide moiety of HA0 between influenza A and B (Table 1), coupled to the observation that the protective response against influenza B appears to target this region of the cleavage site (Table 2), suggested a further experiment, in which we challenged mice, vaccinated with either the B/HA0 or the A/H3/HA0 conjugate, with influenza B virus. Although to a lower degree than mice vaccinated with the homologous vaccine, mice vaccinated with the influenza A vaccine were partially protected from mortality (Fig. 7). The reverse, however, was not true, since mice vaccinated with the B/HA0 vaccine were not protected against influenza A virus challenge (data not shown).
FIG. 7.
A/H3/HA0 vaccine protects against challenge with influenza B. Mice (10 animals/group) were vaccinated with the A/H3/HA0 conjugate (1 μg), the B/HA0 conjugate (1 μg), or OMPC, all formulated with 20 μg of QS21, and challenged with LD90 of B/Ann Arbor/4/55. Solid triangles, B/HA0 vaccinated, open triangles, A/H3/HA0 vaccinated, solid circles, OMPC vaccinated.
Anti-HA0 immune response in the human population.
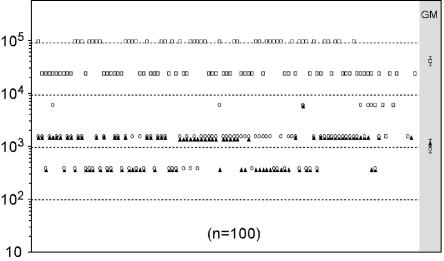
Finally, we investigated if anti-HA0 immunity is present in the human population due to natural exposure to the influenza virus. The analysis was performed by ELISA on a panel of 100 random adult human serum samples, which were tested for positivity for influenza A and B viruses by a commercial diagnostic kit. All subjects were positive for influenza A virus. We examined separately reactivity to A/H3/HA0 and A/H1/HA0 and, as a reference, reactivity to the influenza A virus nucleoprotein (NP) (Fig. 8). While the reactivity to NP ranged between 104 and 105 (geometric mean = 4 × 104), the geometric means of the titers to A/H3/HA0 and A/H1/HA0 were all <103. This level of reactivity is likely nonspecific, since when we analyzed a cohort of 94 sera for reactivity to B/HA0, we found the same geometric mean (≈103), and yet only two subjects were later found positive for influenza B virus infection (their B/HA0 titers were 25,600 and 640, respectively) (data not shown).
FIG. 8.
Prevalence of anti-cleavage site antibodies in the human population. One hundred serum samples from adult humans, all positive for prior exposure to influenza A virus, were tested in ELISA against the A/H1/HA0 (open circle), A/H3/HA0 (solid triangle) peptides, and as a reference antigen to the influenza A nucleoprotein (NP) (open square). The geometric means (GM) with standard errors are shown for each antigen.
In other terms, individuals seronegative for influenza B virus display the same (very low) level of reactivity to the B/HA0 cleavage site as individuals seropositive for influenza A virus do for the A/HA0 cleavage site. Moreover, when we examined the B/HA0 titers of naïve mice surviving challenge with influenza B virus (from the control groups of the challenge experiments), we found that none developed significant titers (GMT < 500) against the cleavage site (data not shown). Although clearly preliminary, these data do suggest that significant immunity against the cleavage site sequence is not usually elicited in the course of natural influenza virus infection. Similar data were reported for M2, i.e., the absence of a significant antibody response to M2 in mice infected with influenza virus (17, 33) and the complete (27) or partial (3) absence of M2 antibodies in humans naturally exposed to influenza A virus.
DISCUSSION
The influenza A and B viruses fluctuate continuously in prevalence (9), and it is not possible to predict which type (A or B) and/or subtype (A/H3 or A/H1) will dominate in a given year in a given geographic area. Accordingly, the conventional inactivated vaccines must be manufactured each year with the World Health Organization-recommended influenza A H1N1, influenza A H3N2, and influenza B strains. To allow enough time for manufacture, designation of the vaccine strains is made well in advance of the influenza season and is often problematic (48). Moreover, the currently available inactivated vaccines have several production limitations and do not allow a strategy of stockpiling vaccine doses, much as recommended for the available anti-influenza drugs (39). Intense research is focused to developing alternatives to the egg-based production system, including DNA vaccines and subunit vaccines produced in cell culture, but each suffers from various limitations (reviewed in reference 20).
The development of a “universal” vaccine is therefore a long-sought goal of influenza research. A universal vaccine should induce an immune response cross-protective against all circulating strains of both influenza A and B viruses. However, because of higher danger associated with influenza A virus infection (59), all the effort so far has been focused solely on the influenza A virus component, and no candidate universal influenza B vaccine is available today. Moreover, the most advanced candidates for influenza A all target the conserved M2 protein (8, 10, 18, 33, 38), a strategy which cannot be extended to influenza B (20).
In this manuscript we report the design and validation in a preclinical animal model of a universal influenza B vaccine. The epitope chosen for our vaccine is the intersubunit cleavage site of the HA0 precursor of the viral HA. Since HA0 cleavage occurs extracellularly in all human influenza viruses, it is an appropriate target for a humoral response. Because of its functional constraints, the epitope is extremely well conserved, providing the vaccine with broad specificity against all circulating influenza B viruses.
We have designed a subunit vaccine in which a peptide encompassing the consensus cleavage site sequence is conjugated to the carrier OMPC (8, 31, 58). Vaccines utilizing this carrier have been used extensively and are well tolerated in humans for carbohydrate epitopes (23).
We show that the proposed HA0 vaccine (i) is highly immunogenic, (ii) completely protects from mortality upon lethal viral challenge with viruses belonging to either one of the currently circulating influenza B lineages, (iii) provides protection from morbidity, as assessed from weight change in infected animals, and (iv) reduces viral replication in the lower respiratory tract of vaccinated animals.
We then show by passive transfer experiments that protection is mediated by antibodies. In addition, we have derived three anti B/HA0 MAbs, which are also able to protect by passive vaccination. These three MAbs all recognize a common epitope, which includes the P1 residue of the scissile bond, and two key amino acids in the fusion peptide moiety, indicating that in our vaccine this epitope is both immunodominant and protective. This finding is encouraging, since it is well established that even very conservative point mutations in the influenza virus fusion peptide dramatically alter the fusion phenotype (12, 43, 54) and detracts from the risk of inducing the formation of escape mutants by vaccination with HA0 conjugates.
Experiments with FcR(−) mice suggest that the B/HA0 antibodies elicited by the vaccine protect through indirect mechanisms, most likely by antibody-dependent cellular cytotoxicity (ADCC) rather than by direct blockage of the HA0 cleavage. NK-dependent ADCC has been reported as the mechanism of protection of the M2-based vaccine (17). Both antigens are abundantly expressed on the membrane of infected cells, with HA about fourfold higher than M2 (24). For the M2 vaccine, it has been argued that ADCC-dependent protection might be less efficacious than blockage of viral entry, which is exploited by the inactivated vaccines (17). In the mouse model, this difference translates into a different ability to prevent morbidity, as measured by the degree of weight loss following viral challenge. The B/HA0 vaccine effectively prevents weight loss, a feature more similar to the inactivated vaccines.
In conclusion, the B/HA0-OMPC conjugate can be proposed for clinical evaluation as a universal influenza B vaccine, with the most immediate use as a substitute for the influenza B virus component of the currently manufactured vaccines: it would provide the distinct advantages of eliciting protection against both circulating lineages of the virus while not requiring annual manufacturing updates.
Can the HA0 approach be extended to influenza A? Our preliminary experiments suggest a positive answer. By using an HA0 peptide from the consensus subtype H3 sequence in the form of an OMPC conjugate, we could induce a protective response against challenge with a homologous as well heterologous subtype virus, although protection against both mortality and morbidity appears considerably less robust than that obtained for influenza B. Our result nevertheless represent an improvement over the original observations with unconjugated HA0 peptide (16, 35), and careful optimization of peptide/carrier/adjuvant combinations might further improve the vaccine.
Although the reasons why the same approach gives better results for influenza B than for influenza A are presently unclear, we speculate that this may depend on the different structure and/or degree of exposure of the fusion peptide region of HA0 in the two viruses. If we map the key residues recognized by all three influenza B-protective MAbs (Table 2) onto the X-ray structure of influenza virus A/HA0 (5), we find that Arg8, Phe11, and Phe17 cannot form a common surface exposed to an antibody. On the other side, this epitope must be accessible in the intact influenza virus B/HA0 protein, since the MAbs are protective in vivo (unpublished data). Vaccination, as we do, with a linear form of HA0 might bias the immune response towards antibodies directed to this particular structure, which would be effective for influenza B but not for influenza A. The intriguing result that vaccination with the A/H3/HA0 conjugate provides some protection against challenge with influenza B virus (Fig. 7) is compatible with this hypothesis, since protection might be mediated by the antibody population directed to the epitope common to influenza A and B viruses. Moreover, the opposite should not occur, and indeed protection against influenza A virus challenge by vaccination with B/HA0 is not observed. Should this hypothesis be true, an A/HA0 vaccine based on a cyclic form of the peptide might be more efficacious.
At this stage, however, our A/HA0 vaccine could be proposed in combination with an M2-based vaccine, such as the M2-OMPC conjugate recently described by our laboratories (8). The presence of two different components against the influenza A virus would provide the potential for synergy and diminish the likelihood of simultaneous antigenic drift of both epitopes driven by an immune pressure apparently new to the human population (Fig. 8).
Further along this line, a triple-component conjugate vaccine (A/HA0, A/M2, and B/HA0) could potentially satisfy all the criteria for a truly universal influenza vaccine.
Finally, given the generality of the fusion mechanism mediated by class I fusion proteins such as HA (6, 52), the maturational cleavage site might be targeted for other viral vaccines.
Acknowledgments
We thank S. Jeffrey for some of the influenza B viral stocks, Y. Rippeon for assistance with vaccine formulations, and F. Bonelli for MS. We also thank J. Katz, Influenza Branch of CDC, who provided the B/Ann Arbor/55, B/Hong Kong/330/2001, and B/Yamanashi/166/1998 viral strains.
REFERENCES
- 1.Abbas, A. K., A. H. Lichtman, and J. S. Pober. 2000. Cellular and molecular immunology, 4th ed. WB Saunders Company, Philadelphia, PA.
- 2.Atherton, E., and R. C. Sheppard. 1989. Solid-phase peptide synthesis, a practical approach. IRL Press, Oxford, England.
- 3.Black, R., P. Rota, N. Gorodkova, H. Klenk, and A. Kendal. 1993. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J. Gen. Virol. 74:143-146. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1986-2002. Annual reports on influenza frequency. Morbid. Mortal. Wkly. Rep.
- 5.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 6.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 7.Evans, T. G., M. J. McElrath, T. Matthews, D. Montefiori, K. Weinhold, M. Wolff, M. C. Keefer, E. G. Kallas, L. Corey, G. J. Gorse, R. Belshe, B. S. Graham, P. W. Spearman, D. Schwartz, M. J. Mulligan, P. Goepfert, P. Fast, P. Berman, M. Powell, D. Francis, and N. A. V. E. Group. 2001. QS-21 promotes an adjuvant effect allowing for reduced antigen dose during HIV-1 envelope subunit immunization in humans. Vaccine 19:2080-2091. [DOI] [PubMed] [Google Scholar]
- 8.Fan, J., X. Liang, M. S. Horton, H. C. Perry, M. P. Citron, G. J. Heidecker, T.-M. Fu, J. Joyce, C. T. Przysiecki, P. M. Keller, V. M. Garsky, R. Ionescu, Y. Rippeon, L. Shi, M. A. Chastain, J. H. Condra, M.-E. Davies, J. Liao, E. A. Emini, and J. W. Shiver. 2004. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 22:2993-3003. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, N. M., A. P. Galvani, and R. M. Bush. 2003. Ecological and immunological determinants of influenza evolution. Nature 422:428-433. [DOI] [PubMed] [Google Scholar]
- 10.Fiers, W., M. De Filette, A. Birkett, S. Neirynck, and W. Min Jou. 2004. A “universal” human influenza A vaccine. Virus Res. 103:173-176. [DOI] [PubMed] [Google Scholar]
- 11.Gamblin, S. J., L. F. Haire, R. J. Russell, D. J. Stevens, B. Xiao, Y. Ha, N. Vasisht, D. A. Steinhauer, R. S. Daniels, A. Elliot, D. C. Wiley, and J. J. Skehel. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838-1842. [DOI] [PubMed] [Google Scholar]
- 12.Han, X., J. H. Bushweller, D. S. Cafiso, and L. K. Tamm. 2001. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 8:715-720. [DOI] [PubMed] [Google Scholar]
- 13.Hatta, M., and Y. Kawaoka. 2003. The NB protein of influenza B virus is not necessary for virus replication in vitro. J. Virol. 77:6050-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helting, T. B., and G. Guthöhrlein. 1981. Process for the isolation of membrane proteins from Neisseria meningitidis and vaccines containing same. June 1981. U.S. patent 4,271,147.
- 15.Hilleman, M. 2002. Realities and enigmas of human viral infuenza:pathogenesis, epidemiology and control. Vaccine 20:3068-3087. [DOI] [PubMed] [Google Scholar]
- 16.Horvath, A., G. K. Toth, P. Gogolak, Z. Nagy, I. Kurucz, I. Pecht, and E. Rajnavolgyi. 1998. A hemagglutinin-based multipeptide construct elicits enhanced protective immune response in mice against influenza A virus infection. Immunol. Lett. 60:127-136. [DOI] [PubMed] [Google Scholar]
- 17.Jegerlehner, A., N. Schmitz, T. Storni, and M. F. Bachmann. 2004. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 172:5598-5605. [DOI] [PubMed] [Google Scholar]
- 18.Jegerlehner, A., A. Tissot, F. Lechner, P. Sebbel, I. Erdmann, T. Kundig, T. Bachi, T. Storni, G. Jennings, P. Pumpens, W. A. Renner, and M. F. Bachmann. 2002. A molecular assembly system that renders antigens of choice highly repetitive for induction of protective B cell responses. Vaccine 20:3104-3112. [DOI] [PubMed] [Google Scholar]
- 19.Kanegae, Y., S. Sugita, A. Endo, M. Ishida, S. Senya, K. Osako, K. Nerome, and A. Oya. 1990. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J. Virol. 64:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemble, G., and H. Greenberg. 2003. Novel generations of influenza vaccines. Vaccine 21:1789-1795. [DOI] [PubMed] [Google Scholar]
- 21.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Klenk, H. D., R. Rott, M. Orlich, and J. Blodorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426-439. [DOI] [PubMed] [Google Scholar]
- 23.Kniskern, P. J., S. Marburg, and R. W. Ellis. 1995. Haemophilus influenzae type b conjugate vaccines. Pharm. Biotechnol. 6:673-694. [DOI] [PubMed] [Google Scholar]
- 24.Lamb, R. A., and R. M. Krug. 1996. Orthomyxoviridae: the viruses and their replication, p. 1353-1395. In P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 25.Lazarowitz, S. G., and P. W. Choppin. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68:440-454. [DOI] [PubMed] [Google Scholar]
- 26.Liu, G., C. Anderson, H. Scaltreto, J. Barbon, and C. R. Kensil. 2002. QS-21 structure/function studies: effect of acylation on adjuvant activity. Vaccine 20:2808-2815. [DOI] [PubMed] [Google Scholar]
- 27.Liu, W., H. Li, and Y. H. Chen. 2003. N-terminus of M2 protein could induce antibodies with inhibitory activity against influenza virus replication. FEMS Immunol. Med. Microbiol. 35:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. D. M. E. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza IV. Elsevier Science, Amsterdam, The Netherlands.
- 29.Marburg, S., D. Jorn, R. L. Tolman, B. Arison, J. McCauley, P. J. Kniskern, A. Hagopian, and P. P. Vella. 1986. Bimolecular chemistry of macromolecules: synthesis of bacterial polysaccharide conjugates with Neisseria meningitidis membrane protein. J. Am. Chem. Soc. 108:5282-5287. [Google Scholar]
- 30.Matsuzaki, Y., K. Sugawara, E. Takashita, Y. Muraki, S. Hongo, N. Katsushima, K. Mizuta, and H. Nishimura. 2004. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J. Med. Virol. 74:132-140. [DOI] [PubMed] [Google Scholar]
- 31.McGaughey, G. B., M. Citron, R. C. Danzeisen, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. Miller, J. Shiver, and M. J. Bogusky. 2003. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 42:3214-3223. [DOI] [PubMed] [Google Scholar]
- 32.Mould, J. A., R. G. Paterson, M. Takeda, Y. Ohigashi, P. Venkataraman, R. A. Lamb, and L. H. Pinto. 2003. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell 5:175-184. [DOI] [PubMed] [Google Scholar]
- 33.Mozdzanowska, K., J. Feng, M. Eid, G. Kragol, M. Cudic, L. Otvos, and W. Gerhard. 2003. Induction of influenza type A virus-specific resistance by immunization of mice with a synthetic multiple antigenic peptide vaccine that contains ectodomains of matrix protein 2. Vaccine 21:2616-2626. [DOI] [PubMed] [Google Scholar]
- 34.Murphy, B. R., and R. G. Webster. 1996. Orthomyxoviruses, p. 1397-1445. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincot-Raven, Philadelphia, Pa.
- 35.Nagy, Z., E. Rajnavolgyi, M. Hollosi, G. K. Toth, G. Varadi, B. Penke, I. Toth, A. Horvath, J. Gergely, and I. Kurucz. 1994. The intersubunit region of the influenza virus haemagglutinin is recognized by antibodies during infection. Scand. J. Immunol. 40:281-291. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa, N., R. Kubota, A. Maeda, and Y. Okuno. 2004. Influenza B virus Victoria group with a new glycosylation site was epidemic in Japan in the 2002-2003 season. J. Clin. Microbiol. 42:3295-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedrud, J. G., X. P. Liang, N. Hague, and M. E. Lamm. 1987. Combined oral/nasal immunization protects mice from Sendai virus infection. J. Immunol. 139:3484-3492. [PubMed] [Google Scholar]
- 38.Neirynck, S., T. Deroo, X. Saelens, P. Vanlandschoot, W. M. Jou, and W. Fiers. 1999. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 5:1157-1163. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson, K. G., J. M. Wood, and M. Zambon. 2003. Influenza. Lancet 362:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oxford, J. S., S. Bossuyt, S. Balasingam, A. Mann, P. Novelli, and R. Lambkin. 2003. Treatment of epidemic and pandemic influenza with neuraminidase and M2 proton channel inhibitors. Clin. Microbiol. Infect. 9:1-14. [DOI] [PubMed] [Google Scholar]
- 41.Palese, P., and A. Garcia-Sastre. 2002. Influenza vaccines: present and future. J. Clin. Investig. 110:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paterson, R. G., M. Takeda, Y. Ohigashi, L. H. Pinto, and R. A. Lamb. 2003. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology 306:7-17. [DOI] [PubMed] [Google Scholar]
- 43.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rota, P. A., M. L. Hemphill, T. Whistler, H. L. Regnery, and A. P. Kendal. 1992. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J. Gen. Virol. 73:2737-2742. [DOI] [PubMed] [Google Scholar]
- 46.Rota, P. A., T. R. Wallis, M. W. Harmon, J. S. Rota, A. P. Kendal, and K. Nerome. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59-68. [DOI] [PubMed] [Google Scholar]
- 47.Sato, M., S. Yoshida, K. Iida, T. Tomozawa, H. Kido, and M. Yamashita. 2003. A novel influenza A virus activating enzyme from porcine lung: purification and characterization. Biol. Chem. 384:219-227. [DOI] [PubMed] [Google Scholar]
- 48.Shaw, M., X. Xu, Y. Li, S. Normand, R. T. Ueki, G. Y. Kunimoto, H. Hall, A. Klimov, N. J. Cox, and K. Subbarao. 2002. Reappearance and global spread of variants of influenza B/Victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology 303:1-8. [DOI] [PubMed] [Google Scholar]
- 49.Shuler, K. R., R. G. Dunham, and P. Kanda. 1992. A simplified method for determination of peptide-protein molar ratios using amino acid analysis. J. Immunol. Methods 156:137-149. [DOI] [PubMed] [Google Scholar]
- 50.Simonsen, L., K. Fukuda, L. B. Schonberger, and N. J. Cox. 2000. The impact of influenza epidemics on hospitalizations. J. Infect. Dis. 181:831-837. [DOI] [PubMed] [Google Scholar]
- 51.Skehel, J. J., and M. D. Waterfield. 1975. Studies on the primary structure of the influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 72:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 53.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 54.Steinhauer, D. A., S. A. Wharton, J. J. Skehel, and D. C. Wiley. 1995. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J. Virol. 69:6643-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens, J., A. L. Corper, C. F. Basler, J. K. Taubenberger, P. Palese, and I. A. Wilson. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866-1870. [DOI] [PubMed] [Google Scholar]
- 56.Takai, T., M. Li, D. Sylvestre, R. Clynes, and J. V. Ravetch. 1994. FcR gamma chain deletion results in pleiotrophic effector cell defects. Cell 76:519-529. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda.2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179-186. [DOI] [PubMed] [Google Scholar]
- 58.Tolman, R. L., M. A. Bednarek, B. A. Johnson, W. J. Leanza, S. Marburg, D. J. Underwood, E. A. Emini, and A. J. Conley. 1993. Cyclic V3-loop-related HIV-1 conjugate vaccines. Synthesis, conformation and immunological properties. Int. J. Peptide Protein Res. 41:455-466. [DOI] [PubMed] [Google Scholar]
- 59.Webby, R. J., and R. G. Webster. 2003. Are we ready for pandemic influenza? Science 302:1519-1522. [DOI] [PubMed] [Google Scholar]
- 60.Zhirnov, O. P., I. V. Vorobjeva, A. V. Ovcharenko, and H. D. Klenk. 2003. Intracellular cleavage of human influenza A virus hemagglutinin and its inhibition. Biochemistry 68:1020-1026. [DOI] [PubMed] [Google Scholar]