Abstract
It has been shown previously that the nonstructural protein NS1 of influenza virus is an alpha/beta interferon (IFN-α/β) antagonist, both in vitro and in experimental animal model systems. However, evidence of this function in a natural host has not yet been obtained. Here we investigated the role of the NS1 protein in the virulence of a swine influenza virus (SIV) isolate in pigs by using reverse genetics. The virulent wild-type A/Swine/Texas/4199-2/98 (TX/98) virus and various mutants encoding carboxy-truncated NS1 proteins were rescued. Growth properties of TX/98 viruses with mutated NS1, induction of IFN in tissue culture, and virulence-attenuation in pigs were analyzed and compared to those of the recombinant wild-type TX/98 virus. Our results indicate that deletions in the NS1 protein decrease the ability of the TX/98 virus to prevent IFN-α/β synthesis in pig cells. Moreover, all NS1 mutant viruses were attenuated in pigs, and this correlated with the amount of IFN-α/β induced in vitro. These data suggest that the NS1 protein of SIV is a virulence factor. Due to their attenuation, NS1-mutated swine influenza viruses might have a great potential as live attenuated vaccine candidates against SIV infections of pigs.
Swine influenza is an acute respiratory disease of swine caused by type A influenza viruses. Its severity depends on many factors, including host age, virus strain, and secondary infections (11). Influenza A viruses are segmented negative-strand RNA viruses and can be isolated from a number of other animal host species, including birds, humans, horses, whales, and mink. Although whole influenza viruses rarely cross the species barrier, gene segments can cross this barrier through the process of genetic reassortment, or genetic shift. Since pigs support the replication of both human and avian influenza A viruses (25), they have been postulated to play an important role in interspecies transmission by acting as a “mixing vessel” for reassortment between viruses specific to different host species (41). This may lead to the generation of novel influenza viruses capable of crossing the species barrier to humans. There are three subtypes of swine influenza viruses (SIVs) currently circulating in pigs in the United States: H1N1, H3N2, and H1N2 (22, 23, 34, 35, 53, 54, 57). Before 1998, mainly “classical” H1N1 SIVs were isolated from swine in the United States (5, 20). In 1998, SIVs of the subtype H3N2 were isolated in multiple states in the United States. These viruses were generated by reassortment between human, avian, and classical swine viruses (57). They are undergoing rapid evolution, and genetic characterization of swine viruses in the last several years has shown that the H3N2 viruses have undergone further reassortment with the classical H1N1 viruses, resulting in novel H1N2 and H1N1 SIVs (52). The ongoing reassortment of swine influenza viruses has implications both for the future surveillance of U.S. swine herds and for the efficacy of current vaccines.
Pathogenicity of influenza viruses is dependent on multiple virus and host factors. Among the host factors that fight virus infections, the alpha/beta interferon (IFN-α/β) system represents a powerful antiviral innate defense mechanism which was established relatively early in the evolution of eukaryotic organisms (15). The antiviral IFN-α/β system involves three major steps: (i) detection of viral infection and IFN-α/β secretion, (ii) binding of IFN-α/β to its receptors and transcriptional induction of IFN-α/β-stimulated genes, and (iii) synthesis of antiviral enzymes and proteins. Most viruses, however, have acquired specific genetic information encoding IFN-α/β antagonist molecules, which effectively block one or more steps of the antiviral IFN-α/β system. Influenza A viruses express a nonstructural protein in infected cells, the NS1 protein, which counteracts the cellular IFN-α/β response (17).
The NS1 protein of influenza A virus binds to double-stranded RNA (dsRNA), prevents dsRNA-mediated activation of protein kinase R (2, 19, 29), and prevents the synthesis of IFN-α/β (10, 15, 49). These properties have been mapped to the amino-terminal domain of the NS1 protein (51). In addition, the carboxy-terminal domain of the NS1 also contributes to its IFN-antagonistic properties, possibly by enhancing NS1 stability and dimerization (32, 50) and by attenuating host gene cell expression (33). The interplay of the influenza virus NS1 protein with the antiviral immune defense of the cell is likely to play a major role in the virulence of the virus and most likely also regulates virus replication in the host. Several studies have shown a role of the NS1 protein in contributing to enhanced virulence in animal models (12, 49). However, these studies were performed mostly with mouse-adapted influenza viruses, and definitive proof of NS1 function in a natural host is lacking. Here we show that mutations in the NS1 of an H3N2 SIV resulted in attenuation of viral replication both in vitro and in vivo. Attenuation correlated with a loss of NS1 potency in inhibiting IFN-α/β synthesis during viral infection. These results demonstrate that the NS1 protein of SIV is a virulence factor in its natural host mediated via its IFN-α/β antagonist properties.
MATERIALS AND METHODS
Cells and viruses.
Pig kidney-15 (PK-15), swine testis, and Madin-Darby canine kidney (MDCK) type II cells were grown in minimal essential medium (MEM) supplemented with 10% fetal bovine serum. Baby hamster kidney, 293T, and A549 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. The influenza A/swine/Texas/4199-2/98 (TX/98, H3N2 subtype) virus was obtained from the repository at St. Jude Children's Research Hospital, Memphis, TN. Influenza viruses were grown in the allantoic cavities of embryonated chicken eggs or in MDCK cells. Viral stocks were prepared in 7-day-old embryonated chicken eggs, which lack the ability to produce IFN-α/β (43, 44). Recombinant vesicular stomatitis virus expressing green fluorescent protein (VSV-GFP) (48) was grown and titrated in baby hamster kidney cells. This virus was used in IFN bioassays and was kindly provided by John Hiscott.
Construction of plasmids.
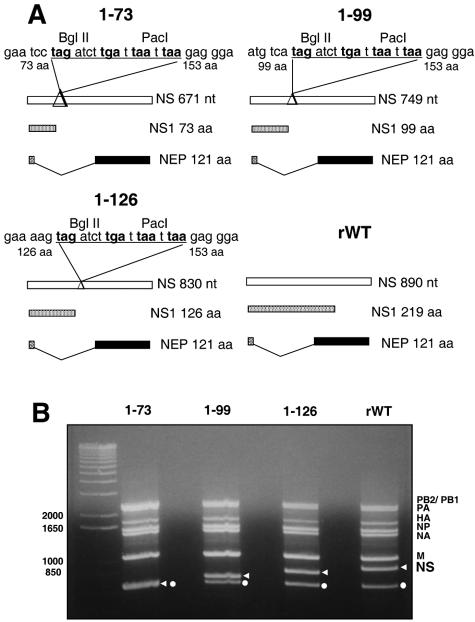
Ten-day-old embryonated eggs were infected with TX/98 virus, and total RNA was extracted by using TRIzol reagent (Invitrogen). The eight reverse genetics plasmids pHW-Sw-PB2, pHW-Sw-PB1, pHW-Sw-PA, pHW-Sw-HA, pHW-Sw-NP, pHW-Sw-NA, pHW-Sw-M, and pHW-Sw-NS were constructed by reverse transcription-PCR (RT-PCR) amplification of single viral RNA segments and cloning of the resulting cDNAs into the vector pHW2000 (21). In this system, influenza viral cDNA is inserted between the RNA polymerase I (pol I) promoter and terminator sequences. This entire RNA pol I transcription unit is flanked by an RNA pol II promoter and a polyadenylation site. The orientation of the two promoters allows the synthesis of negative-sense viral RNA and positive-sense mRNA from one viral cDNA template. pHW-Sw-NS-73, pHW-Sw-NS-99, and pHW-Sw-NS-126 are derivatives of pHW-Sw-NS. They contain a deletion in the NS1 sequence, plus the insertion of four stop codons in the three frames after this deletion (Fig. 1A). While the nuclear export protein (NEP) open reading frame is not altered in these constructs, the NS1 open reading frames encode only the first 73, 99, and 126 amino acids (aa) of the wild-type NS1 protein, respectively. Primer sequences used in the construction of these plasmids can be provided upon request.
FIG. 1.
Generation of plasmid-derived Sw/Tx/98 influenza viruses with mutated NS1 proteins. (A) Schematic diagram of the wild-type and mutated NS influenza virus gene segments. The NS gene segment was modified to create mutated NS1 genes encoding 73, 99, and 126 aa, respectively. NS1 mutations did not affect the sequence of the NEP. Underlined sequences were introduced to generate stop codons (in bold). (B) RT-PCR analysis of the rWT and NS1 deletion mutant viruses. Influenza viral RNA segments were amplified using primers specific for the noncoding regions of all eight influenza virus segments. Arrows indicate the decreasing size of the NS segment. Dots indicate a product corresponding to the 3′ end of the HA gene segment due to internal binding of the forward primer. Size markers (in nucleotides) are indicated on the left.
Transfection-mediated recovery of recombinant SIV.
Rescue of influenza viruses from plasmid DNA was performed as previously reported using an eight-plasmid system (21). To generate the recombinant wild-type (rWT) TX/98 virus, 1.0 μg of each of the eight plasmids was transfected into 293T-MDCK cocultures using the TransIT LT-1 reagent (Panvera, Madison, WI). The NS1-truncated mutant viruses were generated in the same way but substituting the pHW-Sw-NS plasmid by the corresponding mutant one to recover the 1-73, 1-99, or 1-126 virus mutants. The resulting viruses were passaged and cloned by plaque purification into MDCK cells. Virus stocks were grown in 7-day-old embryonated chicken eggs. The identity of the recombinant SIVs was verified by gel electrophoretic analyses of RT-PCR products derived from viral RNA using oligonucleotides specific for the common noncoding regions. The wild-type NS gene or the deleted versions were further confirmed by sequencing.
Virus growth curves.
To analyze viral replication, confluent PK-15 cells were infected at the indicated multiplicity of infection (MOI) and incubated for different periods of times at 37°C in MEM containing 0.3% bovine albumin (MEM/BA) and 5% allantoic fluid. Virus titers were determined by plaque assay on MDCK cells in MEM/BA supplemented with 1 μg/ml of TPCK trypsin. Titers were expressed as PFU per ml.
Bioassay to measure IFN production.
Levels of IFN secreted by virus-infected cells were determined as previously described (10, 36), with some variations. Confluent PK-15 cells seeded in 22-mm dishes were either mock treated or infected with rWT, 1-73, 1-99, or 1-126 viruses at an MOI of 2. Following infection, cells were incubated with MEM containing 0.3% bovine serum albumin and 5% allantoic fluid, and at different time points postinfection (p.i.) supernatants were harvested. Viruses present in the supernatant were UV inactivated by placing samples on ice 6 in. below an 8-W UV lamp (Fisher) for 15 min with constant stirring. New PK-15 cells were seeded in 96-well plates the day before and incubated with the UV-inactivated supernatants for 24 h. The preincubated PK-15 cells were then infected with VSV-GFP (MOI = 0.1). The cells expressing GFP were visualized by fluorescence microscopy 16 h p.i. The amounts of IFN present in the supernatants were estimated based on their abilities to induce an antiviral state in PK-15 cells and inhibit VSV-GFP replication, compared with known amounts of recombinant human IFN-β (Calbiochem).
Analysis of IFN-β and TNF-α mRNA by RT-PCR.
PK-15 cells were infected at an MOI of 2 and, at 24 h p.i., total RNA was extracted using the Absolutely RNA RT-PCR miniprep kit (Stratagene). RT was performed by using oligo(dT) as the primer. PCR was conducted using specific pairs of primers for swine IFN-β (5-SWIFNB+, GGCCATGGCTAACAAGTGCATCC, 3-SWIFNB, CCGGTCAGTTCCGGAGGTAATC) and tumor necrosis factor alpha (TNF-α) (5′SW-TNFA, ATGAGCACTGAGAGCATG; 3′SW-TNFA, TCACAGGGCAATGATCCC) mRNAs (GenBank accession numbers M86762 and X57321). As control, specific primers for β-actin (10) were used to amplify a 550-bp fragment of swine β-actin. The products were sequenced and confirmed to be derived from the expected mRNAs.
Metabolic labeling.
Confluent PK-15 cells seeded in 22-mm dishes were either mock infected or infected with rWT, 1-73, 1-99, or 1-126 TX/98 viruses at an MOI of 2. Cells were incubated in MEM/BA at 37°C for various times and subsequently labeled for 2 h with 10 μCi of [35S]Met-Cys in MEM lacking Met-Cys. Cells were washed with ice-cold phosphate-buffered saline and lysed, and total cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were visualized by autoradiography.
Antisera.
Antibodies against the first 73 amino acids of NS1 TX/98 were raised as follows. We overproduced the protein in Escherichia coli BL21(DE3) cells from plasmid pGEX-6P-1-(1-73)Sw. This plasmid encodes a protein consisting of glutathione S-transferase (GST) fused to the N terminus (aa 1 to 73) of NS1Tx/98. After expression, the 1-73NS1TX/98 protein was cleaved with the PreScission protease (Amersham) from the GST fusion to inject rabbits (Cocalico Biologicals). Antisera raised against PR8 virus were used to detect NPTX/98.
Western blot analysis.
Dishes (diameter, 22 mm) of confluent PK-15 cells were mock infected or infected with the rWT TX/98 and the different deletion mutants at an MOI of 2. Every 2 h p.i., cells were lysed. Cell lysates were subjected to Western blot analysis by using anti-1-73NS1TX/98 or anti-PR8 antibodies.
Infection of pigs.
Out-bred specific-pathogen-free pigs were obtained from a commercial hog farm in Iowa. Sera from these pigs were confirmed to be negative for the presence of hemagglutination inhibition antibodies against H3N2 (Sw/TX/98 and Sw/CO/99) and H1N1 (Sw/IA/30) SIV. Groups of four to five 4-week-old pigs were housed in separate isolation rooms. The methods for intratracheal infection (using 1 ml of virus solution containing 1 × 105 PFU per pig), clinical observation, necropsy, and macroscopic lung lesion scoring of the pigs were performed as described recently (38). At necropsy, the nasal turbinates, trachea, and right cardiac lobe were removed and formalin fixed for further histopathologic evaluation. Blood, bronchoalveolar fluid (BALF), and nasal swabs were also collected during necropsy. All animal experiments were conducted at the National Animal Disease Center in compliance with the Institutional Animal Care and Use Committee.
Virus titrations from nasal swabs and BALF.
Nasal swabs and BALF were tested to determine virus load. Ten-fold serial dilutions were prepared in McCoy's medium without serum and supplemented with 5 μg/ml trypsin. MDCK cells were inoculated with the dilutions and incubated with medium plus trypsin in microtiter plates at 37°C for 72 h. The plates were examined for cytopathic effects after 72 h. Virus titers were calculated by the Reed and Muench method (37).
Histopathological evaluation.
The nasal turbinates and trachea samples were stained with hematoxylin and eosin and examined under the microscope for epithelial changes and subepithelial inflammation. The lungs were examined for bronchiolar epithelial changes and peribronchiolar inflammation in large, medium, and small or terminal bronchioles. The alveoli were also evaluated for inflammatory changes. Because lesions were found most consistently in medium-sized airways, data obtained from the medium bronchioles were used for comparisons (38). Lesion severity was scored by the distribution or extent of lesions within the sections examined as follows: 0, no airways affected; 1, only a few isolated airways affected; 2, approximately 50% of airways affected; 3, greater than 75% of airways affected. One trained examiner was utilized for evaluation of tissue sections. The examiner was unaware of which group of animals the tissues were derived from.
HI assay.
Hemagglutination inhibition (HI) titers were measured in sera from pigs inoculated with recombinant influenza A viruses by determining the highest serum dilution with the ability to inhibit hemagglutination of chicken red blood cells by WT TX/98 virus.
Statistical analysis.
Mean body temperatures, extent of gross and histopathologic changes, and virus replication in infected and control groups were compared by using the two-sided Student's t test. Probability (P) values of <0.05 were considered to indicate a statistically significant difference between groups.
RESULTS
Generation of plasmid-derived Sw/TX/98 viruses encoding truncated NS1 proteins.
To generate SIV using reverse genetics, we cloned the eight viral RNAs from TX/98 virus into ambisense expression plasmids for viral rescue (21). Upon transfection of these plasmids in 293T-MDCK-cocultured cells, we recovered infectious viruses. The parental (WT) and the plasmid-derived wild-type (rWT) SIVs grew to similar titers in MDCK cells and embryonated eggs (unpublished data). When inoculated into pigs, the rWT virus produced similar lesions as the parental WT virus (unpublished data). We also generated three NS1-truncated SIV mutants encoding NS1 proteins of 73, 99, and 126 aa and compared them to the wild-type full-length, 219-aa-long NS1 protein (Fig. 1A). RT-PCR and sequencing analyses confirmed the presence of the truncated NS1 genes in the rescued virus preparations (Fig. 1B and unpublished data).
NS1 mutant Sw/TX/98 viruses are replication attenuated in pig epithelial cells.
In order to investigate the multicycle growth properties of the mutant NS1 viruses, confluent pig cells (PK-15) were infected at a low MOI (MOI = 0.001). Supernatants from infected cells were titrated at different time points p.i. by plaque assay on MDCK cells. The growth kinetics of the NS1 deletion mutants in PK-15 cells were clearly different compared to the wild-type virus. Interestingly, the 1-126 mutant virus was the most compromised in growth, followed by the 1-99 and the 1-73 mutants (Fig. 2A). Plaque sizes in MDCK cells correlated with the growth differences in PK-15 cells (Fig. 2C). These results indicate that deletions in the NS1 protein of Sw/TX/98 virus result in attenuation of viral growth, both in PK-15 and MDCK cells. In contrast, when NS1 mutant viruses were grown at a high MOI (MOI = 2) in PK-15 cells, major differences in growth kinetics were not detected (Fig. 2B).
FIG. 2.
Characterization of plasmid-derived wild-type and NS1 mutant Sw/TX/98 viruses in tissue culture. (A) Multicycle growth curves in PK-15 cells. (B) Single-cycle growth curves in PK-15 cells. (C) Plaque size in MDCK cells at 3 days p.i.
NS1 mutant Sw/TX/98 viruses induce IFN-α/β in pig epithelial cells.
The NS1 protein of influenza virus has previously been shown to act as an IFN-α/β antagonist (17). In order to see whether the observed attenuation of growth in vitro of the NS1-truncated viruses was directly correlated with the ability of these viruses to inhibit the IFN-α/β system, the induction of IFN in cells infected with the recombinant Sw/TX/98 viruses was investigated. Supernatants of infected PK-15 cells were used to determine the levels of secreted IFN-α/β in a bioassay based on inhibition of VSV-GFP replication (Fig. 3A). For this purpose, PK-15 cells were infected (MOI = 2) with rWT and NS1 mutant SIVs and supernatants were collected for IFN determinations every 2 h for 10 h. The results are shown in Fig. 3B. Supernatants from mock-infected cells caused no inhibition of GFP expression by VSV-GFP in PK-15 cells. In contrast, VSV-GFP replication was completely abolished in cells pretreated with the supernatant of 1-126 virus-infected cells at 10 h p.i. IFN-α/β was barely detected by this assay in supernatants of cells infected with rWT virus at any time point analyzed. All NS1-truncated viruses induced detectable levels of IFN-α/β by 6 h p.i., with 1-126 virus being the stronger IFN-α/β inducer, followed by 1-99 and 1-73 viruses. Similar experiments were performed using other virus-infected swine testis and human (A549) cell lines, with basically identical results (unpublished data). The level of IFN-α/β present in the supernatants of virus-infected PK-15 cells at 10 h p.i. was quantified by performing serial dilutions of the samples and comparing their antiviral activities with standard amounts of recombinant human IFN-β. The amount of IFN-α/β induced by the rWT, 1-126, 1-99, and 1-73 viruses corresponded to approximately 100 U/ml, 10,000 U/ml, 4,000 U/ml, and 1,000 U/ml, respectively.
FIG. 3.
Induction of IFN-α/β in PK-15 cells infected with plasmid-derived wild-type and NS1 mutant Sw/TX/98 viruses. (A) Schematic representation of the IFN bioassay. (B) Time course of IFN-α/β synthesis. Fresh PK-15 cells were treated for 24 h with UV-inactivated supernatants (harvested at different times p.i. as indicated) from PK-15 cells which were infected with the indicated viruses, followed by VSV-GFP infection. Sixteen hours p.i., cells expressing GFP were visualized by fluorescence microscopy. (C) RT-PCR analysis of TNF-α-, IFN-β-, and β-actin-specific mRNAs in virus-infected PK-15 cells.
These results also correlated with the ability of the NS1 mutant viruses to induce expression of IFN-β mRNA. Specifically, RT-PCR analysis of IFN-β and TNF-α transcripts revealed the induction of the expression of the mRNA of these cytokines in PK-15 cells infected with NS1 mutant viruses, especially in 1-126 virus-infected cells (Fig. 3C). Overall, our data indicate that the potency of inhibition of IFN production by the recombinant Sw/TX/98 viruses is as follows: rWT > 1-73 > 1-99 > 1-126.
NS1 mutant Sw/TX/98 proteins are detected at lower levels.
In order to compare the levels of expression of the viral proteins in the different viruses, we labeled virus-infected PK cells (MOI = 2) with [35S]Met-Cys metabolically for 2 hours at different time points p.i. (only one time point, 8 to 10 h, is shown as a representative sample). The gel revealed no differences in the kinetics of NP protein synthesis among the 1-73, 1-99, and 1-126 mutant viruses compared to rWT (Fig. 4A). As previously shown with other strains of influenza viruses, truncations in the NS1 protein resulted in reduced expression of the viral proteins M1 and hemagglutinin (HA) in virus-infected cells (12-14, 39). We were not able to detect the NS1 mutant proteins produced by the 1-73, 1-99, and 1-126 mutant viruses with the [35S]Met-Cys labeling method (Fig. 4A). In addition, antibodies against the first 73 aa of the NS1 protein of Sw/TX/98 were produced. Cells were infected with the rWT or the deletion mutants at an MOI of 2. At different times p.i. (steady state), cells were lysed and the cell extracts were analyzed by Western blotting; only one time point (10 h p.i.) is shown as a representative sample (Fig. 4B). Western analysis revealed that the levels of the NS1-truncated proteins are considerably reduced in comparison to WT NS1, with the 1-126 mutant showing the weakest signal. As a control, no differences were found in the expression of NP protein between these viruses.
FIG. 4.
Levels of rWT and mutant NS1 TX/98 proteins determined by metabolic labeling and Western blotting. (A) Metabolic labeling with [35S]Met-Cys of PK-15 cells infected with the rWT and deleted mutants (MOI = 2), from 8 to 10 h p.i. (B) Immunoblot with antibodies specific for NS1 and NP in PK-15 cells infected with rWT and the different mutants at 10 h p.i.
NS1 mutant Sw/TX/98 viruses are attenuated in pigs.
Forty seven 4-week-old outbred pigs were purchased from a commercial hog farm in Iowa. Groups of 10 4-week-old outbred pigs were intratracheally infected with 105 PFU of rWT and NS1 mutant Sw/TX/98 viruses. Seven animals were mock infected with medium only. The mean rectal temperature increased to ≥39.7°C in the rWT group 2 to 5 days p.i. and in the 1-73, 1-99, and 1-126 groups predominantly on days 2 and 3 p.i. (unpublished data). Additional clinical signs like nasal secretion, depression, sneezing, coughing, and dyspnea were observed starting on day 2 p.i. throughout day 5 p.i., with maximal signs on days 3 and 4 p.i. in the rWT virus-infected group. In the 1-99 and 1-73 virus-infected cohorts, only nasal secretions were found, primarily on days 3 to 5 p.i. In the 1-126 virus-infected pigs, besides elevated body temperature on days 2 and 3 p.i., nasal discharge was observed only on day 5 p.i. in about 50% of the pigs.
During necropsy at day 5 p.i., the percentage of each lung surface with macroscopic lesions was estimated. As shown in Fig. 5A, the mock-infected and the 1-126-infected pigs had no or minimal macroscopic lung lesions. Pigs infected with the rWT Sw/Tx/98 virus showed a significantly higher percentage of macroscopic lung lesions than pigs infected with the NS1 deletion mutant viruses 1-73 (P = 0.018), 1-99 (P = 0.008), or 1-126 (P = 0.0006). In general, the gross lesions we observed were marked, plum-colored, consolidated areas on individual lobes. The diaphragmatic lobes were less involved than the other lobes. The mediastinal lymph nodes were usually hyperemic and enlarged.
FIG. 5.
Infection of 4-week-old pigs with plasmid-derived wild-type and NS1 mutant Sw/TX/98 viruses on day 5 p.i. (A) Mean percentage (± standard error of the mean [SEM]) of lung surface with macroscopic lesions. (B) Microscopic lesions (± SEM) in the medium-sized bronchioles. (C) Virus titers (± SEM) in BALF.
Attenuation of epithelium in nasal turbinates was mild, focal, and inconsistently present in all groups; subepithelial lymphocyte infiltration in nasal turbinates was nearly universal and without correlation to overlying epithelial damage. Tracheal lesions (epithelial attenuation and subepithelial inflammation) were present at all levels of the trachea in all rWT-infected pigs but inconsistently present in pigs infected with the various NS1 mutants.
Virus damage to bronchiolar epithelium varied from acute necrosis and sloughing of epithelial cells, to attenuation of the epithelial layer as remaining cells spread out to cover the defect resulting from loss of necrotic cells, to irregular reactive proliferation of the epithelial lining of the airway. Peribronchiolar inflammation of each airway tended to correspond to damage to the epithelial lining. Thickening of alveolar walls (interstitial pneumonia) and accumulation of leukocytes in alveolar lumens were minimal and were most evident in lungs with extensive bronchiolar damage. Consistent with previous studies of SIV infection in swine, in pigs infected with rWT virus in this study bronchiolar damage rarely affected all airways in the sections examined. Lesions were observed most consistently in medium-sized airways; thus, data obtained for the medium bronchioles were used for comparison of rWT and NS1 mutant viruses (Fig. 5B).
Mock-infected animals as well as 1-126 virus-infected pigs showed no or minimal lesions, whereas moderate to severe lesions were detected in animals infected with the rWT and the 1-99 and 1-73 Sw/TX/98 viruses (Fig. 4A and B, 6, and 7). All animals infected with the parental wild type and some infected with the 1-73 and 1-99 viruses had high scores, reflecting widespread damage to bronchioles. Most animals infected with either 1-73 or 1-99 virus had a moderate score, reflecting distribution of bronchiolar damage that was intermediate between the extensive damage in rWT-infected pigs and the minimal damage in pigs infected with 1-126. The lungs of animals infected with the NS1 deletion mutant 1-126 virus were nearly devoid of lesions. As in the macroscopic lung lesions, 1-126- and 1-99-infected pigs showed significantly less damage than rWT-infected pigs (1-126, P < 0.0001; 1-99, P = 0.0002). Interestingly, microscopic lesions in bronchioles of pigs infected with the 1-73 mutant virus were also significantly less than with the parental rWT virus (P = 0.02).
FIG. 6.
Histopathological examination of lungs from pigs infected with plasmid-derived wild-type and NS1 mutant Sw/TX/98 viruses. Magnification, ×200. (Mock) Medium-sized bronchiole from the lung of a mock-infected control pig. (rWT) Severe acute necrotizing bronchiolitis and interstitial pneumonia characteristic of the widespread lesion induced by infection with rWT TX/98 virus. (1-99) One normal bronchiole and one affected bronchiole in early recovery from infection with deletion mutant 1-99 virus. (1-126) Normal bronchiole and surrounding alveoli from the lung of a pig inoculated with the 1-126 virus. Bar, 50 μm.
FIG. 7.
Histopathological examination of lungs from pigs infected with plasmid-derived wild-type and NS1 mutant Sw/TX/98 viruses. Magnification, ×500. (Mock) Epithelial lining of a larger bronchiole from the lung of a mock-infected control pig. (rWT) Epithelial lining from a larger bronchiole from the lung of a pig infected with TX/98 rWT virus. (1-99) Large bronchiole and smaller branching airway from the lung of a pig infected with deletion mutant 1-99 virus. (1-126) Normal epithelial lining from the lung of a pig inoculated with 1-126 virus. Bar, 200 μm.
Virus titers in the BALF were also analyzed at day 5 p.i. All viruses replicated in the respiratory tract of pigs, and the virus titers in animals infected with parental rWT virus were significantly higher (P < 0.01) than with the NS1 deletion mutants (Fig. 5C). This finding correlates with significantly less-extensive gross and microscopic disease observed in pigs infected with NS1 deletion mutants versus wild-type Sw/Tx/98 viruses.
Nasal swabs were taken on days 4 and 5 p.i. from five pigs each infected with rWT, 1-73, 1-99, and 1-126 viruses. All five pigs infected with rWT virus were virus positive at days 4 and 5 p.i., whereas no virus was recovered from the pigs infected with 1-99 and 1-126 viruses (starting dilution for virus titration, 1:25). Virus was recovered from pigs infected with the 1-73 virus on day 4 (1/5) and 5 (3/5) p.i.
When we analyzed the presence of antibodies against wild-type Sw/Tx/98 virus in sera from infected pigs by the HI assay, we found that rWT as well as the attenuated 1-99 and 1-126 NS1 mutant viruses induced detectable HI antibodies in four out of four animals by day 8 p.i., with average HI titers of 80 (rWT), 30 (1-99), and 45 (1-126) at day 14 p.i. Thus, 1-99 and 1-126 viruses appear to be immunogenic despite their lower levels of replication in pigs. Similarly, a mouse-adapted influenza virus strain (A/PR/8/34 virus) containing NS1 mutations was also immunogenic in mice despite limited replication in lungs (12, 49, 50).
DISCUSSION
Nonspecific innate immune responses to viruses include the production of cytokines, particularly IFN-α/β. However, most viruses have developed different mechanisms to evade the host IFN-mediated antiviral response (27). In the case of influenza A virus, the NS1 protein appears to have a prominent role in inhibiting the IFN system by attenuating the production of IFN-α/β during viral infection (8, 16, 51). However, these studies have mostly been conducted in vitro or using rodent models of influenza virus infection. While informative, it remained to be demonstrated whether the IFN-antagonistic properties of the NS1 protein have a role in the pathogenicity of influenza virus in humans or other natural hosts. To investigate the role of this viral protein in the pathogenicity of influenza virus in a natural host, we used a swine model of influenza virus infection. Pigs are a natural host for influenza virus infections, and swine influenza disease is closely related to human influenza (11, 24, 40, 55). The disease consists of an acute febrile respiratory disease characterized by fever, apathy, anorexia, and labored breathing. The gross lesions found in uncomplicated swine influenza are mainly those of a viral pneumonia and are most often limited to the apical and cardiac lobes of the lungs, although in severe cases more than a half of the lung may be affected. In our studies we have used a naturally occurring strain of swine influenza virus, influenza A/Swine/Texas/4199-2/98 virus, that has undergone only a very limited number of passages in vitro.
We first generated a recombinant wild-type TX/98 SIV from plasmid DNA. The recombinant wild-type virus induced clinical symptoms and respiratory lesions indistinguishable from the parental virus in experimentally infected pigs (unpublished data). Three recombinant TX/98 SIVs encoding partially carboxy-terminally truncated NS1 proteins of 73, 99, and 126 aa in length were then generated. All NS1-truncated SIVs were attenuated in replication in vitro and in vivo, and the level of attenuation correlated with the amounts of IFN-α/β induced by the respective NS1-mutated viruses, demonstrating a role of the NS1 of SIV in virulence in its natural host. These data support the concept that highly virulent influenza virus strains, such as the 1918 “Spanish” influenza virus and the H5N1 Hong Kong viruses isolated in 1997, express NS1 proteins with strong IFN-antagonistic properties (18, 45).
Using mouse-adapted influenza virus strains, the length of the NS1 protein has been shown to be correlated with attenuation of replication in vitro and in mice (12, 49, 50). However, in the case of Sw/TX/98 virus, the degree of attenuation did not correlate with the length of the NS1 protein, since the mutant SIVs encoding the shorter NS1 proteins (1-73 and 1-99) were significantly more virulent than the 1-126 virus. All NS1 mutant SIVs express the dsRNA-binding domain of NS1, located within the first 73 amino acids (6, 7), which mediates inhibition of IFN synthesis (49, 50), and lack the NS1 effector domain involved in general inhibition of gene expression (31). Interestingly, the wild-type NS1 protein of Sw/TX/98 virus has only 219 amino acids and lacks the poly(A)-binding protein binding domain that has been shown to be involved in inhibition of host cell mRNA polyadenylation (26). In addition, the NS1 proteins from the 1-126 and 1-99 viruses, but not the 1-73 virus, contain a domain that has been shown to bind to eukaryotic initiation factor 4GI and stimulate translation of viral mRNAs (1, 4). Another function that has recently been shown is that the carboxy-terminal domain of the NS1 protein of influenza virus stabilizes NS1 dimerization, which is necessary for its IFN-antagonistic function (50). A possible explanation for the different degree of virulence of the NS1 mutant SIVs is that truncated Sw/TX/98 NS1 proteins of 73 and 99 aa are more stable than the truncated NS1 of 126 aa. Interestingly, by metabolic labeling we could not detect the truncated NS1 in 1-126 virus-infected cells (Fig. 4A), despite the fact that this mutant NS1 protein retains all the methionines and cysteines of the wild-type NS1 protein. By Western blotting we have determined that while the expression level of NP is similar in cells infected with the WT and the mutant viruses, the amount of the 1-126 NS1 protein is very low. This suggests that a truncated Sw/TX/98 virus NS1 protein of 126 amino acids is highly unstable in infected cells. Why this truncated protein is less stable in the SIV TX/98 virus than the same-length protein in WSN (50) needs to be further studied. Alternatively, the mutations inserted in the NS gene might have differentially affected the levels of replication, transcription, splicing, or stability of the NS-derived RNAs, affecting the levels of NS1 and maybe also of NEP expression. We do not favor this explanation, since our deletion strategy did not affect any known RNA cis-acting signal in the NS gene. In any case, we conclude from our studies that not only the functional domains present in the NS1-deleted proteins but also the absolute amount of the mutated NS1 proteins present in infected cells are determinants of virulence, i.e., the ability to antagonize the host IFN response. This also appears to be the case when NS1 truncations are made in the context of equine influenza A viruses (P. Palese, personal communication).
The NS1 mutant SIVs not only induced higher levels of IFN-α/β, but also of TNF-α, a cytokine that has also been shown to possess antiviral activity against influenza viruses (46, 47). Although the mechanism of NS1-mediated inhibition of TNF-α expression is unclear at the moment, TNF-α inhibition might also contribute to the attenuated phenotype of the NS1 mutant viruses. Finally, we cannot exclude that the loss of additional NS1 functions might be partly responsible for the phenotype of the NS1 mutant SIVs. Interestingly, it has recently been found that NS1 is able to inhibit RNA silencing in insects and plants (3, 9, 28), and this may indicate that animals may have also evolved antiviral small inhibitory RNA systems that are inhibited by the NS1 protein of influenza virus.
Swine influenza is widespread and endemic in pig populations worldwide and is part of the porcine respiratory disease complex. The high morbidity associated with swine influenza has serious economic consequences (24). In addition to the severe disease that the virus causes in swine, pigs may act as an intermediate host and “mixing vessel” for genetic reassortment between human and avian viruses (42), facilitating the generation of novel human influenza virus strains and the initiation of influenza pandemics. Although killed SIV vaccines are available, live attenuated vaccines may afford better and faster protection. For instance, live attenuated vaccines based on cold adaptation in tissue culture have been developed for both human and equine influenza viruses (30, 56). NS1-truncated influenza virus mutants have been proven attenuated and immunogenic in mice (49) and in pigs (this study). Future studies aimed towards a more detailed immunogenic characterization of mutant NS1 SIVs in pigs are planned to evaluate the potential efficacy of these viruses as vaccines against swine influenza.
Acknowledgments
We acknowledge Peter Palese for critical advice and discussions and John Hiscott for kindly providing VSV-GFP virus. We also gratefully acknowledge Deb Adolphson, Richard Cadagan, and Deb Clouser for excellent technical assistance.
This work was partially supported by NIH grants to A.G.-S. A.S. is a recipient of a postdoctoral fellowship from the Ministerio de Educación, Cultura y Deporte from Spain.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Aragón, T., S. de La Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucher, E., H. Hemmes, P. de Haan, R. Goldbach, and M. Prins. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85:983-991. [DOI] [PubMed] [Google Scholar]
- 4.Burgui, I., T. Aragon, J. Ortin, and A. Nieto. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, T. M., V. S. Hinshaw, Y. Kawaoka, B. C. Easterday, and R. G. Webster. 1991. Influenza viral infection of swine in the United States 1988-1989. Arch. Virol. 116:261-265. [DOI] [PubMed] [Google Scholar]
- 6.Chien, C. Y., R. Tejero, Y. Huang, D. E. Zimmerman, C. B. Rios, R. M. Krug, and G. T. Montelione. 1997. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat. Struct. Biol. 4:891-895. [DOI] [PubMed] [Google Scholar]
- 7.Chien, C. Y., Y. Xu, R. Xiao, J. M. Aramini, P. V. Sahasrabudhe, R. M. Krug, and G. T. Montelione. 2004. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 43:1950-1962. [DOI] [PubMed] [Google Scholar]
- 8.Dauber, B., G. Heins, and T. Wolff. 2004. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J. Virol. 78:1865-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgadillo, M. O., P. Saenz, B. Salvador, J. A. Garcia, and C. Simon-Mateo. 2004. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 85:993-999. [DOI] [PubMed] [Google Scholar]
- 10.Donelan, N., C. F. Basler, and A. García-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding defective NS1 protein induces high levels of IFN-β and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Easterday, B. C. 1980. Animals in the influenza world. Philos. Trans. R. Soc. London B 288:433-437. [DOI] [PubMed] [Google Scholar]
- 12.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enami, M., and K. Enami. 2000. Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system. J. Virol. 74:5556-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falcon, A. M., R. M. Marion, T. Zurcher, P. Gomez, A. Portela, A. Nieto, and J. Ortin. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78:3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 16.García-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A virus and other negative strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 17.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 18.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. García-Sastre. 2002. Cellular transcriptional profiling in influenza A virus infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinshaw, V. S., W. J. Bean, Jr., R. G. Webster, and B. C. Easterday. 1978. The prevalence of influenza viruses in swine and the antigenic and genetic relatedness of influenza viruses from man and swine. Virology 84:51-62. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karasin, A. I., J. Landgraf, S. Swenson, G. Erickson, S. Goyal, M. Woodruff, G. Scherba, G. Anderson, and C. W. Olsen. 2002. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J. Clin. Microbiol. 40:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 24.Kay, R. M., S. H. Done, and D. J. Paton. 1994. Effect of sequential porcine reproductive and respiratory syndrome and swine influenza on the growth and performance of finishing pigs. Vet. Rec. 135:199-204. [DOI] [PubMed] [Google Scholar]
- 25.Kida, H., T. Ito, J. Yasuda, Y. Shimizu, C. Itakura, K. F. Shortridge, Y. Kawaoka, and R. G. Webster. 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75:2183-2188. [DOI] [PubMed] [Google Scholar]
- 26.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 27.Levy, D. E., and A. García-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 28.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. García-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 30.Maassab, H. F., J. R. LaMontagne, and D. C. DeBorde. 1998. Live influenza virus vaccines, p. 435-457. In S. A. Plotkin and E. A. Mortimer (ed.), Vaccines. Saunders, Philadelphia, Pa.
- 31.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 32.Nemeroff, M. E., X. Y. Qian, and R. M. Krug. 1995. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212:422-428. [DOI] [PubMed] [Google Scholar]
- 33.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 34.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199-210. [DOI] [PubMed] [Google Scholar]
- 35.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, M. S., A. García-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77:9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50% endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 38.Richt, J. A., K. M. Lager, B. H. Janke, R. D. Woods, R. G. Webster, and R. J. Webby. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 41:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvatore, M., C. F. Basler, J.-P. Parisien, C. M. Horvath, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. García-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholtissek, C. 1995. Molecular evolution of influenza viruses. Virus Genes. 11:209-215. [DOI] [PubMed] [Google Scholar]
- 41.Scholtissek, C. 1994. Source for influenza pandemics. Eur. J. Epidemiol. 10:455-458. [DOI] [PubMed] [Google Scholar]
- 42.Scholtissek, C., V. S. Hinshaw, and C. W. Olsen. 1998. Influenza in pigs and their role as the intermediate host, p. 137-145. In K. G. Nicholson, R. G. Webster, and A. J. Hay (ed.), Textbook of influenza. Blackwell Science, Oxford, England.
- 43.Sekellick, M. J., W. J. Biggers, and P. I. Marcus. 1990. Development of the interferon system. I. In chicken cells development in ovo continues on time in vitro. In Vitro Cell. Dev. Biol. 26:997-1003. [DOI] [PubMed] [Google Scholar]
- 44.Sekellick, M. J., and P. I. Marcus. 1985. Interferon induction by viruses. XIV. Development of interferon inducibility and its inhibition in chick embryo cells “aged” in vitro. J. Interferon Res. 5:651-667. [DOI] [PubMed] [Google Scholar]
- 45.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 46.Seo, S. H., E. Hoffmann, and R. G. Webster. 2004. The NS1 gene of H5N1 influenza viruses circumvents the host anti-viral cytokine responses. Virus Res. 103:107-113. [DOI] [PubMed] [Google Scholar]
- 47.Seo, S. H., and R. G. Webster. 2002. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J. Virol. 76:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stojdl, D. F., B. D. Lichty, B. R. tenOever, J. M. Paterson, A. T. Power, S. Knowles, R. Marius, J. Reynard, L. Poliquin, H. Atkins, E. G. Brown, R. K. Durbin, J. E. Durbin, J. Hiscott, and J. C. Bell. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263-275. [DOI] [PubMed] [Google Scholar]
- 49.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. García-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., C. F. Basler, B. R. G. Williams, R. H. Silverman, P. Palese, and A. García-Sastre. 2002. Functional replacement of the carboxy-terminal two thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 76:12951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents the activation of NF-κB and induction of type I IFN. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webby, R. J., K. Rossow, G. Erickson, Y. Sims, and R. Webster. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res. 103:67-73. [DOI] [PubMed] [Google Scholar]
- 53.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webby, R. J., and R. G. Webster. 2001. Emergence of influenza A viruses. Philos. Trans. R. Soc. London B 356:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webster, R. G. 2002. The importance of animal influenza for human disease. Vaccine 20:S16-S20. [DOI] [PubMed] [Google Scholar]
- 56.Youngner, J. S., P. Whitaker-Dowling, T. M. Chambers, K. E. Rushlow, and R. Sebring. 2001. Derivation and characterization of a live attenuated equine influenza vaccine virus. Am. J. Vet. Res. 62:1290-1294. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]