Abstract
Venezuelan equine encephalitis (VEE) and eastern equine encephalitis (EEE) viruses are important, naturally emerging zoonotic viruses. They are significant human and equine pathogens which still pose a serious public health threat. Both VEE and EEE cause chronic infection in mosquitoes and persistent or chronic infection in mosquito-derived cell lines. In contrast, vertebrate hosts infected with either virus develop an acute infection with high-titer viremia and encephalitis, followed by host death or virus clearance by the immune system. Accordingly, EEE and VEE infection in vertebrate cell lines is highly cytopathic. To further understand the pathogenesis of alphaviruses on molecular and cellular levels, we designed EEE- and VEE-based replicons and investigated their replication and their ability to generate cytopathic effect (CPE) and to interfere with other viral infections. VEE and EEE replicons appeared to be less cytopathic than Sindbis virus-based constructs that we designed in our previous research and readily established persistent replication in BHK-21 cells. VEE replicons required additional mutations in the 5′ untranslated region and nsP2 or nsP3 genes to further reduce cytopathicity and to become capable of persisting in cells with no defects in alpha/beta interferon production or signaling. The results indicated that alphaviruses strongly differ in virus-host cell interactions, and the ability to cause CPE in tissue culture does not necessarily correlate with pathogenesis and strongly depends on the sequence of viral nonstructural proteins.
Alphaviruses are a group of widely distributed human and animal pathogens that includes almost 30 currently known species (39). In natural conditions, alphaviruses are transmitted by mosquitoes that develop lifelong chronic infection (42). The presence of virus in the mosquito salivary glands mediates infection of vertebrates that serve as amplifying hosts, producing high-titer viremia until the virus is cleared by their immune system (14). In accordance with their life cycle, alphaviruses develop different infections in tissue culture. They cause cytopathic infection in commonly used cell lines of mammalian and avian origin, in which the death of the infected cells occurs within 24 to 48 h postinfection (10, 19). In contrast, mosquito-derived cell lines remain persistently or chronically infected during many passages without displaying obvious cytopathic effect (CPE) (37).
The alphavirus genome consists of a single-stranded RNA molecule of positive polarity approximately 11.7 kb in length (21, 38, 40). It contains a domain of nonstructural proteins (nsP1 to nsP4) encoded by the 5′ two-thirds of the genome. These proteins are translated directly from the genomic RNA and form, together with cellular proteins, the RNA-dependent RNA polymerase (RdRp) essential for viral genome replication and transcription of a subgenomic RNA. The latter serves as a template for translation of all of the structural proteins comprising viral particles. In spite of a high level of sequence similarity in the structural and nonstructural proteins, alphaviruses significantly differ in their ability to cause disease. In humans, the Old World alphaviruses, such as Sindbis virus (SIN), Semliki Forest virus (SFV), and O'nyong nyong virus, develop self-limited febrile illness characterized by fever, skin rashes, and arthritis (15). Some of the New World viruses, including the Venezuelan equine encephalitis (VEE), eastern equine encephalitis (EEE), and western equine encephalitis (WEE) viruses, cause severe encephalitis in humans and animals that can result in death or neurological sequelae (5, 15, 17, 24). Pathogenesis of the alphaviruses is likely to be determined by many factors that control virus-host interactions, and our understanding of the processes is far from complete.
SIN is one of the least pathogenic but most intensively studied members of the alphavirus genus. It can efficiently replicate in many commonly used vertebrate cell lines. Within a few hours postinfection, it critically affects the intracellular environment by downregulating cellular macromolecular synthesis (10, 11). This inhibition of transcription and translation of cellular mRNAs, high-level expression of virus-specific proteins, and development of apoptosis accompany cell death (10, 19, 25, 26).
SIN structural proteins are dispensable for RNA replication. Viral genomes lacking structural genes (replicons) can efficiently replicate themselves, and their replication and/or accumulation of their encoded nonstructural proteins results in cell death (3, 10). SIN, SFV, and VEE replicons have become important tools for studying the mechanism of alphavirus RNA replication and different aspects of virus-host cell interactions (3, 27, 33). Previous studies showed that SIN replicons expressing selectable markers can accumulate adaptive mutations in the nsP2-encoding gene (9, 32). These mutations made replicons noncytopathic and capable of persisting in some vertebrate cell lines for an unlimited number of passages. Colocalization of the mutations leading to a noncytopathic phenotype in the same short fragment of nsP2 suggested that, in addition to being an RdRp component, this protein may play a critical role(s) in virus-host cell interactions.
Compared to SIN, replication and virus-host cell interactions of other alphaviruses, particularly VEE, EEE, and WEE, are less well characterized. However, it is expected that differences in pathogenesis are caused not only by the structures of the viral envelope glycoproteins that determine tissue specificity but also by viral nonstructural proteins and/or RNA replication. To further understand the differences in the pathogenesis of alphaviruses on the molecular and cellular levels, we designed EEE- and VEE-based replicons and assayed their replication as well as their abilities to cause CPE and to interfere with other viral infections. Our results demonstrated that VEE and EEE replicons were less cytopathic than similar wild-type (wt) SIN-based constructs. They readily established persistent replication in BHK-21 cells and appeared to cause fewer changes than SIN in the intracellular environment that can lead to cell death. VEE TC-83 strain-based replicons required a mutation in the 5′ untranslated region (UTR) and acquired additional mutations in nsP2 or nsP3 genes when they were forced to persist in the cells with intact alpha/beta interferon (IFN-α/β) production and signaling. These adapted replicons may be potentially useful for the development of cell lines for screening antiviral drugs and for expressing heterologous proteins.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, Mo.). NIH 3T3 cells were obtained from the American Type Tissue Culture Collection (Manassas, Va.). HeLa cells were kindly provided by Eugene Agapov (Washington University, St. Louis). These cell lines were maintained at 37°C in alpha minimum essential medium supplemented with 10% fetal bovine serum (FBS) and vitamins. Huh-7 cells were kindly provided by Charles M. Rice (Rockefeller University, New York) and were propagated in Dulbecco's minimal essential medium supplemented with 10% FBS.
Plasmid constructs.
The high-copy-number plasmid encoding the EEE nonstructural proteins from the genome of North American strain Florida 91 was provided by Michael Parker (U.S. Army Medical Research Institute for Infectious Diseases). The high-copy-number plasmid carrying the VEE TC-83 strain genome was described elsewhere (20). TC-83 is a live attenuated vaccine strain of VEE that is available for vaccination of laboratory workers and military personnel. It was developed by serial passaging of the virulent, subtype IAB Trinidad donkey (TRD) VEE strain in cell culture (2). SINrep/L/Pac and SINrep/Pac replicon-encoding plasmids were described elsewhere (9). They were previously referred to as pSINrep19/Pac and pTSG/Pac, respectively. The names were changed to make them consistent with other names of the constructs developed in this work. Standard recombinant DNA techniques were used for all plasmid constructions. All of the required sequence modifications were carried out using PCR-based mutagenesis, and sequences were verified by sequencing of the cloned fragments. The genetic material from the VEE TC-83 strain and the EEE North American strain Florida 91 was cloned into a pToto1101 plasmid (34) to replace the entire SIN genome. Maps and sequences are available from the authors upon request. The construct, pEEErep/Pac, contained the promoter for SP6 RNA polymerase, followed by nucleotides (nt) 1 to 7594 of the EEE genome, a synthetic TCTAGAGCTTACC sequence that came with a puromycin acetyltransferase (Pac)-encoding cassette, a 764-nt-long sequence containing the entire gene of the selectable marker Pac, and a 356-nt-long sequence of the EEE 3′ UTR followed by poly(A) and a NotI restriction site. pVEErep/Pac contained the promoter for SP6 RNA polymerase, followed by nt 1 to 7561 of the VEE TC-83 genome, a synthetic TCTAGAGCTTACC sequence, a 676-nt-long sequence encoding the entire Pac gene, and a 330-nt-long sequence of VEE TC-83 that included the 3′ end of the E1-encoding gene and the 3′ UTR followed by poly(A) and an MluI restriction site. p5′VEErep/Pac carried essentially the same replicon with a single substitution: the A in the third position of the 5′ UTR was replaced by G. pEEErep/SEAP/Pac also contained an SP6 promoter, followed by nt 1 to 7594 of the EEE genome, a synthetic TCTAGGTGAGC sequence that came with a secreted alkaline phosphatase (SEAP)-encoding cassette, a 1597-nt-long sequence carrying the entire sequence of the SEAP gene derived from pSEAP2-Basic (Clontech), nt 7183 to 7594 of the EEE genome that encoded the subgenomic promoter, a synthetic TCTAGAGCTTACC sequence, a 764-nt-long Pac gene, and a 356-nt-long sequence of the EEE 3′ UTR followed by poly(A) and a NotI restriction site. pEEErep/GFP/Pac had a structure similar to that of pEEErep/SEAP/Pac, but the SEAP gene was replaced by a green fluorescent protein (GFP) coding sequence. p5′VEErep/SEAP/Pac contained the promoter for SP6 RNA polymerase, followed by nt 1 to 7561 of the VEE TC-83 genome (with A3 in the 5′ UTR replaced by G), a synthetic TCTAGGTGAGC sequence, a 1597-nt-long sequence carrying the entire SEAP gene, nt 7501 to 7561 of the VEE genome (carrying the subgenomic promoter), a synthetic TCTAGAGCTTACC sequence, a 676-nt-long sequence carrying the entire Pac gene, and a 330-nt-long sequence of VEE TC-83 that included the 3′ end of the E1-encoding gene and the 3′ UTR followed by poly(A) and an MluI restriction site. p5′VEErep/GFP/Pac had a similar structure, but the SEAP gene was replaced by a GFP coding sequence. p5′VEE/SIN carried the promoter for SP6 RNA polymerase, followed by nt 1 to 7535 of the VEE TC-83 genome (with A3 in the 5′ UTR replaced by G), nt 7601 to 11486 of the SIN genome carrying the SIN structural genes (with T7626 replaced by C to preserve the secondary structure of the 5′ UTR in the subgenomic RNA), a 273-nt-long fragment of the VEE TC-83 genome containing the 3′ end of the E1 coding sequence, the 3′ UTR, poly(A), and an MluI restriction site.
RNA transcriptions.
Plasmids were purified by centrifugation in CsCl gradients. Before the transcription reaction, the viral, replicon, or helper genome- coding plasmids were linearized using the restriction sites located downstream of the poly(A) sequence. RNAs were synthesized by SP6 RNA polymerase in the presence of cap analog using previously described conditions (34). The yield and integrity of transcripts were evaluated by gel electrophoresis under nondenaturing conditions followed by analysis on an Alpha Innotech imaging system. Aliquots of transcription reactions were used for electroporation without additional purification.
RNA transfections.
BHK-21 cells were electroporated using previously described conditions (28). For testing the colony-forming activity of replicons in different experiments, 1 or 4 μg of in vitro-synthesized RNAs was transfected into 5 × 106 cells. Different aliquots of electroporated cells were seeded into 100-mm tissue culture dishes. For the selection of Purr cells, puromycin was added to the medium at 24 h postelectroporation. To rescue 5′VEE/SIN virus, 1 μg of in vitro-synthesized viral genome RNAs was electroporated into the BHK-21 cells, and they were seeded into 100-mm dishes and incubated until the CPE was observed (between 20 and 24 h posttransfection). This virus stock was used for all of the experiments without additional passaging.
Infectious center assay.
In standard experiments, 1 μg of in vitro-synthesized, full-length RNA transcripts of viral genomes was used per electroporation. Tenfold dilutions of electroporated BHK-21 cells were seeded in six-well Costar plates containing subconfluent naïve cells. After 1 h of incubation at 37°C in a 5% CO2 incubator, cells were overlaid with 2 ml of minimum essential medium containing 0.5% Ultra-Pure agarose supplemented with 3% FBS. Plaques were stained with crystal violet after 2 days' incubation at 37°C.
Viral replication analysis.
BHK-21 cells were seeded at a concentration of 5 × 105 cells/35-mm dish. After 4 h of incubation at 37°C, monolayers were infected with the multiplicity of infection (MOI) indicated in figure legends for 1 h, washed three times with phosphate-buffered saline, and overlaid with 1 ml of complete medium. At the indicated times postinfection, media were replaced by fresh media, and virus titers in the harvested samples were determined by plaque assay on BHK-21 cells as previously described (22).
RNA analysis.
Cells were infected at the MOIs indicated in the figure legends with viruses or virus-specific RNAs. RNAs were labeled with [3H]uridine as described in the legends and were isolated from the cells by TRIzol reagent, as recommended by the manufacturer (Invitrogen). They were denatured with glyoxal in dimethyl sulfoxide and analyzed by agarose gel electrophoresis using the previously described conditions (3).
Development of EEE and VEE replicon-containing cell lines expressing alkaline phosphatase.
Four micrograms of in vitro-synthesized RNAs was transfected into 5 × 106 BHK-21 cells. Different aliquots of electroporated cells were seeded into 100-mm tissue culture dishes. For the selection of Purr cells, puromycin was added to the media at 24 h postelectroporation. Colonies of Purr cells were randomly selected and expanded, and stocks of the cells were used for a second selection of Purr colonies and further propagation. This additional cell cloning was expected to make a more homogeneous cell population. Alkaline phosphatase activity in the media was measured using a Great EscAPe SEAP detection kit (BD Biosciences) according to the manufacturer's instructions.
Identification of the adaptive mutations in VEE replicons.
Four micrograms of in vitro-synthesized RNAs was transfected into 5 × 106 Huh-7 cells. Cells were seeded into 100-mm tissue culture dishes, and Purr colonies were selected for 2 weeks. Total RNA was isolated from all of the expanded colonies, and the amount equivalent to 106 cells was transfected into Huh-7 followed by selection in the presence of puromycin. RNA was isolated from three randomly selected Purr cell clones, and RNAs were isolated by TRIzol using the procedure recommended by the manufacturer (Invitrogen). Overlapping fragments approximately 1,000 nt long were synthesized by using VEE TC-83-specific primers with a minimal number of cycles applied (usually between 15 and 20 cycles of PCR). After purification on agarose gels, fragments were sequenced without cloning. Mutation-containing fragments were cloned into pRS2 (an analog of pUC19), sequenced (to confirm the consensus sequence), and cloned into a 5′VEErep/Pac genome to confirm their effect on the replication of VEE-specific RNAs. DNA fragments representing the 5′ terminal sequence of virus isolates were synthesized using a commercially available FirstChoice RLM-rapid amplification of cDNA ends kit according to the procedures recommended by the manufacturer (Ambion). Fragments were purified by agarose gel electrophoresis and cloned into the plasmid pRS2. Multiple, independent clones were sequenced to determine variations in the 5′ ends of the genomes.
RESULTS
VEE and EEE replicons are less cytopathic than wt SIN-based constructs.
In order to compare the in vitro replication of VEE and EEE replicons with similar SIN-based (SINrep) constructs (3), we created cDNA copies of both replicons in low-copy-number plasmids. The use of low-copy-number plasmids made replicon- and entire viral genome-coding plasmids stable during propagation in Escherichia coli. Original cloned genetic material from the VEE TC-83 strain and EEE North American strain Florida 91 was recloned under control of the SP6 promoters into a pToto1101 plasmid to replace the SIN genome (see Materials and Methods for details). The final constructs contained the 5′ and 3′ cis-acting elements required for replication, the nsP1 to nsP4 genes, and the selectable marker Pac gene cloned under control of viral subgenomic promoters (Fig. 1). Two variants of the VEE replicon were generated. One of these (VEErep/Pac) contained the TC-83-derived 5′ UTR followed by TC-83 nonstructural proteins, whereas the second (5′VEErep/Pac) differed in a single position: the adenine in the third position of the 5′ UTR was replaced by guanine. This mutation made the replicon's 5′ UTR identical to that found in the epizootic, highly pathogenic VEE TRD strain. The mutation in this position was previously shown to play a critical role in determining the interferon sensitivity of VEE (20, 44). Equal amounts of the in vitro-synthesized replicons' RNAs (between 1 and 4 μg in different experiments) were transfected into BHK-21 cells, and 24 h later, selection with puromycin was applied. The previously described noncytopathic SINrep/L/Pac replicon (9) (with the nsP2 P726→L mutation) and cytopathic, wt SIN replicon SINrep/Pac were used in these experiments as reference controls for comparing the efficiencies of Purr colony formation.
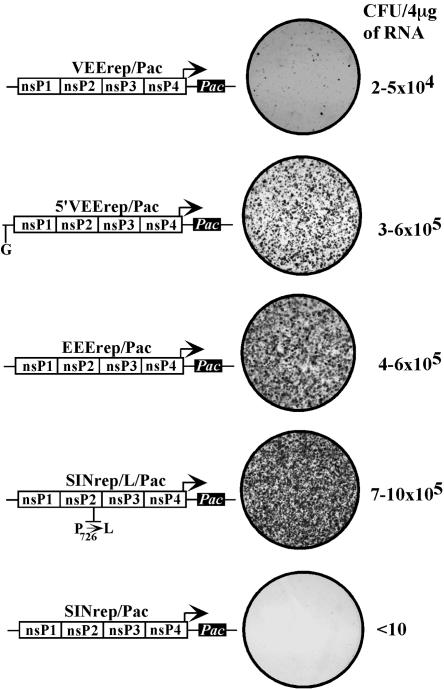
FIG. 1.
Schematic representation of replicons and their ability to persistently replicate in BHK-21 cells. A detailed description of the replicon constructs is presented in Materials and Methods. Arrows indicate the positions of the subgenomic promoters. The positions of the mutations in the VEE 5′ UTR and SIN nsP2 coding gene are indicated. BHK-21 cells were transfected by 4 μg of each in vitro-synthesized replicon RNA using electroporation, and different dilutions of the cells were seeded into 100-mm dishes. Puromycin selection (10 μg/ml) was applied 24 h posttransfection. After 5 days of incubation under puromycin, dishes containing 1% of electroporated cells were stained with crystal violet. The efficiency of focus formation was calculated based on the number of foci in the dishes containing fewer electroporated cells.
Surprisingly, this assay revealed that both VEE- and EEE-based replicons easily established persistent replication. EEErep/Pac formed Purr colonies with an efficiency similar to that found for the mutated SIN replicon SINrep/L/Pac. The replicon-containing cells had the same morphology as naïve BHK-21 cells and demonstrated similar growth rates (Fig. 1 and data not shown). The experiments also demonstrated that compared to EEErep/Pac, the VEErep/Pac replicon was noticeably more cytopathic, and between 10- and 20-fold fewer cells formed Purr colonies. At the early time point, within 1 to 2 days after transfection with VEErep/Pac, all of the cells were Purr, but the majority of them eventually died within the next few days, most likely due to changes in cell metabolism caused by the replication of virus-specific RNAs. The surviving cells exhibited morphological changes and slower growth (Fig. 1). The TRD-specific single point mutation in the 5′ UTR of 5′VEErep/Pac (A3→G) (20) was critical for the noncytopathic phenotype of the VEE replicon. This single nucleotide change decreased the cytopathicity of the 5′VEE replicon, reduced its negative effect on cell growth (compare the sizes of colonies in Fig. 1), and strongly increased the efficiency of forming Purr colonies to the level of noncytopathic SINrep/L/Pac or EEErep/Pac. In agreement with our previous data (9), the control SINrep/Pac replicon, containing nonstructural genes derived from wt SIN, was highly cytopathic and formed Purr foci 5 orders of magnitude less efficiently than both EEErep/Pac and 5′VEErep/Pac constructs (fewer than 10 colonies of Purr cells per μg of transfected RNA).
Taken together, the results indicated that, compared to wt SIN- and wt SFV-derived constructs (27, 29, 30, 32), replication of EEE- and VEE-based replicons was less cytopathic. The latter replicons were capable of establishing persistent replication in a high percentage of transfected BHK-21 cells. In addition, the replacement of A3 by G in the 5′ UTR of the VEE TC-83-based replicon had a strong impact on its ability to persistently replicate in the BHK-21 cell line.
Replication of VEE- and EEE-based replicons.
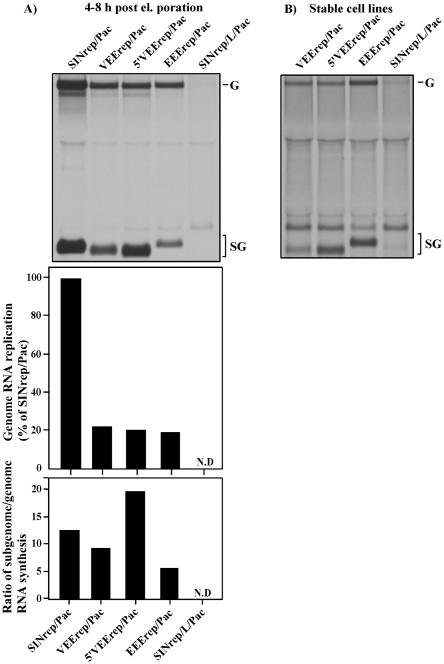
To compare the efficiencies of RNA replication, 4 μg of each replicon (5′VEErep/Pac, VEErep/Pac, EEErep/Pac, SINrep/Pac, and SINrep/L/Pac) was transfected into BHK-21 cells. The RNA replication and transcription of the subgenomic, Pac-encoding RNA was tested by metabolic RNA labeling with [3H]uridine in the presence of dactinomycin, both between 4 and 8 h posttransfection and after a few passages of the Purr cells (Fig. 2). RNAs were isolated, denatured, resolved by gel electrophoresis, and detected by autoradiography. VEE and EEE replicons demonstrated high levels of replication of the genome RNAs and transcription of the subgenomes (Fig. 2A) at between 4 and 8 h postelectroporation. At this time point, they replicated fivefold less efficiently than did SINrep/Pac but dramatically better than the noncytopathic SINrep/L/Pac, whose RNAs could be visualized only after 10-fold-longer exposure of the film or by metabolic labeling of the RNA after superinfection with the wt SIN virus, whose replicative machinery performed additional RNA amplification (data not shown). All of the SINrep/Pac-transfected cells and a large fraction of cells transfected with VEErep/Pac developed profound morphological changes and died within the next 48 h. The surviving cells continued to support the replication of VEErep/Pac, 5′VEErep/Pac, and EEErep/Pac RNAs. They were resistant to puromycin and capable of growth. After a few passages, RNA replication became at least ∼10-fold lower than after electroporation (Fig. 2B), indicating that persistent replication was most likely associated with a lower level of viral nonstructural proteins and replicative complexes present in the cells. During both the initial (acute) and persistent phases, we did not detect significant differences between the replication levels of VEErep/Pac and 5′VEErep/Pac RNAs, but as described above, they displayed significant differences in the ability to cause CPE that were likely a result of the mutation in the 5′ UTR. The same mutation had a pronounced effect on transcription of the subgenomic RNA (Fig. 2), with the VEE TRD-specific sequence of the 5′ UTR leading to a higher level of subgenome synthesis. This difference in transcription of the subgenomic RNA suggested that the balance between replication and transcription could be determined, in part, by competition for the RdRp between the 3′ end and subgenomic promoters in the minus-strand intermediate.
FIG. 2.
Analysis of RNA replication and transcription of the subgenomic RNA in the cells transfected with different replicons. (A) BHK-21 cells were transfected with 4 μg of the in vitro-synthesized replicons' RNA, and equal numbers of electroporated cells (∼106 cells) in six-well Costar plates were labeled with [3H]uridine (20 μCi/ml) in the presence of dactinomycin (1 μg/ml) between 4 and 8 h posttransfection. RNAs were isolated and analyzed by agarose gel electrophoresis as described in Materials and Methods. Lanes contain RNAs from 3 × 105 cells. The gel was fluorographed and exposed for 60 h. Levels of replicon genome RNA accumulation relative to that of the SINrep/Pac replicon and molar ratio of the subgenome-to-genome RNA synthesis were determined by excision of radiolabeled bands and liquid scintillation counting. N.D indicates that RNA replication was below the detection limit of the procedure applied. (B) RNA replication in the cells containing persisting replicons. After 6 days of selection with puromycin, 106 Purr BHK-21 cells containing different replicons were labeled with [3H]uridine (20 μCi/ml) in the presence of dactinomycin (1 μg/ml) for 8 h. RNAs were isolated and analyzed by agarose gel electrophoresis. Lanes contain RNAs from 3 × 105 cells. The gel was fluorographed and exposed for 6 days. G and SG indicate positions of replicons' genomic and subgenomic RNAs, respectively.
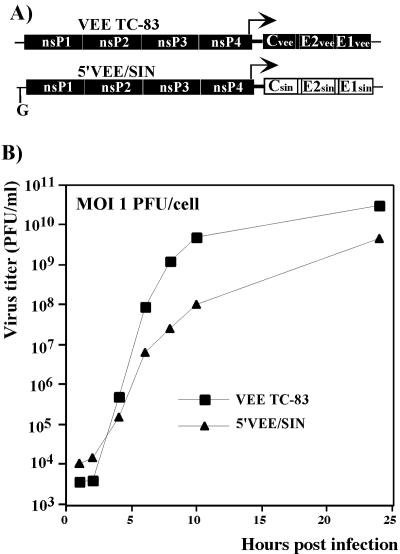
The low cytotoxicity of the VEE- and EEE-derived replicons, compared to wt SIN constructs (3, 9, 10) that we previously studied, was an unexpected phenomenon. Thus, to rule out the possibility that this interesting finding was not a result of spontaneous mutations introduced during cloning procedures, the Pac gene in the 5′VEErep/Pac construct was replaced by SIN structural genes (see Materials and Methods for details). The 5′VEE/SIN chimera was constructed instead of VEE TC-83 virus with the (A3→G) mutation in the 5′ UTR, because this chimeric virus was not pathogenic for mice of any age after either intracutaneous or subcutaneous inoculation and could be used in BSL2 conditions, while VEE TC-83 with the mutated 5′ UTR required BSL3-plus conditions due to a strong positive effect of the 5′ UTR mutation on virus pathogenicity (20, 36, 44). EEE/SIN chimeras were not tested in our studies for two reasons. First, the replication level of EEErep/Pac was very similar to that found for 5′VEErep/Pac. Second, there were safety concerns about creating the EEE/SIN chimera because of the possibility of recreating a WEE-like virus (16, 43) that would also require a higher biosafety containment that is unavailable for our work. The in vitro-synthesized 5′VEE/SIN RNA exhibited the same infectivity (PFU/μg of RNA) as control SIN Toto1101 RNA in the infectious center assay (data not shown), indicating that no mutations were required for virus viability and for making it capable of causing CPE. The chimeric virus caused plaque formation and was capable of efficient growth in BHK-21 cells, but it replicated to noticeably lower final titers than plasmid-derived VEE TC-83 (Fig. 3). Thus, lower cytopathicity is a natural feature of VEE TC-83-based replicons with a TRD-derived 5′ UTR, and the same RNA replication level was sufficient for virus production and plaque formation.
FIG. 3.
Schematic representation of viral genomes and analysis of virus growth. (A) Both genomes encoded the same nonstructural proteins derived from VEE TC-83. 5′VEE/SIN had a 5′ UTR derived from the VEE TRD strain. A detailed description of the chimeric viral genome is presented in Materials and Methods. Solid boxes and open boxes indicate VEE genome- and SIN genome-derived sequences, respectively. (B) BHK-21 cells in six-well Costar plates (5 × 105 cells/well) were infected with the indicated viruses at an MOI of 1 PFU/cell. At the indicated times, the media were replaced, and virus titers were determined as described in Materials and Methods.
VEE and EEE replicons can be used for stable production of heterologous proteins.
Considering the need to develop cell lines for screening the antiviral drugs capable of downregulating the replication of VEE and EEE, we designed double subgenomic replicons expressing the selectable marker gene (Pac) and either a gene of secreted alkaline phosphatase (SEAP) or a GFP-encoding gene (Fig. 4A). Stable Purr cell lines expressing SEAP or GFP were easily established after transfection of in vitro-synthesized RNAs into BHK-21 cells. 5′VEErep/SEAP/Pac- and EEErep/SEAP/Pac-containing cells secreted SEAP at rates of 50 and 100 ng/106 cells/h, respectively (Fig. 4B). Secretion was not linear in these experiments due to cell growth. GFP expression was also readily detectable in the cells transfected by either VEErep/GFP/Pac or EEErep/GFP/Pac replicons. The expression appeared to be almost 10-fold higher than that found for the previously described noncytopathic SINrep/L/GFP/Pac replicon containing a P726→L mutation in nsP2 (Fig. 4A and C). The stability of heterologous protein expression was tested by passaging the EEErep/GFP/Pac- and 5′VEErep/GFP/Pac-containing cells for 10 passages (using a 1:10 to 1:20 dilution at each passage). In the presence of puromycin, the percentage of GFP-negative cells did not noticeably increase and remained below 1 to 2% of the cell population (data not shown). Passaging of the EEErep/GFP/Pac-containing cells in the absence of the drug caused an accumulation of GFP-negative cells. However, essentially all of these GFP-negative cells were sensitive to puromycin and died after reapplication of the drug (Fig. 5). This was an indication that a GFP-negative phenotype was most likely a result of lower replication or complete loss of virus-specific RNAs in the absence of puromycin. However, the possibility of the efficient elimination of both Pac and GFP genes together cannot be completely ruled out.
FIG. 4.
Schematic representation of the double subgenomic replicons expressing Pac and GFP or SEAP and analysis of protein expression. (A) The designed replicons encoded the nonstructural proteins and homologous cis-acting elements (5′ UTR, 3′ UTR, subgenomic promoters, and 5′ UTRs in the subgenomic RNAs) derived from VEE, EEE, and SIN viruses (see Materials and Methods for details). All of the constructs had one subgenomic promoter driving the expression of the Pac gene and a second promoter driving the expression of either GFP or SEAP. Mutations in the 5′ UTR of the VEE TC-83-based replicons and in the nsP2 gene of the SIN-based replicon are indicated. (B) Analysis of alkaline phosphatase expression in the cells carrying persistently replicating VEE and EEE replicons. We seeded 2 × 105 replicon-containing cells into six-well Costar plates. After 4 h of incubation at 37°C, cells were washed with phosphate-buffered saline and further incubated in 2 ml of complete growth medium. At the indicated time points, 200-μl aliquots of medium were taken, and the same volume was added to the wells. Alkaline phosphatase activity was analyzed as described in Materials and Methods. (C) Analysis of GFP expression in the cells carrying persistently replicating VEE, EEE, and SIN replicons with adaptive mutation in nsP2. Cells were analyzed without fixation by flow cytometry on a FACS Vantage (Becton Dickinson).
FIG. 5.
Stability of GFP expression by the persistently replicating EEErep/GFP/Pac replicon. Previously selected Purr cells were passaged (1:10, approximately every 48 h) in the absence of puromycin in the medium. At the indicated times, the percentage of GFP-negative cells was calculated by examination of several random fields on the inverted fluorescent microscope. Puromycin was added back to the medium after 18 days of passaging in drug-free medium.
Both EEErep/GFP/Pac and 5′VEErep/GFP/Pac were also capable of persistent replication in mosquito C710 cells. Similar to BHK-21 cells, in the puromycin-containing media, C710 cells expressed GFP for at least 10 passages without noticeable accumulation of GFP-negative cells (data not shown).
Persistently replicating VEE and EEE replicons do not interfere with replication of heterologous viruses.
VEE- and EEE-specific RNA replication in BHK-21 cells and the ability to express heterologous proteins suggested that they would be useful for gene expression and trans-complementation of defects in the replication of other viruses. One critical problem in the latter application could be the interference of the replication of the alphavirus-specific RNAs with the replication of other, particularly slower-replicating, viruses. To evaluate this type of interference, we infected cells carrying 5′VEErep/Pac and EEErep/Pac replicons with a number of viruses used in our research projects: West Nile virus (WNV), the MP-12 strain of Rift Valley fever virus (RVFV), and different homologous and heterologous alphaviruses. Both WNV and RVFV replicated in the replicon-containing cell lines as efficiently as in naïve BHK-21 cells (Fig. 6), thereby indicating that in this cell type, replication of VEE- and EEE-specific RNAs did not interfere with the replication of WNV and RVFV belonging to other families. Heterologous alphaviruses replicated in the 5′VEErep/Pac- and EEErep/Pac-containing cells as efficiently as in naïve BHK-21 cells. However, the replication of homologous viruses was detectably less effective. Growth rates and final titers of homologous viruses were 10- to 100-fold lower, indicating a significant level of interference caused by the replicons. These results correlated with the previously published data concerning the superinfection exclusion (18). Thus, in spite of more efficient replication than previously selected, noncytopathic, SIN-based constructs, VEE- and EEE-based replicons can be used for trans-complementation experiments with heterologous viruses, including other members of the alphavirus genus.
FIG. 6.
Analysis of replicons' interference with different viral infections. Cells carrying persistently replicating EEErep/Pac or 5′VEErep/Pac replicons were infected with RVFV MP12 (upper left panel), WNV (upper right panel), VEE TC-83 and SIN Toto1101 (lower left panel), and EEE NA Florida 91 and SIN Toto1101 (lower right panel). BHK-21 or Purr cells (5 × 105) carrying the indicated replicons in six-well Costar plates were infected with viruses at the MOIs indicated in the figure. At the indicated times, media were replaced and virus titers were determined as described in Materials and Methods.
VEE genome-based replicons can accumulate adaptive mutations.
As did SIN replicons, the VEE- and EEE-based constructs demonstrated an inability to establish persistent replication in cell lines that possessed no defects in IFN-α/β induction or signaling and that were capable of developing the antiviral response (data not shown). In our previous study, the SIN replicons with the adaptive mutations in nsP2 could replicate indefinitely in a very limited number of cell lines with known defects in IFN signaling. SIN with mutated nsP2 caused persistent infection in IFN-competent cells when grown in the presence of IFN-specific antibodies (1, 9, 11). The originally constructed VEErep/Pac, 5′VEErep/Pac, and EEErep/Pac had very similar ranges of cell types that supported persistent replication. However, after transfection of the Huh-7 cells with in vitro-synthesized 5′VEErep/Pac RNA, we managed to select a very limited number (less than 50) of Purr colonies. This pool of cells resistant to puromycin was expanded, and isolated total RNA was used to retransfect naïve Huh-7 cells. In spite of a low concentration of replicon-specific RNAs in this sample compared to the samples of the in vitro-synthesized replicon, foci of Purr cells were readily selected after the repeated transfection. These cell colonies were expected to contain replicons with adaptive mutations that would allow them to persistently replicate in the Huh-7 cells. The nonstructural genes and 5′ UTR of RNAs obtained from three randomly selected, large colonies were sequenced to identify adaptive mutations. A single mutation was identified in each of the three RNAs: nsP2 Q739→L, nsP2 P773→S, and nsP3 L121→P (Fig. 7). The rest of the nsP1-4 coding sequence and the 5′ UTR in each cell clone-derived RNA was identical to the transfected RNA (20). Interestingly, application of the same selection method to VEErep/Pac and EEErep/Pac replicons was unsuccessful. Both replicons transfected Huh-7 cells equally efficiently and made initially virtually 100% of cells resistant to puromycin. However, transfections of Huh-7 cells with EEErep/Pac made cells incapable of growth, and all of them were dead within the next 2 weeks of selection. In repeated transfection experiments, we selected a few foci of Huh-7 cells with persistently replicating VEErep/Pac, but these likely adapted replicons were present in the cells at very low concentrations. Compared to 5′VEErep/Pac-containing cells, at least 10 more PCR cycles were required to detect VEE-specific sequences in cells transfected with VEErep/Pac, and these replicons were not further investigated.
FIG. 7.
Sequence alignment and adaptive mutations detected in the nsP2 and nsP3 coding sequence of VEE replicons (panels A and B, respectively) and the mutations found in other alphaviruses and replicons that affect their ability to cause CPE. VEE, Venezuelan equine encephalitis virus (21); EEE, eastern equine encephalitis virus (41); SIN, Sindbis virus; SFV, Semliki Forest virus (40). Residues identical to those in the VEE sequence are indicated by dashes. All of the mutations are highlighted. The mutated amino acids in viral proteins are indicated by black boxes. Corresponding amino acids in the proteins of other alphaviruses are indicated by shaded boxes.
Despite more than four orders of magnitude higher efficiency of Purr colony formation activity in BHK-21 cells than Huh-7 cells, we still considered the possibility that the 5′VEErep/Pac replicon required adaptive mutations for persistent replication in both cell lines. To test this, a population of 5′VEErep/Pac-carrying cells was cloned after 2 weeks postelectroporation, and replicons from three randomly selected colonies were sequenced. No mutations were detected in all of the nonstructural genes or the 5′ UTR of the 5′VEErep/Pac replicons persisting in BHK-21 cells. Taken together, the data indicated that the mutations in the replicons were required only for persistence in IFN-α/β-competent Huh-7 cells and that they accumulated more efficiently in the VEE replicons than in the EEE-derived ones.
The amino acid substitutions detected in 5′VEErep/Pac nsP2 were located close in primary sequence to mutated loci that we previously described for SIN replicons with reduced abilities to cause CPE in BHK-21 cells (6, 9). These 5′VEErep/Pac nsP2 mutations were additionally investigated. Both mutations found were transferred into 5′VEErep/Pac (named 5′VEErep/S/Pac and 5′VEErep/L/Pac), and Huh-7 cells were transfected by in vitro-synthesized RNAs. In contrast to the original 5′VEErep/Pac, which was capable of forming very few foci during puromycin selection, electroporation of either 5′VEErep/S/Pac or 5′VEErep/L/Pac replicons made all of the cells drug resistant (Fig. 8B). We detected no differences in the growth of the replicon-transfected cells and untransfected Huh-7 cells propagated in puromycin-free medium and did not observe replicon-induced changes in cell morphology.
FIG. 8.
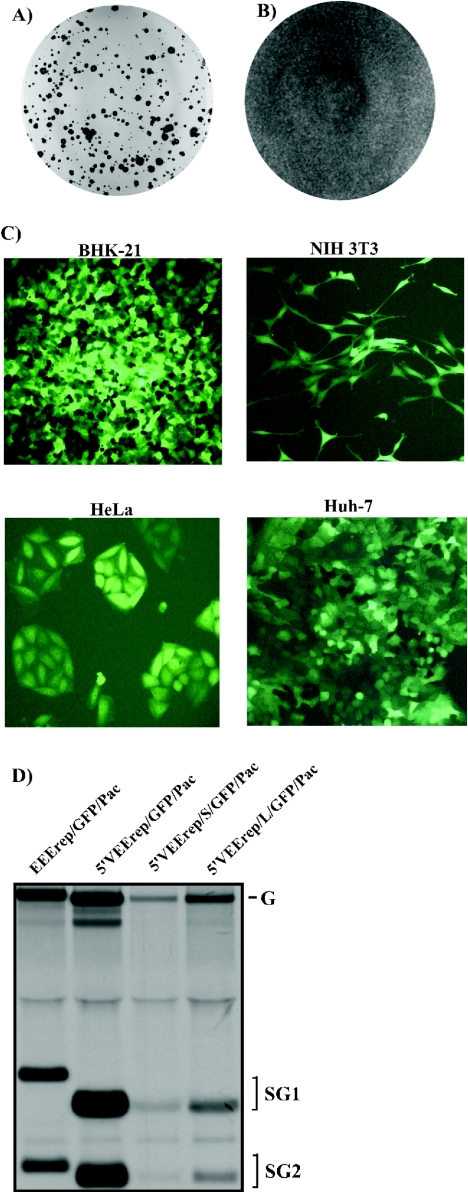
Analysis of replicons' efficiency to persistently replicate in different cell lines. (A) Huh-7 cells were transfected with 12 μg of 5′VEErep/Pac RNA, followed by puromycin selection. Colonies of Purr cells were stained after 20 days of drug treatment. (B) Huh-7 cells were transfected with 12 μg of 5′VEErep/S/Pac RNA (containing the P773→S mutation in nsP2) and treated with puromycin for 10 days. Wedetected no cell death due to CPE caused by replication of virus-specific RNAs or drug treatment. (C) Different Purr, GFP-expressing cell lines were generated by transfection of 4 μg of 5′VEErep/S/GFP/Pac RNA followed by puromycin selection. (D) BHK-21 cells were transfected with 4 μg of in vitro-synthesized replicons' RNA, and equal numbers of electroporated cells (∼106 cells) in six-well Costar plates were labeled with [3H]uridine (30 μCi/ml) in the presence of dactinomycin (1 μg/ml) at 3 to 7 h posttransfection. RNAs were isolated and analyzed by agarose gel electrophoresis as described in Materials and Methods. Lanes contain RNAs from 2.5 × 105 cells. G and SG indicate positions of replicons' genomic and subgenomic RNAs, respectively.
Another group of experiments was performed with 5′VEErep-based replicons expressing GFP and containing the adaptive mutations described above (5′VEErep/S/GFP/Pac and 5′VEErep/L/GFP/Pac). In contrast to the originally designed 5′VEErep/GFP/Pac, both constructs were capable of persistent replication and retained the ability to express high levels of heterologous protein (GFP), not only in BHK-21 cells but also in Huh-7, HeLa, and NIH 3T3 cells (Fig. 8C). Replicons with the introduced mutations demonstrated 5- to 10-fold-lower levels of replication than the parental construct 5′VEErep/Pac (Fig. 8D), suggesting that their noncytopathic phenotype could be at least partially explained by less efficient replication and/or less efficient production of viral nonstructural proteins in the transfected cells. During the first 24 to 48 h postelectroporation, the originally constructed 5′VEErep/GFP/Pac replicon arrested cell growth and caused changes in cell morphology, but this was not the case when the transfections were performed using replicons with the adaptive mutations in nsP2. Thus, 5′VEErep-derived constructs with mutated nsP2 have the ability to persist in a number of cell lines other than BHK-21 cells and accordingly may be useful in a variety of trans-complementation experiments.
DISCUSSION
The VEE- and EEE-based replicons reported in this study differed from similar constructs previously described for SIN and SFV. The efficient replication and pathogenicity of VEE and EEE in small animal models (4, 13, 31) suggested that the ability of these viruses to cause disease would correlate with efficient viral RNA replication, fast CPE development, and suppression of critical cellular functions, including transcription and translation of cellular mRNAs. This could lead to strong interference with the development of an innate immune response and interference with the production of IFN-α/β by the infected cells (11, 12). However, EEE- and VEE TC-83-based replicons expressing the dominant selectable marker Pac gene were less cytopathic than previously described SIN Toto1101- and SFV-derived constructs (3, 27), and they readily established persistent replication in BHK-21 cells. Based on the high efficiency of Purr focus formation, which was comparable to that found for SIN replicons with adaptive mutations in the 726 position of nsP2, these RNAs did not contain or require adaptive mutations for persistent replication in this cell line. The adaptive mutations were also not detected in the 5′VEErep/Pac replicons isolated from the randomly selected colonies. However, the direct sequencing of the PCR fragments did not rule out the possibility of the presence of multiple RNA quasispecies in the samples. It must be noted that VEErep/Pac and EEErep/Pac efficiently replicated until 24 to 48 h postelectroporation and within that time arrested cell growth. At later time points, the levels of replicon-specific RNA production in the replicon-carrying cells were lower but still detectable by metabolic RNA labeling with [3H]uridine. The low level of replication of virus-specific RNAs at the persistent stage compared to the initial, more efficient replication can be explained by a currently accepted hypothesis pertaining to the regulation of alphavirus RNA replication (23, 35, 39). According to this hypothesis, the nonstructural protein nsP2 is a critical factor that controls the number and mode of functioning of the replicative complexes. Within the first few hours after infection or transfection of the alphavirus-specific RNAs, nsP2 is present in the cells in an unprocessed (P123) or partially processed form required for the enzyme complexes to synthesize the minus-strand RNA intermediates. Then, after the nsP2-dependent processing of the P123 precursor, the replicative complexes are transformed into the mature form, capable of plus-strand but not minus-strand RNA synthesis. At this time, nsP2 is at least partially released from the protein complexes and can be detected not only in sites of virus-specific RNA synthesis but also distributed in the cytoplasm and nucleus. Thus, the early time point of replication can be characterized by a low concentration of cleaved nsP2 and a high concentration of the replicative complexes. At later times, the processed nsP2 accumulates in the cytoplasm and interferes with the formation of the new P123-containing replicative complexes by trans cleaving of newly synthesized polyproteins. We speculate that the persistent replication of the virus-specific RNAs is based on establishing a balance between the concentration of nsP2 and the number of replicative complexes in the cells. The continuous presence of nsP2 in the cytoplasm makes it impossible for replication at the persistent stage to reach the same levels as that found during the early, acute stage of replication.
The high cell survival rate was a strong indication that neither inhibition of transcription nor inhibition of translation of cellular RNAs in the transfected cells reached a level that could induce cell death. One of the reasons for the less cytopathic phenotype of VEErep/Pac and EEErep/Pac could be their less efficient initial replication compared to the SINrep/Pac (Fig. 2A) or SFV-based (data not shown) replicons. The reduced level of RNA replication in the cells containing VEE and EEE replicons could lead to less efficient induction of PKR-dependent (45) and PKR-independent (12) translational shutoffs, inefficient inhibition of transcription of cellular mRNA and rRNAs (19), and poor apoptosis induction. This, in turn, could increase the survival of the transfected BHK-21 cells before the replication of virus-specific RNAs slowed at 2 to 3 days posttransfection (at the persistent stage of replication). However, it must be noted that VEErep and EEErep replication at the early points posttransfection was manyfold higher than replication of the previously described noncytopathic SIN replicon (SINrep/L/Pac).
Previously, we demonstrated that the replication of SIN replicons with adaptive mutations in nsP2 (SINrep/L/Pac and others) induced IFN-α/β secretion, and its release arrested the replication of virus-specific RNAs and eliminated replicons from Huh-7 or NIH 3T3 cells (11). The EEErep/Pac replicon also caused efficient IFN-α/β production that stopped its replication in the cell lines having no defects in IFN-α/β production or signaling (data not shown). As a result, the EEErep/Pac-transfected cells died within 1 week of selection if puromycin was present in the medium. Alternatively, the drug could be removed from the medium after 3 days of selection, which provided enough time to eliminate all of the untransfected cells. In the drug-free medium, cells resumed efficient growth, but replicons were no longer detected (data not shown), indicating that similar to our previously published data, the released IFN-α/β induced the antiviral response in the replicon-carrying cells and stopped RNA replication (11). In multiple experiments, we failed to select colonies of Huh-7 or other cells (except BHK-21) with persistently replicating EEE genome-based replicons, suggesting that EEErep/Pac replication was either more toxic for cells or, most likely, more sensitive to the autocrine effect of IFN-α/β.
Based on the previously published data of other research groups (36, 44), we expected replication of VEE-based replicons to be more resistant to IFN-α/β. However, the initially designed construct VEErep/Pac showed significant cytopathicity. Only introducing a single point mutation into the 5′ UTR, making this sequence identical to that of the 5′ UTR of the epizootic VEE strain TRD, the 5′VEErep/Pac replicon converted into less cytopathic and capable of efficient Purr focus formation in BHK-21 cells. This 5′ UTR mutation might also contribute to increased replicon resistance to the autocrine effect of IFN-α/β, because a critical role of the third nucleotide in VEE pathogenesis has been previously documented by other research groups (20, 44). A mutation in the same position affected the resistance of VEE TRD to IFN-α/β and strongly reduced the pathogenicity of the virus (36, 44). The TC-83-specific 5′ UTR was also found to drive translation of the nonstructural proteins in vitro more efficiently than the TRD 5′ UTR (44), indicating that replacement of the third nucleotide, destabilizing a predicted secondary structure of the RNA, could have an effect on accumulation of the VEE-specific nonstructural proteins. In our experiments, we did not detect convincing differences in the replication levels of VEE replicons with TRD- or TC-83-derived 5′ UTRs, but we can speculate that a lower cytopathic effect of 5′VEErep/Pac compared to VEErep/Pac in BHK-21 cells was a result of a less efficient accumulation of VEE nsPs (at least during the first hours posttransfection). However, this hypothesis needs further experimental support. The importance of the 5′ UTR for alphavirus pathogenesis is also supported by the results with SIN (7). The mutation of nt 8 in the SIN 5′ UTR had a strong effect on the SIN neuroinvasive phenotype.
In spite of reducing the cytotoxicity of replicons for BHK-21 cells, the TRD-specific mutation was not sufficient to make them capable of persisting in the IFN-competent cell lines. However, additional adaptive mutations in nsP2 (Q739→L or P773→S) or nsP3 (L121→P) further reduced cytotoxicity of the 5′VEErep/Pac construct and strongly increased its ability to persistently replicate. The nsP2 mutants gained the ability to persist not only in BHK-21 cells but also in the tested IFN-competent cell lines. These replicons remained resistant to an autocrine effect of IFN-α/β, which could be readily detected in the medium of the NIH 3T3 cells carrying the 5′VEErep/L/Pac or 5′VEErep/S/Pac mutant replicon (data not shown), and became significantly less cytopathic. It was noted that in the medium of NIH 3T3 cells carrying VEE replicons with adaptive mutations, the concentration of IFN-α/β could reach a critical point (∼800 IU/ml), at which replicons were no longer resistant to the IFN-induced antiviral state, and all of the cells could die within a few hours due to the loss of puromycin resistance (data not shown).
The intriguing feature of the mutations accumulating in nsP2 of 5′VEErep/Pac was in their localization near the mutations that we previously found in noncytopathic SIN replicons or less cytopathic SIN virus (6, 9, 32) and the mutation that another group found in noncytopathic SFV replicons (32). The adaptive mutations have now been detected in replicons derived from three viruses that belong to different antigenic groups exhibiting strong heterogeneity in the nonstructural genes and demonstrating very different pathogenicity levels. However, in all three cases, the mutations in the same short peptide strongly affected the ability of these self-replicating RNAs to cause CPE, and this effect did not correlate with the level of RNA replication (32). The colocalization of the mutations may be an indication that the carboxy-terminal domain of nsP2 has functions in addition to those necessary for helicase and protease activity described for this protein. We speculate that this domain may play a critical role in interaction with host proteins during virus replication. The L121→P adaptive mutation in nsP3 also made one of the 5′VEErep/Pac replicons noncytopathic for Huh-7 cells. In a previous work, we described mutations in SIN nsP2 and nsP3 that synergistically affected RNA promoter binding (8), suggesting a direct or indirect interaction between these two proteins. The present data also indicate possible interplay between VEE nsP2 and nsP3 that is critical for the replicon's ability to cause CPE. However, we cannot rule out the possibility that mutations in these proteins affect completely different processes during replication of VEE-specific RNAs, leading to development of the same noncytopathic phenotype. Based on present data and the results of other research groups (30, 32), we believe that adaptive mutations that make alphavirus replicons capable of persistent, noncytopathic replication can appear in other parts of the replicons' genome. However, the carboxy-terminal part of nsP2 can likely tolerate multiple mutations leading to the same noncytopathic phenotype. Thus, such mutations can occur more frequently in this genome fragment of VEE, SIN, and EEE than in other parts of their genomes. Indeed, in another project, we sequenced the nsP2 and nsP3 fragments (the same as described in this report) of 5′VEErep/Pac replicons derived from the other three clones of Purr Huh-7 cells. No mutations were found, indicating that the noncytopathic phenotype was due to changes in different parts of the replicons' genome.
In conclusion, we have shown that self-replicating RNAs derived from different alphaviruses are very diverse in terms of their ability to cause CPE and to persistently replicate in vertebrate cells. (i) Replicons derived from wt SIN are cytopathic, strongly affect transcription and translation of cellular mRNAs, and require the mutations in nsP2 to become noncytopathic. They are incapable of persisting in cells which can produce IFN-α/β and have no defects in IFN-α/β signaling. (ii) Replicons derived from wt EEE of a North American strain are less cytopathic and require no additional mutations to persist in BHK-21 cells which have defects in IFN signaling. (iii) VEE TC-83-based replicons require a mutation in the 5′ UTR to attain a less cytopathic phenotype making them capable of persisting at least in BHK-21 cells. However, they also need an adaptive mutation in nsP2 or nsP3 to make them even less cytopathic for cells with no defects in IFN-α/β response and to make them capable of persistent replication in these cell lines.
This information will allow us to expand our understanding of profound differences in the biology of different alphaviruses and to further increase the application of alphavirus replicons in the production of heterologous proteins and development of recombinant vaccines. The adaptive mutations that occurred in nsP2 and nsP3 may be useful for developing attenuated VEE viruses, in which combinations of mutations might reduce to a very low level the possibility of reversion to wt virulence. Attenuation of VEE TRD by applying such mutations is now under investigation. Finally, the stable replicon-containing cell lines expressing alkaline phosphatase may be useful for screening compounds for their ability to suppress the replication of VEE and EEE.
Acknowledgments
O.P. and E.V. contributed equally to this work.
We thank Peter Mason and Scott Weaver for critical reading of the manuscript and Frank Scholle for help in the experiments with WNV.
This work was supported by Public Health Service grants AI053135 and AI50537 from the National Institute of Allergy and Infectious Diseases. S.P. was supported by K08AI059491.
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Prägai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berge, T. O., I. S. Banks, and W. D. Tigertt. 1961. Attenuation of Venezuelan equine encephalomyelitis virus by in vitro cultivation in guinea pig heart cells. Am. J. Hyg. 73:209-218. [Google Scholar]
- 3.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles, P. C., E. Walters, F. Margolis, and R. E. Johnston. 1995. Mechanism of neuroinvasion of Venezuelan equine encephalitis virus in the mouse. Virology 208:662-671. [DOI] [PubMed] [Google Scholar]
- 5.Dal Canto, M. C., and S. G. Rabinowitz. 1981. Central nervous system demyelination in Venezuelan equine encephalomyelitis infection. J. Neurol. Sci. 49:397-418. [DOI] [PubMed] [Google Scholar]
- 6.Dryga, S. A., O. A. Dryga, and S. Schlesinger. 1997. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology 228:72-83. [DOI] [PubMed] [Google Scholar]
- 7.Dubuisson, J., S. Lustig, Y. Akov, and C. M. Rice. 1997. Genetic determinants of Sindbis virus neuroinvasiveness. J. Virol. 71:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fayzulin, R., and I. Frolov. 2004. Changes of the secondary structure of the 5′ end of the Sindbis virus genome inhibit virus growth in mosquito cells and lead to accumulation of adaptive mutations. J. Virol. 78:4953-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov, I., and S. Schlesinger. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorchakov, R., E. Frolova, B. R. Williams, C. M. Rice, and I. Frolov. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78:8455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grieder, F. B., N. L. Davis, J. F. Aronson, P. C. Charles, D. C. Sellon, K. Suzuki, and R. E. Johnston. 1995. Specific restrictions in the progression of Venezuelan equine encephalitis virus-induced disease resulting from single amino acid changes in the glycoproteins. Virology 206:994-1006. [DOI] [PubMed] [Google Scholar]
- 14.Griffin, D. E. 1986. Alphavirus pathogenesis and immunity, p. 209-250. In S. Schlesinger and M. J. Schlesinger (ed.), The togaviridae and flaviviridae. Plenum Press, New York, N.Y.
- 15.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, New York, N.Y.
- 16.Hahn, C. S., S. Lustig, E. G. Strauss, and J. H. Strauss. 1988. Western equine encephalitis virus is a recombinant virus. Proc. Natl. Acad. Sci. USA 85:5997-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven, New York, N.Y.
- 18.Johnston, R. E., K. Wan, and J. H. R. Bose. 1974. Homologous interference induced by Sindbis virus. J. Virol. 14:1076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kääriäinen, L., and M. Ranki. 1984. Inhibition of cell functions by RNA virus infections. Annu. Rev. Microbiol. 38:91-109. [DOI] [PubMed] [Google Scholar]
- 20.Kinney, R. M., G.-J. Chang, K. R. Tsuchiya, J. M. Sneider, J. T. Roehrig, T. M. Woodward, and D. W. Trent. 1993. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J. Virol. 67:1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinney, R. M., B. J. B. Johnson, J. B. Welch, K. R. Tsuchiya, and D. W. Trent. 1989. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 170:19-30. [DOI] [PubMed] [Google Scholar]
- 22.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leon, C. A. 1975. Sequelae of Venezuelan equine encephalitis in humans: a four year follow-up. Int. J. Epidemiol. 4:131-140. [DOI] [PubMed] [Google Scholar]
- 25.Levine, B., J. E. Goldman, H. H. Jiang, D. E. Griffin, and J. M. Hardwick. 1996. Bcl-2 protects mice against fatal alphavirus encephalitis. Proc. Natl. Acad. Sci. USA 93:4810-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine, B., Q. Huang, J. T. Isaacs, J. C. Reed, D. E. Griffin, and J. M. Hardwick. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739-742. [DOI] [PubMed] [Google Scholar]
- 27.Liljeström, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. BioTechnology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 28.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundstrom, K., A. Abenavoli, A. Malgaroli, and M. U. Ehrengruber. 2003. Novel Semliki Forest virus vectors with reduced cytotoxicity and temperature sensitivity for long-term enhancement of transgene expression. Mol. Ther. 7:202-209. [DOI] [PubMed] [Google Scholar]
- 30.Lundstrom, K., C. Schweitzer, J. G. Richards, M. U. Ehrengruber, F. Jenck, and C. Mülhardt. 1999. Semliki Forest virus vectors for in vitro and in vivo applications. Gene Ther. Mol. Biol. 4:23-31. [Google Scholar]
- 31.Paessler, S., P. Aguilar, M. Anishchenko, H. Q. Wang, J. Aronson, G. Campbell, A. S. Cararra, and S. C. Weaver. 2004. The hamster as an animal model for eastern equine encephalitis—and its use in studies of virus entrance into the brain. J. Infect. Dis. 189:2072-2076. [DOI] [PubMed] [Google Scholar]
- 32.Perri, S., D. A. Driver, J. P. Gardner, S. Sherrill, B. A. Belli, T. W. Dubensky, Jr., and J. M. Polo. 2000. Replicon vectors derived from Sindbis virus and Semliki Forest virus that establish persistent replication in host cells. J. Virol. 74:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239:389-401. [DOI] [PubMed] [Google Scholar]
- 34.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spotts, D. R., R. M. Reich, M. A. Kalkhan, R. M. Kinney, and J. T. Roehrig. 1998. Resistance to alpha/beta interferons correlates with the epizootic and virulence potential of Venezuelan equine encephalitis viruses and is determined by the 5′ noncoding region and glycoproteins. J. Virol. 72:10286-10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stollar, V. 1980. Togaviruses in cultured arthropod cells, p. 584-621. In R. W. Schlesinger (ed.), The togaviruses—biology, structure, replication. Academic Press, Inc., New York, N.Y.
- 38.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 39.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takkinen, K. 1986. Complete nucleotide sequence of the non-structural protein genes of Semliki Forest virus. Nucleic Acids Res. 14:5667-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volchkov, V. E., V. A. Volchkova, and S. V. Netesov. 1991. Complete nucleotide sequence of the Eastern equine encephalomyelitis virus genome. Mol. Genet. Mikrobiol. Virusol. 5:8-15. (In Russian.) [PubMed] [Google Scholar]
- 42.Weaver, S. C., and A. D. Barrett. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2:789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver, S. C., W. Kang, Y. Shirako, T. Rumenapf, E. G. Strauss, and J. H. Strauss. 1997. Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J. Virol. 71:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, L. J., J. G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 75:3706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, S., and R. J. Kaufman. 1997. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J. Biol. Chem. 272:1291-1296. [DOI] [PubMed] [Google Scholar]