Abstract
Alfalfa mosaic virus (AMV) RNAs 1 and 2 encode the replicase proteins P1 and P2, respectively, whereas RNA 3 encodes the movement protein and the coat protein (CP). When RNAs 1 and 2 were transiently expressed from a T-DNA vector (R12 construct) by agroinfiltration of Nicotiana benthamiana, the infiltrated leaves accumulated minus-strand RNAs 1 and 2 and relatively small amounts of plus-strand RNAs. In addition, RNA-dependent RNA polymerase (RdRp) activity could be detected in extracts of the infiltrated leaves. After transient expression of RNAs 1 and 2 with the 3′-untranslated regions (UTRs) of both RNAs deleted (R1Δ/2Δ construct), no replication of RNAs 1 and 2 was observed, while the infiltrated leaves supported replication of RNA 3 after inoculation of the leaves with RNA 3 or expression of RNA 3 from a T-DNA vector (R3 construct). No RdRp activity could be isolated from leaves infiltrated with the R1Δ/2Δ construct, although P1 and P2 sedimented in a region of a glycerol gradient where active RdRp was found in plants infiltrated with R12. RdRp activity could be isolated from leaves infiltrated with constructs R1Δ/2 (3′-UTR of RNA 1 deleted), R1/2Δ (3′-UTR of RNA 2 deleted), or R1Δ/2Δ plus R3. This demonstrates that the 3′-UTR of AMV RNAs is required for the formation of a complex with in vitro enzyme activity. RNAs 1 and 2 with the 3′-UTRs deleted were encapsidated into virions by CP expressed from RNA 3. This shows that the high-affinity binding site for CP at the 3′-termini of AMV RNAs is not required for assembly of virus particles.
RNA-dependent RNA polymerase (RdRp) proteins of many plus-strand RNA viruses have been expressed in heterologous systems, including Escherichia coli (15, 23–25, 31, 35, 42, 61), Saccharomyces cerevisiae (17, 36), insect cells (4, 14, 30, 43, 63), and mammalian cells (26). The availability of in vitro active RdRp has greatly facilitated research into the mechanisms of plus-strand virus replication. Mutant RNA polymerases can be expressed despite their inability to support viral replication. In this way, insight can be gained into the roles of the various domains of viral replicase proteins.
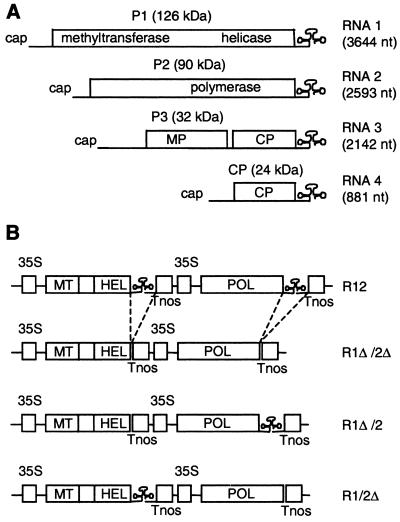
Alfalfa mosaic virus (AMV) (for reviews, see references 6 and 18) is a tripartite plus-strand RNA virus belonging to the family Bromoviridae (Fig. 1A). The AMV replication complex consists of two viral proteins, P1 and P2, and possibly host proteins. P1, encoded by RNA 1, has homologies to known methyltransferase domains in its N-terminal half and homologies to helicase domains in its C-terminal half (12, 21). P2, encoded by RNA 2, contains the RNA polymerase domain characterized by the GDD motif (for a review, see reference 33). AMV RNA 3 codes for the viral movement protein (P3) and the coat protein (CP), which is translated from a subgenomic mRNA 4. All three genomic RNAs of AMV contain 5′- and 3′-untranslated regions (UTRs), which are highly structured and presumed to contain important cis-acting regulatory elements for viral replication and translation (32, 52, 54, 55, 58, 59).
FIG. 1.
Schematic representation of AMV RNAs and T-DNA constructs. (A) Organization of the AMV genome. The 3′-UTR of the RNAs can be folded into a linear array of hairpins that represents a CP-binding site or into a TLS that is recognized by the RdRp. The TLS conformation is indicated. (B) Inserts in the T-DNA vector that were transiently expressed in plants. The R12 construct contains full-length DNA copies of RNAs 1 and 2, each flanked by the CaMV 35S promoter (35S) and nos terminator (Tnos). In the other constructs, the 3′-UTR of RNA 1 (1Δ) and/or RNA 2 (2Δ) is deleted. Domains of P1 and P2 with methyltransferase motifs (MT), helicase motifs (HEL), and polymerase motifs (POL) are indicated. nt, nucleotides.
The 3′-terminal 145 bases of the 3′-UTRs of the AMV RNAs are homologous and can be folded into either a linear array of stem-loop structures (22) or a pseudoknot resembling tRNA (tRNA-like structure [TLS]) (32). When folded into the linear array of stem-loop structures, the RNA contains several high-affinity CP-binding sites (16, 41) whereas the TLS conformation is specifically recognized by the RdRp (32). A mixture of the three genomic RNAs of AMV is infectious only when each RNA is complexed with a few molecules of CP (reviewed in references 6 and 18). It has been proposed that the 3′-UTR acts as a molecular switch that regulates the transition from translation to replication of the parental RNAs (32). In this model, CP bound to inoculum RNAs would force the 3′-UTR into the CP-binding conformation to enhance translation and/or to prevent premature initiation of minus-strand RNA synthesis. Subsequently, CP has to dissociate from the parental RNA to allow the formation of the TLS and the initiation of minus-strand RNA synthesis. In a later step of the replication cycle, de novo-synthesized CP could shut off minus-strand synthesis by binding to the 3′-UTR of progeny RNAs.
In protoplasts inoculated with AMV RNAs 1 and 2, minus-strand RNA accumulation is detectable only when CP is present in the inoculum (27). However, when RNAs 1 and 2 are expressed from the 35S promoter in transgenic plants (R12 plants), CP is not required for minus-strand RNA synthesis (49). It was proposed that the poly(A) tail of the nuclear AMV transcripts may compensate for a putative role of CP in translation of the inoculum RNAs (49). When R12 plants were inoculated with RNA 3, RNAs 1 and 2 started to coreplicate with RNA 3 (49). Tobacco plants that express the P1 and P2 proteins from nuclear transgenes that are flanked by incomplete 5′- or 3′-UTRs (P12 plants) support the replication of AMV RNA 3 but no replication of the transgenic 5′-truncated RNA 1 or 3′-truncated RNA 2 is observed (47). Minus-strand synthesis in RNA 3-infected P12 protoplasts requires neither CP in the inoculum nor expression of the CP gene in RNA 3 (27). However, de novo-synthesized CP is required for asymmetric accumulation of plus-strand AMV RNA in P12 protoplasts (56) and CP stimulates the accumulation of plus-strand RNA 4 in an in vitro RdRp assay (9). Using immunoprecipitation studies and the two-hybrid assay, interactions between P1 and P2 are detectable but no interaction of these proteins with CP was observed (51).
Until now, AMV RdRp could be partially purified from either AMV-infected tobacco plants (39) or the transgenic P12 plants (9). In vitro, these RdRp preparations supported minus-strand synthesis on plus-strand AMV templates as well as subgenomic plus-strand RNA synthesis on a minus-strand RNA 3 template. RdRp complexes of a number of other plus-strand RNA viruses have been isolated from infected plants (for references, see reference 34). The RdRp of Bamboo mosaic virus (24) and that of Tobacco vein mottling virus (15) have been isolated from Escherichia coli. The RdRp of Turnip yellow mosaic virus was expressed in insect cells (14), although no replicase activity was shown in vitro. Brome mosaic virus (BMV) RdRp has been isolated from recombinant S. cerevisiae (37).
In this work we developed a novel method for the transient expression of a plant virus RdRp in planta. Expression of P1 and P2 of AMV by the agroinfiltration technique (5) resulted in the assembly of an RdRp complex that was active both in vivo and in vitro. The 3′-UTR of the AMV RNAs was required for the formation of a functional RdRp complex, and the 3′-UTRs of the three genomic RNAs were found to be equivalent in this function. Although the 3′-UTRs contain high-affinity binding sites for CP, these binding sites were found to be dispensable for encapsidation of the viral RNAs.
MATERIALS AND METHODS
DNA constructs.
cDNA 1 of AMV, cloned between the Cauliflower mosaic virus (CaMV) 35S promoter and the terminator sequence of the nopaline synthase gene (Tnos), was cut from pCA17T (29) using KpnI and PvuII. This fragment was cloned in pBluescript-SK(+) (pBS). Therefore, pBS was restricted with BamHI, which created an overhang that was made blunt using T4 DNA polymerase, and KpnI. The resulting construct was termed pBS-R1. cDNA 2 of AMV, cloned between the CaMV 35S promoter and Tnos, was cut from pCA27T (29) using SmaI and PvuII. This fragment was cloned in pUC19, which had been restricted with KpnI and SphI and treated with T4 DNA polymerase to make the overhangs blunt. The resulting construct was termed pUC-R2. Subsequently, both cDNA 1 and cDNA 2 were cloned in the binary vector pMOG800 (20). cDNA 1 was inserted using KpnI and SstI, and cDNA 2 was inserted using SstI and HindIII. The resulting construct was termed pMOGR12 (Fig. 1B). pMOG800 constructs containing either cDNA 1 or cDNA 2 were termed pMOGR1 and pMOGR2, respectively.
The 3′-UTRs of cDNAs 1 and 2 were deleted by PCR-mediated site-directed mutagenesis. The 3′-terminal 166 nucleotides of the coding sequence of cDNA 1 were amplified using primers pCo1 (5′CAATAAATGGCCCATGCCATG3′) and pCo2 (5′CTCGAGACGCGTATGTCAGAAATTATGATTATAGC3′). Tnos was amplified with primers pCo3 (5′CATACGCGTCTCGAGGATCGTTCAAACATTTGG3′) and pCo9 (5′GCATGCGAGCTCGATCGATCTAGTAACATAGATGACACC3′). Since pCo2 and pCo3 had been designed to have a complementary linker sequence of 15 nucleotides, the two fragments could be fused by PCR using pCo1 and pCo9. The resulting fragment was exchanged with the NcoI-SstI fragment of pBS-R1, yielding pBS-R1Δ. The 3′-terminal 143 nucleotides of the coding sequence of cDNA 2 were amplified using primers pCo5 (5′GAATCCCTAGGTAAGATC3′) and pCo6 (5′CTCGAGACGCGTATGTCAAGCTCGGCGTG3′). Tnos was amplified using primers pCo3 and pCo7 (5′CATGATTACGCCAAGCTTG3′). Due to a complementary sequence in pCo6 and pCo3, the two fragments could be fused by PCR using pCo5 and pCo7. The resulting fragment was exchanged with the BglII-HindIII fragment of pUC-R2, yielding pUC-R2Δ. All PCR-derived sequences in pBS-R1Δ and pUC-R2Δ were sequenced (T7 sequencing kit; Pharmacia). Finally, R1Δ and R2Δ were cloned in pMOG800 and either pMOGR2 or pMOGR1. R1Δ was inserted using KpnI and SstI, and R2Δ was inserted using SstI and HindIII. The resulting constructs were termed pMOG R1Δ/2Δ, pMOG R1Δ/2, and pMOG R1/2Δ (Fig. 1B).
C189S was introduced into cDNA 1 by PCR-mediated site-directed mutagenesis. Fragments were amplified with primers pCo12 (5′CATCTGCATGCTTTGCGGCTGCCCATC3′) plus pCo25 (5′CCTTGAGCTTTTCTGAAACGTATCC3′) and with primers pCo24 (5′GGATACGTTTCAGAAAAGCTCAAGG3′) plus pCo13 (5′GACTAGCTCCCAAATTGGGCTCG3′). Due to complementarity of pCo24 and pCo25, the two fragments could be fused by PCR using pCo12 and pCo13. The resulting fragment was cloned in pGEMT and sequenced (T7 sequencing kit). The SspI-SstI fragment of the cloned PCR fragment was ligated to the SspI-SalI fragment of cDNA 1, and the ligation product was cloned in pBS restricted with SstI and SalI. The SalI-BglII fragment of this clone was exchanged with the corresponding fragment of pBS-R1. Finally, R1C189S was cloned in pMOGR2 using SstI and HindIII. The resulting construct was termed pMOG R12C189S.
cDNA 3, cloned between the CaMV 35S promoter and Tnos, was cut from pCa32T (29) using PvuII and KpnI. This fragment was cloned in pMOG800 to yield pMOGR3. Therefore, pMOG800 was restricted with XhoI, which created an overhang that was made blunt using T4 DNA polymerase, and KpnI.
Agrobacterium-mediated transient expression.
All pMOG constructs were transformed to Agrobacterium tumefaciens strain LBA 4404 by electroporation. pMOG800 was generally used as a negative control. Cultures of 5 ml were grown for 48 h at 28°C in LC medium containing 50 μg of kanamycin per ml and 50 μg of rifampin per ml. Of this culture, 1 ml was grown overnight at 28°C in 100 ml of minimal A medium [46 mM K2HPO4, 33 mM KH2PO4, 7.5 mM (NH4)2SO4, 1.5 mM C6H5Na3O7 · 2H2O, 1 mM MgSO4, 0.2% glucose, 100 μM CaCl2], containing 10 mM N-morpholinoethanesulfonic acid (MES) (pH 5.6), 40 μM 3′, 5′-dimethoxy-4′-hydroxyacetophenone (acetosyringone), 50 μg of kanamycin per ml, and 50 μg of rifampin per ml. Subsequently, the cells were pelleted and resuspended in MMA (10 mM MgCl2, 10 mM MES [pH 5.6], 200 μM acetosyringone) to a final optical density at 600 nm (OD600) of 0.5 to 1.0. When two A. tumefaciens strains were coinfiltrated, the OD600 of each strain was at least 0.5 and the OD600 of the culture was between 1.0 and 1.5. The cells were kept at 23°C for 1 to 3 h prior to infiltration. N. benthamiana leaves were infiltrated using a syringe without a needle. The plants were kept humid at 23°C under mild light conditions (1,000 lux). One day after infiltration, the plants were transferred to the greenhouse. If necessary, leaves were inoculated with P12 virus 2 days after infiltration, as described previously (47). P12 virus consists of virions containing RNA 3 and virions containing RNA 4. The plants were kept in the greenhouse. Plants were inoculated with wild-type AMV as described previously (60).
Isolation and analysis of virions, virion RNA, and total RNA.
Virions and RNA were extracted from leaves 2 days after infiltration of A. tumefaciens or 5 days after subsequent inoculation of P12 virus. Virions were extracted from 500 mg of tissue as decribed by Van Vloten-Doting and Jaspars (60). Total RNA was extracted from 500 mg of tissue essentially as described by Van der Kuyl et al. (53). The particles were analyzed by Northern blot hybridization. Per slot, virions from 25 mg of tissue were loaded. Particles containing RNA 1 were labeled using a random-primed 32P-labeled probe of nucleotides 856 to 3086 of RNA 1. Particles containing RNA 2 were labeled using a random-primed 32P-labeled probe of nucleotides 280 to 1206 of RNA 2. Virion and total RNA was denatured by dimethyl sulfoxide-glyoxal treatment. RNA from 5 mg of leaf tissue was loaded per slot. Northern blot hybridizations were performed using digoxigenin-labeled (Boehringer Mannheim) riboprobes specific for plus- or minus-strand AMV RNAs 1, 2 and 3. All Northern blots were performed with Nylon membranes (Boehringer Mannheim).
RdRp isolation and analysis.
RdRp was isolated from approximately 10 g of leaf tissue 2 days after infiltration, essentially as described previously (38). In short, leaves were homogenized and large debris and nuclei were centrifuged. The supernatant was subsequently centrifuged at 30,000 × g for 20 min. To obtain template-dependent RdRp, the 30,000 × g pellet (P30) was solubilized using high salt and detergent. The isolate was further purified on a glycerol gradient. The gradients were fractionated in 18.5 fractions of 2 ml. Per fraction, 15 μl was analyzed in an in vitro RdRp assay essentially as described previously (13). Products of this assay were extracted with phenol-chloroform, precipitated, treated with nuclease S1, again extracted with phenol-chloroform, precipitated, and finally run on a 1.5% agarose gel.
The template RNAs used in the RdRp assays were transcribed using T7 RNA polymerase (13). Plus-strand AMV RNA 3 was transcribed from pAL3 (28). Minus-strand RNA 3 was transcribed from pT71-301 (52). Therefore, pAL3 and pT71-301 were linearized using PstI, creating overhangs that were made blunt using T4 DNA polymerase.
Protein analysis.
From each fraction of the glycerol gradients, 7.5 μl was analyzed by Western blotting (50). The Western blot analyses were performed with Hybond-P polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). Rabbit polyclonal antibodies directed against the C-terminal amino acids 1100 to 1120 of P1 (57) and the N terminus of P2 (51) were used.
RESULTS
Transient expression of P1 and P2 in N. benthamiana.
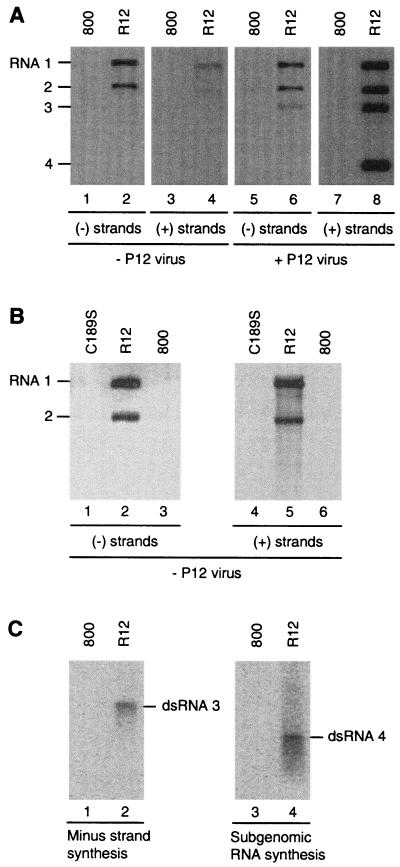
A. tumefaciens was transformed with a binary vector containing cDNAs 1 and 2 each flanked by the CaMV 35S promoter and nos terminator. This A. tumefaciens strain will be referred to below as R12 (Fig. 1B). The empty vector pMOG800 was also transformed to A. tumefaciens (referred to as 800) to be used as a negative control. At 2 days after infiltration of an R12 suspension, total-RNA extracts of the infiltrated leaves contained detectable levels of plus- and minus-strand AMV RNAs 1 and 2 (Fig. 2A, lanes 2 and 4). The minus-strand RNAs resulted from transcription of plus-strand templates by the AMV RdRp. As observed in R12 plants expressing RNAs 1 and 2 from transgenes (49), transient expression of RNAs 1 and 2 from the 35S promoter obviated the requirement for CP to initiate minus-strand RNA synthesis. The plus-strand RNAs could result from transcription of the R12 construct by the host polymerase II, from transcription of AMV minus strands by the RdRp, or both.
FIG. 2.
Viral RNA accumulation and RdRp activity induced in agroinfiltrated leaves. (A) Viral RNA accumulation. Leaves were infiltrated with A. tumefaciens suspensions containing the empty T-DNA vector (800; odd lane numbers) or the R12 construct (R12; even lane numbers). Two days after infiltration, half of the leaves were inoculated with virus particles containing RNAs 3 and 4 (P12 virus; lanes 5 to 8). RNA was extracted from the leaves 2 days (lanes 1 to 4) or 7 days (lanes 5 to 8) after infiltration and analyzed by Northern blot hybridization using probes detecting minus-strand (lanes 1, 2, 5, 6) or plus-strand (lanes 3, 4, 7, 8) AMV RNAs. The positions of RNAs 1 to 4 are indicated in the left margin. (B) Viral RNA accumulation after infiltration of a replication-deficient mutant. Leaves were infiltrated with A. tumefaciens suspensions containing the empty T-DNA vector (800; lanes 3 and 6), the R12C189S construct (C189S; lanes 1 and 4), or the R12 construct (R12; lanes 2 and 5). RNA was extracted from the leaves 2 days after infiltration and analyzed by Northern blot hybridization using probes detecting minus-strand (lanes 1 to 3) or plus-strand (lanes 4 to 6) AMV RNAs. The positions of RNAs 1 and 2 are indicated in the left margin. (C) RdRp activity. Leaves were infiltrated with A. tumefaciens suspensions containing the empty T-DNA vector (800; lanes 1 and 3) or the R12 construct (R12; lanes 2 and 4). Two days after infiltration, the leaves were homogenized and RdRp was solubi- lized from the 30,000 × g membrane fraction and purified by glycerol gradient centrifugation. The RdRp was assayed in vitro for minus-strand RNA 3 synthesis using a plus-strand RNA 3 template (lanes 1 and 2) and subgenomic plus-strand RNA 4 synthesis using a minus-strand RNA 3 template (lanes 3 and 4). Radiolabeled products were analyzed by gel electrophoresis. The positions of double-stranded RNA 3 and 4 products are indicated in the right margins.
To distinguish between these options, a conserved cysteine residue in the proposed methyltransferase domain of P1 was mutated to serine and the mutation was introduced into the R12 construct (R12C189S). Mutation of the homologous residue in the methyltransferase protein of Semliki Forest virus (nsP1) reduced the methyltransferase and guanylyltransferase activities of nsP1 to undetectable levels (1). Furthermore, C189S completely abolished AMV replication (A. C. Vlot, A. Menard, and J. F. Bol, unpublished result), and no minus-strand RNAs 1 and 2 were detected in total-RNA extracts of leaves 2 days after infiltration of R12C189S (Fig. 2B, lane 1). Compared to the R12-derived plus strand RNAs, plus-strand RNAs 1 and 2 could hardly be detected when R12C189S was expressed (compare lanes 4 and 5), unless films were exposed for very long times (data not shown). Since no minus-strand synthesis was detected, even on films that were exposed for a long time, the traces of plus-strand RNAs were attributed to transcription by the host Pol II.
Therefore, after infiltration of R12, the Pol II transcripts of this construct are used as templates for transcription of minus strands by the AMV RdRp. As in R12 plants, the accumulation of plus-strand RNA 1 and 2 could then be the result of protection of the Pol II transcripts from degradation after minus-strand synthesis or of transcription of plus strands by the AMV RdRp on the minus-strand templates (49). In both cases, plus-strand accumulation would be dependent on and coincide with minus-strand accumulation.
Two days after agroinfiltration of R12, leaves were inoculated with virus particles containing RNAs 3 and 4 (P12 virus). These particles are purified from RNA 3-inoculated P12 plants and do not contain RNAs 1 or 2 (47). Five days after inoculation of P12 virus, the R12-infiltrated leaves were found to contain minus-strand RNAs 1, 2, and 3 (Fig. 2A, lane 6) and plus-strand RNAs 1, 2, 3, and 4 (lane 8). P12 virus inoculation of leaves infiltrated with the empty vector did not result in accumulation of viral RNA (lanes 5 and 7). Virus accumulation obtained by this procedure in R12-infiltrated leaves was about 10-fold lower than the accumulation obtained by inoculation of leaves with wild-type virus (result not shown). The observation that the accumulation of plus-strand RNAs 1 and 2 strongly increased after inoculation of the infiltrated leaves with P12 virus (compare lanes 4 and 8) is in line with the previous conclusion that CP is required for asymmetric accumulation of plus-strand AMV RNAs (53, 56).
When the protocol for the purification of RdRp from infected leaves (38) was applied to leaves agroinfiltrated with R12, an enzyme activity was obtained that was able to both transcribe a plus-strand RNA 3 template into full-length minus-strand RNA 3 (Fig. 2C, lane 2) and synthesize subgenomic RNA 4 on a minus-strand RNA 3 template (lane 4) in vitro. This enzyme was called R12-RdRp. Like the enzyme purified from transgenic P12 plants (P12-RdRp), R12-RdRp does not contain CP, which is present in RdRp purified from plants infected with wild-type virus (39, 51). It was reported that plus-strand RNA 4 synthesis in vitro by P12-RdRp was strongly stimulated by addition of CP to the RdRp assay mixture (9). However, CP did not significantly stimulate RNA 4 synthesis by the purified R12-RdRp (P. C. J. Haasnoot, F. T. Brederode, R. C. L. Olsthoorn and J. F. Bol, unpublished result). From the results presented in Fig. 2, we conclude that transient expression of full-length RNAs 1 and 2 in leaves of N. benthamiana results in the assembly of an RdRp that supports AMV RNA synthesis in vivo and in vitro.
Expression of 3′-UTR deletion mutants.
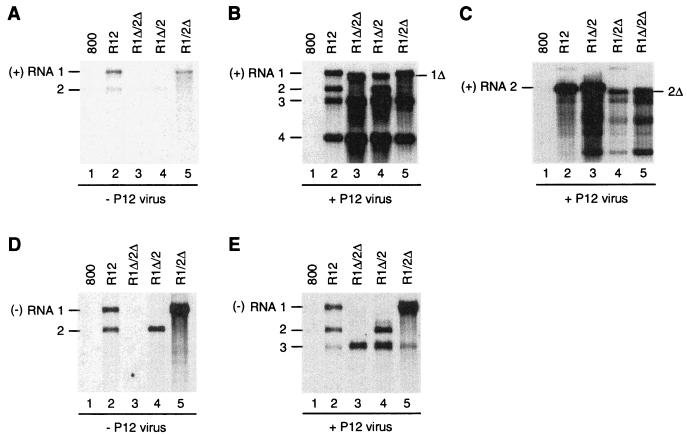
As a first step toward defining cis-acting elements required for the in vivo assembly of a functional replication complex, 3′-UTR deletion mutations were engineered in cDNAs 1 and 2 in pMOGR12 (Fig. 1B). Binary vector constructs containing RNA 1-Δ3′-UTR and RNA 2-Δ3′-UTR (R1Δ/2Δ), RNA 1 and RNA 2-Δ3′-UTR (R1/2Δ), or RNA 1-Δ3′-UTR and RNA 2 (R1Δ/2) were transformed to A. tumefaciens and infiltrated into N. benthamiana leaves. Figure 3 shows the accumulation of viral plus-strand RNAs (Fig. 3A to C) and minus-strand RNAs (Fig. 3D and E) in the infiltrated leaves before (Fig. 3A and D) and after (Fig. 3B, C, and E) inoculation of the leaves with P12 virus. In several experiments, total RNA extracts of the infiltrated leaves contained small amounts of plus-strand AMV RNAs 1 and 2 (Fig. 3A, lanes 2, 4, and 5), coinciding with the accumulation of corresponding minus strands (Fig 3D, lanes 2, 4, and 5). Construct R1Δ/2Δ did not induce the synthesis of minus-strand RNAs (Fig. 3D, lane 3); however, after inoculation of P12 virus the R1Δ/2Δ-infiltrated leaves supported the accumulation of minus-strand RNA 3 (Fig. 3E, lane 3) and plus-strand RNAs 3 and 4 (Fig. 3B, lane 3; the origin of RNA 1Δ seen in this lane is discussed below). This demonstrates that the truncated RNAs 1 and 2 are translated into functional replicase proteins. Leaves infiltrated with the R1Δ/2 or R1/2Δ constructs showed the accumulation of minus-strand RNAs corresponding to wild-type RNA 2 and wild-type RNA 1, respectively, but not of minus-strand RNAs corresponding to the encoded truncated RNAs (Fig. 3D, lanes 4 and 5). These data are consistent with the previous conclusion that the 3′-UTRs of the AMV RNAs contain the promoters for minus-strand RNA synthesis (59). After inoculation of P12 virus, R1Δ/2- or R1/2Δ-infiltrated leaves showed the accumulation of minus-strand RNAs 2 and 3 or 1 and 3, respectively (Fig. 3E, lanes 4 and 5) and the accumulation of corresponding plus-strand RNAs (Fig. 3B, lanes 4 and 5).
FIG. 3.
Accumulation of viral RNA in leaves agroinfiltrated with 3′-UTR deletion mutants. Leaves were infiltrated with A. tumefaciens suspensions containing the empty T-DNA vector (800) or constructs R12, R1Δ/2Δ, R1Δ/2, or R1/2Δ, as indicated above the lanes. Two days after infiltration, some of the leaves were inoculated with virus particles containing RNAs 3 and 4 (B, C, and E); the other leaves were not inoculated (A and D). RNA was extracted from the leaves 2 days (A and D) or 7 days (B, C, and E) after infiltration and analyzed by Northern blot hybridization using probes detecting plus-strand RNAs 1 to 4 (A and B), plus-strand RNA 2 (C), or minus-strand RNAs 1, 2, and 3 (D and E). The positions of plus-strand RNAs 1 to 4 and minus-strand RNAs 1, 2, and 3 are indicated in the left margins; the positions of truncated RNAs are indicated in the right margins.
Lanes 3 and 4 of Fig. 3B show the accumulation of an RNA species that migrates slightly faster than RNA 1 and probably corresponds to RNA 1Δ. Since the accumulation of this RNA is not paralleled by the synthesis of a corresponding minus strand (Fig. 3E, lanes 3 and 4), the truncated RNA 1 probably represents a Pol II transcript of cDNA 1Δ in the T-DNA vector. A similar transcript of cDNA 2Δ would migrate very close to RNA 3. To reveal such a transcript, the blot in Fig. 3C was hybridized with a probe detecting only RNA 2-specific sequences. Accumulation of RNA 2Δ was clearly detectable in leaves infiltrated with R1/2Δ or R1Δ/2Δ after inoculation of P12 virus (Fig. 3C, lanes 4 and 5). Before inoculation of P12 virus, accumulation of RNAs 1Δ and 2Δ was barely detectable (Fig. 3A, lanes 3 to 5). Possibly, the transiently expressed truncated RNAs were stabilized by the RNA 3-encoded CP that was expressed after inoculation of P12 virus.
The 3′-UTR of AMV is not required for encapsidation.
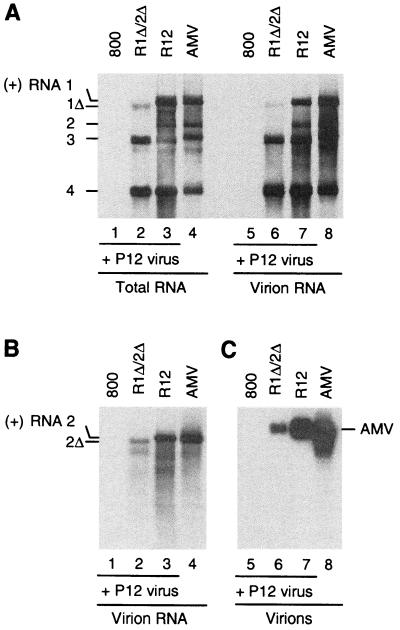
To find whether the putative protection of the 3′-truncated RNAs 1 and 2 by CP was mediated by the formation of virus particles, virions were isolated from infiltrated leaves that had been inoculated with P12 virus. Part of the leaf material was used to isolate total RNA as a control. In the blot in Fig. 4A, the RNAs in the total-RNA extract and the extract from virions were visualized using a probe corresponding to RNAs 1, 2, and 3. Extracts of leaves infiltrated with R12 and inoculated with P12 virus (Fig. 4, lanes 3 and 7) and extracts of leaves inoculated with wild-type AMV (lanes 4 and 8) served as controls. On infiltration of R1Δ/2Δ, accumulation of RNA 1Δ was detectable (Fig. 4A, lane 2) and part of this RNA was apparently encapsidated into virions (lane 6). Since RNA 2Δ migrates close to RNA 3, the blot was hybridized to an RNA 2-specific probe in Fig. 4B, showing that the truncated RNA 2 expressed from the R1Δ/2Δ construct was at least partially encapsidated into virus particles (Fig. 4B, lane 2).
FIG. 4.
Analysis of the encapsidation of RNAs 1 and 2 with 3′-UTR deletions. Leaves were infiltrated with A. tumefaciens suspensions containing the empty T-DNA vector (800; lanes 1 and 5) or constructs R1Δ/2Δ (lanes 2 and 6) or R12 (lanes 3 and 7). In addition, noninfiltrated leaves were inoculated with wild-type AMV (lanes 4 and 8). The agroinfiltrated leaves were inoculated with virus particles containing RNAs 3 and 4 2 days after infiltration. Some of the leaves were used to extract total RNA (panel A, lanes 1 to 4), whereas the remainder of the leaves was used to extract virus particles (panel A, lanes 5 to 8; panels B and C, lanes 1 to 8). Extraction was done 7 days after infiltration of the leaves or 5 days after inoculation of leaves with wild-type AMV. Total RNA (panel A, lanes 1 to 4) and RNA extracted from virus particles (panel A, lanes 5 to 8; panel B, lanes 1 to 4) were analyzed by Northern blot hybridization using a probe detecting plus-strand RNAs 1 to 4 (panel A, lanes 1 to 8) or plus-strand RNA 2 (panel B, lanes 1 to 4). In addition, purified virus particles were run in an agarose gel and analyzed by Northern blot hybridization using a probe detecting coding sequences of RNAs 1 and 2 (panel C, lanes 5 to 8). Lanes 4 and 8 of all panels were loaded with 10 times less material than the standard amount used to load all other lanes. The positions of full-length and truncated AMV RNAs are indicated in the left margin; the position of AMV particles is indicated in the right margin.
It has been shown that the four types of AMV virions containing RNAs 1 to 4 migrate as a single band when run in an agarose gel (56). Figure 4C shows a Northern blot of an agarose gel run with virions isolated from leaves that were infiltrated with R12 or R1Δ/2Δ and inoculated with P12 virus and with virions isolated from leaves that were infected with wild-type AMV. The blot was hybridized to probes corresponding to the coding sequences of RNAs 1 and 2. It is clear that 3′-truncated RNAs 1 and 2 that are transiently expressed from the R1Δ/2Δ construct are encapsidated into virions. RNA extracted from leaves infiltrated with the R12 construct and inoculated with P12 virus was infectious to N. benthamiana, whereas RNA extracted from leaves infiltrated with the R1Δ/2Δ construct and inoculated with P12 virus was not (result not shown). Moreover, RNAs 1 and 2 produced in the R1Δ/2Δ-infiltrated leaves were not detectable on Northern blots using a probe corresponding to the 3′-UTR of these RNAs (result not shown). These data demonstrate that the 3′ deletions in RNAs 1 and 2 had not been restored by recombination with RNA 3.
One 3′-UTR is required to induce extractable RdRp activity.
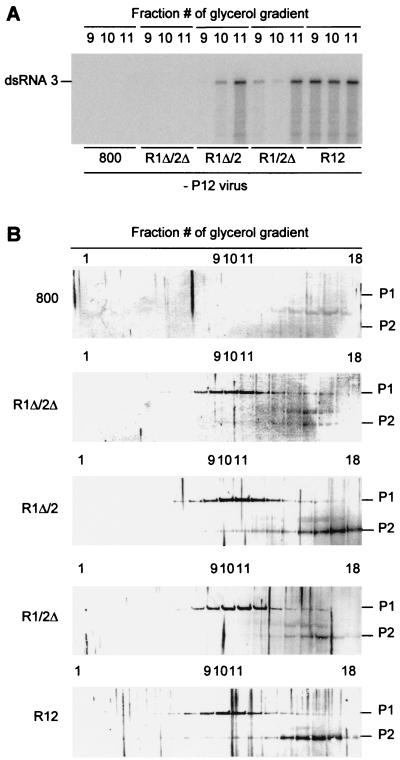
In Fig. 2C it was shown that RdRp activity could be detected in vitro in extracts of R12-infiltrated leaves. To analyze a possible role of the 3′-UTR sequences of RNAs 1 and 2 in the formation of this enzyme, leaves were infiltrated with R1Δ/2Δ, R1Δ/2, and R1Δ/2Δ, as well as with R12 and the empty vector as controls. The leaves were homogenized 2 days after infiltration, after which RdRp was solubilized from the 30,000 × g membrane fraction and further purified in a glycerol gradient. RdRp activity was detected in the gradient fractions by measuring the conversion of an AMV RNA 3 template into a double-stranded radiolabeled product (9). When the fractions are numbered 1 to 18 from the bottom to the top of a gradient, maximum RdRp activity is detected in fractions 9 to 11 under standard conditions. Figure 5A shows the synthesis of double-stranded RNA 3 by enzyme activities in fractions 9, 10, and 11 of glycerol gradients run with extracts of the infiltrated leaves. RdRp activity could be detected in isolates from leaves infiltrated with R12 but not in isolates from leaves infiltrated with R1Δ/2Δ, although the two types of leaves support RNA 3 replication in vivo with similar efficiencies (Fig. 3B and E). However, RdRp activity was detectable in extracts from leaves infiltrated with constructs R1Δ/2 or R1/2Δ (Fig. 5A). This demonstrates that a single 3′-UTR, either from RNA 1 or from RNA 2, is sufficient to permit the in vivo formation of an RdRp complex that is active in vitro.
FIG. 5.
Accumulation of replicase proteins and RdRp activity in agroinfiltrated leaves. Leaves were infiltrated with A. tumefaciens suspensions containing the empty T-DNA vector or constructs R1Δ/2Δ, R1Δ/2, R1/2Δ, or R12 as indicated at the bottom of panel A and in the left margin of panel B. Two days after infiltration, the leaves were homogenized, after which RdRp was solubilized from the 30,000 × g membrane fraction and sedimented in a glycerol gradient. (A) In vitro RdRp assay. Samples from fractions 9, 10, and 11 of the glycerol gradients were used in RdRp assays with plus-strand AMV RNA 3 as a template (fraction 1 is at the bottom of the gradient). Radiolabeled products were analyzed by gel electrophoresis. The position of double-stranded RNA 3 is indicated in the left margin. (B) Western blot analysis of the protein composition of glycerol gradient fractions. Fractions 1 (bottom) to 18 (top) were analyzed with antisera to P1 and P2 proteins. The positions of P1 and P2 are indicated in the right margin.
To find whether RdRp activity in the gradient fractions was correlated with the presence of P1 and P2, the fractions were analyzed by Western blotting using P1- and P2-specific antisera. The distribution of P1 and P2 in the gradient run with the extract of R12-infiltrated leaves (Fig. 5B) was similar to the distribution obtained in the gradient run with an extract of AMV-infected leaves (51). P1 cosediments with the enzyme activity, whereas P2 is present in the fractions with enzyme activity but most prominently in the top fractions of the gradient. The distribution of P1 and P2 in the gradients run with extracts from leaves infiltrated with R1Δ/2Δ, R1Δ/2, or R1/2Δ was largely similar to the distribution in the gradient run with the extract of R12-infiltrated leaves. Apparently, expression of RNA 2Δ into P2 was slightly less efficient than expression of full-length RNA 2. However, this difference cannot explain the observation that the extract of R1Δ/2Δ-infiltrated leaves did not exhibit RdRp activity while the extract of R1/2Δ-infiltrated leaves did. We conclude that when both 3′-UTRs are absent in the transiently expressed RNAs, P1 and P2, which are present in gradient fractions 9 to 11, do not form an active enzyme complex.
All AMV 3′-UTRs facilitate activation of the RdRp-complex.
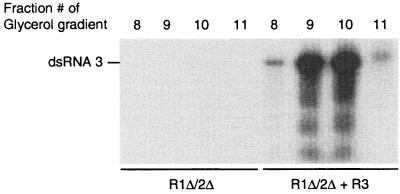
The observation that leaves infiltrated with R1Δ/2Δ efficiently support RNA 3 replication indicates that the transiently expressed P1 and P2 form an active RdRp after inoculation of the leaves with P12 virus. However, it appeared to be difficult to find the right conditions for the isolation of this enzyme. In an alternative approach, we coinfiltrated leaves with a mixture of two A. tumefaciens strains, one containing the R1Δ/2Δ construct and one containing a construct with full-length cDNA of RNA 3 flanked by the 35S promoter and nos terminator (R3 construct). When infiltration was done with a mixture of the R12 and R3 A. tumefaciens strains, accumulation of RNAs 1 to 4 occurred at levels similar to those in plants inoculated with wild-type virus (result not shown). Figure 6 shows the RdRp activity in fractions 8 to 11 of glycerol gradients run with extracts of leaves infiltrated with R1Δ/2Δ or with R1Δ/2Δ and R3. The observation that RNA 3 replication induced RdRp activity indicates that the 3′-UTRs of RNAs 1, 2, and 3 are equivalent in their function in RdRp activation.
FIG. 6.
Induction of RdRp activity by replication of RNA 3. Leaves were infiltrated with an A. tumefaciens suspension containing construct R1Δ/2Δ or a mixture of A. tumefaciens suspensions containing constructs R1Δ/2Δ and R3 as indicated at the bottom of the lanes (R3 expresses full-length RNA 3). Two days after infiltration, the leaves were homogenized, after which RdRp was solubilized from the 30,000 × g membrane fraction and sedimented in a glycerol gradient. Samples of fractions 8, 9, 10, and 11 of the gradients were assayed for RdRp activity using plus-strand AMV RNA 3 as a template. Radiolabeled products were analyzed by gel eclectrophoresis. The position of double-stranded RNA 3 is indicated in the left margin.
DISCUSSION
Expression of AMV RdRp by agroinfiltration.
In the past, A. tumefaciens-mediated agroinfection of plants has been used to initiate virus infections locally as an alternative to mechanical inoculation. In the present work, we developed the agroinfiltration technique for the transient expression of genes in plants into a novel method for the expression and isolation of viral RdRp from a natural host of the virus but in a nonviral background. Previously, AMV RdRp could be isolated only from AMV-infected plants (39) or transgenic P12 plants (47). In theory, transgenic plants could be used to express mutant RdRps for in vitro studies, but in practice this is not feasible due to the large genetic variability between primary transformants and the long periods required to generate transformed plants. Moreover, the amount of RdRp per gram of leaf tissue that can be isolated from transgenic P12 plants is 30-fold smaller than the amount isolated from agroinfiltrated leaves. By agroinfiltration, the T-DNA from the A. tumefaciens vector is transferred to a large proportion of the cells of the infiltrated leaves and the accumulation of viral RNAs and proteins does not involve cell-to-cell movement of virus material. This is indicated by the observation that there is no increase in the accumulation of minus-strand RNAs 1 and 2 in R12-infiltrated leaves on inoculation of the leaves with RNA 3-containing particles (compare lanes 2 and 6 of Fig. 2). In addition, RNA 3 accumulation in infiltrated leaves is independent of replication of RNAs 1 and 2 (Fig. 3). On average, the accumulation of viral RNAs in R12-infiltrated leaves inoculated with RNA 3-containing virus particles was several times lower than the wild-type levels of virus accumulation that were obtained by infiltration of a mixture of the R12 and R3 A. tumefaciens strains. We will analyze a possible role of cell-to-cell movement in this accumulation by using an R3 construct expressing a defective movement protein gene. In addition, it will be interesting to analyze virus accumulation induced by agroinfiltration in a nonhost of AMV such as Arabidopsis thaliana. This would make Arabidopsis genetics available to studies on a wide range of viruses.
Transient expression of AMV RdRp in agroinfiltrated leaves provided further insight into the role of CP in AMV replication. R12-infiltrated leaves permit a study of RNA replication in vivo in the absence of CP and act as source for the purification of highly active and stable RdRp that is devoid of CP. RNA 3 replication in R12-infiltrated leaves could be initiated by inoculation of virus particles containing RNA 3, by inoculation of RNA 3 transcribed with T7 RNA polymerase (data not shown), or by coinfiltration of the R3 construct. This demonstrates that CP is not required to initiate RNA replication in agroinfiltrated leaves. Moreover, our results corroborate previous conclusions that CP is not involved in minus-strand AMV RNA synthesis (9, 27, 56) whereas it causes an approximately 100-fold increase of plus-strand RNA accumulation in infected protoplasts (53, 56). Different CP mutants that were defective in the assembly of detectable virions induced either high or low levels of plus-strand RNA accumulation (56), indicating that CP stimulated RNA synthesis rather than preventing its degradation. However, our observation that accumulation of the nonreplicatable RNAs 1Δ and 2Δ strongly increased after expression of CP in agroinfiltrated leaves suggests a role for CP in the protection of plus-strand RNA from degradation. Possibly, in a normal infection cycle, CP is required in a step prior to minus-strand synthesis, in encapsidation of virus RNA, and in cell-to-cell and long-distance movement (27, 48, 56). It has been proposed that the step prior to minus-strand RNA synthesis involves translation of the genomic RNAs (32). Recent evidence supports the notion that the poly(A) tail of transgenically or transiently expressed AMV RNAs can substitute for the early function of CP (L. Neeleman and J. F. Bol, unpublished results).
The 3′-UTR of AMV is not required for encapsidation.
The 3′-terminal 39 nucleotides of the AMV RNAs contain a high-affinity binding site for CP (16, 41). Peptides corresponding to the N-terminal 26 amino acids of CP bind specifically to this RNA sequence and can substitute for CP in the inoculum to initiate infection (3). Mutation of Arg-17 into Ala interfered with the binding of N-terminal peptides or full-length CP to the 3′ end of the RNAs and with initiation of infection (2, 62). However, this mutation did not interfere with the role of CP in virion assembly (48). Our observation that RNAs 1Δ and 2Δ are encapsidated into virions also supports the notion that binding of CP to the 3′ termini of AMV RNAs plays a role in the initiation of infection but not in the assembly of virus particles. In addition to the 3′ termini, CP has been found to bind to several internal sites in AMV RNAs (64), which may act as the origin of assembly of the virus particles. Within the Bromoviridae family, a coding sequence of BMV RNA 1 was found to be required for encapsidation whereas the 3′-UTRs of the RNAs were not (10).
Possible role of the 3′-UTR of AMV in assembly of an active RdRp complex.
In AMV-infected protoplasts, P1 and P2 accumulate in the vacuolar membrane (51). Active RdRp can be solubilized from the 30,000 × g membrane fractions of homogenates of AMV-infected or R12-infiltrated leaves, and P1 and P2 in the two extracts show a similar distribution in glycerol gradients. From gradient fractions containing RdRp activity, P1 and P2 can be coimmunoprecipitated, indicating that they are part of a single enzyme complex (51). When the 3′-UTRs of RNAs 1 and 2 were both deleted, in leaves infiltrated with R1Δ/2Δ, P1 and P2 appeared to be present in a complex with similar sedimentation properties to P1 and P2 in extracts from R12-infiltrated leaves; however, no RdRp activity could be detected in vitro (Fig. 5). This is in line with previous findings that BMV replication proteins expressed in S. cerevisiae from 1a and 2a genes devoid of flanking UTRs do not form a complex capable of RNA synthesis in vitro (37), although 1a and 2a are targeted to the endoplasmic reticulum, where BMV replication complexes are assembled (7, 40). Infection of 1a- and 2a-expressing yeast cells with BMV RNA 3 induced extractable RdRp activity, and deletion analysis indicated that both the 3′-UTR and the intercistronic region of BMV RNA 3 are required for activity of an RdRp isolate in vitro (37). In addition, extractable in vitro RdRp activity paralleled minus-strand synthesis in vivo (37). The BMV RNA 3 intercistronic region, as well as the 5′-UTRs of BMV RNAs 1 and 2, contain box B motifs corresponding to the TΨC stem-loop of host tRNAs (8, 37, 44). A sequence similar to the box B motif of BMV has been found in the 5′-UTR of AMV RNA 3 (54).
The box B motif is required for 1a-induced stabilization of the BMV RNAs, and it has been proposed that this stabilization is the result of recruitment of the RNAs to the replication complex whereby translation would be inhibited (8, 44). Indeed, it was found that box B is required for membrane association of the BMV RNAs (8). It is possible that the intercistronic region of RNA 3 was found to be required for activation of the BMV RdRp in yeast due to its function in template recruitment, since minus-strand RNA synthesis seems to be necessary to isolate an in vitro active RdRp complex. Similarly, the 3′-UTR of BMV RNA 3 and the 3′-UTRs of the AMV RNAs could be required for activation of the RdRp due to their function as minus-strand promoters.
We showed that a 3′-UTR of either AMV RNA 1, 2, or 3 is required for the induction of AMV replicase activity, although we cannot rule out the possibility that other sequences in the viral RNAs, for instance the 5′-UTR or subgenomic promoter region of RNA 3, are involved as well. Furthermore, the 3′-UTRs of the three AMV RNAs were found to be equivalent in their function in RdRp activation. Although we have strong indications that P1 and P2 expressed from RNAs lacking a 3′-UTR are assembled in membrane-bound protein complexes, we speculate that either the presence of a 3′-UTR or minus-strand synthesis is required for a final step in the assembly or stabilization of the AMV RdRp, which could result in activation of the enzyme. Similarly, it has been shown that RNA synthesis is required for the formation of the poliovirus replication complex, although viral replicase proteins induce membrane proliferation in the absence of viral RNA (11).
For BMV and Bovine viral diarrhea virus, it was proposed that viral replication complexes are stabilized by a transition that occurs between the phase of initiation and elongation of RNA synthesis in vitro (19, 45, 46). In vivo, the assembly of an RdRp may occur in a two-step process: recruitment of viral RNA, viral replicase proteins, and possibly host proteins to the endomembrane in the cell, where replication complexes are established, and subsequent stabilization of the RdRp after the initiation of RNA synthesis on the 3′-UTR of the RNA template. We will use the agroinfiltration system to further study the sequence elements in the noncoding and coding sequences of AMV RNAs that are required for assembly of the RdRp in plants and to study functional domains in P1 and P2 by mutational analyses.
REFERENCES
- 1.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of semlike forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansel-McKinney P, Scott S W, Swanson M, Ge X, Gehrke L. A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO J. 1996;15:5077–5084. [PMC free article] [PubMed] [Google Scholar]
- 3.Baer M L, Houser F, Loesch-Fries L S, Gehrke L. Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 1994;13:727–735. doi: 10.1002/j.1460-2075.1994.tb06312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 5.Bendahmane A, Kanyuka K, Baulcombe D C. The Rx gene from potato controls separate virus resistance and cell death. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bol J F. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J Gen Virol. 1999;80:1089–1102. doi: 10.1099/0022-1317-80-5-1089. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Ahlquist P. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Noueiry A, Ahlquist P. Brome mosaic virus protein 1a recruits viral RNA 2 to RNA replication through a 5′ proximal RNA 2 signal. J Virol. 2001;75:3207–3219. doi: 10.1128/JVI.75.7.3207-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Graaff M, Man in't Veld M R, Jaspars E M J. In vitro evidence that the coat protein of alfalfa mosaic virus plays a direct role in the regulation of plus and minus-RNA synthesis: implications for the life cycle of alfalfa mosaic virus. Virology. 1995;208:583–589. doi: 10.1006/viro.1995.1189. [DOI] [PubMed] [Google Scholar]
- 10.Duggal R, Hall T C. Identification of domains in brome mosaic virus RNA-1 and coat protein necessary for specific interaction and encapsidation. J Virol. 1993;67:6406–6412. doi: 10.1128/jvi.67.11.6406-6412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger D, Teterina N, Ehrenfeld E, Bienz K. Formation of the poliovirus replication complex requires coupled viral translation, vesicle formation, and viral RNA synthesis. J Virol. 2000;74:6570–6580. doi: 10.1128/jvi.74.14.6570-6580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4729. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haasnoot P C J, Brederode F T, Olsthoorn R C L, Bol J F. A conserved hairpin structure in alfamovirus and bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA. 2000;6:708–716. doi: 10.1017/s1355838200992471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hericourt F, Blanc S, Redeker V, Jupin I. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem J. 2000;349:417–425. doi: 10.1042/0264-6021:3490417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong Y, Hunt A G. RNA polymerase activity catalyzed by a potyvirus-encoded RNA-dependent RNA polymerase. Virology. 1996;226:146–151. doi: 10.1006/viro.1996.0639. [DOI] [PubMed] [Google Scholar]
- 16.Houser-Scott F, Baer M L, Liem K F, Cai J-M, Gehrke L. Nucleotide sequence and structural determinants of specific binding of coat protein or coat protein peptides to the 3′ untranslated region of alfalfa mosaic virus RNA 4. J Virol. 1994;68:2194–2205. doi: 10.1128/jvi.68.4.2194-2205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 18.Jaspars E M J. Genome activation in alfamo- and ilarviruses. Arch Virol. 1999;144:843–863. doi: 10.1007/s007050050551. [DOI] [PubMed] [Google Scholar]
- 19.Kim M, Zhong W, Hong Z, Kao C C. Template nucleotide moieties required for de novo initiation of RNA synthesis by a recombinant viral RNA-dependent RNA polymerase. J Virol. 2000;74:10312–10322. doi: 10.1128/jvi.74.22.10312-10322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoester M, van Loon L C, van den Heuvel J, Hennig J, Bol J F, Linthorst H J M. Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc Natl Acad Sci USA. 1998;95:1933–1937. doi: 10.1073/pnas.95.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E V, Gorbalenya A E, Purdy M A, Rozanov M N, Reyes G R, Bradley D W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-stranded RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koper-Zwarthoff E C, Brederode F T, Walstra P, Bol J F. Nucleotide-sequence of the 3′-noncoding region of alfalfa mosaic virus RNA 4 and its homology with the genomic RNAs. Nucleic Acids Res. 1979;7:1887–1900. doi: 10.1093/nar/7.7.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai V C H, Kao C C, Ferrari E, Park J, Uss A S, Wright-Minogue J, Hong Z, Lau J Y N. Mutational analysis of bovine diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Cheng Y, Huang Y, Tsai C, Hsu Y, Meng M. Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of bamboo mosaic virus. J Virol. 1998;72:10093–10099. doi: 10.1128/jvi.72.12.10093-10099.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez Vazquez A, Martin Alonso J M, Casais R, Boga J A, Parra F. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J Virol. 1998;72:2999–3004. doi: 10.1128/jvi.72.4.2999-3004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubinski J M, Ransone L J, Dasgupta A. Primer-dependent synthesis of covalently linked dimeric RNA molecules by poliovirus replicase. J Virol. 1987;61:2997–3003. doi: 10.1128/jvi.61.10.2997-3003.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neeleman L, Bol J F. cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology. 1999;254:324–333. doi: 10.1006/viro.1998.9568. [DOI] [PubMed] [Google Scholar]
- 28.Neeleman L, van der Kuyl A C, Bol J F. Role of alfalfa mosaic virus coat protein gene in symptom formation. Virology. 1991;181:687–693. doi: 10.1016/0042-6822(91)90902-n. [DOI] [PubMed] [Google Scholar]
- 29.Neeleman L, van der Vossen E A G, Bol J F. Infection of tobacco with alfalfa mosaic virus cDNAs sheds light on the early function of the coat protein. Virology. 1993;196:883–887. doi: 10.1006/viro.1993.1551. [DOI] [PubMed] [Google Scholar]
- 30.Neufeld K L, Richards O C, Ehrenfeld E. Expression and characterization of poliovirus proteins 3BVPg, 3Cpro, and 3Dpol in recombinant baculovirus-infected Spodoptera frugiperda cells. Virus Res. 1991;19:173–188. doi: 10.1016/0168-1702(91)90044-v. [DOI] [PubMed] [Google Scholar]
- 31.Oh J, Ito T, Lai M M C. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J Virol. 1999;73:7694–7702. doi: 10.1128/jvi.73.9.7694-7702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsthoorn R C L, Mertens S, Brederode F T, Bol J F. A conformational switch at the 3′-end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Reilly E K, Kao C C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 34.Plante C A, Kim K H, Pillai-Nair N, Osman T A M, Buck K W, Hemenway C L. Soluble, template-dependent extracts from Nicotiana benthamiana plants infected with potato virus X transcribe both plus- and minus-strand RNA templates. Virology. 2000;275:444–451. doi: 10.1006/viro.2000.0512. [DOI] [PubMed] [Google Scholar]
- 35.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price B D, Roeder M, Ahlquist P. DNA-directed expression of functional flock house virus RNA 1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J Virol. 2000;74:11724–11733. doi: 10.1128/jvi.74.24.11724-11733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadt R, Jaspars E M J. Purification and characterization of brome mosaic virus RNA dependent RNA polymerase. Virology. 1990;178:189–194. doi: 10.1016/0042-6822(90)90393-6. [DOI] [PubMed] [Google Scholar]
- 39.Quadt R, Rosdorff H J M, Hunt T W, Jaspars E M J. Analysis of the protein composition of the alfalfa mosaic virus RNA-dependent RNA polymerase. Virology. 1991;182:309–315. doi: 10.1016/0042-6822(91)90674-z. [DOI] [PubMed] [Google Scholar]
- 40.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reusken C B E M, Bol J F. Structural elements of the 3′-terminal coat protein binding site in alfalfa mosaic virus RNAs. Nucleic Acids Res. 1996;24:2660–2665. doi: 10.1093/nar/24.14.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sankar S, Porter A G. Expression, purification, and properties of recombinant encephalomyocarditis virus RNA-dependent RNA polymerase. J Virol. 1991;65:2993–3000. doi: 10.1128/jvi.65.6.2993-3000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steffens S, Thiel H J, Behrens S E. The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro. J Gen Virol. 1999;80:2583–2590. doi: 10.1099/0022-1317-80-10-2583. [DOI] [PubMed] [Google Scholar]
- 44.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA 3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun J, Adkins S, Faurote G, Kao C C. Initiation of (−) strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: Synthesis of oligoribonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 46.Sun J, Kao C C. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: transition from initiation to elongation. Virology. 1997;233:63–73. doi: 10.1006/viro.1997.8583. [DOI] [PubMed] [Google Scholar]
- 47.Taschner P E M, van der Kuyl A C, Neeleman L, Bol J F. Replication of an incomplete alfalfa mosaic virus genome in plants transformed with viral replicase genes. Virology. 1991;181:445–450. doi: 10.1016/0042-6822(91)90876-d. [DOI] [PubMed] [Google Scholar]
- 48.Tenllado F, Bol J F. Genetic dissection of the multiple functions of alfalfa mosaic virus coat protein in viral RNA replication, encapsidation, and movement. Virology. 2000;268:29–40. doi: 10.1006/viro.1999.0170. [DOI] [PubMed] [Google Scholar]
- 49.Thole V, Garcia M, van Rossum C M A, Neeleman L, Brederode F T, Linthorst H J M, Bol J F. Alfalfa mosaic virus RNAs 1 and 2 expressed in transgenic plants start to replicate only after infection of the plants with RNA 3. J Gen Virol. 2001;82:25–28. doi: 10.1099/0022-1317-82-1-25. [DOI] [PubMed] [Google Scholar]
- 50.Towbin H, Staehelin R, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Heijden M W, Carette J E, Reinhoud P J, Haegi A, Bol J F. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J Virol. 2001;75:1879–1887. doi: 10.1128/JVI.75.4.1879-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Kuyl A C, Langereis K, Houwing C J, Jaspars E M J, Bol J F. cis-acting elements involved in replication of alfalfa mosaic virus RNAs in vitro. Virology. 1990;176:346–354. doi: 10.1016/0042-6822(90)90004-b. [DOI] [PubMed] [Google Scholar]
- 53.Van der Kuyl A C, Neeleman L, Bol J F. Role of alfalfa mosaic virus coat protein in regulation of the balance between plus and minus strand RNA synthesis. Virology. 1991;185:496–499. doi: 10.1016/0042-6822(91)90807-n. [DOI] [PubMed] [Google Scholar]
- 54.Van der Vossen E A G, Bol J F. Analysis of cis-acting elements in the 5′ leader sequence of alfalfa mosaic virus RNA 3. Virology. 1996;220:539–543. doi: 10.1006/viro.1996.0345. [DOI] [PubMed] [Google Scholar]
- 55.Van der Vossen E A G, Neeleman L, Bol J F. Role of the 5′ leader sequence of alfalfa mosaic virus RNA 3 in replication and translation of the viral RNA. Nucleic Acids Res. 1993;21:1361–1367. doi: 10.1093/nar/21.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Vossen E A G, Neeleman L, Bol J F. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology. 1994;202:891–903. doi: 10.1006/viro.1994.1411. [DOI] [PubMed] [Google Scholar]
- 57.Van Pelt-Heerschap H. Immunochemical analysis of the alfalfa mosaic virus gene products. Ph.D. thesis. 1987. Leiden, The Netherlands. [Google Scholar]
- 58.Van Rossum C M A, Neeleman L, Bol J F. Comparison of the role of 5′-terminal sequences of alfalfa mosaic virus RNAs 1, 2, and 3 in viral RNA replication. Virology. 1997;235:333–341. doi: 10.1006/viro.1997.8707. [DOI] [PubMed] [Google Scholar]
- 59.Van Rossum C M A, Reusken C B E M, Brederode F T, Bol J F. The 3′untranslated region of alfalfa mosaic virus RNA 3 contains a core promoter for minus-strand RNA synthesis and an enhancer element. J Gen Virol. 1997;78:3045–3049. doi: 10.1099/0022-1317-78-11-3045. [DOI] [PubMed] [Google Scholar]
- 60.Van Vloten-Doting L, Jaspars E M J. The uncoating of alfalfa mosaic virus by its own RNA. Virology. 1972;48:699–708. doi: 10.1016/0042-6822(72)90154-7. [DOI] [PubMed] [Google Scholar]
- 61.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 62.Yusibov V, Loesch-Fries L S. Functional significance of three basic N-terminal amino acids of alfalfa mosaic virus coat protein. Virology. 1998;242:1–5. doi: 10.1006/viro.1997.8973. [DOI] [PubMed] [Google Scholar]
- 63.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuidema D, Jaspars E M J. Comparative investigations on the coat protein binding sites of the genomic RNAs of alfalfa mosaic virus and tobacco streak viruses. Virology. 1984;135:43–52. doi: 10.1016/0042-6822(84)90115-6. [DOI] [PubMed] [Google Scholar]