Abstract
G-protein coupled receptors (GPCRs) are encoded by nonabundant mRNAs, and it is difficult to detect them reliably with the highly parallel methods that are in general use. Because of this, we developed and validated a sensitive, specific, semi-quantitative method for detecting these transcripts. We have used the method to profile GPCR transcripts in white blood cells (WBCs)–B, CD4, CD8, NK, and dendritic cells; monocytes, and macrophage-like monocytes treated with granulocyte-macrophage colony-stimulating factor–as well as platelets. On average, the white cells studied expressed 160 receptor mRNAs (range, 123–206). Platelets made 69. Some, but far from all, of the receptors we found have been detected earlier. We believe our data should stimulate studies of receptor function and contribute to drug development.
Subject terms: Drug development, Receptor pharmacology
Background & Summary
In 2007 we published a detailed description of a method that could be used to profile GPCR mRNAs in cell and tissue samples1. The technique is based on multiplex PCR with panels of specific fluorescent primers. The products that are produced are hybridized to arrays of target DNAs and the slides are then scanned. Two amounts of mRNA are used in the reactions: 10 ng and 100 ng. Products that are detected when 10 ng are employed represent relatively abundant mRNAs. Ones that are only detected when 100 ng are used are rarer.
We have profiled many mRNA samples at this point and believe that the results represent a useful starting point for studies of cell-cell interactions. In addition, they may reveal potential targets for drug development. For example, our profiling data suggested that hematopoietic stem cells (HSCs) make vasopressin receptors, and it appears that activating these increases RBC production and could represent a novel treatment for anaemia2.
In this paper, we share our white blood cell and platelet GPCR profiles.
Methods
All primary white blood cells were obtained from healthy volunteers. All donors provided informed consent that left-over blood donation products can be used for scientific purposes.
B cells
Human Peripheral Blood Mononuclear Cells (PBMCs) were isolated by Ficoll-Hypaque density gradient centrifugation, incubated in the presence of a CD19 monoclonal antibody (PharMingen) solution for 15 min, and washed in FACS staining buffer–Phosphate Buffered Saline (PBS) without Ca2+/Mg2+ plus 1% BSA. Using a FACStarPlus (Becton Dickinson), the cells in the CD19-positive gate were sorted. Reanalysis of the sorted population by flow cytometry indicated a population purity of greater than 97%.
CD4 and CD8 T cells and NK cells
PBMCs from single donors were resuspended in PBS with 2% heat-inactivated fetal calf serum and 2 mM EDTA. CD4+ T cells, CD8+ T cells and NK cells were isolated by negative selection using StemCell Human enrichment kits (StemCell Technologies, Cambridge, MA) as specified by the manufacturer. The cells were shown to be greater than 95% pure using flow cytometry and anti-human CD3, CD4, CD8, CD56, and CD14-20 (Lin-) antibodies.
Monocytes, macrophages, and immature dendritic cells
CD14+ monocytes from single donors were isolated from PBMCs of unidentified healthy volunteers using immunomagnetic negative selection (Monocyte isolation kit StemCell Technology Vancouver, Canada) as specified by the manufacturer. Isolated monocytes were used as is or cultured to differentiate into macrophages or immature dendritic cells. To differentiate unpolarized macrophages, monocytes were cultured in six-well plates for 6–8 days and fed every 2 days with RPMI 1640 (Bio-Whittaker, Walkersville, Md.) supplemented with 10% heat-inactivated fetal calf serum (HyClone Laboratories, Ogden, Utah), 5% heat-inactivated human serum (Sigma, St. Louis, Mo.), and macrophage colony-stimulating factor (0.1 μg/ml) (Peprotech, Frederick, Md.)3. To differentiate immature dendritic cells4, monocytes were cultured for 6 days at 1 × 106 cells/ml in 6 well plates in the in the presence of granulocyte-macrophage colony-stimulating factor (GMCSF, 100 ng/ml) and IL4 (50 ng/ml) both from R & D in RPMI 1640, 10% FCS.
Platelets
Human platelet packs were obtained from 5 healthy donors at the University Medical Center Hamburg-Eppendorf Blood Bank. The platelets were centrifuged twice for 10 minutes at 120 x g, and then washed in modified Tyrode’s buffer (137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 3.3 mM NaH2PO4, 5.55 mM glucose and 20 mM HEPES, pH 7.4), pelleted again, and suspended in PBS. The final platelet preparation appeared to be free from contaminating leukocytes based on automated cell counting on a COULTER® Ac·T diffTM Analyzer (Beckman Coulter).
mRNA preparation
Total RNA was made from the cell samples described above using Ultraspec RNA kits (Biotecx Laboratories Inc., Houston, TX) according to the manufacturer’s instructions. After extraction the RNA was analyzed by spectrometer to determine the concentration and verify a 260/280 ratio of approximately 2.0.
GPCR transcript profiling
Optimization of assay parameters and detection of GPCR mRNAs was performed as described in Hansen, et al.1 (Fig. 1). In brief, RNA was purified from the platelets and cells and contaminating DNA was removed with DNase. Eight sets of 50 primer pairs were used to amplify the GPCR mRNAs present 10 ng and 100 ng of each RNA sample. These reactions were run in triplicate. Aminoallyl-modified dUTP was incorporated into the amplicons. After the PCR reactions were performed, the products of 4 reactions (200 potential products) were pooled. Thus, two pools (x3) were created for each of the two concentrations of RNA studied (10 ng and 100 ng), then the products were labelled by coupling the NHS ester of Cy5 to the amino-modified bases that had been incorporated into the amplicons. The final labelled products were hybridized to glass-slide arrays printed with both plus and minus sense oligonucleotides so that the products of balanced or unbalanced reactions generate signals. Finally, the arrays were read, and the signals were analyzed1. Receptor mRNAs are reported to be present below if all three replicates were positive. Nearly all the RNAs detected in 10 ng of template were also found in 100 ng. Fewer transcripts were detected in 10 ng of template than in 100 ng reflecting the fact that only abundant receptor RNAs are detectable when the smaller amounts of template are used.
Fig. 1.
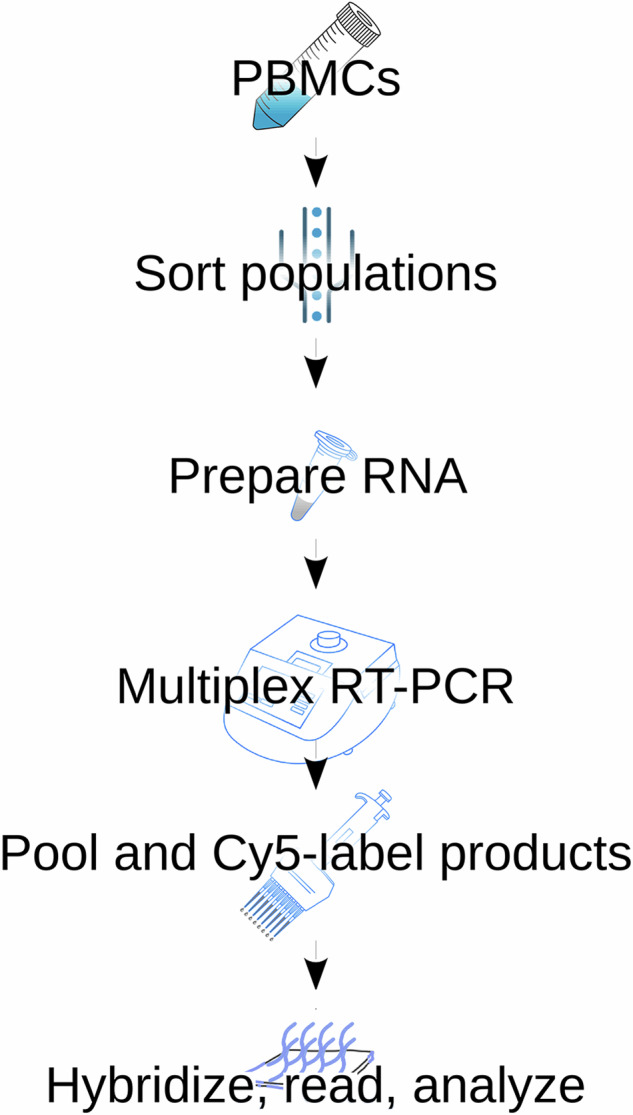
Experimental workflow. PBMCs were collected from healthy donors and sorted into different cell populations. RNA was prepared and its quality and concentration assayed according to standardized techniques. A total of 100 ng per reaction were used as input material to amplify GPCR transcripts in eight separate reactions each containing 50 GPCR specific primer pairs and aminoallyl-dUTP. Separate reactions with 10 ng of input material were used to determine those receptors expressed at higher levels. PCR reactions were pooled and Cy5 labelled on the amino-modified nucleotides. Labelled PCR products were hybridized to custom GPCR arrays that contained plus and minus sense oligonucleotides. Arrays were processed and scanned, and their signal extracted analyzed.
Analyzes
A GPCR expression call was made if amplification was detected in 3 out of 3 independent technical replicates (e.g. separate PCR amplification, labeling and hybridization). Because we used two amounts of input material per reaction, we defined GPCRs as being expressed if they were amplified in 100 ng of input material and expressed at high levels if they were also amplified in 10 ng of input material. A table of GPCR expression calls was assembled in Excel with the results of the triplicate amplifications and subsequent analyzes were performed using pivot tables. A harmonized GPCR symbol space was produced by iterative manual curation of ours and Groot-Komerlink et al.5 datasets using the web-based ToppGene6 tool and the HGNC database (genenames.org)7. GPCR symbols are current as of March 2024.
Data Records
We assayed 382 GPCR mRNAs. Their names (based on HUGO) are listed in the G protein coupled receptor transcripts in human immune cells and platelets database8, which includes all the assay data.
As noted, the PCR reactions were done in triplicate with two amounts of template-10 ng and 100 ng and all the receptors detected in 10 ng of template were also detected in 100 ng. Those that were only detected in 100 ng represent less abundant mRNAs than those that are also detected in 10 ng of template.
In the various white blood cells (WBCs) we studied, we detected a total of 265 GPCR transcripts. One hundred and sixty-two of these appeared to be relatively abundant (detected in 10 ng of RNA) in at least one cell type, and 103 were relatively rare (detected exclusively in 100 ng of RNA).
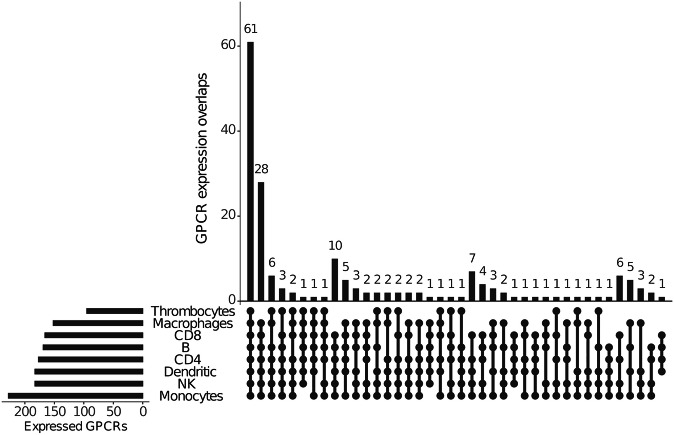
Sixty-one GPCR mRNAs were detected in all the 100 ng samples (Table 1a and Fig. 2). If platelets are excluded, the number increases to 89 (Table 1b). TRPA1 was expressed at high levels in all the RNAs except the one from platelets. One hundred and two of the RNAs were detected in 100 ng of 7 of the 8 samples (Table 2a). Only 11 of these were detected in 10 ng (Table 2b). Some relatively abundant receptors appear to be unique to each sample studied (Table 3).
Table 1.
GPCR mRNAs that are expressed by all cell types assayed.
| a. GPCR mRNAs detected in all WBCs and platelets. |
| ADGRE4P, C5AR2, CCRL2, CXCR2, CXCR3, CXCR4, CYSLTR1, F2R, F2RL1, F2RL2, FFAR3, GPR108, GPR132, GPR135, GPR146, GPR155, GPR174, GPR183, GPR19, GPR20, GPR21, GPR27, GPR37, GPR55, GPR68, GPR75, GPR82, GPR83, GPR84, GPR89, GRM2, GRM4, HCAR2, HCAR3, HTR2B, LGR4, LPAR2, LPAR3, LPAR5, LTB4R, LTB4R2, MC1R, NPBWR1, NTSR1, OPN3, OXER1, P2RY11, P2RY8, PTAFR, PTGIR, RRH, RXFP3, S1PR1, S1PR2, S1PR4, S1PR5, SMO, TBXA2R, TM7SF3, VN1R1, XCR1 |
| b. GPCR mRNAs detected in all WBCs. |
| ACKR3, ADGRA2, ADGRB1, ADGRD1, ADGRE4P, ADGRE5, ADRA2B, ADRB1, C5AR2, CCRL2, CELSR2, CXCR2, CXCR3, CXCR4, CYSLTR1, F2R, F2RL1, F2RL2, FFAR3, FZD1, FZD2, FZD3, FZD6, GIPR, GPR108, GPR132, GPR135, GPR142, GPR146, GPR151, GPR155, GPR157, GPR161, GPR174, GPR183, GPR19, GPR20, GPR21, GPR25, GPR27, GPR3, GPR35, GPR37, GPR55, GPR65, GPR68, GPR75, GPR82, GPR83, GPR84, GPR89, GRM2, GRM4, HCAR2, HCAR3, HTR2B, LANCL2, LGR4, LPAR2, LPAR3, LPAR5, LPAR6, LTB4R, LTB4R2, MC1R, NPBWR1, NTSR1, OPN3, OPRL1, OXER1, P2RY11, P2RY4, P2RY8, PTAFR, PTGIR, RRH, RXFP3, S1PR1, S1PR2, S1PR4, S1PR5, SLC52A1, SMO, TAS2R14, TAS2R5, TBXA2R, TM7SF3, VN1R1, XCR1 |
Fig. 2.
Analysis of individual GPCR expression overlaps. Bars indicate the number of GPCR shared by the cell types indicated by connected black dots in the cell matrix. The total amount of GPCRs expressed by each cell type is indicated by the horizontal bars in the lower left quadrant.
Table 2.
GPCR mRNAs that are expressed by most cell types assayed.
| a. GPCR mRNAs found in at least 7 out of 8 cell types. |
| ACKR2, ACKR3, ADGRA2, ADGRB1, ADGRD1, ADGRE1, ADGRE4P, ADGRE5, ADORA2B, ADRA2B, ADRB1, C3AR1, C5AR2, CCRL2, CELSR2, CXCR2, CXCR3, CXCR4, CXCR5, CYSLTR1, F2R, F2RL1, F2RL2, FFAR1, FFAR2, FFAR3, FZD1, FZD2, FZD3, FZD6, GIPR, GPR108, GPR132, GPR135, GPR142, GPR146, GPR151, GPR155, GPR157, GPR161, GPR174, GPR183, GPR19, GPR20, GPR21, GPR25, GPR27, GPR3, GPR35, GPR37, GPR55, GPR65, GPR68, GPR75, GPR82, GPR83, GPR84, GPR89, GRM2, GRM4, HCAR2, HCAR3, HTR2B, LANCL2, LGR4, LPAR1, LPAR2, LPAR3, LPAR5, LPAR6, LTB4R, LTB4R2, MC1R, MRGPRE, NPBWR1, NTSR1, OPN3, OPRL1, OXER1, P2RY11, P2RY12, P2RY4, P2RY8, PTAFR, PTGER3, PTGIR, RRH, RXFP3, S1PR1, S1PR2, S1PR4, S1PR5, SLC52A1, SMO, TACR2, TAS2R14, TAS2R5, TBXA2R, TM7SF3, TPRA1, VN1R1, XCR1 |
| b. GPCR mRNAs present at high levels in at least 7 out of 8 cell types. |
| FFAR3, GPR108, GRM2, GRM4, LPAR2, LPAR5, P2RY8, S1PR4, TBXA2R, TPRA1, VN1R1 |
Table 3.
GPCRs that are uniquely expressed by specific cells, bold ones are at high levels.
| B | ADGRF1, GPR18, NPFFR1, GCGR, MRGPRX4 |
| CD4 | DRD2, FZD6, OPRD1, SLC52A1, SSTR3, BRS3, CXCR6, MRGPRD |
| CD8 | CELSR2, LPAR3, TAAR8 |
| Dendritic Cells | CMKLR1, FPR3, GPR153, GPR161, HCAR1, HRH1, P2RY14, P2RY6, HCRTR2, HTR1D, MAS1L, RHO |
| Macrophages | ADRA2C, CRHR2, GPR156 |
| Monocytes | ADGRD1, ADGRE1, ADGRE2, ADGRE3, ADORA2B, ADRB1, ADRB2, C3AR1, C5AR1, CCR2, CCR3, CCRL2, CHRM4, CNR2, CX3CR1, F2RL1, FPR2, GPBAR1, GPR142, GPR34, GPR89, GPRC5B, GRM3, LANCL1, LANCL2, MRGPRE, NTSR1, P2ry13, PTGDR2, SSTR5, ADGRL3, APLNR, FZD5, GPR141, GPR160, GRM6, HTR1E, HTR1F, HTR7, MC5R, MRGPRX3, MTNR1A, OXGR1, PROKR2, PTGFR |
| NK | ADGRB2, ADGRG5, ADGRG7, CCR9, GHSR, GPR52, MC3R, P2RY4 |
| Thrombocytes | OXTR, P2RY12 |
Technical Validation
Technical validation of the assay method we used can be found in the paper by Hansen et al.1. The false positive rate appears to be 1–3%.
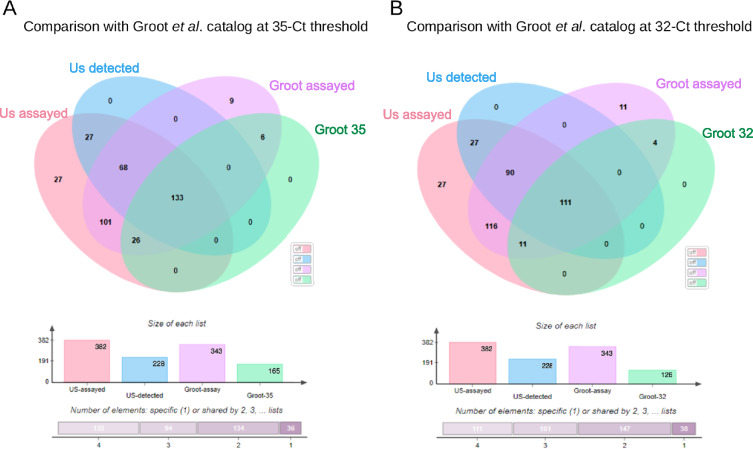
We looked for studies like ours in the literature, but only two have been published that might help validate our results. One of those papers cultured the monocytes before assaying thus we compared our results with Groot-Kormelink et al.5 who used q-PCR to assay 365 individual GPCR transcripts in monocytes isolated from 5 subjects (Fig. 3). Recall that we studied 382 receptors. Groot-Kormelink et al.5 looked at 25 mRNAs that we omitted, but only 11 of these are annotated as GPCR transcripts (Table 4).
Fig. 3.
GPCR expression detection method comparison. (A) Venn analysis was used to compare the sensitivity of our method compared to the Groot et al. study at a Ct threshold of 35 cycles. Us-assayed means the GPCRs assayed in our study. Us-detected, the GPCRs detected in our 100 ng input assay. Groot-assayed, the total amount of GPCRs assayed by Groot-Kormelink et al. Groot-35 the GPCRs detected by TaqMan QPCR assays at a 35-cycle threshold in that study. (B) Venn analysis was used to compare the sensitivity of our methods compared to the Groot et al. study at a Ct threshold of 32 cycles. Us-assayed, the GPCRs assayed in our study. Us-detected, the GPCRs detected in our 100 ng input assay. Groot-assayed, the total amount of GPCRs assayed by Groot-Kormelink et al. Groot-32 the GPCRs detected by TaqMan QPCR assays at a 32-cycle threshold in that study.
Table 4.
Extra genes in Groot-Kormelink et al. monocyte dataset. Bona-fide GPCRs in bold fonts.
| SYMBOL | Description | EntrezID |
|---|---|---|
| LONP2 | LONP2 (lon peptidase 2, peroxisomal) | 83752 |
| SENP3 | SENP3 (SUMO specific peptidase 3) | 26168 |
| HDAC3 | HDAC3 (histone deacetylase 3) | 8841 |
| GPR137 | GPR137 (G protein-coupled receptor 137) | 56834 |
| PTPN22 | PTPN22 (protein tyrosine phosphatase non-receptor type 22) | 26191 |
| OR2A4 | OR2A4 (olfactory receptor family 2 subfamily A member 4) | 79541 |
| MATK | MATK (megakaryocyte-associated tyrosine kinase) | 4145 |
| TRBV5-4 | TRBV5-4 (T cell receptor beta variable 5-4) | 28611 |
| PHGDH | PHGDH (phosphoglycerate dehydrogenase) | 26227 |
| NPY6R | NPY6R (neuropeptide Y receptor Y6 (pseudogene)) | 4888 |
| OR2C3 | OR2C3 (olfactory receptor family 2 subfamily C member 3) | 81472 |
| GCNT2 | GCNT2 (glucosaminyl (N-acetyl) transferase 2 (I blood group)) | 2651 |
| OR7C2 | OR7C2 (olfactory receptor family 7 subfamily C member 2) | 26658 |
| VN1R5 | VN1R5 (vomeronasal 1 receptor 5 (gene/pseudogene)) | 317705 |
| ADGRF4 | ADGRF4 (adhesion G protein-coupled receptor F4) | 221393 |
| GPR26 | GPR26 (G protein-coupled receptor 26) | 2849 |
| VN1R10P | VN1R10P (vomeronasal 1 receptor 10 pseudogene) | 387316 |
| VN1R2 | VN1R2 (vomeronasal 1 receptor 2) | 317701 |
| GNRHR2 | GNRHR2 (gonadotropin releasing hormone receptor 2 (pseudogene)) | 114814 |
| TAS1R1 | TAS1R1 (taste 1 receptor member 1) | 80835 |
| OR7E5P | OR7E5P (olfactory receptor family 7 subfamily E member 5 pseudogene) | 219445 |
| PCDH15 | PCDH15 (protocadherin related 15) | 65217 |
| SLC26A7 | SLC26A7 (solute carrier family 26 member 7) |
We assayed 42 receptor mRNAs that they neglected. Using a threshold of 32 cycles as the cutoff for “expression”, they found 126 GPCR transcripts in monocytes and we found 228 transcripts in 100 ng of RNA. We both detected 111 of the receptor RNAs. They found 4 that we omitted and 11 that we assayed but did not detect. We assayed 54 receptor RNAs that they did not look for and detected half of them. We failed to detect 11 of the receptors that they found, and they did not see 9 of the receptors that we saw. If we increase their threshold for expression to 35 cycles, they detected 165 GPCR mRNAs, 133 of which we also saw. We did not assay 6 of the receptors they found and did not detect 26 of them. Perhaps this should have been detected. The comparison between both assays is summarized in Fig. 3. The assays that both groups used were difficult and time consuming. We looked at single cell samples. Had both of us studied more cells, we might have seen additional overlap that may have been more extensive. The non-abundant receptor mRNAs that we did detect may or may not have been translated. This would have to be determined for one receptor at a time. On the other hand, their presence may hint that they have significant roles. We did not study cells harvested from non-normal patients. This might have revealed additional transcripts or a shift in the abundance of those we detected. The full comparison between the studies and results are summarized in the G protein coupled receptor transcripts in human immune cells and platelets database8 in Excel file format.
Acknowledgements
The project was supported by the NIH/NIDCR DIR intramural project # ZIA DE000714 and # ZIC DC000086. The authors want to thank Lynn-Vitale Cross for her help with preparing Tables and lists.
Author contributions
A.H. assayed the GPCRs. F.L., K.H., J.K., C.C. and E.Ma provided the cells that were studied, and descriptions of the purification methods used. DM did the informatics. M.J.B. and E.Me designed the study and wrote the manuscript with help from all the other authors.
Funding
Open access funding provided by the National Institutes of Health.
Code availability
No custom code was used in data analysis. The data are shown in excel sheets in pivot tables and manual curation.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Arne Hansen, Daniel Martin.
These authors jointly supervised this work.
Contributor Information
Daniel Martin, Email: Daniel.martin@nih.gov.
Eva Mezey, Email: mezeye@nih.gov.
References
- 1.Hansen, A. et al. Sensitive and specific method for detecting G protein-coupled receptor mRNAs. Nat Methods4, 35–37, 10.1038/nmeth977 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Mayer, B. et al. Vasopressin stimulates the proliferation and differentiation of red blood cell precursors and improves recovery from anemia. Sci Transl Med910.1126/scitranslmed.aao1632 (2017). [DOI] [PMC free article] [PubMed]
- 3.Cota, M. et al. Selective inhibition of HIV replication in primary macrophages but not T lymphocytes by macrophage-derived chemokine. Proc Natl Acad Sci USA97, 9162–9167, 10.1073/pnas.160359197 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinelli, E. et al. HSV-2 infection of dendritic cells amplifies a highly susceptible HIV-1 cell target. PLoS Pathog7, e1002109, 10.1371/journal.ppat.1002109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groot-Kormelink, P. J., Fawcett, L., Wright, P. D., Gosling, M. & Kent, T. C. Quantitative GPCR and ion channel transcriptomics in primary alveolar macrophages and macrophage surrogates. BMC Immunol13, 57, 10.1186/1471-2172-13-57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res37, W305–311, 10.1093/nar/gkp427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seal, R. L. et al. Genenames.org: the HGNC resources in 2023. Nucleic Acids Res51, D1003–D1009, 10.1093/nar/gkac888 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen, A. et al. G protein coupled receptor transcripts in human immune cells and platelets database. figshare10.6084/m9.figshare.c.7194810.v1 (2024). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
Data Availability Statement
No custom code was used in data analysis. The data are shown in excel sheets in pivot tables and manual curation.