Abstract
CD38 has emerged as a potential therapeutic target for patients with systemic lupus erythematosus (SLE) but it is not known whether CD38 alters CD4+ T cell function. Using primary human T cells and CD38-sufficient and CD38-deficient Jurkat T cells, we demonstrate that CD38 shifts the T cell lipid profile of gangliosides from GM3 to GM2 by upregulating B4GALNT1 in a Sirtuin 1-dependent manner. Enhanced expression of GM2 causes ER stress by enhancing Ca2+ flux through the PLCγ1-IP3 pathway. Interestingly, correction of the calcium overload by an IP3 receptor inhibitor, but not by a store-operated calcium entry (SOCE) inhibitor, improves IL-2 production by CD4+ T cells in SLE. This study demonstrates that CD38 affects calcium homeostasis in CD4+ T cells by controlling cell membrane lipid composition that results in suppressed IL-2 production. CD38 inhibition with biologics or small drugs should be expected to benefit patients with SLE.
Subject terms: Systemic lupus erythematosus, Autoimmunity, CD4-positive T cells, Interleukins, Lymphocyte activation
CD38 has been reported to be upregulated on T cells in lupus with unclear functional consequences. Here the authors show that CD4+CD38+ T cells from lupus patients have altered calcium homeostasis accompanied by alteration in cell membrane lipid composition and a suppression of IL-2.
Introduction
Systemic lupus erythematosus (SLE) causes organ injury through autoantibodies, immune complexes, inflammatory cytokines and autoreactive immune cells1. The multitude of pathways which have been identified to contribute to the expression of the disease has stalled the development of efficacious therapeutics to control disease activity and organ damage2.
The ectonucleotidase CD38 has been reported to be elevated in immune cells in various autoimmune diseases including SLE3–5, rheumatoid arthritis6 and systemic sclerosis7. CD38, a transmembrane protein, displays ectoenzymatic activity and is widely accepted as a key regulator of extracellular and intracellular NAD+ levels8,9. The NAD reduction resulting from the high expression of CD38 causes cell senescence10 and aging11. We reported previously that increased expression of CD38 on CD8+ T cells attenuates cytotoxicity12. Recently, case series studies have reported that blocking of CD38 with the monoclonal antibody daratumumab delivered clinical benefit in two patients with refractory SLE13–15. Although multiple reports have shown a strong correlation between the expression of CD38 in immune cells and disease activity in patients with SLE, nothing is known on how CD38 affects the function of CD4+ T cells.
CD38-deficient mice are resistant to high fat diet-induced obesity, while CD38-sufficient mice accumulate fat in the liver, muscle and fat tissues16. CD38-sufficient mice also had higher expression of key genes related to lipid synthesis compared to CD38 total knock-out mice during differentiation of adipocytes17. Lipid metabolism, besides being critical for cell surface membrane structures, it is important in T cell activation because it serves as a source of energy18,19.
Lipid microdomains are enriched with cholesterol and gangliosides20 and represent unique platforms on the plasma membrane to bring together receptors and molecules important for the initiation of signaling. Previously, we had reported that GM1 gangliosides detected by cholera toxin B (CTB) staining are upregulated in the lipid microdomains and changes in ganglioside containing lipid rafts are linked to abnormal T cell signaling in SLE T cells21. Inhibition of all ganglioside synthesis by a glucosylceramide synthase (GCS) inhibitor decreased double-stranded DNA antibody in SLE22. Depletion of plasma cholesterol with statins and plasma membrane cholesterol with methyl-beta cyclodextrin also limits the production of inflammatory cytokines including IL-6 and IL-1023. Thus, the modification of lipid microdomains represents an effective approach to correct inflammatory signals in SLE.
In this communication we present evidence that the activity of CD38 which is increased in CD4+ T cells isolated from patients with SLE, enriches the cell membrane with the ganglioside GM2 which accounts for increased Ca2+ entry and endoplasmic reticulum (ER) stress. Control of ganglioside distribution on the T cell membrane and of early signaling events through CD38 may offer therapeutic options for patients with SLE.
Results
Increased expression of CD38 on the surface membrane of SLE CD4+ T cells correlates with increased lipid microdomain formation
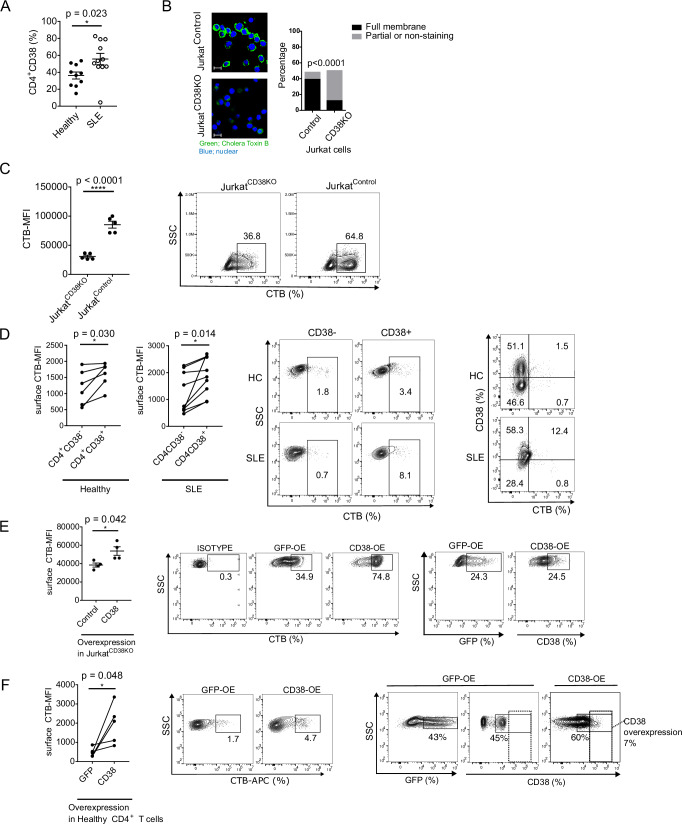
The ectonucleotidase CD38 has been reported to be expressed at increased levels in T, B and NK cells in patients with SLE3–5,24 but the mechanisms responsible for altering CD4+ T cell function are not known. Previously, we linked the expanded CD8+CD38+ T cell population in patients with SLE to suppressed expression of genes responsible for cytotoxic activity and increased rates of infections12. We now interrogated the expression of CD38 in CD4+ T cells and whether they alter their function. First, we established, in a new cohort of patients, that the CD4+CD38+ T cell subpopulation among CD4+ cells is expanded in patients with SLE (Fig. 1A, Fig. S1A). CD38 controls fatty acid synthesis in adipocytes17 and SLE T cells display aggregated lipid microdomains on their cell surface21. Accordingly, we considered that the formation of lipid microdomains on the cell surface membrane of CD4+ T cells is controlled by CD38. To gain evidence that CD38 is involved in the formation of lipid microdomains, we used Cas9-expressing Jurkat human T cells (JurkatControl) and Jurkat cells lacking CD38 prepared using Crispr/Cas9 (JurkatCD38KO) T cells12. Indeed, JurkatCD38KO cells displayed significantly less staining with CTB, which binds to the lipid microdomain component ganglioside GM1, compared to JurkatControl cells as determined by fluorescence microscopy (Fig. 1B) and flow cytometry analysis (Fig. 1C). These data suggest that the expression levels of GM1 on the surface membrane is linked to the expression and/or function of CD38. CTB binding was increased on the surface of CD4+CD38+ T cells from both healthy participants and patients with SLE compared to CD4+CD38- T cells (Fig. 1D). Staining with CTB in CD4+CD38+ T cells was comparable between healthy participants and SLE patients. We also observed increased levels of CTB staining intracellularly in SLE CD4+CD38+ compared to CD4+CD38- T cells (Fig. S1B). To further document the contribution of CD38 to the increased expression of GM1 we force-expressed CD38 in JurkatCD38KO cells (Fig. 1E) and in CD4+ T cells from healthy participants (Fig. 1F), and indeed, in both settings CTB binding was increased significantly. These results suggest that CD38 upregulates the expression of the CTB-binding to GM1. To examine the distribution of CD38 in CD4+ T cells we determined its expression in each major subpopulation of CD4+ T cells including naïve, central memory (CM), effector memory (EM) and terminally differentiated effector memory (TEM) cells in healthy participants and patients with SLE (Figure S1C). In both healthy and SLE participants, the naïve population was significantly higher, EM cells were significantly lower and CM and TEM were similar in CD4+CD38+ T cells compared to CD4+CD38- T cells (Figure S1C). These results indicate that each subpopulation distributed similarly in CD4+CD38+ T cells both in SLE and healthy participants. CD38 expression in naïve and EM cells was higher in SLE derived CD4+ T cells compared to healthy CD4+ T cells (Fig. S1D). When we analyzed the distribution of various T cell subsets among CD4+CD38+ T cells we found that naïve, CM and TEM cells were comparable, however, the EM population was significantly lower in SLE patients than in healthy participants (Figure S1E). The observed distribution of CD4+CD38+ among T cell subsets suggests that the expression of CD38 in SLE CD4+ T cells is not due to cell activation. To further explore this aspect, we investigated the metabolic phenotype of CD38+CD4+ T cells. A glycolysis stress test using sorted CD4+CD38+ and CD4+ CD38 - T cells from HC and SLE patients (n = 5) showed no differences between the two cell subsets and between the two groups (Figure S1F).
Fig. 1. Increased expression of CD38 on the surface membrane of SLE CD4+ T cells correlates with increased lipid microdomain formation.
A Flow cytometry analysis of CD38 expression (%) in CD4+ T cells derived from healthy participants (n = 10) and patients with SLE (n = 11). B Immunofluorescence analysis of cholera toxin B (CTB) binding in JurkatCD38KO and JurkatControl cells. CTB-FITC is evaluated as surface staining. Green is CTB and Blue is DAPI (left panel), scale bar = 20 μm. The number of cells with full, partial or non-staining with CTB on the cell surface were calculated (right panel, 3 independent experiments). C Flow cytometry analysis of CTB binding on the surface membrane comparing JurkatCD38KO and JurkatControl cells (n = 5, 3 independent experiments, cumulative data (left panel) and a representative plot (right panel)). D Flow cytometry analysis of surface CTB binding comparing CD4+CD38- and CD4+CD38+ derived from healthy participants (n = 6) and patients with SLE (n = 9). Cumulative data (left panel) and representative dot plots (middle and far right panel). E Surface CTB expression was compared between control-vector and CD38 overexpressing-vector in JurkatCD38KO cells (n = 4 biological replicates). Cumulative data (left panel) and representative dot plots of CTB expression after overexpression (middle panel) and representative dot plots of transfection efficacy of GFP-control vector and CD38 overexpression vector (far right panels). F Surface CTB binding was compared between control-vector and CD38 overexpressing-vector in CD4+ T cells from healthy participants (n = 5. Cumulative data, left panel) and representative dot plots of CTB expression after overexpression (middle panel) and representative dot plots of transfection efficacy of GFP-control vector and CD38 overexpression vector (far right panels). Dotted quadrant shows the increased CD38 expression (7%) in transfected compared to GFP-control transfected cells). MFI mean fluorescence intensity. Data analysis using unpaired 2-tailed Student’s t test with Welch’s correction (A), unpaired 2-tailed Student’s t test (B, C, E) or paired 2-tailed Student’s t test (D, F). All data are represented as mean ± SEM. *p < 0.05, ****p < 0.0001. Source data are provided as a Source Data file.
We also examined whether CD38 expression on CD4+ T cells from patients with SLE correlates with disease activity. We found that increased expression of CD38 correlated positively with SLE Disease Activity Index (SLEDAI) and high ds-DNA antibody titers (Table S1), which is consistent with a previous report3. With regard to the lipid profiles, serum LDL-cholesterol trended higher in SLE patients who had high CD38+CD4+ T cell numbers, while serum HDL-cholesterol trended lower (Table S1).
CD38 positive cells have skewed lipid raft profile towards the α-series of monosialogangliosides
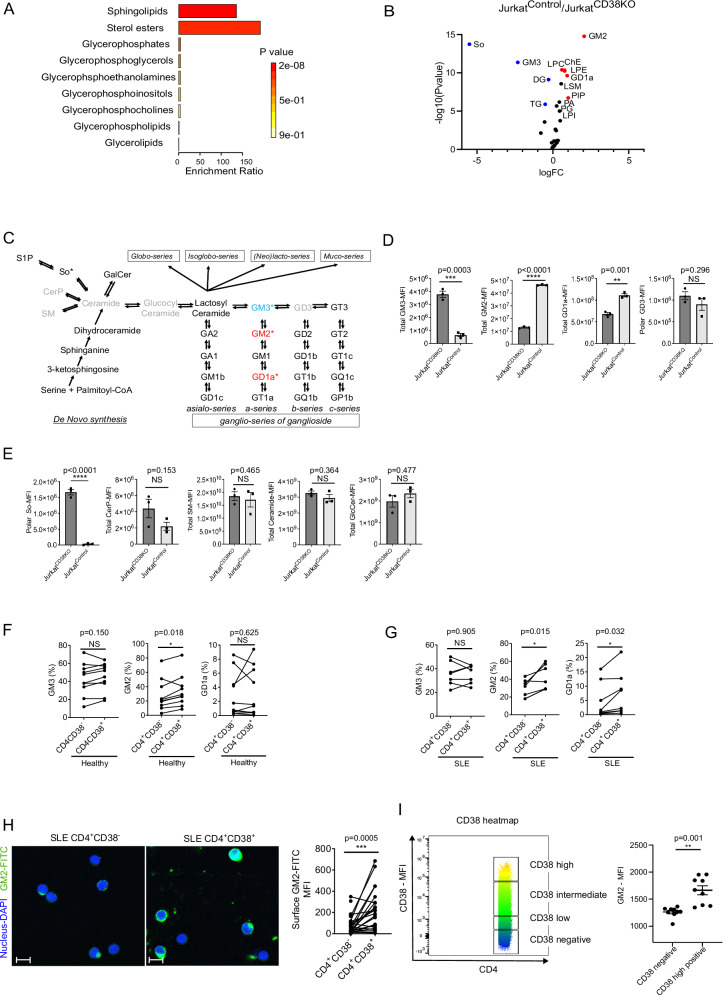
The observed differences in CTB staining between CD38- and CD38+ T cells suggested that CD38 may widely alter lipid component composition on the surface membrane of CD4+ T cells. To explore in a precise manner the global lipid profile affected by CD38, we performed lipidomic analysis in JurkatCD38KO and JurkatControl cells. Samples for lipidomic analysis were acquired both from the hydrophobic phase containing non-polar lipids represented by cholesterol and triglycerol and from the aqueous phase containing glycosphingolipids and other small organic lipids. Both phases were submitted separately to LS-MS/MS and the peak area from each set of data were combined as “total” lipid distribution (data from hydrophobic phase + data from aqueous phase). Subsequently, the data were analyzed by MetaboAnalyst 5.0 as described in the Methods Section. The lipid profile analysis showed differences between CD38-deficient and -sufficient Jurkat cells (Fig S2A, B). Notably, the most significant change appeared in the profile of sphingolipids, which includes sphingomyelins and glycosphingolipids, including gangliosides, the sialylated glycosphingolipids (Fig. 2A). We recorded (volcano plot analysis, Fig. 2B) significant differences between the two groups in the expression of several gangliosides (GM3, GM2 and GD1a), which represent only small fractions of total lipids, but not in major lipid constituents such as cholesterol-ester (ChE) and triglycerides (TG). When we plotted the expression levels of individual components, we found significant changes in the levels of α-series of monosialogangliosides with a shift from GM3 to GM2 and GD1a (p = 0.0003, p < 0.0001 and p = 0.001, respectively in Figs. 2C, D). There were no differences between the expression levels of GD3 (p = 0.29, Fig. 2C, D). Ceramide, the core structure of gangliosides and the hydrophobic backbone which anchors gangliosides into the plasma membrane, was also expressed at comparable levels in both JurkatCD38KO and JurkatControl cells (Fig. 2E). Besides converting to glycosphingolipids (GSLs) through glucosylceramide (GlcCer), ceramide is also converted to sphingosine-1-phosphate (S1P), Ceramide-1-phosphate (CerP), or sphingomyelin (SM)25. However, the branch metabolites of Ceramide, CerP and SM, were expressed at comparable levels in JurkatCD38KO and JurkatControl cells and S1P was not detectable in either group (Fig. 2E), indicating that the direction of the metabolic process of ceramide is skewed towards ganglioside synthesis in a CD38-dependent manner in this Jurkat system.
Fig. 2. CD38 skews lipid raft profile into α-series monosialoganglioside.
A, B Enrichment analysis (A) and volcano plot (B) of lipidomics analysis comparing JurkatCD38KO (n = 3) and JurkatControl (n = 3). The threshold was set as 1.5 for fold change (FC) and 0.05 for p-value in the ratio of JurkatControl/JurkatCD38KO. C Schematics of ceramide and ganglioside synthesis pathway. The ganglioside pathway is composed of the sequential addition of sugars and/or sialic acids. *; statistically significant in the lipidomics data. Red indicates higher expression and blue a lower expression, gray color shows no statistically significant alteration in expression. D Changes in the levels of lipids in the ganglioside series from the lipidomics data comparing JurkatCD38KO and JurkatControl cells (n = 3 independent data points, biological replicates). E Changes in the levels of lipids in the ceramide synthesis from the lipidomics data comparing JurkatCD38KO and JurkatControl cells (n = 3 independent data points, biological replicates). F, G Changes of the lipids in the group of α-series of gangliosides comparing CD4+CD38− and CD4+CD38+ T cells from healthy participants (F, n = 10) and from patients with SLE (G, n = 7). H Fluorescence microscopic analysis of GM2 expression on CD4+ T cells. CD4+CD38− and CD4+CD38+ T cells were sorted by Aria and subsequently stained for GM2 (FITC-green) and DAPI (blue), scale bar = 20 μm. Representative images (left panel). Quantification of GM2 expression as mean fluorescence intensity (25 cells per group from n = 5 biological replicates, right panel). I Flow cytometry-based quantification of GM2 and CD38 in CD4+ cells. The heatmap of CD38 expression gated in CD38 negative, CD38 low-positive, CD38 intermediate-positive and CD38 high-positive based on the expression of CD38 (left panel). Right panel shows GM2 MFI in CD38 negative and CD38 high-positive cells (n = 9 biological replicates). Data analysis using unpaired 2-tailed Student’s t test D, E unpaired 2-tailed Student’s t test with Welch’s correction (I) or paired 2-tailed Student’s t test (F–H). All data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Source data are provided as a Source Data file.
Next, we addressed whether this phenotype can be recapitulated using human primary cells. Flow cytometry analysis following intracellular staining confirmed increased expression of GM2 and GD1a positive JurkatControl cells and CD4+CD38+ T cells from healthy (Fig. 2F and S2C) and SLE samples (Fig. 2G and S2D) compared to their CD4+CD38- counterparts. No difference was observed between HC and SLE patients. Gangliosides are expressed primarily in the outer leaflets of the plasma membrane26 and the distribution of gangliosides is not random at the cell surface, but rather, they cluster together with cholesterol to form lipid microdomains27 and one of α-series ganglioside, GM1, serves as a lipid microdomain marker28. GM1 could not be detected in our lipidomics studies probably because its expression in mass spectrometry determinations is much lower than that which is detectable by flow cytometry22. We also confirmed that GM2 was significantly elevated on the surface membrane of CD4+CD38+ T cells compared to CD4+CD38- T cells in patients with SLE (Fig. 2H). Furthermore, we found that CD38high CD4+ T cells expressed the highest GM2 levels compared to CD38-, CD38low and CD38intermediate in patients with SLE (Fig. 2I). Collectively, these results demonstrate that CD4+CD38+ T cells display GM2 rich lipid microdomains on the cell surface membrane compared to CD4+CD38- T cells.
B4GALNT1 promotes GM2 dominance in lipid microdomains in CD4+CD38+ cells
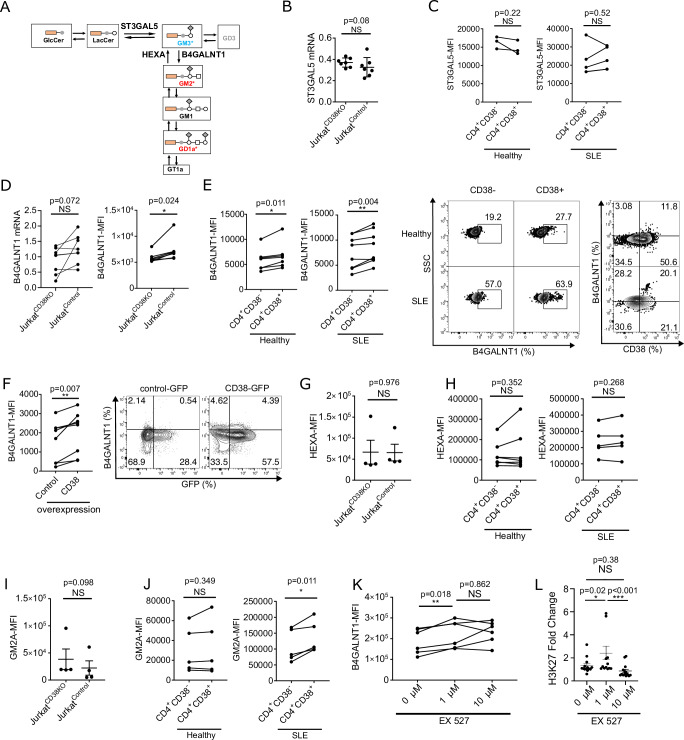
Next, we investigated the identity of the key enzyme responsible for the CD38-dependent skewing of GM3 to GM2 in the α-series of gangliosides. Gangliosides are synthesized in a stepwise manner as illustrated in Fig. 3A. The synthetic pathway from GM3 to GM2, and subsequently to GD1a, is promoted by the GM2 synthase (B4GALNT1; beta-1,4-N-acetylgalactosaminyl transferase-1). B4GALNT1 is a key enzyme of this process transferring N-acetylgalactosamine (GalNAc) to GM3 and GD3. First, we quantified the levels of ST3GAL5, the GM3 synthase from lactoceramide, but we did not find a difference between CD38- and CD38+ Jurkat cells at the RNA level (Fig. 3B) and in CD38- and CD38+ CD4+ cells from healthy participants or patients with SLE at the protein level (Fig. 3C). These data indicates that the synthetic activity from lactoceramide (LacCer) to GM3 was similar between CD4+CD38- and CD4+CD38+ T cells. Though mRNA levels of B4GALNT1 were comparable between Jurkatcontrol and JurkatCD38KO cells (Fig. 3D, left panel), the protein levels were significantly upregulated in JurkatControl compared to JurkatCD38KO cells (Fig. 3D, right panel). Protein levels of B4GALNT1 were also increased in CD4+CD38+ T cells from healthy participants and patients with SLE compared to CD4+CD38- T cells (Fig. 3E). Finally, we confirmed that overexpression of CD38 in CD4+ T cells from healthy participants induced the expression of B4GALNT1 (Fig. 3F). These data suggest that the biosynthesis of GM2 from GM3, catalyzed by B4GALNT1, is associated with higher CD38 expression.
Fig. 3. B4GALNT1 promotes GM2 dominance in lipid microdomains in CD4+CD38+ cells.
A Schematic depicting the α-series of ganglioside synthesis pathway. B,C Quantitative PCR analysis comparing JurkatCD38KO and JurkatControl (B, n = 7) and Flow cytometry analysis comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (C, n = 3, left panel) or from patients with SLE (C, n = 5, right panel) of ST3GAL5. D Quantitative PCR analysis (left panel, n = 8) and flow cytometry analysis (right panel, n = 7) of B4GALNT1 expression comparing JurkatCD38KO and JurkatControl cells (n = 7, 3 independent experiments). E Flow cytometry analysis of B4GALNT1 comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (n = 7, left panel) and from patients with SLE (n = 8, middle panel). Representative dot plots of B4GALNT1 expression and representative dot plots of B4GALNT1 versus CD38 expression in CD4+CD38- and CD4+CD38+ T cells from a healthy participants and a SLE patient (right panels). F Flow cytometry analysis of B4GALNT1 after transfection with a control vector or a CD38-overexpression plasmid in CD4+ cells from healthy participants (n = 8). Cumulative data (left panel) and representative dot plots (right panel). G Flow cytometry analysis of HEXA expression comparing JurkatCD38KO and JurkatControl cells (n = 4, 3 independent experiments). H Flow cytometry analysis of HEXA expression comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (n = 7) and from patients with SLE (n = 5). I, J Flow cytometry analysis of GM2A expression in JurkatCD38KO and JurkatControl cells (I, n = 4, 3 independent experiments) and in CD4+CD38- and CD4+CD38+ T cells from healthy participants (J, n = 5, left panel) and patients with SLE (J, n = 5, right panel). K Flow cytometry analysis of B4GALNT1 expression. CD4+ T cells from healthy participants were isolated from whole blood and treated with indicated concentrations of the SIRT1 inhibitor (EX527) at 37 °C overnight (n = 5). L ChIP assay analysis of H3K27 (fold change) on B4GALNT1. A ChIP assay was performed in CD4+ T cells isolated from whole blood of healthy participants treated with the indicated concentrations of the SIRT1 inhibitor (EX527) at 37 °C overnight (n = 3), using an H3K27 antibody. Data analysis using unpaired 2-tailed Student’s t test (B, G, I) or paired 2-tailed Student’s t test (C–F, H, J) or 1-way ANOVA with Tukey-Kramer test (K, L). All data are represented as mean ± SEM. *p < 0.05, **p < 0.01. Source data are provided as a Source Data file.
Next, we assessed whether increased levels of GM2 resulted from decreased degradation to GM3, a process controlled by beta-hexosaminidase (HEXA). HEXA hydrolyzes GM2 in the presence of the GM2 ganglioside activator (GM2A)29. The expression of HEXA was comparable between JurkatCD38KO and JurkatControl cells (Fig. 3G) and between CD4+CD38- and CD4+CD38+ T cells from healthy participants and patients with SLE (Fig. 3H). Also, we did not find differences in the expression of GM2A between JurkatCD38KO and JurkatControl (Fig. 3I) or between CD4+CD38+ and CD4+CD38- T cells from healthy participants and SLE patients (Fig. 3J).
B4GALNT1 is elevated in CD4+CD38+ T cells through SIRT1
Although the precise mechanisms which regulate the expression of B4GALNT1 are not clear, histone acetylation has been reported to be important30. Accumulation of histone acetylation levels (H3K9, H3K14, or both) at the promoter region (+38/ +187) of the B4GALNT1 gene increases mRNA expression levels of B4GALNT1 in human renal cell carcinoma cell lines30. CD38 acetylates target genes by degrading NAD+, thus decreasing the deacetylating activity of the NAD+-dependent Sirtuin1 (SIRT1). It is widely accepted that SIRT1 regulates the acetylation of histones found in the promoter region of multiple genes31. To confirm whether acetylation affects the levels of B4GALNT1 in a SIRT1-dependet manner, CD4+ T cells from healthy participants were cultured in the presence of the selective SIRT1 inhibitor, EX527. Flow cytometry analysis showed a significant increase of B4GALNT1 expression in cells cultured in the presence of 1 μM of SIRT1 inhibitor (Fig. 3K). ChIP assay analysis showed a significant increase of histone acetylation levels (H3K27) at the promoter region ( + 57 to +178) of the B4GLANT1 gene after treatment with the SIRT1 inhibitor (Fig. 3L). These data suggest that the levels of B4GALNT1 in CD4+ T cells are increased when SIRT1 activity is inhibited.
CD38 is colocalized with lipid microdomain components LAT and GM2
One of the critical roles of lipid microdomains is to serve as platforms to initiate T cell signaling (Fig. 4A). The earliest T cell receptor signaling events involve tyrosine phosphorylation of ITAMs, especially on CD3ζ, mediated by the Src tyrosine kinase LCK, followed by the phosphorylation of ZAP70 and tyrosine phosphorylation of Linker for Activation of T cells (LAT). LAT, an essential T cell specific adapter protein32, loses its mobility and localizes to the lipid rafts to promote sustained cell signaling26,33. Phosphorylated (p) LAT binds PLCγ1 and Grb2 and other adapter proteins including SLP-7634.
Fig. 4. CD4+CD38+ T cells display enhanced early CD3/TCR signaling response through the CD38-B4GALNT1-GM2 pathway.
A Schematic of calcium signaling in T cells through T cell receptor. B Representative confocal images of CD4+ T cells from healthy participants stained for nucleus (blue), lipid rafts (CTB, green), CD38 (orange) and GM2 (red) acquired at an original magnification of 62X (left panel), scale bar represents 10 μm. Scatter plot of colocalization score of CD38 within and outside of LR in CD4+ T cells of healthy individuals (n = 5, for each 5 patients 5 images analyzed with Imaris). C Representative confocal images of CD4+ T cells from healthy participants stained for nucleus (blue), LAT (green), CD38 (orange), and GM2 (red) acquired at an original magnification of 62X (left panel) scale bar represents 10μm. Scatter dot plots of colocalization scores of CD38 with LAT or LAT alone (middle panel); and GM2 colocalization with LAT in CD4+CD38+ and CD4+CD38- T cells (right panel) of healthy individuals (n = 5, for each 5 patients 5 images analyzed with Imaris). D Flow cytometry analysis of pCD3ζ (healthy n = 5, SLE n = 5), pZAP70 (healthy n = 7, SLE n = 6), pLCK (healthy n = 6, SLE n = 6) and pPLCγ1 (Thr783) (healthy n = 7, SLE n = 7), comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (top row) and SLE patients (bottom row), with representative dot plots of pPLCγ1 (right panel). Cells were coated with 20 μg/mL of anti-CD3ε mAb (OKT3 clone) in 50 µl of PBS for 30 min on ice. To induce phosphorylation of downstream signaling molecules we added 10 μg of crosslinker (goat-anti-mouse) and incubated the cells for 2 min at 37 °C. E Flow cytometry analysis of pCD3ζ (Healthy n = 5, SLE n = 5). Representative histogram comparing CD4+CD38+ T cells from healthy participants and CD4+CD38+ T and CD4+CD38- T cells from patients with SLE (left panel) and cumulative data (right panel) comparing CD4+CD38+ T cells from healthy participants and CD4+CD38+ T cells from patients with SLE. Cells were coated with 20 μg/mL of anti-CD3ε mAb (OKT3 clone) in 50 µl of PBS for 30 min on ice. To induce phosphorylation of downstream signaling molecules we added 10 μg of crosslinker (goat-anti-mouse) and incubated the cells for 2 min at 37 °C. Data analysis using unpaired 2-tailed Student’s t test with Welch’s correction (B, C), paired 2-tailed Student’s t test (D) or unpaired 2-tailed Student’s t test (E). All data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001. Source data are provided as a Source Data file.
First, we investigated whether CD38 is localized within lipid microdomain components after activation of CD4+ T cells. When CD4+CD38+ T cells were preactivated with CD3/28 antibodies, CD38 colocalized with with CTB (Fig. 4B) and LAT, a raft-associated protein (Fig. 4C). Next, we examined the difference in GM2 abundance in the lipid rafts of CD38+ and CD38- in CD4+ T cells from healthy participants. We confirmed that CD4+CD38+ T cells express significantly more GM2 colocalized with LAT compared to CD4+CD38- T cells (Fig. 4C). These results suggest that CD38 expression leads to increased expression of GM2 that is localized within the lipid microdomains.
CD4+CD38+ T cells display enhanced early CD3/TCR signaling response through the CD38-B4GALNT1-GM2 pathway
To examine how the presence of GM2 in the lipid microdomains of CD4+CD38+ cells contributes to T cell signaling, we measured the phosphorylation levels of CD3ζ, ZAP70, LCK, and PLCγ1 by flow cytometry. Compared to CD4+CD38- T cells, CD4+CD38+ T cells had significantly higher levels of pCD3ζ and comparable levels of pZAP70 and pLCK (Fig. 4D) both in healthy participants and SLE, and significantly higher pPLCγ1 only in SLE compared to healthy participants (Fig. 4D). When we compared the levels pCD3ζ in CD4+CD38+ T cells from healthy participants and patients with SLE, its levels were higher in healthy participants compared to SLE (Fig. 4E), while the levels of pPLCγ1 were higher in SLE T cells (Fig. 4D). These results are in agreement with previous reports that the levels of CD3ζ are low in SLE T cells22 while pPLCγ1 localizes in the lipid rafts of T cells21. The levels of the pSyk, pSHP2, pJNK, pAKT and pS6 signaling molecules were significantly increased in CD4+CD38+ T cells compared to CD4+CD38- T cells only in SLE (Figure S3A) and are in agreement with previous reports on total SLE T cells23,35–37. Only pPLCγ1, pJNK and pAKT were significantly higher in SLE CD4+CD38+ T cells compared to healthy participants (Fig. S3B). In patients with SLE, we observed that total PLCγ1 levels were increased in CD4+CD38+ T cells compared to CD4+CD38- T cells (Fig. S3C).
Next, we investigated the role of CD38 in T cell metabolism. We observed that OXPHOS activity was not different between JurkatControl and JurkatCD38KO but the glycolytic activity was significantly decreased in JurkatControl compared to JurkatCD38KO cells (Fig. S4A–C).
Since we observed a link between CD38 expression and B4GALNT1 expression, we next asked whether there is also a difference in cell signaling when B4GALNT1 is over expressed. We found that even a slight increase in the levels of B4GALNT1, led to an increase in the levels of GM2 (Fig. S5A) and of pPLCγ1 in SLE CD4+ T cells (Fig. S5B).
Increased calcium flux in CD4+CD38+ T cells
pPLCγ1 hydrolyzes phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) into inositol-1,4,5-trisphosphate (IP3) and releases Ca2+ from endoplasmic reticulum (ER) through the IP3 receptor (IP3R), as shown in Fig. 5A (green quadrant). Elevated calcium flux has been observed in patients with SLE, which was further enhanced by crosslinking with a CTB antibody38,39. Accordingly, we assessed whether the GM2 dominant lipid microdomains in CD4+CD38+ T cells affect calcium fluxes. Firstly, we measured the calcium signaling after CD3 stimulation with Ca2+ sufficient medium in CD4+CD38- and CD4+CD38+ T cells from healthy participants and patients with SLE. Calcium influx was significantly increased in CD4+CD38+ T cells from healthy participants compared to CD4+CD38- T cells (Fig. 5B). Furthermore, calcium flux was even higher in CD4+CD38+ T cells from patients with SLE compared to that from healthy participants (Fig. 5C), while there was no difference in the calcium flux in CD4+CD38- T cells between healthy participants and patients with SLE (Fig. 5D).
Fig. 5. Increased calcium entry in CD4+CD38+ cells through CRAC channels.
A Schematics of calcium signaling in T cells. B Intracellular calcium kinetics comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (n = 3, summary figure in left panel). CD4+ T cells loaded with Fluo-4 were stimulated with anti-CD3 and a cross-linker in calcium-sufficient RPMI medium. PMA and ionomycin were added as a positive control. Quantification of peak MFI of Fluo-4 after anti-CD3 stimulation (n = 5, cumulative data in right panel). C Same experimental conditions as (B) comparing CD4+CD38+ T cells from healthy participants and patients with SLE. Intracellular calcium kinetics (n = 3, summary figure in left panel). Quantification of peak MFI of Fluo-4 after anti-CD3 stimulation (cumulative data in the right panel, n = 4). D Same experimental conditions as (B) comparing CD4+CD38- T cells from healthy participants and from patients with SLE. Intracellular calcium kinetics (n = 3, summary figure in left panel). Quantification of peak MFI of Fluo-4 after anti-CD3 stimulation (cumulative data in the right panel, n = 6). E Thapsigargin-induced intracellular calcium kinetics comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (Representative figure in left panel) and quantification of peak MFI of Fluo-4 after 1 mM Ca2+ addition (Summary scatter plot in right panel, n = 3). CD4+ T cells loaded with Fluo-4 were stimulated with 1 μM of thapsigargin in HBSS to induce passive Ca2+ release from ER (ER-Ca2+). F Calcium kinetics focusing on ER-Ca2+ presented in Figure E. Representative figure (left panel) and quantification of peak MFI of Fluo-4 after 1 μM of thapsigargin addition (Cumulative data in the right panel, n = 8). CD4+ T cells loaded with Fluo-4 were stimulated with anti-CD3 in HBSS to measure release of ER-Ca2+. G Anti-CD3-induced intracellular calcium kinetics comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (Representative figure in left panel) and quantification of peak MFI of Fluo-4 after 1 mM Ca2+ addition (Summary scatter plot in the right panel, n = 4 biological replicates). CD4+ T cells loaded with Fluo-4 were stimulated with anti-CD3 in HBSS to release of ER-Ca2+ which is induced by TCR-IP3 pathway. Data analysis using unpaired 2-tailed Student’s t test (C, D) or paired 2-tailed Student’s t test (B, E–G). All data are represented as mean ± SEM. *p < 0.05, **p < 0.01. Source data are provided as a Source Data file.
To inquire whether B4GALNT1 accounts for the higher calcium flux in in CD4+CD38+ T cells, we measured calcium flux in T cells from SLE patients after B4GALNT1 overexpression. Although only a slight elevation of B4GALNT1 protein was achieved (Fig. S5A), a significant increase of calcium influx (Fig. S5C) was observed, following anti-CD3 stimulation in a Ca2+-sufficient medium. These results indicate that lipid composition alteration led by B4GALNT1, causes higher calcium signaling in SLE T cells possibly by affecting proximal T cell signaling.
As shown in Fig. 4D, CD4+CD38+ T cells displayed higher Ca2+ flux by enhancing proximal TCR signaling. pPLCγ1-generated IP3 stimulates IP3R expressed by the ER to release and empty the Ca2+ stores. When Ca2+ is depleted, stromal interaction molecule 1 (STIM1) in the ER membrane interacts with Orai1, the pore subunit of the calcium release activated channels (CRAC) on plasma membrane. This interaction allows bulk entry of Ca2+, known as Store-Operated Calcium Entry (SOCE)40 (Fig. 5A, red quadrant). Therefore, we next measured the calcium entry through SOCE activity in CD4+CD38- and CD4+CD38+ T cells from healthy participants. We assessed two conditions: first, we used thapsigargin, a sarco/endoplasmic reticulum ATPase (SERCA) inhibitor which passively releases the ER-Ca2+ by inhibiting replenishment of Ca2+ through SERCA on ER membrane; second, we used a CD3 antibody to release ER-Ca2+ through TCR signaling and the downstream production of IP3 by pPLCγ1. After each stimulation, 1 mM of Ca2+ was added to activate SOCE. CD4+CD38+ T cells showed significantly higher SOCE activity (Fig. 5E). Thus, CD4+CD38+ T cells had higher ability to activate SOCE in response to thapsigargin. Since CD4+CD38+ T cells showed slight, but significantly higher, ER-Ca2+ release after stimulation with thapsigargin compared to CD4+CD38- T cells (Fig. 5F), there is a possibility that CD4+CD38+ T cells may have higher baseline of ER-Ca2+ storage which may cause more Ca2+ to be released after stimulation with thapsigargin. This high activity of SOCE was also confirmed by the anti-CD3 induced ER-Ca2+ depletion through IP3R (Fig. 5G).
Interestingly, when B4GALNT1 was overexpressed in CD4+ T cells of patients with SLE, SOCE activity was increased only when stimulated with anti-CD3 (Fig. S5D), but not with thapsigargin (Fig. S5E). These results indicate that enhanced SOCE activity by B4GALNT1 is due to Ca2+ depletion from ER through the PLCγ1-IP3R pathway. Our data also shows that simple overexpression of B4GALTN1 does not contribute to higher amounts of stored ER-Ca2+.
We considered that CD38 through its enzymatic activity may produce second messengers including cyclic ADP ribose (cADPR)41 and nicotinic acid adenine dinucleotide phosphate (NAADP) from NAD+ 9. NAADP releases Ca2+ from lysosomes and is involved in a short, early Ca2+ peak after anti-CD3 stimulation, while cADPR releases Ca2+ from endoplasmic reticulum (ER) and contributes to sustained Ca2+ after anti-CD3 stimulation42, but there is no clear evidence that CD38 induces Ca2+ flux through the production of cADPR and NAADP in mammalian cells. To test whether the production of cADPR and NAADP in CD4+CD38+ T cells contributes to the noted increased calcium flux, we incubated CD4+ T cells from healthy participants with 8-Bromo-cADP-ribose and NED-19, the antagonist for cADPR and NAADP, respectively and compared Ca2+ flux in CD4+CD38+ cells after anti-CD3 stimulation in HBSS (Fig. S6A). However, neither of the antagonists decreased Ca2+ release (Fig. S6B) or calcium entry (Figure S6C), suggesting that the effect of cADPR and NAADP in CD38 positive T cells is not responsible for the drastic differences of calcium flux between CD4+CD38- and CD4+CD38+ T cells presented in Fig. 5B, E, F.
Calcium flux in CD4+CD38+ cells leads to ER stress in T cells
SOCE is so far the main mechanism found to be responsible for the maintenance of sustained Ca2+ elevation43. Indeed, Ca2+ was maintained at high levels for at least 4 hours through SOCE activity after stimulation with thapsigargin (Fig. S6D). Notably, CD4+CD38+ T cells from SLE patients had higher and sustained SOCE activity compared to their counterparts from healthy participants (Fig. S6D). CD4+CD38+ T cells in patients with SLE showed significantly higher ER-Ca2+ release after stimulation with thapsigargin compared to those from healthy participants (Fig. S6E).
The maintenance of sustained Ca2+ is important for the translocation of transcriptional factors to activate T cell functions44,45. However, overall cytokine productions including IL-2 were rather significantly suppressed in CD4+CD38+ T cells compared to CD4+CD38- T cells from patients with SLE and healthy participants (Fig. 6A for IL2 and Fig. S6F and S6G for others). The production of IFNγ and TNF were comparable between CD4+CD38+ T cells from patients with SLE and healthy participants (Fig. S6H).
Fig. 6. Calcium flux in CD4+CD38+ T cells leads to ER stress in T cells.
A Scatter plot of flow cytometry analysis of IL-2 comparing CD4+CD38- and CD4+CD38+ T cells from patients with SLE (n = 10, left panel) and from healthy participants (n = 6, middle panel) and representative dot plot of IL-2 production in healthy participants and SLE patients in CD38- and CD38+ in CD4+T cells (right panel). CD4+ T cells were stimulated with PMA/Ionomycin for 6 h with brefeldin and intracellular staining for IL-2 was performed. B Cumulative data (left panel) and representative plots (right panel) of flow cytometry analysis of IL-2 positive CD4+CD38+ T cells from patients with SLE (n = 3) and from healthy participants (n = 4). CD4+ T cells were stimulated with PMA/Ionomycin for 6 h with brefeldin and intracellular staining for IL-2 was performed. C Cumulative data of flow cytometry analysis of IL-2 production in total CD4+ T cells comparing healthy participants (n = 6) and patients with SLE (n = 5). Cells were stimulated with PMA/Ionomycin for 5 h and intracellular staining for IL-2 was performed. D Pathway analysis of RNA-seq data comparing JurkatControl and JurkatCD38KO cells (n = 3 from each group). Bar graphs represent GO Cellular Component 2018 pathway that was upregulated in JurkatControl compared to JurkatCD38KO cells with adjusted p values < 0.05 and Fold Change > 1.5 (2-sided tests). E Gene set enrichment plot of the genes upregulated in response to ER stress calculated by GSEA (gene set from GO:1990440 was used). F Flow cytometry analysis of IRE1 comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (n = 6, left panel) and from patients with SLE (n = 6, right panel). G Flow cytometry analysis of spliced XBP1 comparing CD4+CD38- and CD4+CD38+ T cells from healthy participants (n = 5, left panel) and from patients with SLE (n = 5, middle panel) and representative dot plot (right panel). H Representative electron microscope images of ER morphology in CD4+CD38- and CD4+CD38+ T cells from patients with SLE and healthy participants. The arrow points to a normal strand of ER in a CD4+CD38- cell of a patient with SLE and the star marks point to dilated strands of ER in the cytoplasm and adjacent to the nuclear membrane (inset) in a CD4+ CD38+ cell from another patient with SLE. Cells from healthy individuals (low panels) show no ER changes. 3 independent experiments were performed. Scale bars, 500 nm. Data analysis using unpaired 2-tailed Student’s t test with Welch’s correction (B, C) or paired 2-tailed Student’s t test (A, F, G). All data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
The production of IL-2 was even lower in CD4+CD38+ T cells derived from SLE compared to healthy participants (Fig. 6B), and in CD4+ T cells from SLE patients compared to those from healthy participants (Fig. 6C). Decreased IL-2 production is considered as one of key features in the pathogenesis of SLE46,47 because it is responsible for the impaired regulatory T cell function46 and proliferation48. IL-2 was even more suppressed following overexpression of B4GALNT1 (Fig. S7A). ATAC-seq showed widely limited chromatin accessibility (only 8 genes were significantly accessible in CD4+CD38+ T cells compared to 332 genes in CD4+CD38- T cells) in CD4+CD38+ T cells compared to CD4+CD38- T cells in SLE (Fig. S7B, right panel), which is consistent with the feature of decreased cytokine production in CD4+CD38+ T cells. The chromatin accessibility was not apparently different between CD4+CD38+ T cells and CD4+CD38- T cells in healthy participants (Fig. S7B, left panel). This suggests that CD4+CD38+ T cells in SLE are transcriptionally less active when Ca2+ flux is enhanced. Therefore, we hypothesized that CD4+CD38+ T cells may be stressed or exhausted under sustained high Ca2+ load through altered lipid microdomain composition.
To understand how CD38 alters T cell function, we performed RNA-seq and compared Jurkat CD38KO and JurkatControl cells. The pathway analysis revealed that JurkatControl had significantly altered ER-related gene expression (Fig. 6D). Specifically, genes related to ER stress response49 were upregulated in JurkatControl compared to JurkatCD38KO cells (Fig. 6E). These data suggest that CD38 causes ER stress linked to Ca2+ overload. It has been previously reported that in the tumor environment the ER stress-XBP1 pathway reduces T cell function and decreases IFNγ production by inducing CD8+ cell exhaustion50 or limiting mitochondrial respiration51. It is known that impaired calcium homeostasis as well as hypoxia, oxidative stress and other forms of cell stimulation cause ER stress52. Therefore, we considered that ER stress caused by the Ca2+ overload in CD4+CD38+ T cells may lead to the impaired T cell function. To demonstrate this, we measured ER stress markers involved in three major ER stress pathways: PKR-like endoplasmic reticulum kinase (PERK), Activating Transcription Factor 6 (ATF6) and Inositol-Requiring Enzyme (IRE1). Among these ER stress markers, IRE1 is the most conserved one and cleaves mRNA of X-box binding protein (XBP)−1 into spliced XBP-1 (sXBP-1). Although IRE1 expression was comparable (Fig. 6F), spliced XBP1 (XBP1s), a downstream factor of Inositol-requiring enzyme 1 (IRE1) was significantly increased in CD4+CD38+ T cells compared to CD4+CD38- T cells derived from patients with SLE but not in CD4+ T cells from healthy participants (Fig. 6G). B4GALNT1 overexpression increased XBP1 expression (Fig. S7C), suggesting that ER stress in CD4+CD38+ T cells is linked to B4GALNT1. The PERK-ATF4 pathway was also upregulated in CD4+CD38+ T cells from SLE compared to CD4+CD38- T cells (Fig. S7D). The ATF6 pathway was comparable between the two groups (Fig. S7D). Using transmission electron microscopy, we found that CD4+CD38+ cells from patients with SLE exhibited mild to moderate dilation of the ER in the cytoplasm and adjacent to the nuclear membrane, in contrast to CD4+CD38- T cells, which had normal appearing ER (Fig. 6H). CD4+CD38- and CD4+CD38+ T cells from healthy individuals also did not show significant ER dilation (Fig. 6H). Because ER dilation is a morphologic indication of ER stress (Chavez-Valdez et al., 2016), its presence in the CD4+ CD38+ cells of patients with SLE supports that they are in a condition of ER stress.
Inhibition of proximal calcium flux restores IL-2 production by CD4+CD38+ cells
Next, to determine whether regulation of Ca2+ through the proximal TCR signaling affects ER stress, we treated CD4+ T cells from patients with SLE with the IP3 receptor inhibitor 2-Aminoethyl diphenylborinate (2-APB) which is known to inhibit IP3-provoked intracytoplasmic Ca2+ increases42. 2-APB significantly decreased the expression of XBP1s (Fig. 7A), suggesting that blocking Ca2+ signaling prevents cellular ER stress. Calcium entry through SOCE following anti-CD3 stimulation was inhibited with 2-APB and the selective SOCE inhibitor YM-58483 (Fig. 7B). YM-58483 decreased cytosolic Ca2+ more dramatically (Fig. 7B, right panel) compared to 2-APB (Fig. 7B, left panel). Consistent with our hypothesis, IL-2 was restored by inhibiting the IP3 receptor with 2-APB in patients with SLE (Fig. 7C). The effect of 2-APB did not change IL-2 production in healthy participants (Fig. 7C), indicating that the IP3 pathway is dysregulated in SLE. Interestingly, the SOCE inhibitor, which more dramatically shut down cytosolic Ca2+ (Fig. 7B, right panel), did not restore IL2, but rather significantly decreased IL-2 production both in healthy and in SLE cells (Fig. 7D). These results suggest that selective inhibition of Ca2+ through IP3 signaling mediated Ca2+ entry, not through SOCE, is pathogenetically important in CD4+CD38+ T cells in SLE. Finally, we also checked whether modulation of lipid microdomain by knocking down B4GALNT1 restores IL-2 production. Although only a slight downregulation of B4GALNT1 was achieved in CD4+ T cells from patients with SLE, our data already show that Ca2+ entry induced with anti-CD3 was significantly decreased (Figure S7E). Importantly, knock-down of B4GALNT1 even restored IL-2 expression (Figure S7F). Collectively, modulation of IP3 signaling or B4GALNT1-depenedent microdomain correct calcium entry and restore IL-2 production in CD4+ T cells from patients with SLE.
Fig. 7. Inhibition of proximal calcium flux restores IL-2 production in CD4+CD38+ cells.
A Scatter plot of flow cytometry analysis of spliced XBP1 in CD4+ T cells treated with 2-APB (IP3 receptor inhibitor) in indicated concentration following overnight incubation (n = 5 biological replicates). B SOCE activity of CD4+ T cells treated with 2-APB (left panel) or with YM-58483 (SOCE inhibitor) (right panel). CD4+ T cells loaded with Fluo-4 were stimulated with anti-CD3 in HBSS to release ER-Ca2+ and subsequently added 1 mM Ca2+ to activate SOCE. Peak MFI of Fluo-4 after 1 mM Ca2+addition is shown (scatter plot of n = 6 biological replicates in each experiment). C Flow cytometry analysis of IL-2 in CD4+ T cells from healthy participants (left panel) or patients with SLE (right panel) treated with 2-APB overnight at indicated concentration. Cells were stimulated with PMA/Ionomycin for 5 h (n = 6 in healthy participants and n = 5 in patients with SLE, Paired T-test). D Same experimental condition as (C) except for the treatment was done with YM-58483, not with 2-APB (n = 6 in healthy participants and n = 5 in patients with SLE). Data analysis using paired 2-tailed Student’s t test (A, B, D) and 1-way ANOVA with Tukey-Kramer test (C). All data are represented as mean ± SEM. *p < 0.05, **p < 0.01; ***p < 0.001, ****p < 0.0001. Source data are provided as a Source Data file.
Discussion
Lipid microdomains dynamically assemble cholesterol and sphingolipids. We and others have reported that lipid microdomains represented by CTB staining are aggregated on the surface membrane of T cells from patients with SLE and are involved in the aberrant CD3/TCR-initiated signaling21,22,53.
Here we have presented information on the composition of these lipid microdomains and the metabolic processes that are responsible for their formation. Specifically, we have revealed a further role of CD38 in determining lipid microdomain composition through the β-1,4-N-Acetylgalactosaminyl transferaseB4GALNT1 responsible for the synthesis of the monosialoganglioside GM2. By utilizing lipidomics analysis of Jurkat CD38-sufficient and -deficient cells we recorded the dominance of GM2 ganglioside in the plasma membrane of CD38 sufficient cells. GM2-enriched CD4+CD38+ T cells displayed enhanced proximal T cell signaling but less cytokine production including IL-2. Previously we have shown that SLE T cells mobilize the calcium/calmodulin kinase IV (CaMK4) to the nucleus to suppress the production of IL-2 through the transcriptional repressor CREMα54.
In the current study we linked the increased calcium response to ER stress and suppression of IL-2 production. In brief, we have provided evidence for the following mechanistic link: CD38>decreased NAD+>decreased Sirtuin1>increased B4GALNT1>increased GM2>increased calcium response through IP3R>increased ER stress>suppressed cytokine production.
The regulation of lipid microdomain formation and composition have not been clearly elucidated. Different profiles of gangliosides characterize distinct T cell subset function55,56 and association with molecules involved in the early signaling events57. Lipid microdomains have been recognized by the proxy binding of CTB and have been reported aggregated on the surface membrane of SLE T cells21. We found that the GM2 present on the surface of CD4+CD38+ cells comes from GM3 through the increased action of B4GALNT1. Previously it was reported that the Liver X receptor β (LXR β), which reduces cholesterol content by efflux, increases CTB staining in T cells from patients with SLE22 and that LXR β regulates the glycosphingolipid:cholesterol balance by upregulating UGCG, the synthase which converts ceramide to glucosylceramide58. These data support the concept that elevated levels of gangliosides are involved in the pathogenesis of SLE. By focusing on the expanded CD4+CD38+ subpopulation in patients with SLE, we were able to identify the dominance of GM2 of the alpha ganglioside series over other gangliosides and other lipids. Earlier focus on the contribution of cholesterol synthesis and the putative therapeutic effect of statins in an effort to normalize lipid content and distribution of the surface membrane of immune cells in SLE59 has not yielded the expected benefit60–62. Targeting either CD38 or the synthesis of GM2 which localizes preferentially within the lipid microdomains20 could prove of therapeutic value in SLE.
The involvement of B4GALNT1 in development of disease is not clear but there are reports that it is involved in carcinogenesis63,64 and lysosomal disorders65. Our data indicate a role for B4GALNT1 in promoting GM2 synthesis on the surface membrane of CD4+CD38+ cells, the alteration of the composition of lipid microdomains and the enhancement of calcium flux. Previously, the increased calcium flux in SLE T cells was attributed to a “rewiring” of the CD3 complex which involved the replacement of the CD3ζ/ZAP70 complex with the FcRγ/complex66 and aggregates of lipid rafts defined by CTB staining21 in which additional molecules like Kv1.3 were found translocated67. pPLCγ1 was found to be linked to the early calcium response and to depend on B4GALNT1. Lipid microdomains can recruit molecules important for the early signaling response. Our finding that increased content of GM2 through the action of B4GALNT1 increased cell surface membrane fluidity may explain facile translocation of signaling-relevant molecules to the lipid microdomains.
To complete the study of calcium dynamics, we evaluated the activity of SOCE and found it increased in CD4+CD38+ T cells compared to CD4+CD38- T cells after thapsigargin or TCR/CD3 stimulation and found it higher in patients with SLE compared to healthy participants. This activity depends on the TCR/CD3 stimulation-initiated depletion of calcium from the ER and not in the thapsigargin-induced depletion in a B4GALNT1-dependent manner. We observed higher calcium flux in CD38+CD4+ T cells from HC and SLE compared to CD38-CD4+ T cells. The CD38+CD4+ T cells from HC have higher CD3ξ which can recruit more pPLCγ1. It is possible that the recorded changes in the cell membrane lipid composition, due to higher B4GALNT1 through CD38, also affects the ER membrane leading to increased sensitivity of IP3 receptor. Further experiments will be needed to demonstrate this. The role of SOCE activity in T cell malfunction in autoimmune diseases has not been studied. Our study suggests that chronic deletion of the ER stores in calcium accounts for the increased SOCE activity in SLE CD4+CD38+ cells and correction of steps involved in the aberrant early signaling response rather than the activity of SOCE may help normalize SLE T cell function.
Our findings in CD4+CD38+ cells from patients with SLE appear to be in contradiction with the reported importance of SOCE in the activity of the transcription factor NFAT in T cell activation68–70. We asked if this discrepancy was due to ER stress, which induces exhaustion and loss of T cell function in the tumor environment, and whether a similar condition existed in SLE T cells due to chronic stimulation. Indeed, we found that CD4+CD38+ T cells display ER stress by demonstrating increased expression of XBP1s, as shown by pathway analysis of the RNA- and ATAC-seq data, and by providing ultrastructural evidence of ER dilation. ER stress may reflect a key event in the exhaustion or senescence phenotype of CD38high cells as described in previous reports10,11.
Finally, we show that inhibition of the proximal T cell signaling by a IP3 receptor inhibitor restores IL-2. Decreased IL-2 production is considered as one of the key features in SLE pathogenesis47,71, responsible for the impaired regulatory T cell function46 and cytotoxic cell responses72,73. Low-dose IL-2 administration to people with various autoimmune diseases restores Treg cell function and improves clinical outcomes74. Our study demonstrated that targeting the CD38/GM2 axis can restore IL-2 production in SLE and can be exploited clinically. Most importantly, we provide evidence that correction of altered proximal T cell calcium signaling which is mediated by GM2-enriched microdomains, but not direct inhibition of SOCE, is effective in reversing low IL-2 production. Our observation that IP3 receptor inhibition restores IFNγ, as well as the report in two patients with SLE treated with the CD38 antibody daratumumab further support to consider CD38-instigated aberrant signaling and cytokine production therapeutically13. It will be interesting to investigate whether the beneficial effects of daratumumab treatment in SLE patients are due to a depletion of CD38+ lymphocytes or CD38 function inhibition.
There are several limitations of our studies. Several of the experiments were performed using fresh ex vivo T cells in which manipulation, that is silencing or overexpression, of a number of molecules was not perfect. Several of the small drug inhibitors that we used may not have been highly specific. We have studied CD38 expression in T cells from the angle of considering this molecule as an ectonucleotidase and relied on its ability to degrade NAD and influence the activity of NAD-dependent enzymes. Yet, we have significantly advanced our previous observations that CD3ζ is decreased in SLE T cells38 and that lipid rafts are aggregated in these cells21 and that their manipulation can alter disease in lupus-prone mice53. We still do not know why CD38 is increased in SLE T cells and whether autoantibodies or inflammatory cytokines present in the sera of patients with SLE are responsible. Our study having demonstrated the presence of an aberrant CD38 > NAD + >Sirtuin1 > B4GALNT1 > GM2 > IP3R > ER stress>cytokine pathway, offers novel targets for the treatment of patients with SLE.
Methods
Experimental model and cohort details
Human samples
All SLE patients fulfilled the American College of Rheumatology classification criteria75. Patients with SLE were recruited from the Division of Rheumatology at Beth Israel Medical Center and provided consent, as approved by the institutional review board. As controls, age- and sex-matched healthy individuals were recruited (Supplementary Table 2).
Disease activity score for the patients with SLE was measured using the SLE Disease Activity Index (SLEDAI) scoring system76. The experiments except for those in Fig. 1A have been performed with frozen T cells from healthy participants and patients with SLE. As for healthy participants, we purchased whole blood from Boston Children’s Donor Center, which is remaining in the leukoreduction system chambers of apheresis instruments after routine platelet collection (“Trima collars”).
Cell lines
Jurkat human CD4+ T cell line (Clone: E6-1, #TIB-152, ATCC) was purchased from, authenticated and tested for mycoplasma by ATCC and maintained in RPMI1640 with L-glutamine (# 10040CV, Corning), 1% of Penicillin/Streptomycin (#30002CI, Corning) and 10% of FBS (#35-011-CV, Corning). CD38 knocked-down Jurkat cell line (JurkatCD38KO) was generated as previously described {Katsuyama, 2020 #25}. Cas9-expressing Jurkat cells were used as control (JurkatControl). B4GALNT1 knocked-down Jurkat cell line was generated by Crispr/Cas9. Briefly, 1 μM of pooled three different gRNA targeting B4GALNT1-exon (ACCGGGATGTGTGCGTAGCG, ACGGCGCAAGAGGTAGCCGG, TGACCGGGATGTGTGCGTAG) was electroporated into 1 million of Cas9-expressing Jurkat cells {Katsuyama, 2020 #25} by Amaxa nucleofector (#VCA-1003, Lonza).
Method details
Cell isolation and sorting
Total T cells or CD4+ T cells were enriched by RosetteSep human CD4+ Enrichment Cocktail (#15062, STEMCELL Technologies) from peripheral blood from the study participants. Freshly isolated or frozen T cells were used in further experiments. CD4+CD38- and CD38+ T cells were sorted from human primary cells by FACSAria Cell Sorter (BD) or MoFlo Astrios EQ Cell Sorter (Beckman Coulter).
Electroporation
For overexpression assays, 1–5 million primary cells were electroporated with 1.25 μg of CD38- or GFP- overexpressing control plasmid (Sino Biological) or B4GALNT1- overexpressing plasmid (ORIGENE) using the Amaxa nucleofector (Human T Cell Nucleofector kit (#VCA-1002, Lonza), the program U-014). For Jurkat cells we used the Cell Line Nucleofector kit (program X-001).
Flow cytometry
The phenotype of T cell subpopulations was measured by flow cytometry, using fluorochrome-conjugated antibodies against CD3 (Clone: UCHT1, #300439, Biolegend), CD4 (Clone: RPA-T4, #300528, Biolegend), CD8 (Clone: HIT8a, #300907, Biolegend), CD38 (Clone: HIT2, #303516, Biolegend), CTB (C1655, Sigma Aldrich), CD45RA (Clone:HI100, #304118, Biolegend), CCR7 (Clone:G043H7, #353212, Biolegend), IL-2 (Clone: MQ1-17H12, #500326, Biolegend), IFNγ (Clone: 4S.B3, 502521, Biolegend), TNF (Clone: MAb11, #502913, Biolegend), IL-4 (Clone: MP4-25D2, #500811, Biolegend), IL-6 (Clone: MQ2-13A5, #501107, Biolegend), IL-10 (Clone:JES3-9D7, #50-7108-42, eBioscience), IL-13 (Clone: JES10-5A2, #501903, Biolegend), IL-17 (Clone; eBio64DEC17, #17-7179-42, eBioscience), XBP1s (Clone: 143 F, #647504, Biolegend) with proper isotypes as controls (PE/Cy7 Isotype control, Clone:MOPC-21, #400126, Biolegend). For intracellular staining, cells were fixed and permeabilized using a Cytofix/Cytoperm kit (#554714, BD Biosciences) to stain cytoplasmic proteins or using a True Nuclear Transcription Factor buffer set (#424401, Biolegend) to stain B4GALNT1 (Clone:M02, #AT1255a, Abcepta, 1:50), HEXA (Clone: #MAB6237-SP, R&D, 1:50), ST3GAL5 (Clone:aa389-418, #LS-C161530, LSBio,1:50), GM3 (Clone: GMR6, #A2582, TCI, 1:50), GM2A (Clone:06, # 13246-MM06, Sino Biological, 1:50), GM2 (Clone:06, #A2575, TCI, 1:50), GD1a (Clone: GD1a-1, #MAB5606Z, TCI, 1:50), pCD3ζ (Clone: 6B10.2, # 644103, Biolegend, 1:50), pZAP70 (Clone: 1503310 #683703, Biolegend, 1:50), pPLCγ1 (Clone: 3H1C10, # NBP2-52533, Novus, 1:50), pPLCγ1-Tyr783 (FITC, PLCG1Y783-C4, #MA5-28029, Invitrogen), PLCγ1 (#LS-C669421, LSBio), pLCK (Clone: SRRCHA, #46-9076-41, eBioscience), pZAP70/Syk (Clone:, #12-9006-41, eBioscience), pSHP1 (Clone:P1C1-A5, #656603, Biolegend), pSHP2 (Clone: L99-921, #L99-921, BD), pJNK (Clone:N9-66, #562480, BD), pMAPK (Clone: 4NIT4KK, #12-9078-42, eBioscience), pERK (Clone:4B11B6, #675503, Biolegend), pAKT (Clone:D25E6, #13038S, CST), pS6 (Clone: D57.2.2E, #4851S, CST). Goat anti-mouse IgG (APC, Poly4053, #405308, Biolegend), ATF4 (Clone: SD20-92, #MA5-32364, 1:50), ATF6 (Clone: 2358 C, # IC71527S, Novus Biologicals, 1:50), IRE1 (polyclonal, #A00683-1, Boster, 1:200) and PERK (polyclonal, #A01992-2, Boster,1:400) with subsequent secondary antibodies. All the antibodies conjugated with fluorescence were used at a concentration of 1:100 in PBS containing 2% FBS. When indicated, cells were stimulated with plate-coated 5 µg/ml of anti-CD3/28 (CD3, Clone: OKT-3, #BE0001-2, Bioxcell; CD28, Clone:CD28.2, #302934, Biolegend) or with 25 ng/ml of Phorbol 12-myristate 13-acetate (PMA, #P1585-1MG, Sigma) and 1 μM of Ionomycin (#19657-5MG, Sigma) for 4 hours. Cell viability was determined by gating with negative Zombie-Aqua (#423102, Biolegend, 1:500) or Zombie-Nir (#423106, Biolegend, 1:500). Data were acquired with a CytoFLEX LX Flow Cytometer (Beckman Coulter).
For the quantification of GM2 in SLE patients, cells stained for CD3 (PeCy7, Clone: UCHT1, #300426, Biolegend), CD4 (FITC, Clone: RPA-T4, #300506, Biolegend), CD38 (APC, Clone: HIT2, #303510,Biolegend), GM2 (monoclonal antibody IgM unconjugated, Clone: MK1-16, #A2576, TCI chemicals), Livedead (Zombie UV Dye, #77474, Biolegend) in PBS, for 1 h at 4 °C. Cells were then washed in PBS. Finally, goat anti-mouse IgM cross adsorbed secondary antibody (DyLight 550, #SA5-10151, Invitrogen) was added for an hour at 4 °C.
Unless otherwise specified flow cytometry antibodies were used at 1:100 dilution.
Phospho-Flow
Cells were coated with 20 μg/mL of anti-CD3ε mAb (Clone: OKT3, #317726, Biolegend) in 50 µl of PBS for 30 min on ice. To start stimulation, 10 μg of crosslinker (goat-anti-mouse, #AP124, Sigma) were added and the cells were incubated at 37 °C for 2 min. Reaction was stopped by diluting with 1 ml of ice-cold PBS. Cells were then fixed, stained with appropriate markers mentioned in method section of flow cytometry. As positive control, cells were treated with 4 mM of Na3VO4 (#S6508-10G, Sigma) to maximize tyrosyl phosphorylation and confirmed the highest fluorescent signal in each phospho-flow antibody.
Immunofluorescence
For surface staining, 0.5 M of cells were fixed using 4% paraformaldehyde (#15710, EMS) for 20 min, at room temperature. Subsequently FITC conjugated CTB (#C1655, Sigma) was added. For ganglioside staining, cells were stained with primary antibodies of mouse IgM GM3 (#A2583, Tokyo Chemical Industry,1:50) or mouse IgM GM2 (#A2576, Tokyo Chemical Industry, 1:50) overnight, at 4 °C. DyLight 488 anti-mouse IgM (#VJ3099414, Invitrogen, 1:100) was used as secondary antibody. The images were captured using a Zeiss LSM 880 Inverted Live-cell Laser Scanning Confocal Microscope.
For the colocalization of GM2-CTB-CD38 CD4+ T cells were isolated from healthy donor fresh blood with RosetteSep human CD4+ Enrichment Cocktail (#15062, STEMCELL Technologies). Cells were activated with PMA (25 ng/ml) and Ionomycin (1 µM) for 3 h at 37 °C.
Cells were then washed and stained with CTB-FITC (#C1655-250UG, Sigma), CD38-Biotin (Clone: HIT2, #303518, Biolegend), GM2-IgM monoclonal antibody IgM unconjugated, clone MK1-16, #A2576, TCI chemicals), CD4-PB (Clone: RPA-T4, #300521, Biolegend) over night at 4 °C. The next day, cells were washed in PBS and stained for 4 h at 4 °C with secondary antibodies Streptavidin-Alexa Fluor 647 (#405237, Biolegend) and Goat anti-mouse IgM-DyLyght550 (#SA5-10151, Invitrogen).
Cells were then washed and resuspended in PBS and CD4+CD38- and CD4+CD38+ T cells were sorted with MoFlo Astrios EQ Cell Sorter (Beckman Coulter). Finally, sorted cells were mounted on coverslips using ProLong Diamound antifade mountant with DAPI (#P36962, Invitrogen) mounting solution. Images were acquired with upright confocal microscope (Zeiss LSM880) 63× and processed with Zen Blue (version 2.3) and Imaris (version 9.9.0). For the colocalization of GM2-LAT-CD38 we followed the same protocol but we incubated overnight and stained for LAT using Rabbit IgG LAT (#PA5-78472, Invitrogen, 1:100) with secondary goat anti-Rabbit antibody (#F2765, Invitrogen, 1:100).
Lipidomics
The samples for lipidomics were prepared following the protocol (Metabolomics.2017.13:30). Briefly, 10 million JurkatControl (n = 3) and JurkatCD38KO (n = 3) cells were washed with PBS. The cells were resuspended in 20 times volume of chloroform:methanol (2:1). The cell suspension was agitated by shaking for 20 min at room temperature. Twenty percent volume of water was added and the samples were vortexed for 1 min. After settling for 10 min, the samples were centrifuged for 10 min at 1000 × g to separate into three phases (upper, middle and lower). The upper aqueous phase containing small organic polar molecules and the lower phase containing non-polar lipids were separately harvested in two different days and evaporated under vacuum using a SpeedVac at 37 °C. The dried-out lipid samples were kept in −80 °C and resuspended in LS/MS grade isopropanol:acetonitrile methanol just prior to LC/MS/MS analysis. Lipid extracts were subjected to LC-MS/MS using C18 reversed-phase chromatography coupled to a hybrid QExactive Plus/HF Orbitrap mass spectrometer (Thermo Fisher Scientific). Lipid identification has done by both accurate precursor ion mass and fragmentation featured. For quantification, we used Lipid-Search. The area of each lipid ions in the same lipid class from each sample group (JurkatControl #1-#3 and JurkatCD38KO #1-#3) were averaged to submit to MetaboAnalyst 5.0. The name of each lipid classes was standardized with the ID conversion tool in MetaboAnalyst 5.0. Several ambiguous lipid classes were manually corrected based on HMDB. Then the data were processed with built-in parameter optimization procedures as described in MetaboAnalyst 5.0 to perform enrichment analysis, cluster analysis (heatmap) and univariate analysis (volcano plot).
Transmission electron microscopy
Freshly isolated T cells were used for ultrastructural evaluation. In order to sort sufficient numbers of CD4+CD38- and CD38+ T cells from human primary cells, we used blood from “Trima Collars” and FACSAria. The cells were fixed in 2.5% glutaraldehyde (#16200, EMS), 2% paraformaldehyde, in 0.1 M cacodylate buffer pH 7.4 (modified Karnovsky’s fixative) for at least 1 h at room temperature and then at 4 °C overnight. Cells were washed in 0.1 M cacodylate buffer pH 7.4, and post-fixed in 1% osmium tetroxide in 0.1 M cacodylate buffer pH 7.4 for 1 hr at 4 °C. Subsequently, cells were washed in distilled water and incubated overnight in 2% aqueous uranyl acetate at 4 °C. After washes in distilled water, cells were dehydrated in ethanol and propylene oxide and embedded in SeaKem LE Agarose. Agarose blocks were sectioned using a Leica Ultracut E ultramicrotome and sections were stained with 2% uranyl acetate and lead citrate. The cells were evaluated using a JEOL 1400 TEM (JEOL, Inc) equipped with a Orius SC1000 digital CCD camera (Gatan, Inc.).
Chromatin immunoprecipitation (ChIP)-qPCR
CD4+ T cells were isolated from healthy donor fresh blood with RosetteSep human CD4+ Enrichment Cocktail (#15062, STEMCELL Technologies). Next, they were cultured for 24 h at 37 °C at 1.5 M/ml in culture medium with the following conditions: Non Treated, 1 µM EX527, 10 µM EX527.
Following incubation cells were collected and 1 million cells per condition were prepared for ChIP.
ChIP experiments were performed with the MAGnify Chromatin Immunoprecipitation System (#49-2024; Life Technologies) following the manufacturer’s instructions. Briefly, 1 × 106 cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The reaction was stopped with glycine for 5 min, and the samples were lysed and sonicated to obtain 200- to 500-bp fragments. Immunoprecipitation was performed with a H3K27 (#ab4729, abcam) or an IgG control (#49-2024; Life Technologies) antibody. Cross-linking was reversed, and DNA was eluted and purified using DNA-purification magnetic beads. Enrichment for the human B4GALANT1 promoter, B4GALANT1 genomic sequence in the samples was quantified by real-time qPCR and was normalized with the input samples. The primers used were the following (Eurofins genomics): Forward:ATCCCTCCCGTCTTGGAA, Reverse:GGGTGCAACAGACTCTTAACT.
RNA isolation and quantitative PCR
RNeasy Mini Kit (#74104, QIAGEN) was used to extract total RNA. The following TaqMan probes (Thermo Fisher Scientific) were used to detect target genes, CD38 Hs01120071_m1, B4GALNT1 Hs01110791_g1, ST3GAL5 Hs01105377_m1, and TBP Hs00427620_m1. Gene expression was assessed by the comparative CT method and normalized to the reference gene TBP. We did not use GAPDH as endogenous control because CD38, a NADase, may affect metabolism and change the expression of GAPDH.
Chemical compounds
Six-Chloro-2,3,4,9-tetrahydro-1H-Carbazole-1-carboxamide as Sirtuin1 inhibitor (EX527, #E7034, Sigma-Aldrich), thapsigargin (#1138/1, Tocris), 2-Aminoethyl diphenylborinate as IP3 receptor inhibitor (2-APB, #17146, Cayman Chemical), YM-58483 as SOCE inhibitor (#Y4895, Sigma), trans-Ned-19 as NAADP antagonist (#17527, Cayman Chemical), and 8-Bromo-cADP-Ribose as cADPR antagonist (8-Br-cADPR, #sc201514, Santa Cruz) were used. Cells were incubated with these inhibitors overnight in culture medium at the indicated concentrations.
RNA sequencing
Total RNA was extracted from JurkatControl and JurkatCD38KO using the RNeasy mini kit (#74104, QIAGEN) and submitted for RNA sequencing to the Molecular Biology Core Facilities at the Dana-Farber Cancer Institute (DFCI). Libraries were prepared using Illumina TruSeq Stranded mRNA sample preparation kits according to the manufacturer’s protocols. Samples were sequenced on an Illumina NextSeq500 run with single-end 75-bp reads. Data analyzes were performed using the VIPER pipeline by the DFCI Core.
Omni-ATAC-seq (assay for transposase accessible chromatin with high-throughput sequencing)
To assay chromatin accessibility, we used 50,000 freshly sorted CD4+CD38- and CD4+CD38+ T cells from two different healthy donors and two different SLE patients. For ATAC-seq cell preparation we followed the Omni-ATAC protocol77. Deep sequencing was performed using PE35-bp reads on Illumina NextSeq500 at the Boston Nutrition Obesity Research Center Functional Genomics Core. After filtering low quality reads and mitochondrial DNA sequences, high quality sequences were mapped to human genome (GRcH37/hg19) using Bowtie278. Low mapping quality, duplicates, and mismatched sequences were further filtered before further analysis. MACS2 (version 2.1.1) was used to call peaks from the aligned results79, and the aligned results were normalized by CPM. To compare differential binding between CD4+CD38- and CD4+CD38+ samples, DEseq2 was used to calculate statistically significant differences in binding regions80, as defined by fold change >1.5 and p-value < 0.05, and R (version 4.2.0) package “EnhancedVolcano” was used for visualization.
Measurement of intracellular Ca2+ by flow cytometry
CD4+ T cells were stained with surface markers, washed and incubated with 4 µM of Fluo-4 (#F14217, Thermo Fisher) in culture medium for 30 min at 37 °C, washed and incubated in medium with 1 mM of CaCl2 for 30 min at 37 °C, then analyzed by Cytoflex LX2. To establish a baseline, events were collected for 45 s and Ca2+ flux was measured for the indicated duration immediately after addition of the agonist anti-CD3 at a final concentration of 2 µg/mL followed by 10 mg/mL goat-anti-mouse crosslinker (Millipore Sigma, #AP124). Ionomycin was used as a positive control at a concentration of 1 µM for 45 s. For depletion of calcium stores from ER, cells were treated with 1 µM thapsigargin in Ca2+-free HBSS and 1 mM of Ca2+ was subsequently added when measuring SOCE activity. Data analysis was performed using the kinetics package of FlowJo software (Tree Star, Inc).
CRISPR editing for CD4+ T cells from patients with SLE
Guide RNAs for CRISPR-mediated gene knock-down were designed using Benchling, and chemically modified synthetic guide RNAs (sgRNAs) were synthesized by Synthego. The following sequences were used for CRISPR editing: 3ʹ-ACCGGGATGTGTGCGTAGCG-5ʹ, 3ʹ-ACGGCGCAAGAGGTAGCCGG-5ʹ, and 3ʹ-TGACCGGGATGTGTGCGTAG-5ʹ for pooled human B4GALNT1 targeting sgRNAs. Nontargeting sgRNA was used as a negative control. To form ribonucleoprotein complexes, a total of 50 pmols of pooled three targeting sgRNAs or nontarget sgRNA were mixed with 20 pmols of Cas9 2NLS nuclease (Streptococcus pyogenes) (New England Biolabs) and incubated for 10 min at room temperature. For B4GALNT1 depletion, sgRNAs targeted B4GALNT1 or negative control sgRNAs were transfected using the Amaxa human T Cell Nucleofector Kit with the T-023 program. CD4+ T cells from patients with SLE were pre-stimulated for overnight with plate-coated anti-CD3 and CD28 (2.5 µg/ml) before transfection to improve transfection efficacy. The cells after transfection were kept stimulated with plate-coated anti-CD3 and CD28 stimulation for another 2 days and harvested for further analysis.
Immunometabolism
0.15 million/well of JurkatControl and JurkatCD38KO cells were applied in 8-well plate (XFp FluxPak, #103022-100, Agilent). Cells were stimulated with anti-CD3/28 (2 μg/ml as a final concentration) during each assay as an acute-stimulation. Glycolysis (XFp Glycolysis Stress Test Kit,#103017-100, Agilent) and Oxidative phosphorylation (XFp Cell Mito Stress Test Kit, #103010-100, Agilent) were analyzed by Seahorse XF HS Mini Analyzer according to manufacturer’s instructors. A glycolysis stress test with human samples was performed on sorted CD4+CD38- T cells and CD4+CD38+ T cells from SLE and from healthy participants (n = 5) under the same experimental condition as Jurkat cells.
Quantification and Statistical analysis
Unpaired 2-tailed Student’s t test or paired 2-tailed Student’s t test or 1-way ANOVA with Tukey-Kramer test were used for statistical comparisons. All data are represented as mean ± SEM. GraphPad Prism (version 7) was used for statistical analysis. P-value is defined as follows; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Suzanne Krishfield, Julianne O’Connell and Christina Ioannidis for collecting patients’ data. Molly Veregge and Linus Tsai for ATAC sample preparation and Kyle Smith in Electron Microscopy Core for EM sample preparation and imaging. Work was supported by grants NIH RO1AI42269 and NIH RO1 AI148161 to GCT.
Author contributions
Conceptualization, E.K. and G.C.T.; Methodology, E.K., M.H., A.S.F. and G.C.T.; Investigation, E.K., A.S.F., A.S., N.Y., M.G.T., M.H.; Formal Analysis, E.K., M.H.; Writing - Original Draft, E.K., M.H. and G.C.T.; Writing – Review & Editing, E.K., V.C.K., M.G.T., M.H. and G.C.T.; Funding Acquisition, G.C.T.; Resources, G.C.T.; Supervision, G.C.T.
Peer review
Peer review information
Nature Communications thanks Sun Jung Kim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The ATACseq data generated in this study have been deposited in the GEO database under accession code: GSE273008. The RNAseq data generated in this study have been deposited in the GEO database under accession code: GSE273009. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.
Competing interests
GCT is on the scientific advisory boards of Cugene, Biolojic, ABPRO, A2 biotherapeutics, COREVITAS. VCK is site PI for clinical trial for BMS, Astra Zeneca and Novartis. VK is advisory board member for Janssen and Aurinia. VK is in the clinical trial adjudication committee member for Cabaletta. ASF currently employed at Abbvie. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Eri Katsuyama, Morgane Humbel.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52617-7.
References
- 1.Tsokos, G. C. Systemic lupus erythematosus. N. Engl. J. Med365, 2110–2121 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Tsokos, G. C. Autoimmunity and organ damage in systemic lupus erythematosus. Nat. Immunol.21, 605–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcocer-Varela, J., Alarcon-Riquelme, M., Laffon, A., Sanchez-Madrid, F. & Alarcon-Segovia, D. Activation markers on peripheral blood T cells from patients with active or inactive systemic lupus erythematosus. Correlation with proliferative responses and production of IL-2. J. Autoimmun.4, 935–945 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. et al. Integrative analysis reveals CD38 as a therapeutic target for plasma cell-rich pre-disease and established rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res Ther.20, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erkeller-Yuksel, F. M., Lydyard, P. M. & Isenberg, D. A. Lack of NK cells in lupus patients with renal involvement. Lupus6, 708–712 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Wang, H., Li, S., Zhang, G., Wu, H. & Chang, X. Potential therapeutic effects of cyanidin-3-O-glucoside on rheumatoid arthritis by relieving inhibition of CD38+ NK cells on Treg cell differentiation. Arthritis Res Ther.21, 220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi, B. et al. Targeting CD38-dependent NAD(+) metabolism to mitigate multiple organ fibrosis. iScience24, 101902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aksoy, P., White, T. A., Thompson, M. & Chini, E. N. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys. Res Commun.345, 1386–1392 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Chini, E. N., Chini, C. C., Kato, I., Takasawa, S. & Okamoto, H. CD38 is the major enzyme responsible for synthesis of nicotinic acid-adenine dinucleotide phosphate in mammalian tissues. Biochem J.362, 125–130 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covarrubias, A. J. et al. Senescent cells promote tissue NAD(+) decline during ageing via the activation of CD38(+) macrophages. Nat. Metab.2, 1265–1283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chini, C. C. S. et al. CD38 ecto-enzyme in immune cells is induced during aging and regulates NAD(+) and NMN levels. Nat. Metab.2, 1284–1304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsuyama, E. et al. The CD38/NAD/SIRTUIN1/EZH2 axis mitigates cytotoxic CD8 T cell function and identifies patients with SLE prone to infections. Cell Rep.30, 112–123.e114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostendorf, L. et al. Targeting CD38 with Daratumumab in refractory systemic lupus erythematosus. N. Engl. J. Med. 383, 1149–1155 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Roccatello, D. et al. Daratumumab monotherapy for refractory lupus nephritis. Nat. Med. 29, 2041–2047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander, T. et al. Sustained responses after anti-CD38 treatment with daratumumab in two patients with refractory systemic lupus erythematosus. Ann. Rheum. Dis.82, 1497–1499 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Barbosa, M. T. et al. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J.21, 3629–3639 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Wang, L. F. et al. CD38 deficiency suppresses adipogenesis and lipogenesis in adipose tissues through activating Sirt1/PPARgamma signaling pathway. J. Cell Mol. Med. 22, 101–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angela, M. et al. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARgamma directs early activation of T cells. Nat. Commun.7, 13683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. et al. Regulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. J. Immunol.192, 3190–3199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prinetti, A., Chigorno, V., Tettamanti, G. & Sonnino, S. Sphingolipid-enriched membrane domains from rat cerebellar granule cells differentiated in culture. A compositional study. J. Biol. Chem.275, 11658–11665 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Krishnan, S. et al. Alterations in lipid raft composition and dynamics contribute to abnormal T cell responses in systemic lupus erythematosus. J. Immunol.172, 7821–7831 (2004). [DOI] [PubMed] [Google Scholar]
- 22.McDonald, G. et al. Normalizing glycosphingolipids restores function in CD4+ T cells from lupus patients. J. Clin. Invest124, 712–724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jury, E. C., Isenberg, D. A., Mauri, C. & Ehrenstein, M. R. Atorvastatin restores Lck expression and lipid raft-associated signaling in T cells from patients with systemic lupus erythematosus. J. Immunol.177, 7416–7422 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Pavon, E. J. et al. Increased CD38 expression in T cells and circulating anti-CD38 IgG autoantibodies differentially correlate with distinct cytokine profiles and disease activity in systemic lupus erythematosus patients. Cytokine62, 232–243 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Hannun, Y. A. & Obeid, L. M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol.19, 175–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanimura, N. et al. Dynamic changes in the mobility of LAT in aggregated lipid rafts upon T cell activation. J. Cell Biol.160, 125–135 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakomori, S., Handa, K., Iwabuchi, K., Yamamura, S. & Prinetti, A. New insights in glycosphingolipid function: “glycosignaling domain,” a cell surface assembly of glycosphingolipids with signal transducer molecules,involved in cell adhesion coupled with signaling. Glycobiology8, xi–xix (1998). [DOI] [PubMed] [Google Scholar]
- 28.Sonnino, S., Mauri, L., Chigorno, V. & Prinetti, A. Gangliosides as components of lipid membrane domains. Glycobiology17, 1R–13R (2007). [DOI] [PubMed] [Google Scholar]
- 29.Hoon, D. S., Irie, R. F. & Cochran, A. J. Gangliosides from human melanoma immunomodulate response of T cells to interleukin-2. Cell Immunol.111, 410–419 (1988). [DOI] [PubMed] [Google Scholar]
- 30.Banerjee, A., Mahata, B., Dhir, A., Mandal, T. K. & Biswas, K. Elevated histone H3 acetylation and loss of the Sp1-HDAC1 complex de-repress the GM2-synthase gene in renal cell carcinoma. J. Biol. Chem.294, 1005–1018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rifai, K. et al. SIRT1-dependent epigenetic regulation of H3 and H4 histone acetylation in human breast cancer. Oncotarget9, 30661–30678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finco, T. S., Kadlecek, T., Zhang, W., Samelson, L. E. & Weiss, A. LAT is required for TCR-mediated activation of PLCgamma1 and the Ras pathway. Immunity9, 617–626 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Lin, J., Weiss, A. & Finco, T. S. Localization of LAT in glycolipid-enriched microdomains is required for T cell activation. J. Biol. Chem.274, 28861–28864 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Shah, K., Al-Haidari, A., Sun, J. & Kazi, J. U. T cell receptor (TCR) signaling in health and disease. Signal Transduct. Target Ther.6, 412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enyedy, E. J. et al. Fc epsilon receptor type I gamma chain replaces the deficient T cell receptor zeta chain in T cells of patients with systemic lupus erythematosus. Arthritis Rheum.44, 1114–1121 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Wang, J. et al. Inhibition of SHP2 ameliorates the pathogenesis of systemic lupus erythematosus. J. Clin. Invest126, 2077–2092 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai, Z. W. et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. Lancet391, 1186–1196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liossis, S. N., Ding, X. Z., Dennis, G. J. & Tsokos, G. C. Altered pattern of TCR/CD3-mediated protein-tyrosyl phosphorylation in T cells from patients with systemic lupus erythematosus. Deficient expression of the T cell receptor zeta chain. J. Clin. Invest101, 1448–1457 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassilopoulos, D., Kovacs, B. & Tsokos, G. C. TCR/CD3 complex-mediated signal transduction pathway in T cells and T cell lines from patients with systemic lupus erythematosus. J. Immunol.155, 2269–2281 (1995). [PubMed] [Google Scholar]
- 40.Prakriya, M. et al. Orai1 is an essential pore subunit of the CRAC channel. Nature443, 230–233 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Howard, M. et al. Formation and hydrolysis of cyclic ADP-ribose catalyzed by lymphocyte antigen CD38. Science262, 1056–1059 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Guse, A. H. et al. Regulation of calcium signalling in T lymphocytes by the second messenger cyclic ADP-ribose. Nature398, 70–73 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Feske, S. Calcium signalling in lymphocyte activation and disease. Nat. Rev. Immunol.7, 690–702 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Kar, P. et al. Dynamic assembly of a membrane signaling complex enables selective activation of NFAT by Orai1. Curr. Biol.24, 1361–1368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kar, P. & Parekh, A. B. Distinct spatial Ca2+ signatures selectively activate different NFAT transcription factor isoforms. Mol. Cell58, 232–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharabi, A. et al. Regulatory T cells in the treatment of disease. Nat. Rev. Drug Discov.17, 823–844 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Linker-Israeli, M. et al. Defective production of interleukin 1 and interleukin 2 in patients with systemic lupus erythematosus (SLE). J. Immunol.130, 2651–2655 (1983). [PubMed] [Google Scholar]
- 48.Willerford, D. M. et al. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity3, 521–530 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Bouman, L. et al. Parkin is transcriptionally regulated by ATF4: evidence for an interconnection between mitochondrial stress and ER stress. Cell Death Differ.18, 769–782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma, X. et al. Cholesterol induces CD8(+) T cell exhaustion in the tumor microenvironment. Cell Metab.30, 143–156.e145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song, M. et al. IRE1alpha-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature562, 423–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim, I., Xu, W. & Reed, J. C. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov.7, 1013–1030 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Deng, G. M. & Tsokos, G. C. Cholera toxin B accelerates disease progression in lupus-prone mice by promoting lipid raft aggregation. J. Immunol.181, 4019–4026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Juang, Y. T. et al. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J. Clin. Invest115, 996–1005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagafuku, M. et al. CD4 and CD8 T cells require different membrane gangliosides for activation. Proc. Natl Acad. Sci. USA109, E336–E342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, Y. et al. Lowering glycosphingolipid levels in CD4+ T cells attenuates T cell receptor signaling, cytokine production, and differentiation to the Th17 lineage. J. Biol. Chem.286, 14787–14794 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barbat, C. et al. p56lck, LFA-1 and PI3K but not SHP-2 interact with GM1- or GM3-enriched microdomains in a CD4-p56lck association-dependent manner. Biochem J.402, 471–481 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waddington, K. E. et al. LXR directly regulates glycosphingolipid synthesis and affects human CD4+ T cell function. Proc. Natl Acad. Sci. USA118, e2017394118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fatemi, A. et al. Atorvastatin effect on systemic lupus erythematosus disease activity: a double-blind randomized clinical trial. Clin. Rheumatol.33, 1273–1278 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Costenbader, K. H. et al. A pravastatin dose-escalation study in systemic lupus erythematosus. Rheumatol. Int27, 1071–1077 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Knapp, A. C., Huang, J., Starling, G. & Kiener, P. A. Inhibitors of HMG-CoA reductase sensitize human smooth muscle cells to Fas-ligand and cytokine-induced cell death. Atherosclerosis152, 217–227 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Kwak, B., Mulhaupt, F., Myit, S. & Mach, F. Statins as a newly recognized type of immunomodulator. Nat. Med6, 1399–1402 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Liang, Y. J. et al. Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells. Proc. Natl Acad. Sci. USA110, 4968–4973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuyama, Y., Dohi, T., Morita, H., Furukawa, K. & Oshima, M. Enhanced expression of GM2/GD2 synthase mRNA in human gastrointestinal cancer. Cancer75, 1273–1280 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Gondre-Lewis, M. C., McGlynn, R. & Walkley, S. U. Cholesterol accumulation in NPC1-deficient neurons is ganglioside dependent. Curr. Biol.13, 1324–1329 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Tsokos, G. C., Nambiar, M. P., Tenbrock, K. & Juang, Y. T. Rewiring the T-cell: signaling defects and novel prospects for the treatment of SLE. Trends Immunol.24, 259–263 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Nicolaou, S. A. et al. Differential calcium signaling and Kv1.3 trafficking to the immunological synapse in systemic lupus erythematosus. Cell Calcium47, 19–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feske, S. et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature441, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Kaufmann, U. et al. Calcium signaling controls pathogenic Th17 cell-mediated inflammation by regulating mitochondrial function. Cell Metab.29, 1104–1118.e1106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaw, P. J. & Feske, S. Physiological and pathophysiological functions of SOCE in the immune system. Front Biosci. (Elite Ed.)4, 2253–2268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katsuyama, E. et al. Correction: downregulation of miR-200a-3p, targeting CtBP2 complex, is involved in the hypoproduction of IL-2 in systemic lupus erythematosus-derived T cells. J. Immunol.201, 1104 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Gillis, S. & Smith, K. A. Long term culture of tumour-specific cytotoxic T cells. Nature268, 154–156 (1977). [DOI] [PubMed] [Google Scholar]
- 73.Gillis, S., Baker, P. E., Ruscetti, F. W. & Smith, K. A. Long-term culture of human antigen-specific cytotoxic T-cell lines. J. Exp. Med. 148, 1093–1098 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kolios, A. G. A., Tsokos, G. C. & Klatzmann, D. Interleukin-2 and regulatory T cells in rheumatic diseases. Nat. Rev. Rheumatol.17, 749–766 (2021). [DOI] [PubMed] [Google Scholar]
- 75.Tan, E. M. et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum.25, 1271–1277 (1982). [DOI] [PubMed] [Google Scholar]
- 76.Bombardier, C., Gladman, D. D., Urowitz, M. B., Caron, D. & Chang, C. H. Derivation of the SLEDAI. a disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum.35, 630–640 (1992). [DOI] [PubMed] [Google Scholar]
- 77.Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol.9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ATACseq data generated in this study have been deposited in the GEO database under accession code: GSE273008. The RNAseq data generated in this study have been deposited in the GEO database under accession code: GSE273009. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.