Abstract
Respiratory syncytial virus (RSV) produces intense pulmonary inflammation, in part through its ability to induce chemokine synthesis in infected airway epithelial cells. RANTES (regulated upon activation, normally T-cell expressed and presumably secreted) is a CC chemokine which recruits and activates monocytes, lymphocytes, and eosinophils, all cell types present in the lung inflammatory infiltrate induced by RSV infection. In this study, we analyzed the mechanism of RSV-induced RANTES promoter activation in human type II alveolar epithelial cells (A549 cells). Promoter deletion and mutagenesis experiments indicate that RSV requires the presence of five different cis regulatory elements, located in the promoter fragment spanning from −220 to +55 nucleotides, corresponding to NF-κB, C/EBP, Jun/CREB/ATF, and interferon regulatory factor (IRF) binding sites. Although site mutations of the NF-κB, C/EBP, and CREB/AP-1 like sites reduce RSV-induced RANTES gene transcription to 50% or less, only mutations affecting IRF binding completely abolish RANTES inducibility. Supershift and microaffinity isolation assays were used to identify the different transcription factor family members whose DNA binding activity was RSV inducible. Expression of dominant negative mutants of these transcription factors further established their central role in virus-induced RANTES promoter activation. Our finding that the presence of multiple cis regulatory elements is required for full activation of the RANTES promoter in RSV-infected alveolar epithelial cells supports the enhanceosome model for RANTES gene transcription, which is absolutely dependent on binding of IRF transcription factors. The identification of regulatory mechanisms of RANTES gene expression is fundamental for rational design of inhibitors of RSV-induced lung inflammation.
Respiratory syncytial virus (RSV) is an enveloped, negative-sense single-stranded RNA virus (18). Since its isolation, RSV has been identified as a leading cause of epidemic respiratory tract illness in children in the United States and worldwide. In fact, RSV is so ubiquitous that it will infect 100% of children before the age of 3 (15). In infants and young children, RSV is the most common etiologic agent of bronchiolitis and is also responsible for 50% of pneumonia cases in children up to 2 years of age (38). Each year approximately 100,000 children are hospitalized with RSV disease, with an estimated annual cost close to $300 million in the United States alone (15, 17).
The main targets of RSV infection are respiratory epithelial cells. In bronchiolitis and pneumonia, RSV antigen can be identified in epithelial cells from throughout the lower respiratory tract, with less virus found in lungs of children with bronchiolitis than in lungs of children with pneumonia, where large amounts of viral antigen are detected. Necrosis of the airway epithelium is associated with mononuclear cell infiltration, mainly peribronchial and perivascular in bronchiolitis, and between the interalveolar walls, leading to alveolar filling, in pneumonia (reviewed in reference 38). Moreover, the presence of cell-specific inflammatory mediators in nasopharyngeal secretions and in tracheobronchial aspirates of children with bronchiolitis suggests that RSV infection triggers the migration to the airways and local activation of eosinophil and basophil leukocytes (11, 13).
Much of the cellular response at sites of tissue inflammation is controlled by gradients of chemotactic factors that direct leukocyte transendothelial migration and movement through the extracellular matrix. The composition of this cellular response is dependent on the discrete target cell selectivity of these chemotactic molecules. Chemokines, a family of small chemotactic cytokines, regulate the migration and activation of leukocytes and therefore play a key role in inflammatory and infectious processes of the lung (29). RANTES (regulated upon activation, normal T-cell expressed and presumably secreted) is a member of the CC branch of the chemokine family and is strongly chemotactic for T lymphocytes, monocytes, basophils, and eosinophils (2), all cell types which are present or activated in the inflammatory infiltrate that follows RSV infection of the lung.
Recent in vivo studies have shown elevated RANTES concentrations in nasal washes of children infected with RSV (11a, 42, 44). Several reports have shown that epithelial cells are a major source of RANTES in the lung (3, 16). This observation is particularly relevant to viral infections, since respiratory epithelial cells are the primary targets of viruses that enter the airways. We have recently demonstrated in an in vitro model that RSV is a potent stimulus for RANTES production in cultured human nasal, bronchial, and alveolar epithelial cells (34). Synthesis of RANTES was dose and time dependent and required replicating virus.
Mechanisms for inducible RANTES gene expression following viral infections have not been fully elucidated. Thomas et al. (45) have shown that the transcription factor NF-κB plays an important role in RSV-inducible RANTES production in airway epithelial cells. Lin et al. have recently identified the essential role played by interferon (IFN) regulatory factor 3 IRF-3 and the cooperation existing between IRF-3 and NF-κB in activation of RANTES transcription following infection with Sendai virus (14, 26). However, a complete analysis of the promoter cis regulatory elements and nuclear factors involved in regulation of RANTES gene transcription following viral infection of epithelial cells has not been done. Therefore, we have investigated the mechanisms of RSV-induced RANTES transcription in A549 cells, a cell line retaining features of human alveolar type II epithelial cells, which is a widely accepted model for studying RSV-epithelial cell interactions (7, 9, 10, 12, 20). Our results indicate that RSV-induced RANTES transcription requires multiple cis regulatory elements corresponding to the NF-κB and NF-IL6 binding sites and to the cyclic AMP-responsive element (CRE) and IFN-stimulated response element (ISRE). The NF-IL6 site binds nuclear proteins in a constitutive manner, while all of the other three cis regulatory elements of the promoter bind transcription factors in an RSV-inducible manner. Identification of the molecular mechanisms involved in RANTES gene expression is fundamental for developing strategies to modulate the inflammatory response associated with RSV infection of the lung.
MATERIALS AND METHODS
RSV preparation.
The human Long strain of RSV (A2) was grown in HEp-2 cells and purified by centrifugation on discontinuous sucrose gradients as described elsewhere (47). The virus titer of the purified RSV pools was 7.5 to 8.5 log10 PFU/ml in a methylcellulose plaque assay. No contaminating cytokines, including interleukin-1 (IL-1), tumor necrosis factor, IL-6, IL-8, granulocyte-macrophage colony-stimulating factor, and IFN were found in these sucrose-purified viral preparations (36). Lipopolysaccharide, assayed using the limulus hemocyanin agglutination assay, was not detected. Virus pools were aliquoted, quick-frozen on dry ice-alcohol, and stored at −70°C until used.
Cell culture and infection of epithelial cells with RSV.
A549 and 293 cells, a human embryonic kidney epithelial cell line (American Type Culture Collection, Manassas, Va.), were maintained in F12K and minimal essential medium respectively, containing 10% (vol/vol) fetal bovine serum 10 mM glutamine, penicillin (100 IU/ml), and streptomycin (100 μg/ml). Cell monolayers were infected with RSV at a multiplicity of infection (MOI) of 1 (unless otherwise stated) as described elsewhere (12). An equivalent amount of a 20% sucrose solution was added to uninfected A549 cells as a control.
Northern blotting.
Total RNA was extracted from control and infected A549 cells by the acid guanidium thiocyanate-phenol-chloroform method (40). Twenty micrograms of RNA was fractionated on a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to a radiolabeled RANTES cDNA (a generous gift from A. Krensky, Stanford, Calif.), as previously described (4). After washing, the membrane was exposed for autoradiography on Kodak XAR film at −70°C, using intensifying screens. The membrane was stripped and reprobed for β-actin, as an internal control for equal loading of the samples.
Plasmid construction.
5′-deletion constructs of the human RANTES promoter were produced by PCR using as template the full-length human RANTES promoter from nucleotides (nt) −974 to +55 relative to the mRNA start site, designated +1 (a generous gift from A. Krensky), cloned into the pGL2 vector (pGL2-974). Upstream primers, incorporating a unique KpnI restriction site, were designed to produce 5′ deletions at nt −400, 300, 220, 195, 150, and 120, while the downstream oligonucleotide, containing a unique HindIII restriction site, was designed to hybridize from nt +30 to +55. The PCR products were restricted with KpnI and HindIII, gel purified, cloned into the luciferase reporter gene vector pGL2 (Promega, Madison, Wis.), and named pGL2-400, pGL2-300, pGL2-220, pGL2-195, pGL2-150, and pGL2-120.
Site-directed mutations of the RANTES promoter were introduced by the PCR overlap extension mutagenesis technique (7), using pGL2-220 as the template and mutagenic primers identical to the oligonucleotides used in electrophoretic mobility shift assay (EMSA) (Table 1), excluding the 5′ GATC sequence. The site mutations introduced in the promoter correspond to mutations shown to be able to disrupt binding of the relevant transcription factor in EMSA. Mutations of the ISRE, NF-κB1, and NF-κB2 binding sites corresponded to the ones previously described (26, 31). Mutations of the CRE and NF-IL6 sites were introduced by replacing a purine with a noncomplementary pyrimidine.
TABLE 1.
cis regulatory elements of the RANTES promoter analyzed in this study
| cis-responsive elements | Positiona | Oligonucleotide used in EMSAb
|
|
|---|---|---|---|
| Type | Sequence | ||
| NF-κB1 RANTES | −71 to −53 | WT | 5′GATCCATTTT GGAAACTCCCCTTAT 3′ |
| 3′ TAAAACCTTTGAGGGGAATATCTAG 5′ | |||
| MUT | 5′GATCCATTTT GGcAcCTtaaCg TAT 3′ | ||
| 3′TAAAACCgTgGAattGcATATCTAG 5′ | |||
| NF-κB2 RANTES | −57 to −39 | WT | 5′GATCCATTAGGGGATGCCCCTCAT 3′ |
| 3′TAAAACCgTgGAattGcATATCTAG 5′ | |||
| MUT | 5′GATCCATTAcGccATGCatCTCAT 3′ | ||
| 3′TAATgCggTACGtaGAGTACTAG 5′ | |||
| NF-IL6 | −120 to −100 | WT | 5′GATCCAGTTTTGTGCAATTTCGA 3′ |
| 3′TCAAAACACGTTAAAGCTCTAG 5′ | |||
| MUT | 5′GATCCAGTTgTt Tta ccTTTCGA 3′ | ||
| 3′TCAAcAaAatggAAAGCTCTAG 5" | |||
| ISRE | −138 to −117 | WT | 5′GATCCATATTTCAG TTTT CTTTT CCGT 3′ |
| 3′TATAAAGTCAAAAGAAAAGGCATCTAG 5′ | |||
| MUT | 5′GATCCATATTTCAGTaaaCTaaaCCGT 3′ | ||
| 3′TATAAAGTCAtttGA ttt GGCATCTAG 5′ | |||
| CRE | −210 to −180 | WT | 5′GATCCAAAGAGGAAACTGATGAGCTCACTC TAGAT 3′ |
| 3′TTT CTCC TTTGACTACTC GAGTGAGATCTATCTAG 5′ | |||
| MUT | 5′GATCCAAAGAGGAAACT tcT tAtagacCgCTAGAT 3′ | ||
| 3′TTT CTCC TTT GAagAaTatctg GcGATCTATCTAG 5′ | |||
Relative to the transcription start site.
Mutated nucleotides are in lowercase. WT, wild type; MUT, mutant.
The phosphorylation-defective IκB-α mutant, which has serine residues 32 and 36 replaced with alanine, was produced by PCR using the upstream mutagenic primer 5′-TTTCATGGCGTCCAGGCCGGCGTCGTGGCGGTCGTC-3′ and the downstream mutagenic primer 5′-CGCCACGACGCCGGCCTGGACGCCATGAAAGAC-3′. The PCR product was then ligated into the BamHI/HindIII-restricted pcDNA3 vector.
The eukaryotic expression vector expressing NF-IL6 was produced in two steps. A synthetic oligonucleotide containing the sequence 5′-AGCTTGCCGCCACCATGGGCAACTGCGCGTGGAG-3′ was annealed to 5′-AATTCTCACGCGCAGTTGCCCATGGTGGCGGCA-3′ and ligated into HindIII-EcoRI-digested pGEM7Z (Promega), creating pGEM7Zad. pGEM7Zad contains an NcoI site (underlined) containing an initiation codon (bold) downstream of a consensus Kozak initiator sequence. The coding sequences for the major translation product of NF-IL6 (1) encoding amino acids 24 to 245 was excised as an Nco-EcoRI fragment and cloned into pGEM7Zad. The modified NF-IL6 coding sequences including the ribosomal binding sequences were then excised with HindIII and EcoRI and ligated into pcDNAI, producing pcDNAI-NF-IL6 an expression vector producing human NF-IL6 under control of the cytomegalovirus (CMV) promoter.
All plasmids were purified by ion exchange (Endo-Free Qiagen kit; Qiagen, Chatsworth, Calif.) and sequenced, prior to transfection, by the dideoxy-chain termination method using a Sequenase version 2.0 kit (Amersham International).
Plasmids expressing c-Jun (a generous gift from M. J. Birrer), IRF-1, IRF-3, and IRF-7 dominant negative mutants have been previously described (5, 26, 4).
Cell transfection.
Logarithmically growing A549 cells were transfected in triplicate in 60-mm-diameter petri dishes by using DEAE-dextran as previously described (7). Cells were incubated in 2 ml of HEPES-buffered Dulbecco modified Eagle medium (10 mM HEPES, pH 7.4) containing 20 μl of DEAE-dextran (60 mg/ml; Pharmacia) premixed with 6 μg of RANTES-pGL2 plasmids and 1 μg of CMV-β-galactosidase internal control plasmid. After 3 h, the medium was removed and 0.5 ml of 10% (vol/vol) dimethyl sulfoxide in phosphate-buffered saline was added to the cells for 2 min. Cells were washed with phosphate-buffered saline PBS and cultured overnight in 10% fetal bovine serum–Dulbecco modified Eagle medium. The next morning, cells were infected with RSV; at different time postinfection, cells were lysed to measure independently luciferase and β-galactosidase reporter activity as previously described (4). Luciferase activity was normalized to the internal control β-galactosidase activity. When the dominant negative (DN) expression plasmids were used, 293 or A549 cells were transfected using FuGene 6 (Roche, Indianapolis, Ind.). Two-microgram aliquots of pGL2-220 plasmid and different amounts of the DN mutant expression plasmids were premixed with FuGene6 in a 1:3 (μg/μl) ratio and added to the cells in 3 ml of regular medium. The next morning, cells were infected with RSV; 24 h later, cells were lysed to measure luciferase and β-galactosidase reporter activities. All experiments were performed in duplicate or triplicate.
EMSA.
Nuclear extracts of uninfected and infected A549 cells were prepared using hypotonic/nonionic detergent lysis as previously described (7). Proteins were normalized by protein assay (Bio-Rad, Hercules, Calif.) and used to bind to duplex oligonucleotides corresponding to the RANTES CRE, ISRE, NF-IL6, and NF-κB1 and NF-κB2 wild-type and mutated binding sites. Sequences of the oligonucleotides used for EMSA are shown in Table 1. Nuclear extracts, used for binding to the CRE site, were prepared from control and infected A549 cells that had been serum starved, before and throughout the period of infection, for a total of 36 h.
DNA binding reactions using the CRE, ISRE, and NF-IL6 probes contained 10 to 15 μg of nuclear protein, 5% glycerol, 12 mM HEPES, 80 mM NaCl, 5 mM dithiothreitol (DTT), 5 mM Mg2Cl, 0.5 mM EDTA, 1 μg of poly(dI-dC), and 40,000 cpm of 32P-labeled double-stranded oligonucleotide in a total volume of 20 μl. DNA binding reactions using the NF-κB1 and -2 probes contained 10 to 15 μg of total protein, 5% glycerol, 12 mM HEPES, 80 mM NaCl, 5 mM DTT, 1 μg of poly(dA-dT), and 40,000 cpm of 32P-labeled double-stranded oligonucleotide in a total volume of 20 μl. The nuclear proteins were incubated with the probe for 15 min at room temperature and then fractionated by 6% nondenaturing polyacrylamide gel electrophoresis (PAGE) in Tris-borate-EDTA buffer (22 mM Tris-HCl, 22 mM boric acid, 0.25 mM EDTA [pH 8]). After electrophoretic separation, gels were dried and exposed for autoradiography on Kodak XAR film at −70°C, using intensifying screens. In competition assays, 2 pmol of unlabeled wild-type or mutated competitor was added at the time of probe addition. In the gel mobility supershift assay, commercial antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) against specific transcription factors were added to the binding reactions and incubated on ice for 1 h prior to fractionation by 6% PAGE. For the CRE supershift assay, we used antibodies broadly reactive with different members of the ATF/CREB or AP-1 family. Anti-CREB-1 (sc-186) recognizes also ATF-1 and CREM-1; anti-c-Fos (sc-413) recognizes c-Fos, Fos-B, Fra-1, and Fra-2; anti-c-Jun (sc-44) recognizes c-Jun, Jun-B, and Jun-D. Preimmune serum was used as a control for any nonspecific effects of the immune antisera.
Microaffinity isolation assay.
Microaffinity purification of proteins binding to the RANTES ISRE was performed by a two-step biotinylated DNA-streptavidin capture assay (7). In this assay, duplex oligonucleotides are chemically synthesized containing 5′ biotin on a flexible linker (Genosys, The Woodlands, Tex.). Four hundred micrograms of 12-h-infected A549 cells nuclear extracts were incubated at 4°C for 30 min with 50 pmol of biotinylated ISRE, in the absence or presence of a 10-fold molar excess of nonbiotinylated wild-type or mutated ISRE. The binding buffer contained 8 μg of poly (dI/dC) (as nonspecific competitor) and 5% (vol/vol) glycerol, 12 mM HEPES, 80 mM NaCl, 5 mM DTT, 5 mM Mg2Cl, and 0.5 mM EDTA. One hundred microliters of a 50% slurry of prewashed streptavidin-agarose beads was then added to the sample, which was incubated at 4°C for an additional 20 min with gentle rocking. Pellets were washed twice with 500 μl of binding buffer, and the washed pellets were resuspended in 100 μl of 1× sodium dodecyl sulfate (SDS)-PAGE buffer, boiled, and fractionated on an SDS–10% polyacrylamide gel. After SDS-PAGE separation, proteins were transferred to polyvinylidene difluoride membrane for Western blot analysis.
Western immunoblotting.
Nuclear and cytoplasmic proteins were prepared as previously described (7), fractionated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% milk in Tris-buffered saline–Tween and incubated with rabbit polyclonal antibodies to c-Jun, IRF-3, and IRF-7 (Santa Cruz Biotechnology). For secondary detection, we used a horseradish peroxidase-coupled donkey anti-rabbit antibody in the enhanced chemiluminescence assay (Amersham Life Sciences, Arlington Heights, Il.).
Statistical analysis.
Data from experiments involving multiple samples subject to each treatment were analyzed by the Student-Newman-Keuls t test for multiple pairwise comparisons. Results were considered significantly different at a P value of <0.05.
RESULTS
RSV infection induces RANTES gene expression in A549 cells.
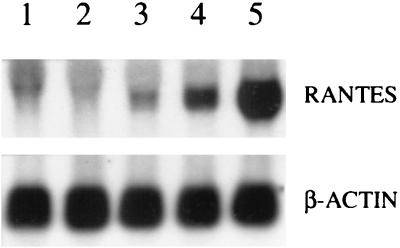
We previously reported that RSV infection of normal human nasal, bronchial, and small alveolar epithelial cells induced RANTES secretion (34, 39). Infection of A549 cells, a lung carcinoma cell line that retains features of well-differentiated lung type II alveolar epithelial cells, also induced increased RANTES protein release, whose synthesis was dose and time dependent and required replicating virus (34). To determine if the increased protein release paralled an increase in the steady-state level of RANTES mRNA, A549 cells were infected with RSV, and total RNA was extracted from control and infected cells at different times postinfection for Northern blot analysis. A small increase in RANTES mRNA expression was first detected at 6 h postinfection, with maximal induction at 24 h (Fig. 1). There was no further increase in mRNA levels at later time points (data not shown). These data indicate that RANTES gene expression and protein secretion are coupled in A549 cells.
FIG. 1.
Northern blot of RANTES mRNA in RSV-infected A549 cells. A549 cells were infected with RSV (MOI of 1) for various lengths of time (lane 1, uninfected cells; lane 2, 3 h postinfection; lane 3, 6 h postinfection; lane 4, 12 h postinfection; lane 5, 24 h postinfection). Total RNA was extracted from control and infected cells, and 20 μg of RNA was fractionated on a 1.2% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to a radiolabeled RANTES or β-actin cDNA probe.
RSV infection induces RANTES promoter activity in a time- and dose-dependent manner.
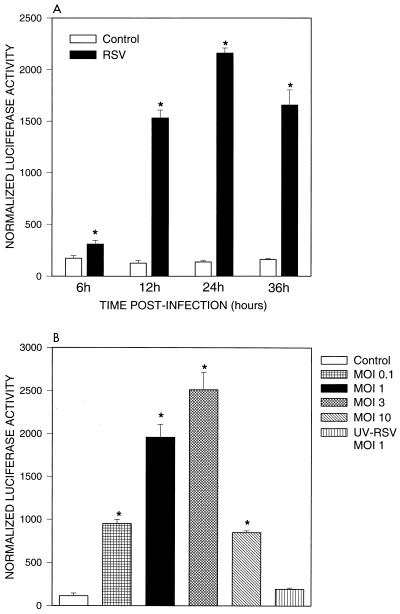
To determine whether RSV infection of alveolar epithelial cells was able to induce RANTES gene transcription, we transiently transfected A549 cells with a construct containing the first 974 nt of the human RANTES promoter linked to the luciferase reporter gene (pGL2-974). A schematic diagram of the promoter construct is shown in Fig. 2. Previous studies have shown that this fragment of the promoter is sufficient to drive regulated luciferase expression in a variety of cell types (30–32). As shown in Fig. 3A, RSV infection of transfected A549 cells induced a time-dependent increase of luciferase activity, compared to uninfected cells, that started at 6 h and peaked around 24 h postinfection, slightly decreasing at 36 h, a kinetic profile identical to that of RSV-induced RANTES mRNA accumulation. The RSV-induced promoter activation was dose dependent, with maximal stimulation seen at an MOI of 3 (Fig. 3B), corresponding to the situation where 99% of the cells are initially RSV infected. Higher MOIs result in lower degree of luciferase activity, perhaps due to the effect of defective interfering particles. UV-inactivated virus was unable to induce RANTES transcription, confirming our previous observations that RANTES secretion requires replicating virus (34).
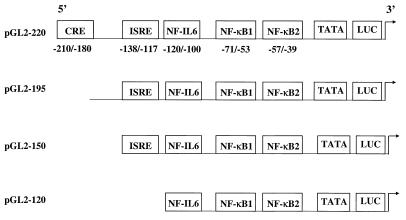
FIG. 2.
Schematic representation of RANTES promoter deletion constructs. Locations of the putative binding sites for CRE, ISRE, NF-IL6, and NF-κB are illustrated. Numbering is relative to the transcription initiation site.
FIG. 3.
RANTES promoter activation following RSV infection. (A) Time course. A549 cells were transiently transfected with pGL2-974 and infected with RSV (MOI of 1). At different times postinfection, cells were harvested to measure luciferase activity. Uninfected plates served as controls. For each plate, luciferase activity was normalized to β-galactosidase reporter activity. Data are expressed as mean ± standard deviation of normalized luciferase activity. ∗, P < 0.01 relative to mock-infected plates. (B) Effects of different MOIs. A549 cells were transiently transfected with pGL2-974 and infected with RSV at MOIs of 0.1, 1, 3, and 10; 24 h after infection, cells were harvested to measure luciferase activity. Uninfected plates served as controls. For each plate, luciferase activity was normalized to β-galactosidase reporter activity. Data are expressed as mean ± standard deviation of normalized luciferase activity. ∗, P < 0.01 relative to mock-infected plates.
Effects of 5′ deletions and site mutations of the RANTES promoter sequence on RSV-inducible activity.
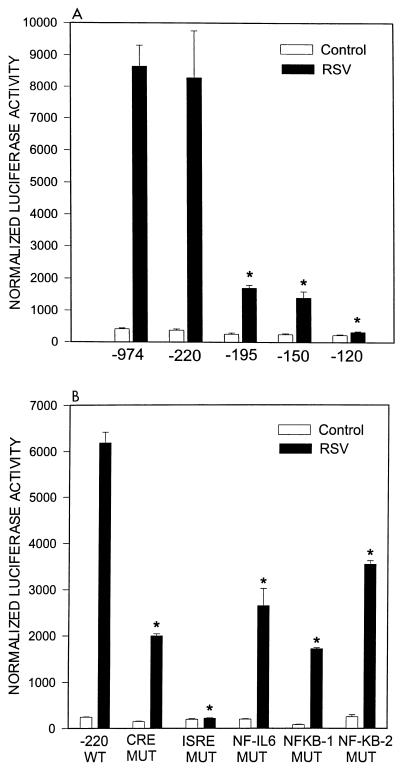
To define the regions of the RANTES promoter involved in regulating gene expression after RSV infection, A549 cells were transiently transfected with plasmids containing serial 5′ to 3′ deletions of the RANTES promoter linked to the luciferase reporter gene. Cells were infected with RSV and harvested at 24 h postinfection (conditions corresponding to peak reporter gene induction) to measure luciferase activity. As shown in Fig. 4A, deletions from nt −974 to −220 did not affect basal or RSV-inducible luciferase activity. Further deletion to nt −195 reduced the basal activity of the promoter by ∼30% and significantly reduced the RSV-induced luciferase activity by ∼60 to 70%, indicating that the sequence between nt −220 and −195 is critically involved in RANTES promoter activation. Deletion to nt −150 did not further change promoter activity, while deletion to nt −120 completely abolished RSV-induced luciferase activity. Computer analysis of the RANTES promoter and very recent studies (23) have identified a CRE site in the region from nt −220 to −190 and an ISRE site in the region from nt −150 to −120 of the promoter. To establish the role of the CRE and ISRE sites of the RANTES promoter in conferring responsiveness to RSV infection, we tested the effects of point mutations of these sites in the context of the minimal RANTES promoter fragment (nt −220) that retains full RSV inducibility. As shown in Fig. 4B, mutation of the CRE site affected both basal activity and RSV inducibility of the promoter to the same extent of deletion of the promoter region spanning nt −220 and −195. Mutation of the ISRE did not affect the basal activity but it completely abolished RSV-induced promoter activation, as did deletion of the promoter region spanning from nt −195 to −150. Together, the data of the 5′-deletion and site mutation analysis indicate that the ISRE is indeed a responsive element required for RSV-induced RANTES activation. Previous studies have indicated that the NF-IL6 and two NF-κB binding sites located between nt −110 and −30 of the promoter can play an important role in RANTES gene transcription (30, 35). Although the 5′-deletion analysis indicates that these sequences are not sufficient by themselves to confer RSV inducibility (Fig. 4A), we have previously shown that RSV is a strong stimulus of NF-IL6 expression and NF-κB nuclear translocation (12, 21). To determine their role, if any, in RANTES expression, we introduced site-directed mutations in each of the binding sites and tested the mutant plasmids for RSV inducibility. The proximal NF-κB site was defined as NF-κB2, while the distal NF-κB site was defined as NF-κB1 (Fig. 2). As shown in Fig. 4B, mutation of NF-κB1 affected the promoter basal activity and greatly reduced RSV-induced promoter activation. Mutation of NF-IL6 and NF-κB2 sites also decreased the RSV-induced luciferase activity, although to a lesser extent.
FIG. 4.
Effects of 5′ deletions and site mutations in the RANTES promoter sequence on RSV-inducible activity. (A) A549 cells were transiently transfected with 5′ deletions of the human RANTES promoter and infected with RSV for 24 h at an MOI of 1. Uninfected plates served as controls. For each plate, luciferase activity was normalized to β-galactosidase reporter activity. Data are expressed as mean ± standard deviation of normalized luciferase activity. ∗, P < 0.01 from pGL2-974. (B) A549 cells were transiently transfected with site-mutated (MUT) plasmids of the pGL2-220 RANTES promoter and infected with RSV for 24 h. Uninfected plates served as controls. For each plate, luciferase activity was normalized to β-galactosidase reporter activity. Data are expressed as mean ± standard deviation of normalized luciferase activity. ∗, P < 0.01 relative to -220 wild type (WT).
Role of CRE and ISRE binding proteins in RANTES promoter inducibility following RSV infection.
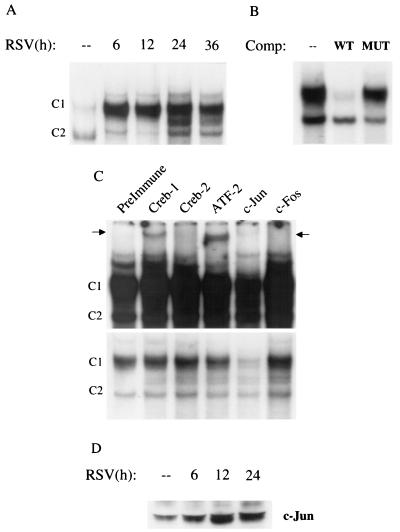
To determine whether RSV infection produced changes in the abundance of DNA binding proteins recognizing the RANTES CRE site, EMSA was performed on nuclear extracts prepared from control and RSV-infected A549 cells. The CRE sites can bind homo- and heterodimeric complexes formed by members of the CREB, ATF, Jun, and Fos transcription factor families. To exclude uncontrolled effects due to the presence of serum on c-Fos and Jun expression (24), serum-starved cells were infected with RSV and harvested at different time points to prepare nuclear extracts. As shown in Fig. 5A, two binding complexes, C1 and C2, were detected in control A549 cells; RSV infection markedly increased C1 binding, detectable at 6 h and persisting for the duration of the experiments. The inducible C1 complex was sequence specific, as demonstrated by its competition by an unlabeled wild-type but not mutant oligonucleotide (Fig. 5B). To determine the composition of the RSV-inducible complex, we performed supershift assays using a panel of antibodies broadly reacting with the different members of CREB, ATF, Fos, and Jun families of transcription factors (see Materials and Methods). The anti-Jun antibody induced the complete disappearance of C1, as shown by the lighter exposure of Fig. 5C, indicating that members of the Jun family are a component of the CRE complex induced by RSV infection. The anti-CREB-1 and anti-ATF-2 antibodies, although they did not cause a reduction of the same complex, induced the appearance of a supershifted band (darker exposure in Fig. 5C), suggesting that these members of the CREB/ATF family are also minor components of the RSV-inducible CRE complex. Activation and therefore nuclear translocation of c-Jun were also confirmed by Western blotting. Increased amounts of c-Jun were detected in nuclear extracts of infected A549 cells, compared to control cells, as shown in Fig. 5D.
FIG. 5.
EMSA of RANTES CRE binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from serum-starved control and RSV-infected A549 cells at the indicated times and used for EMSA. Time following RSV infection is indicated at the top. Two DNA-protein complexes, C1 and C2, are detected in control cells. C1 binding is further increased by RSV infection, while C2 binding is not. (B) Competition (Comp) analysis. Nuclear extracts from 24-h-infected cells were used to bind to the CRE probe in the absence (–) or presence of 2 pmol of wild-type (WT) or mutated (MUT) unlabeled competitor in the binding reaction, as indicated at top. C1 is competed by the wild-type oligonucleotide but not by the mutant one, indicating binding specificity. (C) Supershift interference assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum and anti-Jun, Fos, CREB-1, -CREB-2, and -ATF-2 antibodies. Top, long exposure showing the presence of supershifted bands (indicated by the arrows) induced by the anti-CREB-1 and anti-ATF-2 antibodies; bottom, light exposure showing the disappearance of C1 induced by the anti-Jun antibody. (D) Western blot of c-Jun in A549 cells infected with RSV. A549 cells were infected with RSV (MOI of 1) for various lengths of time. Nuclear extracts were prepared from control and infected cells, and equal amounts of protein were assayed for c-Jun protein.
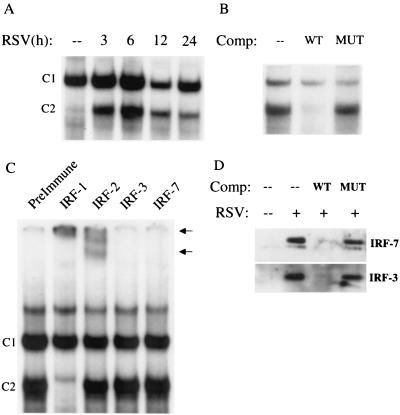
We next determined whether RSV infection produced changes in the abundance of DNA binding proteins recognizing the RANTES ISRE site. As shown in Fig. 6A, a single nucleoprotein complex, C1, was formed in control cells using the ISRE probe, while a second complex, C2, appeared following RSV infection. RSV infection increased the binding of C2 as early as 3 h postinfection, with a peak in binding intensity at 6 h postinfection. The sequence specificity of the ISRE complexes was examined by competition with unlabeled oligonucleotides in EMSA (Fig. 6B). C2 was competed by the wild-type oligonucleotide but not by the mutated one, indicating binding specificity. The RANTES ISRE binds transcription factors belonging to the ISRE family. To determine the composition of the RSV-inducible complex, preimmune serum or antibodies recognizing IRF-1, IRF-2, IRF-3, and IRF-7 were added to the binding reaction. We did not include an antibody to the IFN consensus sequence binding protein since this protein is expressed only in hematopoietic cell types (43). As shown in Fig. 6C, the anti-IRF-1 antibody affected C2 binding, inducing the disappearance of the complex and the appearance of a supershifted band. A supershift was also produced by addition of anti-IRF-2. These data indicate that IRF-1 is a major component of the RSV-inducible complex, although IRF-2 may be also present. Lin et al. (14, 26) have previously shown that IRF-3 and -7 play an important role in virus-induced RANTES gene expression. Since the ability of an antibody to produce a supershift depends on its affinity and the availability of the epitope within the DNA-protein complex, the absence of a supershift cannot be used to exclude the presence of IRF-3 and -7 binding to the ISRE. To independently address this issue, we used a two-step microaffinity isolation-Western blot assay to determine whether IRF-3 and -7 bound to the RANTES ISRE following RSV infection. In this assay, biotinylated ISRE was used to bind nuclear extracts of control and 12-h-infected A549 cells. ISRE binding proteins were captured by the addition of streptavidin-agarose beads and washed, and the presence of bound IRF-3 and -7 was detected by Western blotting. As shown in Fig. 6D, there was no detectable binding of IRF-3 and -7 in control nuclear extracts, but their abundance was greatly increased after RSV infection. IRF-3 and -7 detection was abolished when 10-fold excess ISRE wild-type, but not mutated, oligonucleotide was included as competitor in the initial binding reaction, indicating sequence specificity. These data indicate that RSV infection induces IRF-1, -3, and -7 to bind to the RANTES ISRE.
FIG. 6.
EMSA of RANTES ISRE binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from control and RSV-infected cells at the indicated times and used for EMSA. Time following infection is shown at the top. A single nucleoprotein complex 1 (C1) is formed in control cells, while a second complex (C2) appears following RSV infection. (B) Competition (Comp) analysis. Nuclear extracts from A549 cells infected for 12 h were used to bind to the ISRE probe; 2 pM unlabeled wild-type (WT) or mutated (MUT) competitor was included in the binding reaction, as indicated at the top. C1 is competed by the wild-type oligonucleotide but not by the mutant one, indicating binding specificity. (C) Supershift assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum and anti-IRF-1, -IRF-2, -IRF-3, and -IRF-7 antibodies. The anti-IRF-1 antibody induced the complete disappearance of C2 and the appearance of a supershifted band, which was also induced by the anti-IRF-2 antibody (as indicated by the arrows). (D) Microaffinity isolation-Western blot analysis for IRF-3 and -7 Control and 12-h-infected A549 nuclear extracts were affinity purified using biotinylated ISRE in the absence or presence of nonbiotinylated wild-type (WT) or mutated (MUT) competitor. After capture with streptavidin-agarose beads, complexes were eluted and assayed for IRF-3 and IRF-7 by Western blotting. IRF-3 and -7 are not present in the ISRE complex formed by control nuclear extracts, but they are strongly induced to bind to the ISRE following RSV infection. Binding is competed by the wild-type nonbiotinylated oligonucleotide but not the mutated one, indicating sequence specificity.
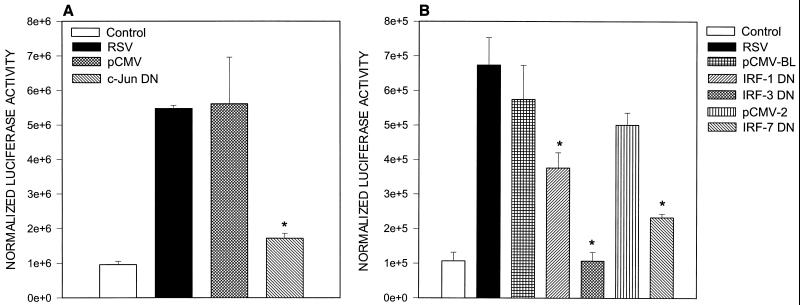
To further investigate the role of CRE and ISRE binding proteins in RSV-induced RANTES promoter activation, we cotransfected 293 cells with pGL2-220 and expression plasmids of DN inhibitors of c-Jun, IRF-1, IRF-3, and IRF-7. The day after transfection, cells were infected with RSV (MOI of 1) and harvested 24 h later to measure luciferase activity. As shown in Fig. 7A, overexpression of c-Jun DN greatly affected RSV-induced luciferase activity, indicating its central role in RANTES promoter induction. Among the IRF proteins, overexpression of IRF-1 and -7 mutants significantly reduced RSV-induced luciferase activity, while IRF-3 completely abolished it, indicating that IRF-3 is the major transactivating factor of the RSV-induced ISRE binding complex (Fig. 7B).
FIG. 7.
Effects of overexpressing DN mutant c-Jun and IRF transcription factors on RSV-induced RANTES gene transcription. 293 cells were transfected with 2 μg of pGL2-220 plasmid alone or cotransfected with either 0.5 μg of c-Jun DN or the empty vector pCMV (A) or with 0.2 μg of IRF-1, -3, and -7 DN or the corresponding empty vector (pCMV-BL for IRF-1 and -3; pCMV2 for IRF-7) (B). Transfected cells were infected with RSV (MOI of 1) and harvested at 24 h postinfection for determination of luciferase activity. Data are expressed as mean ± standard deviation of normalized luciferase activity. ∗, P < 0.01 relative to empty vector.
Role of NF-κB and NF-IL6 in RANTES promoter inducibility following RSV infection.
Since the site mutation analysis of the RANTES promoter had shown that the NF-κB and NF-IL6 binding sites are important in RSV-induced promoter activation, we performed gel shift assays to determine whether RSV infection produced changes in the abundance of DNA binding proteins recognizing these three regions of the RANTES promoter.
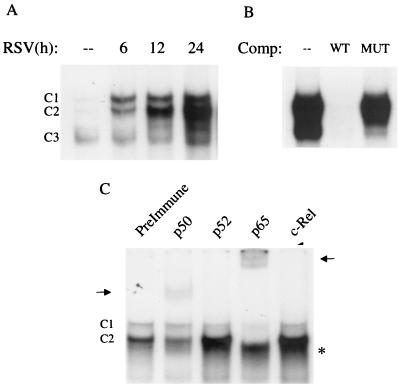
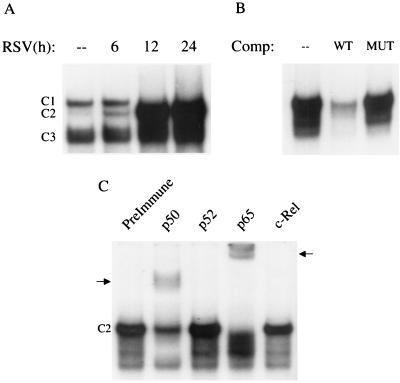
As shown in Fig. 8A, a single nucleoprotein complex (C3) was formed from nuclear extracts of control cells on the NF-κB1 probe, while two other complexes, C1 and C2, were faintly detected. RSV infection markedly increased the binding of C1 and C2 at 6 h postinfection, with a progressive increase in binding intensity of C2 at 12 and 24 h postinfection. The sequence specificity of the different complexes was examined by competition with unlabeled oligonucleotides in EMSA (Fig. 8B). C1 and C2 were competed by the wild-type but not the mutated oligonucleotide, indicating binding specificity. To determine the composition of the inducible complexes, we performed supershift assays, adding specific antibodies to various NF-κB subunits in EMSA. We did not include an antibody to RelB, since we and others have previously shown that this protein is not present in epithelial cells (12, 45). As shown in Fig. 8C, addition of the anti-p50 antibody induced the appearance of a supershifted band, with a concomitant reduction of C2. Addition of the anti-p65 antibody supershifted both C1 and C2 and produced a faster-migrating complex, which could represent a p50 homodimer. Together, these data indicate that C2 is a p50-p65 heterodimer and C1 is a p65 homodimer.
FIG. 8.
EMSA of RANTES NF-κB1 binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from control and RSV-infected cells at the indicated times and used for EMSA. Time following RSV infection is shown at the top. Complexes 1 and 2 (C1 and C2) are RSV inducible; complex 3 (C3) is constitutive. (B) Competition (Comp) analysis. Nuclear extracts from A549 cells uninfected or infected for 12 h were used to bind to the NF-κB1 probe in the absence (–) or presence of 2 pmol of unlabeled wild-type (WT) or mutated (MUT) competitor in the binding reaction, as indicated at the top. C1 and C2 are competed by the unlabeled oligonucleotide, indicating binding specificity. (C) Supershift assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum and anti-p50, -p52, -c-Rel, and -p65 antibodies. Addition of the anti-p50 and anti-p65 antibodies induces the appearance of a supershifted band (indicated by the arrows), with a concomitant reduction of C2 (in the case of anti-p50) or both C1 and C2 (in the case of anti-p65). Addition of the anti-p65 antibody also induces the appearance of a faster-migrating complex, indicated by the asterisks.
Using the NF-κB2 probe, two nucleoprotein complexes (C1 and C3) were formed from nuclear extracts of control cells, while another, C2, was faintly detected. RSV infection markedly increased the binding of C2, starting at 6 h postinfection, while C1 was no longer detectable after 12 h of infection (Fig. 9A). C2 was sequence specific, as shown by competition assay (Fig. 9B). The addition of either anti-p50 or anti-p65/RelA antibody induced the appearance of a supershifted band, with a concomitant reduction of C2, indicating that C2 is a p50-p65 heterodimer (Fig. 9C).
FIG. 9.
EMSA of RANTES NF-κB2 binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from control and RSV-infected cells at the indicated times and used for EMSA. Time following RSV infection is shown at the top. Complex 2 (C2) is RSV inducible; complexes 1 and 3 (C1 and C3) are constitutive. (B) Competition (Comp) analysis. Nuclear extracts from A549 cells uninfected or infected for 12 h were used to bind to the NF-κB1 probe in the absence (–) or presence of 2 pmol of unlabeled WT or MUT competitor in the binding reaction, as indicated at the top. C2 is competed by the unlabeled oligonucleotide, indicating binding specificity. (C) Supershift assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum and anti-p50, -p52, -c-Rel, and -p65 antibodies. Addition of the anti-p50 and anti-p65 antibodies produces a reduction in (in the case of anti-p50) or disappearance of (in the case of anti-p65) C2 and the appearance of a supershifted band (indicated by the arrows).
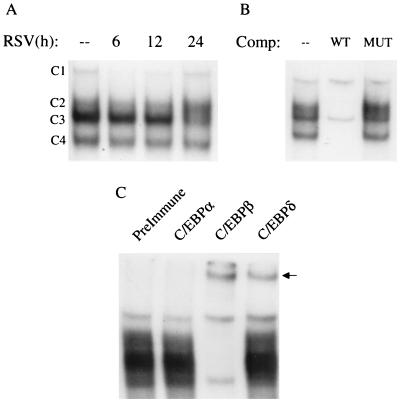
When nuclear extracts from A549 cells were used to bind to the NF-IL6 probe, four complexes (C1, C2, C3, and C4) were formed similarly in control and RSV-infected cells (Fig. 10A). In competition assay, C2, C3, and C4 were competed by the wild-type but not the mutated oligonucleotide, indicating binding specificity (Fig. 10B). In supershift assays using antibodies specific to C/EBPα, C/EBPβ/NF-IL6, and C/EBPδ, which are the C/EBP family members mainly involved in promoter transactivation (37), the addition of anti-C/EBPβ produced the disappearance of the specific complexes and the appearance of a supershifted band, showing that C/EBP-β is the major component of the NF-IL6 nucleoprotein complexes. Since the addition of anti-C/EBPδ also induced the appearance of a supershifted band, it is likely that this protein is also part of the DNA-protein complexes formed on the NF-IL6 probe (Fig. 10C).
FIG. 10.
EMSA of RANTES NF-IL6 binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from control and RSV-infected cells at the indicated times and used for EMSA. Time following RSV infection is shown at the top. Four complexes (C1, C2, C3, and C4) are formed similarly in control and RSV-infected cells. (B) Competition (Comp) analysis. Nuclear extracts from A549 cells uninfected or infected for 12 h were used to bind to the NF-IL6 probe in the absence (–) or presence of 2 pmol of unlabeled WT or MUT competitor. C2, C3, and C4 are competed by the unlabeled oligonucleotide, indicating binding specificity. (C) Supershift assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum, anti-C/EBPα, anti-C/EBPβ, and anti-C/EBPδ. The addition of anti-C/EBPβ produces the disappearance of the specific complexes and the appearance of a supershifted band; the addition of anti-C/EBPδ also induces the appearance of a supershifted band.
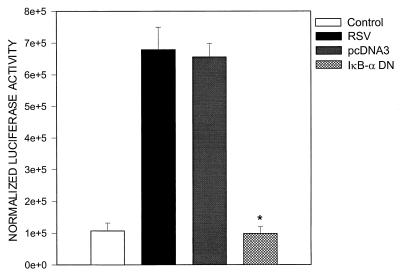
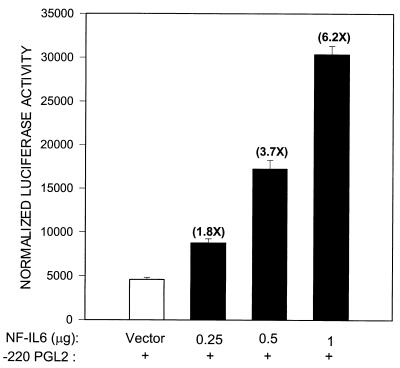
Together, the EMSA data indicate that RSV infection induced significant changes in the composition of proteins binding to the proximal RANTES NF-IL6 and the NF-κB sites. To confirm the role of NF-κB activation in RANTES gene transcription, we cotransfected pGL2-220 with an expression vector encoding a DN mutant of IκB-α, in which serine residues 32 and 36 were changed to alanine, producing a nonproteolyzable form of IκB that blocks NF-κB activation (46). As shown in Fig. 11, overexpression of the IκB-α mutant completely abolished RSV-induced luciferase activity. To better define the role that NF-IL6 plays in RANTES gene transcription, we cotransfected A549 cells with an expression vector containing the NF-IL6 cDNA or the empty vector alone and the pGL2-220 plasmid. As shown in Fig. 12, transfection of increasing amounts of NF-IL6 plasmid in alveolar epithelial cells resulted in a severalfold increase of RANTES promoter activity, confirming its important role in RANTES gene expression.
FIG. 11.
Effect of overexpressing IκB-α DN on RSV-induced RANTES gene transcription. 293 cells were transfected with 2 μg of pGL2-220 plasmid alone or cotransfected with 0.5 μg of either the plasmid expressing IκB-α DN or the empty vector (pcDNA3). Transfected cells were infected with RSV (MOI of 1) and harvested at 24 h postinfection for determination of luciferase activity. Data are expressed as mean ± standard deviation of normalized luciferase activity. ∗, P < 0.01 relative to empty vector.
FIG. 12.
Activation of RANTES transcription by NF-IL6/C/EBPβ overexpression. A549 cells were cotransfected with 2 μg of pGL2-220 plasmid and either different amounts of the NF-IL6 expression plasmid or 1 μg of empty vector; 48 h later, cells were harvested to measure luciferase and β-galactosidase reporter activities. Data are expressed as mean ± standard deviation of normalized luciferase activity.
DISCUSSION
RSV is the most common cause of epidemic respiratory disease in children. It is estimated that 40 to 90% of children with bronchiolitis and 25 to 50% of children with pneumonia are infected with RSV, resulting in 100,000 hospital admissions annually in the United States alone (18). Infants with congenital heart disease infected with RSV can have a significantly more severe illness and a higher mortality rate than normal children (27). RSV is also a significant contributor to illness in adult populations (48). An effective vaccine is not yet available. The mechanisms of RSV-induced acute airway disease and its long-term consequences are largely unknown, but the delicate balance between immunopathology and immunoprotection in the airway mucosa may be altered by an exuberant and unwanted local inflammatory response. Airway infiltration of monocytes and lymphocytes is typically present in RSV infection of the lung (18), and activation of eosinophil and basophil leukocytes has been shown to correlate with the severity of acute RSV disease (11, 13). RANTES is a CC chemokine highly chemoattractant for T lymphocytes, monocytes, eosinophils, and basophils, all cell types present in RSV-induced lung inflammatory infiltrate. We and other groups have recently shown that RANTES is detected in high concentrations in the nasal and bronchoalveolar lavages of RSV-infected children (42, 44; Garofalo, unpublished data) and is strongly expressed in RSV-infected respiratory epithelial cells (34, 39), which are the primary target for viral infection. Therefore, it is likely that RANTES produced by infected epithelial cells plays a pivotal role in the pathogenesis of RSV-induced airway inflammation.
Human RANTES gene expression appears to be differentially regulated depending on the cell type and the stimulus applied (19, 30–32, 35). Induction of transcription is an important level of control of RANTES gene expression and different combinations of cis regulatory elements of the promoter seem to be required for optimal levels of transcription in a variety of cell types, such as monocytes, lymphocytes, and astrocytes, following stimulation with cytokines or phorbol esters (19, 30–32, 35). Mechanisms of inducible RANTES gene expression following viral infections of airway epithelial cells have not been fully investigated. In this study, the results of transient transfections clearly indicate that RANTES gene transcription is activated following RSV infection of A549 cells (Fig. 3). The kinetic of promoter activation mirrors the induction of the endogenous RANTES gene mRNA, suggesting that in alveolar epithelial cells RANTES expression, following RSV infection, is controlled mainly at the level of transcription. A similar result has been reported for human embryonic kidney cells infected with Sendai virus, a paramyxovirus similar to RSV (26). However, increased transcription is not the only mechanism by which RANTES gene expression is regulated in epithelial cells following viral infection. Koga et al. have recently reported that RSV infection of bronchial epithelial cells markedly increased the RANTES mRNA half-life, which was identified as the major mechanism responsible for virus-induced mRNA accumulation (23). Similarly, RSV infection of small alveolar epithelial cells and rotavirus infection of HT-29, an intestinal epithelial cell line, are able to induce RANTES protein secretion and mRNA upregulation without activating RANTES reporter gene transcription more than twofold (A. Casola, unpublished data). These results suggest that RANTES induction can be regulated at both transcriptional and posttranscriptional levels, with a preponderance of one of the two mechanisms dependent on the cell type.
Identification of the regulatory mechanisms involved in RANTES gene transcription is important for a rational design of therapeutic agents that can block its expression in the lung. Therefore, in this study we report for the first time a detailed analysis of the cis regulatory elements required for RANTES promoter activation following RSV infection. Results from 5′-deletion analysis indicate that two different regions are required for RSV-induced promoter activation: the first from −220 to −195 nt, which is responsible for half of the promoter inducibility, and the second from −150 to −120 nt, which is absolutely necessary for promoter induction following RSV infection (Fig. 4). A few studies have investigated the minimal enhancer region of the RANTES promoter required to confer responsiveness to external stimuli like cytokines or phorbol esters. In T-cell and monocytic-like cell lines, the first 500 nt of the RANTES promoter are sufficient to confer full phorbol myristate acetate and ionomycin inducibility, which is lost by further deletions to nt −200 (31). Differently, in phytohemagglutinin-stimulated lymphocytes, a 195-bp fragment of RANTES promoter has the same inducibility of longer fragments, while a 120-bp fragment is no longer inducible (32, 35). In a human astrocytoma cell line, IL-1 stimulation of RANTES promoter requires a region spanning from nt −220 to −120 (30), similar to what we have observed in alveolar epithelial cells infected with RSV. These results clearly indicate that the minimal enhancer region required for RANTES promoter activation differ among cell types and possibly among stimuli, although in none of these studies was there a direct comparison of different stimuli applied to the same cell line.
The region of the promoter spanning nt −220 to −190, which accounts for more than half of RSV inducibility of the RANTES promoter, contains a CRE site. Site-directed mutation experiments clearly demonstrate that this site plays a major role in promoter activation following RSV infection (Fig. 7). Gel shift assays show that the CRE site binds RSV-inducible proteins belonging to the CREB/ATF and Jun families of transcription factors. The virus-inducible complex formed on the CRE site is likely formed by heterodimers of c-Jun and CREB/ATF proteins, since the anti c-Jun antibody almost completely blocked the formation of the DNA complex (Fig. 5). c-Jun is likely the relevant transactivating factor of the CRE complex, since overexpression of a c-Jun DN mutant reduces RANTES transcription similarly to the mutation of the CRE site (compare Fig. 7 and 8A). This is the first time that an important role for the CRE site has been reported for virus-induced RANTES transcription. Miyamoto et al. have recently shown that the CRE site is relevant for RANTES activation in astrocytoma cells stimulated with IL-1, although there was no inducible binding in IL-1-stimulated cells and the composition of the DNA binding complexes was different from that of the RSV-inducible complex formed on the CRE site (30).
Our 5′-deletion analysis shows that a further deletion of the RANTES promoter to nt −120 completely abolishes RSV-induced RANTES transcription. The promoter region spanning nt −138 to −117 contains a functional ISRE site, and site-directed mutation experiments clearly show that this site plays a fundamental role in promoter activation following RSV infection, since the ISRE mutant is no longer RSV inducible. Supershift assays and microaffinity isolation experiments clearly show that IRF-1, -3, and -7 are components of the RSV-inducible complex formed on the RANTES ISRE site (Fig. 6). We have previously reported that RSV infection of A549 cells induces IRF-1 synthesis, nuclear translocation, and binding to a newly identified responsive element of the IL-8 promoter involved in RSV-induced IL-8 transcription (7). IRF-7 synthesis is also induced in RSV-infected A549 cells, while IRF-3 is constitutive (data not shown). Among the IRF proteins, IRF-3 seems to be essential for RSV-induced RANTES transcription, since overexpression of its DN mutant completely abolished RANTES promoter activation (Fig. 8B). Similar results were recently reported by Lin et al., who showed that activation of IRF-3 and -7 and binding to the ISRE site are critical for RANTES gene transcription in Sendai virus-infected cells (26).
We have previously shown that RSV infection of A549 cells is a potent activator of p65/RelA, a member of the NF-κB family, which is absolutely required for RSV-inducible IL-8 gene transcription (12). The present study demonstrates that the NF-κB site plays an important role also in RSV-induced RANTES gene transcription, since mutation of the NF-κB1 site greatly reduces both basal and RSV-induced promoter activity. The composition of the DNA-nuclear complex formed on both RANTES NF-κB1 and NF-κB2 sites is slightly different from the one formed on the NF-κB site of the IL-8 promoter, since it contains p65 and p50 subunits but not c-Rel (12). We have also shown that inhibition of IκB-α degradation, and therefore of NF-κB nuclear translocation, blocks RSV-induced RANTES transcription (Fig. 12). Our results are similar to the data shown by Genin et al. for 293 cells infected with Sendai virus (14). In their model, overexpression of IκB-α DN mutant greatly reduced Sendai virus-induced RANTES gene expression and blocked virus induced binding not only to the NF-κB site but also to the ISRE site. Therefore, disruption of NF-κB and IRF cooperation in RSV-infected cells overexpressing the IκB-α mutant would explain our finding of almost complete inhibition of RSV-induced RANTES transcription.
C/EBPs are basic domain/leucine zipper-containing transcription factors involved in inducible gene expression during acute infectious and inflammatory responses, as well as cell differentiation (1, 6, 8, 25, 28). We have previously shown that RSV infection of A549 cells induces C/EPBβ/NF-IL6 activation and that the NF-IL6 site is important for RSV-induced IL-8 gene transcription (7, 21). This study shows that the RANTES NF-IL6 site, which binds C/EPBβ/NF-IL6 and C/EPBδ, is important in RSV-induced RANTES transcription, similarly to what has been shown for phytohemagglutinin-stimulated lymphocytes (25). A different result was reported for T cells and monocytes stimulated with phorbol esters or lipopolysaccharide (23, 24), in agreement with the fact that C/EBP expression is regulated in a tissue-type-dependent fashion.
Our findings that multiple binding sites contribute to RANTES promoter induction after RSV infection and that cooperation among these different sites is required for full activation of the promoter support the enhanceosome model for RANTES gene transcription. An enhanceosome is a nuclear protein complex assembled at a given enhancer, where various combinations of ubiquitous, signal- and tissue-specific activators allow different interactions with coactivators and with the basal transcriptional machinery, recruiting them to DNA to generate synergistic transcription. In the case of the RANTES gene, cooperation among transcription factors belonging to the C/EBP, NF-κB, IRF, and CREB/AP-1 families is necessary for full transcriptional activation. Expression of the RANTES gene, as well as many other genes, appears to be differentially regulated depending on the cell type and the stimulus applied (19, 30–32, 35). Lin et al. have identified IRF and NF-κB as major players in Sendai virus-induced RANTES expression in kidney epithelial cells (26). We have obtained similar results for RSV-infected alveolar epithelial cells. Therefore, it is likely that viruses infecting epithelial cells require similar subsets of nuclear factors for induction of RANTES transcription. It is possible, however, that other stimuli, like cytokines, as well as viral infection of different cell types may have different mechanisms for induction of RANTES gene transcription. We have recently reported that NF-κB, NF-IL6, IRF, and AP-1 binding sites are necessary for full activation of IL-8 gene transcription in alveolar epithelial cells infected with RSV (7), and cooperation of the same transcription factors, with the exclusion of NF-IL6, is also required for virus-induced IFN-β (33), a gene highly expressed in RSV-infected A549 cells (22). These data suggest the existence of common mechanisms of activation for different virus-induced genes in airway epithelium. Identification of these mechanisms is important to identify novel targets for modulation of virus-induced gene expression in the lung.
ACKNOWLEDGMENTS
This work was supported in part by grants from NIAID (PO1 46004 and AI 15939) and NIEHS (P30ES0 6676). A.C. is a Child Health Research Center Young Investigator (Child Health and Human Development grant HD 27841) and the recipient of the 1998 European Society for Pediatric Infectious Disease Fellowship Award sponsored by Bristol-Myers Squibb. H.H. was supported by a grant from the Fortune Program of the University of Tuebingen, Tuebingen, Germany. A.R.B. is an Established Investigator of the American Heart Association.
We thank Tianshuang Liu for excellent technical assistance.
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam R, Stafford S, Forsythe P, Harrison R, Faubion D, Lett-Brown M A, Grant J A. RANTES is a chemotactic and activating factor for human eosinophils. J Immunol. 1993;150:3442–3448. [PubMed] [Google Scholar]
- 3.Alam R, York J, Boyers M, Stafford S, Grant J A, Lee J, Forsythe P, Sim T, Ida N. Increased MCP-1, RANTES, and MIP-1α in bronchoalveolar lavage fluid of allergic asthmatic patients. Am J Respir Crit Care Med. 1996;153:1398–1404. doi: 10.1164/ajrccm.153.4.8616572. [DOI] [PubMed] [Google Scholar]
- 4.Brasier A R, Jamaluddin M, Casola A, Duan W, Shen Q, Garofalo R P. A promoter recruitment mechanism for tumor necrosis factor-alpha-induced interleukin-8 transcription in type II pulmonary epithelial cells. Dependence on nuclear abundance of RelA, NF-κB1, and c-Rel transcription factors. J Biol Chem. 1998;273:3551–3561. doi: 10.1074/jbc.273.6.3551. [DOI] [PubMed] [Google Scholar]
- 5.Brown P H, Chen T K, Birrer M J. Mechanism of action of a dominant-negative mutant of c-Jun. Oncogene. 1994;9:791–799. [PubMed] [Google Scholar]
- 6.Cardinaux J R, Allaman I, Magistretti P J. Pro-inflammatory cytokines induce the transcription factors C/EBPβ and C/EBPδ in astrocytes. J Neurosci. 1996;16:919–929. [Google Scholar]
- 7.Casola A, Garofalo R P, Jamaluddin M, Vlahopoulos S, Brasier A R. Requirement of a novel upstream response element in RSV induction of interleukin-8 gene expression: stimulus-specific differences with cytokine activation. J Immunol. 2000;164:5944–5951. doi: 10.4049/jimmunol.164.11.5944. [DOI] [PubMed] [Google Scholar]
- 8.Dunn S M, Coles L S, Lang R K, Gerondakis S, Vadas M A, Shannon M F. Requirement for nuclear factor (NF)-κB p65 and NF-interleukin-6 binding elements in the tumor necrosis factor response region of the granulocyte colony-stimulating factor promoter. Blood. 1994;83:2469–2479. [PubMed] [Google Scholar]
- 9.Fiedler M A, Wernke-Dollries K, Stark J M. Inhibition of viral replication reverses respiratory syncytial virus-induced NF-κB activation and interleukin-8 gene expression in A549 cells. J Virol. 1996;70:9079–9082. doi: 10.1128/jvi.70.12.9079-9082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiedler M A, Wernke-Dollries K, Stark J M. Mechanism of RSV-induced IL-8 gene expression in A549 cells before viral replication. Am J Physiol. 1996;271:L963–L971. doi: 10.1152/ajplung.1996.271.6.L963. [DOI] [PubMed] [Google Scholar]
- 11.Garofalo R P, Kimpen J L L, Welliver R C, Ogra P L. Eosinophil degranulation in the respiratory tract during naturally acquired respiratory syncytial virus infection. J Pediatr. 1992;120:28–32. doi: 10.1016/s0022-3476(05)80592-x. [DOI] [PubMed] [Google Scholar]
- 11a.Garofalo, R. P., B. Olszewska-Pazdrak, P. L. Ogra, and R. C. Welliver. Beta-chemokines in nasal secretions of infants with respiratory syncytial virus-induced respiratory infections. Pediatr. Asthma Allergy Immunol., in press.
- 12.Garofalo R P, Sabry M, Jamaluddin M, Yu R K, Casola A, Ogra P L, Brasier A R. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J Virol. 1996;70:8773–8781. doi: 10.1128/jvi.70.12.8773-8781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garofalo R P, Welliver R C, Ogra P L. Concentrations of LTB4, LTC4, LTD4 and LTE4 in bronchiolitis due to respiratory syncytial virus. Pediatr Allergy Immunol. 1991;2:30–37. [Google Scholar]
- 14.Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-κB and IFN-regulatory factor transcription factors. J Immunol. 2000;164:5352–5361. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- 15.Glezen W P, Taber L H, Frank A L. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalo J A, Lloyd C M, Wen D, Albar J P, Wells T N C, Proudfoot A, Martinez-A C, Dorf M, Bjerke T, Coyle J, Gutierrez-Ramos J C. The coordinated action of CC chemokines in the lung orchestrates allergic inflammation and airway hyperresponsiveness. J Exp Med. 1998;188:157–167. doi: 10.1084/jem.188.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groothuis J R, Gutierre K M, Lauer B A. Respiratory syncytial virus infection in children with bronchopulmonary dysplasia. Pediatrics. 1988;82:199–203. [PubMed] [Google Scholar]
- 18.Hall C B, McCarthy C A. Respiratory syncytial virus. In: Mandel G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingston; 1995. p. 1501-1519. [Google Scholar]
- 19.Hiura T S, Kempiak S J, Nel A E. Activation of the human RANTES gene promoter in a macrophage cell line by lipopolysaccharide is dependent on stress-activated protein kinases and the I-κB kinase cascade: implications for exacerbation of allergic inflammation by environmental pollutants. Clin Immunol. 1999;90:287–301. doi: 10.1006/clim.1998.4659. [DOI] [PubMed] [Google Scholar]
- 20.Jamaluddin M, Casola A, Garofalo R P, Han Y, Elliott T, Ogra P L, Brasier A R. The major component of IκBα proteolysis occurs independently of the proteasome pathway in respiratory syncytial virus-infected pulmonary epithelial cells. J Virol. 1998;72:4849–4857. doi: 10.1128/jvi.72.6.4849-4857.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamaluddin M, Garofalo R P, Ogra P L, Brasier A R. Inducible translational regulation of the NF-IL6 transcription factor by respiratory syncytial virus infection in pulmonary epithelial cells. J Virol. 1996;70:1554–1563. doi: 10.1128/jvi.70.3.1554-1563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamaluddin, M., S. Wang, R. P. Garofalo, T. Elliott, A. Casola, S. Baron, and A. R. Brasier. 2001. Am. J. Physiol. 280:248–257. [DOI] [PubMed]
- 23.Koga T, Sardina E, Tidwell R M, Pelletier M, Look D C, Holtman M J. Virus-inducible expression of a host chemokine gene relies on replication-linked mRNA stabilization. Proc Natl Acad Sci USA. 1999;96:5680–5685. doi: 10.1073/pnas.96.10.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamph W W, Wamsley P, Sassone-Corsi P, Verma I M. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 25.Lane M D, Tang Q Q, Jian M S. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Commun. 1999;266:677–683. doi: 10.1006/bbrc.1999.1885. [DOI] [PubMed] [Google Scholar]
- 26.Lin R, Heylbroeck C, Genin P, Pitha P M, Hiscott J. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacDonald N E, Hall C B, Suffin S C, Alexson C, Harris P J, Manning J A. Respiratory syncytial viral infection in infants with congenital heart disease. N Engl J Med. 1982;307:397–400. doi: 10.1056/NEJM198208123070702. [DOI] [PubMed] [Google Scholar]
- 28.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci USA. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller M D, Krangel M S. Biology and biochemistry of the chemokines: a family of chemotactic and inflammatory cytokines. Crit Rev Immunol. 1992;12:17–46. [PubMed] [Google Scholar]
- 30.Miyamoto N G, Medberry P S, Hesselgesser J, Boejlk S, Nelson P J, Krensky A M, Perez H D. Interleukin-1β induction of the chemokine RANTES promoter in the human astrocytoma line CH235 requires both constitutive and inducible transcription factors. J Neuroimmunol. 2000;105:78–90. doi: 10.1016/s0165-5728(00)00195-8. [DOI] [PubMed] [Google Scholar]
- 31.Moriuchi H, Moriuchi M, Fauci A S. Nuclear factor-κB potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J Immunol. 1997;158:3483–3491. [PubMed] [Google Scholar]
- 32.Nelson P J, Ortiz B D, Pattison J M, Krensky A M. Identification of a novel regulatory region critical for expression of the RANTES chemokine in activated T lymphocytes. J Immunol. 1996;157:1139–1148. [PubMed] [Google Scholar]
- 33.Nguyen H, Hiscott J, Pithas P M. The growing family of interferon regulatory factors. Cytokine Growth Ed Rev. 1997;4:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 34.Olszewska-Pazdrak B, Casola A, Saito T, Alam R, Crowe S E, Mei F, Ogra P L, Garofalo R P. Cell-specific expression of RANTES, MCP-1, and MIP-1α by lower airway epithelial cells and eosinophils infected with respiratory syncytial virus J. Virol. 1998;72:4756–4764. doi: 10.1128/jvi.72.6.4756-4764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz B D, Krensky A M, Nelson P J. Kinetics of transcription factors regulating the RANTES chemokine gene reveal a developmental switch in nuclear events during T-lymphocyte maturation. Mol Cell Biol. 1996;16:202–210. doi: 10.1128/mcb.16.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel J A, Kunimoto M, Sim T C, Garofalo R, Eliott T, Baron S, Ruuskanen O, Chonmaitree T, Ogra P L, Schmalstieg F. Interleukin-1 alpha mediates the enhanced expression of intercellular adhesion molecule-1 in pulmonary epithelial cells infected with respiratory syncytial virus. Am J Respir Cell Mol Biol. 1995;13:602–609. doi: 10.1165/ajrcmb.13.5.7576697. [DOI] [PubMed] [Google Scholar]
- 37.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 38.Ruuskanen O, Ogra P L. Respiratory syncytial virus. Curr Probl Pediatr. 1993;2:50–79. doi: 10.1016/0045-9380(93)90003-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito T, Deskin R W, Casola A, Haeberle H, Olszewska B, Ernst P B, Alam R, Ogra P L, Garofalo R. Respiratory syncytial virus induces selective production of the chemokine RANTES by upper airway epithelial cells. J Infect Dis. 1997;175:497–504. doi: 10.1093/infdis/175.3.497. [DOI] [PubMed] [Google Scholar]
- 40.Salkind A R, Nichols J E, Roberts N J., Jr Suppressed expression of ICAM-1 and LFA-1 and abrogation of leukocyte collaboration after exposure of human mononuclear leukocytes to respiratory syncytial virus in vitro. Comparison with exposure to influenza virus. J Clin Investig. 1991;88:505–511. doi: 10.1172/JCI115332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Servant M J, ten Oever B, LePage C, Conti L, Gessani S, Julkunen I, Lin R, Hiscott J. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J Biol Chem. 2001;276:355–363. doi: 10.1074/jbc.M007790200. [DOI] [PubMed] [Google Scholar]
- 42.Sheeran P, Jafri H, Carubelli C, Saavedra J, Johnson C, Krisher K, Sanchez P J, Ramilio M O. Elevated cytokine concentrations in the nasopharyngeal and tracheal secretions of children with respiratory syncytial virus disease. Pediatr Infect Dis J. 1999;18:115–122. doi: 10.1097/00006454-199902000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 44.Teran L M, Seminario M C, Shute J K, Papi A, Compton S J, Low J L, Gleich G J, Johnston S L. RANTES, macrophage-inhibitory protein 1alpha, and the eosinophil product major basic protein are released into upper respiratory secretions during virus-induced asthma exacerbations in children. J Infect Dis. 1999;179:677–681. doi: 10.1086/314618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas H, Friedland J S, Sharland M, Becker S. Respiratory syncytial virus-induced RANTES production from human bronchial epithelial cells is dependent on nuclear factor-κB nuclear binding and is inhibited by adenovirus-mediated expression of inhibitor of κBa. J Immunol. 1998;161:1007–1016. [PubMed] [Google Scholar]
- 46.Traenckner E B-M, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2882. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueba O. Respiratory syncytial virus. I. Concentration and purification of the infectious virus. Acta Med Okayama. 1978;32:265–272. [PubMed] [Google Scholar]
- 48.Walsh E E, Falsey A R, Hennessey P A. Respiratory syncytial and other virus infections in persons with chronic cardioplumonary disease. Am J Respir Crit Care Med. 1999;160:791–795. doi: 10.1164/ajrccm.160.3.9901004. [DOI] [PubMed] [Google Scholar]