Abstract
The assessment of ctDNA has emerged as a minimally invasive avenue for molecular diagnosis and real-time tracking of tumor progression in NSCLC. However, the evaluation of ctDNA by amplicon-based NGS has been not endorsed by all the healthcare systems and remains to be fully integrated into clinical routine practice. To compare tissue single-gene with plasma multiplexed testing, we retrospectively evaluated 120 plasma samples from 12 consecutive patients with advanced non-squamous NSCLC who were part of a prospective study enrolling treatment-naïve patients and in which tissue samples were evaluated using a single-gene testing approach. While the plasma ctDNA detection of EGFR and BRAF mutations had an acceptable level of concordance with the archival tissue (85%), discordance was seen in all the patients in whom ALK alterations were only detected in tissue samples. Among six responders and six non-responders, early ctDNA mutant allelic frequency (MAF) reduction seemed to predict radiologic responses and longer survival, whereas increasing MAF values with the emergence of co-mutations like BRAFV600E, KRASG12V or TP53M237I seemed to be an early indicator of molecular and radiologic progression. This report using an amplicon-based NGS assay on ctDNA underscores the real-life need for plasma and tissue genotyping as complementary tools in the diagnostic and therapeutic decision-making process.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-73046-y.
Keywords: NSCLC, Liquid biopsy, CtDNA, NGS, Monitoring
Subject terms: Lung cancer, Molecular medicine, Oncology
Introduction
Despite the expanding adoption of targeted and immunotherapy-based interventions, the prognosis of patients with advanced non-small-cell lung cancer (NSCLC) remains regrettably grim1. In the era of precision oncology, the introduction of liquid biopsy has enabled a paradigmatic transformation in the care of such patients, offering a promising solution to the limitations of traditional tissue biopsies and establishing itself as a valuable diagnostic tool in current clinical practice2. Beyond its clinical applicability for diagnostic purposes, the integration of liquid biopsy testing holds the potential to serve as a valuable tool in monitoring clinical outcomes and prognostication3,4. Specifically, the assessment of circulating tumor DNA (ctDNA), a part of cell-free DNA (cfDNA) shed from tumor sites into the bloodstream of cancer patients, has emerged as a minimally invasive avenue for molecular diagnosis and real-time tracking of tumor progression at the time of acquired resistance, with ctDNA kinetics holding promise as an indicator of treatment efficacy especially in patients with oncogene-driven NSCLC5. Despite the mounting body of evidence within the scientific literature, the serial monitoring of ctDNA for predicting radiological responses to conventional treatments has been not endorsed by all the healthcare systems and remains to be fully integrated into clinical routine practice6.
Even the most recent clinical trials have only adopted polymerase chain reaction (PCR)- and immunohistochemistry (IHC)-based single-gene testing techniques for assessing the molecular status of tissue samples7–9. Such targeted methodologies employ specific probes to identify known mutations, do not encompass the entire spectrum of oncogene addictions, and thus fail to detect less prevalent yet clinically significant genomic alterations. Furthermore, these methods have limited multiplexing capabilities, thereby constraining the concurrent analysis of other emerging biomarkers10. To address such limitations, the adoption of plasma next-generation sequencing (NGS) proves promising, as it saves tissue while facilitating the sequencing of extensive genomic regions or multiple exons on ctDNA samples11. Despite several research groups have reported results on the prognostic significance of ctDNA in NSCLC while many pan-cancer liquid biopsy panels are commercially available, however, liquid biopsies remain not widely adopted or reimbursed12, while only two hybrid capture-based cfDNA technologies, such as Guardant360® CDx (Guardant Health, Inc.; Redwood, CA, USA) and FoundationOne® Liquid CDx (Foundation Medicine, Inc.; Cambridge, MA, USA), have granted the FDA approval13. Target enrichment, generally achieved by hybrid capture- or amplicon-based approaches, represents a crucial step in the targeted NGS sequencing workflow, significantly influencing the success, efficiency, and accuracy of variant detection14. To date, no multiplex amplicon-based liquid biopsy assays have yet received full FDA approval.
Hence, there is a pressing need for additional data to validate the role of ctDNA by amplicon-based NGS in forecasting and tracking clinical outcomes in the real-life context of lung cancer. This real-world report, presented herein, conducts the diagnostic evaluation along with the retrospective assessment of longitudinal plasma samples by amplicon-based NGS, compared to baseline tissue single-gene testing, to explore the potential of ctDNA as a predictor of response and survival at the time of first disease restaging in treatment-naive patients with advanced NSCLC undergoing standard first-line treatments.
Materials and methods
Patient samples and study design
To compare tissue single-gene with plasma multiplexed testing, we retrospectively evaluated 12 consecutive patients with advanced non-squamous lung cancer who were part of a prospective study enrolling treatment-naïve patients at the Paolo Giaccone University Hospital, Palermo (Italy) and in which formalin-fixed paraffin‐embedded (FFPE) tissue samples were evaluated according to clinical practice using a targeted single-gene testing approach (real time-PCR and IHC for the detection of EGFR/BRAF hotspot mutations and ALK/ROS1 alterations, respectively) by a distinct referring pathology unit, as previously described15. Real time-PCR was performed on FFPE specimens by amplification of 15–30 ng of extracted DNA using the EasyPGX® Ready EGFR and BRAF kits on EasyPGX® qPCR (Diatech Pharmacogenetics), according to the manufacturer’s instructions. These tests allowed the detection of the most clinically relevant hotspot alterations, as reported in Supplementary Table 1. Data were automatically analyzed as positive or negative results using the EasyPGX® analysis software version 4.0.10 (Diatech Pharmacogenetics)16. Paired blood samples were collected at baseline (T0) and following the first radiologic evaluation of disease within 12 ± 1 weeks (T1 or W12) during the treatment course. The collected plasma samples were used to isolate, quantify, and analyze cfDNA using a DNA/RNA-based NGS testing approach both at T0 and T1. All the patients underwent a computerized tomography scan at T0-T1 and were classified as radiologic responders (complete (CR) or partial response (PR)) or non‐responders (stable disease (SD) or progressive disease (PD)) according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.117. Patients with oligo-progressive disease (oligo-PD), defined as limited metastatic areas progressing on first-line treatment and treated using local radiation therapy followed by continued targeted agents according to clinical practice, were labeled as non-responders18. Plasma molecular response or progression was evaluated according to the reduction/clearance or increase/persistence of the maximal ctDNA mutant allelic fraction (MAF), respectively. The study was conducted following the Declaration of Helsinki, and the protocol was approved by The Ethics Committee Palermo I (AIFA code CE 150109).
Plasma separation, cfDNA quantification, and molecular analysis
According to the standard procedure6, blood samples (5 mL) were collected into K2 EDTA tubes for times ranging from 15 min to less than 2 h at room temperature and centrifuged twice (10 min at 1,200 x g; 10 min at 16,000 x g) using a refrigerated centrifuge (4 °C) for plasma collection. The collected plasma samples were stored at -80 °C until further processing or immediately used to extract cfDNA. We extracted cfDNA from 2 mL of plasma using a QIAamp Circulating Nucleic Acid Kit (Qiagen) and quantified it in terms of ng/µl using a Qubit™ dsDNA HS Assay Kit. Namely, 20 ng of isolated cfDNA was analyzed using Oncomine™ Lung cfTNA Research Assay while, according to the manufacturer’s recommendations, we accepted an input range of 1–50 ng of extracted cfDNA to create a successful library. According to the manufacturer’s instructions ant external quality assessment for our laboratory, a contrived analytic positive control was used to monitor each batch for quality assurance. The analytical performance of each sequencing run was inspected by evaluating the technical parameters (reads, medium coverage depth, uniformity of coverage). Quality control check for single nucleotide variant/indel target regions was based on molecular coverage. As regards the detection of fusion and exon skipping amplicons, the panel provided five assays to perform the quality check: two non-fused process control genes (HMBS and TBP) consistently detected in cell-free nucleic acid (cfNA) extracts and other three assays (one with the skipping between exon 13 and 15, and two wild type assays) were used to inform the variant call quality check of fusions and MET exon 14 skipping, respectively. At least one control from each group must have passed a molecular count > 2. The libraries were quantified using an Ion Library TaqMan™ quantification kit on a QuantStudio7 Pro Real-Time PCR System (Applied Biosystems) using Design and Analysis Software v2.4.3. The libraries were diluted to 30 ng and pooled together. The pool was charged on Ion 510 and Ion 520 and Ion 530 Chef reagents (Thermo Fisher Scientific); then, an automatic system (Ion Chef instrument, Thermo Fisher Scientific) was used to automatically charge the Ion 530 chip with the pooled libraries according to the manufacturer’s instructions.
Using 20 ng of cfNAs, the specificity of this kit was 99.0% at 0.1% of the limit of detection (LoD). The data were tested on an amplicon-based sequencing platform Ion Torrent S5™ System. Oncomine TagSeq Lung v2 Liquid Biopsy‐w2.5‐Single Sample was the workflow applied for the analysis of cfNAs samples. To test the reliability of the data for cfNA sequencing, we used the following thresholds: total mapped reads > 3 M, median read coverage Avg 40,000 – Min > 25,000, median molecular coverage > 2500. The data of DNA sequencing were analyzed with Ion Torrent TorrentSuite™ (TS, version 5.18) using the Coverage Analysis and Variant Caller plugins. The LoD of single nucleotide variants/indels detected was calculated by the level of molecular amplicon coverage and displayed for each variant call. Molecular coverage had to be at least 2 with a minimum detection cutoff frequency of 0.035%. To be reported, fusion and exon skipping amplicons must have > 2 molecular counts. The sequencing data were categorized by relevance with the related percentage of allelic frequency as annotated by Ion Reporter Software v5.18 applying the Variant Matrix Summary (5.18) filter chains for default use.
Statistical considerations
The categorical clinical-pathological variables of the population enrolled in the study were described as absolute numbers (N) and percentages (%). To describe the treatment efficacy, progression-free survival (PFS) was computed as the time from treatment start to disease progression or death from any cause; overall survival (OS) was computed as the period from treatment initiation to death from any cause. To assess the diagnostic accuracy of liquid biopsy, contingency tables were constructed to describe the results of overall baseline tissue and plasma testing and subsequently for each gene of interest (EGFR, ALK, and BRAF). The genomic status of tumor tissue was considered as a gold standard whereas ctDNA evaluation was considered as an experimental group. All analyses were performed using SPSS software (ver 27.0). For the diagnostic accuracy analyses, the following definitions were considered: true positive (TP) as the number of patients with a mutation discovered in both tissue and liquid biopsy, true negative (TN) as the number of patients with a mutation not discovered in either the tissue or liquid biopsy, false positive(FP) as the number of patients with a mutation not found in the tissue but found in liquid biopsy, and finally false negative (FN) as the number of patients with a positive tissue biopsy and negative liquid biopsy. Consequently, sensitivity and specificity were calculated as the ratio between TP and the sum of TP and FN × 100 (TP/[TP + FN] × 100) and the ratio between TN and the sum of TN and FP × 100 (TN/[TN + FP] × 100) respectively. Lastly, concordance between ctDNA and tissue was evaluated as ([TP + TN]/ [TP + FN + TN + FN]) × 100.
Results
Among 73 patients prospectively enrolled in the real-world cohort, in this report, we retrospectively focused on consecutive non-squamous lung cancer patients who received baseline single-gene testing on archival tissue and had sufficient circulating biospecimens. Briefly, a total of 120 liquid biopsy plasma samples were collected isolating cfDNA from 12 patients at baseline with paired available plasma samples at disease radiologic re-evaluation. Systemic treatment was performed according to clinical indication and routine practice. Clinical-pathological characteristics of patients included in our analysis are listed in [Supplementary Table 2].
Diagnostic accuracy of NGS plasma ctDNA at baseline
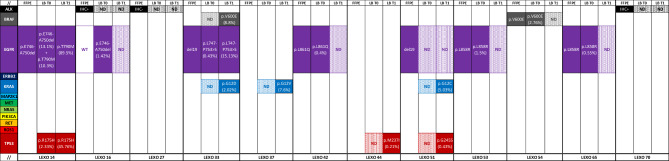
In our patients’ cohort, the molecular landscape determined by tissue single-gene testing identified four distinct profiles: six patients presented with EGFR mutations (LEXO14, LEXO33, LEXO42, LEXO51, LEXO53, and LEXO65), three patients with ALK IHC positivity (LEXO16, LEXO27, and LEXO70), one patient with a BRAF mutation (LEXO54), and two classified as non-oncogene addicted (LEXO37 and LEXO44) ([Table 1], [Fig. 1]).
Table 1.
Predictive molecular pathology of the included patients at baseline (T0) and first disease restaging (T1) undergoing first-line treatments.
| ID | Tissue single-gene testing (RT-PCR, IHC) | Treatment | ctDNA T0 (NGS) |
MAF T0 (%) | ctDNA T1 (NGS) |
MAF T1 (%) | CT SCAN | cfDNA T0 (ng/µl) | cfDNA T1 (ng/µl) |
|---|---|---|---|---|---|---|---|---|---|
| LEXO 14 | p.E746_A750del, EGFR | afatinib |
p.E746_A750del, EGFR p.T790M, EGFR p.R175H, TP53 |
13.1% 10.3% 2.33% |
p.E746_A750del, EGFR; p.T790M, EGFR; p.R175H, TP53 |
0% 89.5% 45.76% |
PD | 0.28 | 0.42 |
| LEXO 16 | ALK+ | alectinib | p.E746_A750del, EGFR | 1.42% | ND | 0% | PD | 0.23 | 0.43 |
| LEXO 27 | ALK+ | alectinib | ND | N.A. | ND | N.A. | PR | 0.47 | 0.34 |
| LEXO 33 | p.E746_A750del, EGFR | osimertinib | p.L747_P753delinsS, EGFR | 0.43% |
p.L747_P753delinsS, EGFR; p.V600E, BRAF; p.G12D, KRAS |
15.13% 8.8% 2.02% |
PD | 0.84 | 0.54 |
| LEXO 37 | - | CT + IO | ND | N.A. | p.G12V, KRAS | 7.60% | PD | 0.63 | 4.01 |
| LEXO 42 | p.L861Q, EGFR | osimertinib | p.L861Q, EGFR | 0.4% | ND | 0% | PR | 0.59 | 0.38 |
| LEXO 44 | - | CT + IO | ND | N.A. | p.M237I, TP53 | 0.21% | PD | 0.37 | 0.78 |
| LEXO 51 | p.E746_A750del, EGFR | osimertinib | ND | N.A. |
p.G12C, KRAS; p.G245S, TP53 |
5.03% 0.43% |
PD | 0.24 | 0.46 |
| LEXO 53 | p.L858R, EGFR | osimertinib | p.L858R, EGFR | 1.5% | ND | 0% | PR | 0.92 | 0.46 |
| LEXO 54 | p.V600E, BRAF |
dabrafenib+ trametinib |
p.V600E, BRAF | 2.76% | ND | 0%. | PR | 0.61 | 0.91 |
| LEXO 65 | p.L858R, EGFR | osimertinib | p.L858R, EGFR | 0.55% | ND | 0% | PR | 0.45 | 0.57 |
| LEXO 70 | ALK+ | alectinib | ND | N.A. | ND | N.A. | PR | 8.07 | 2.74 |
RT-PCR, reverse transcriptase-polymerase chain reaction; IHC, immunohistochemistry; ctDNA, circulating tumor DNA; cfDNA, circulating cell-free DNA; NGS, next-generation sequencing; MAF, mutant allelic frequency; CT, computed Tomography; CHT + IO, platinum doublet chemotherapy (carboplatin and pemetrexed) plus pembrolizumab; -, negative single-gene testing by both RT-PCR and IHC; PD, radiologic progressive disease; PR, radiologic partial response; ND, not detected; N.A., not available.
Fig. 1.
Overview of the predictive molecular pathology characterization of the enrolled patients including the mutant allelic frequencies (in brackets) of liquid biopsy ctDNA variants detected by NGS. FFPE, formalin-fixed, paraffin-embedded tissue; LB, liquid biopsy ctDNA; T0, baseline; T1, disease re-staging; WT, wild-type; N.D., not detected.
At baseline, genomic testing showed a tissue-plasma concordance of 85% in the overall population, with a sensitivity and positive predictive value of 85% whereas presenting with a specificity and negative predictive value of 75%, respectively. According to genomic subgroups, EGFR and BRAF mutations showed the best tissue-plasma concordance (85%) whereas ALK alterations presented with a weaker concordance (75%) ([Supplementary Tables 3–6]).
Baseline amplicon-based NGS testing on ctDNA confirmed the presence of tissue EGFR mutations in all patients except for LEXO51 who presented with intrathoracic disease only ([Figure 1]). Namely, compared to the canonical exon 19 in-frame deletions identified by tissue RT-PCR, LEXO33 presented on plasma a distinct and less frequently detected EGFR variant (L747_P753delinsS), whereas LEXO14 exhibited an additional de novo EGFRT790M along with a TP53 point mutation. Moreover, we successfully detected a classical BRAFV600E both on tissue and plasma. While the plasma ctDNA detection of EGFR and BRAF point mutations had an acceptable level of concordance with the archival tissue, discordance was seen in all the patients in whom ALK alterations were only detected in tissue samples by IHC ([Figure 1], [Table 1] [Supplementary Tables 3-6]).
Prognostic significance of longitudinally monitoring NSCLC using ctDNA
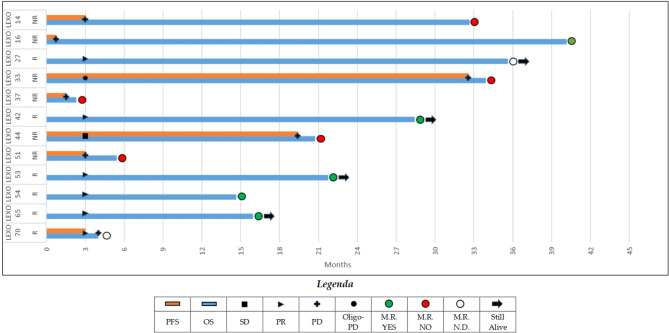
In the overall cohort, we identified six responders and six non-responders according to RECIST 1.1. radiologic evaluation. PFS and OS according to radiologic and molecular response are shown in [Fig. 2].
Fig. 2.
Swimmer plot depicting survival of the included patients according to radiologic (lines) and molecular (circles) response. PFS, progression-free survival; OS, overall survival; SD, stable disease; PR, radiologic partial response; PD, radiologicprogressive disease; M.R., molecular response; N.D., not detected.
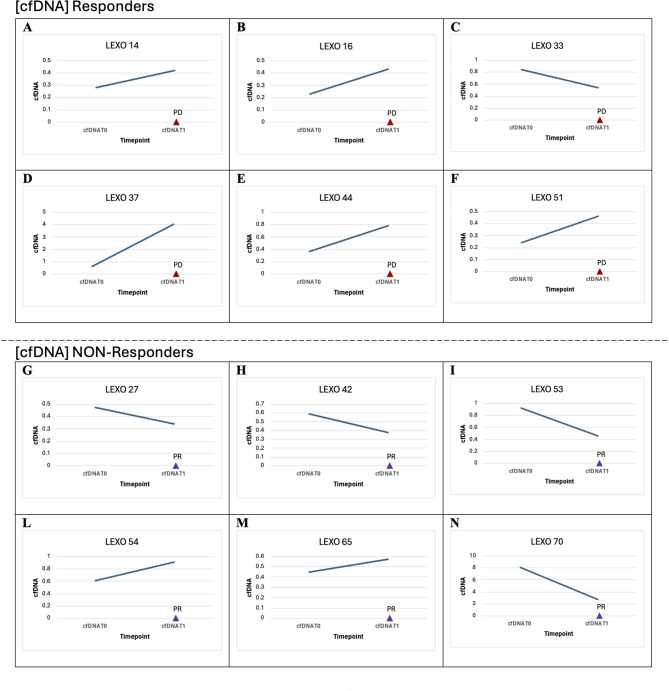
Among radiologic responders, four patients (the EGFR-positive LEXO42, LEXO53, LEXO65 and the BRAF-mutant LEXO54) experienced a detectable ctDNA MAF reduction showing a durable and ongoing response ([Figs. 1 and 2]). Significantly, a ctDNA response was not evaluable in two tissue ALK-positive patients (LEXO27 and LEXO70) that, however, had a favorable radiologic response paralleled by significantly decreasing cfDNA levels ([Figs. 2 and 3]). Of note, patient LEXO70, despite showing a radiologic partial response with no detectable molecular assessment, unfortunately, died soon because of disseminated intravascular coagulation.
Fig. 3.
Dynamics of cfDNA among responders and non-responders according to radiologic restaging. cfDNA, circulating cell-free DNA; T0, baseline; T1, first disease restaging; PD, radiologic progressive disease; PR, radiologic partial response.
Among radiologic non-responders, LEXO14 had a systemic PD on afatinib and received second-line osimertinib whereas LEXO33 on first-line osimertinib experienced an oligo-PD disease that was treated according to clinical practice (Table 1). Intriguingly, in these patients, ctDNA monitoring unveiled increasing on-target allelic frequencies and additional off-target alterations (such as BRAF, KRAS and TP53 point mutations) that, following a sequential single-gene approach, were not initially detected on tissue at baseline ([Figure 1]).
Likewise, three patients (the non-oncogene addicted LEXO37 and LEXO44 together with the EGFR-mutant LEXO51) experienced molecular progression with the detection of additional KRAS and TP53 mutations at T1, progressing on standard treatments and presenting with very poor long-term survival compared to the other cohort patients ([Figs. 1 and 2]).
Considering the tissue ALK-positivity, patient LEXO16 started an ALK inhibitor but rapidly presented a clinically symptomatic and radiologic progression before the planned radiologic restaging at W12. Surprisingly, the retrospective evaluation of plasma ctDNA at baseline revealed a classical EGFR exon 19 E746_A750 deletion that was not previously detected by tissue RT-PCR. Of note, the patient harbored an impressive EGFR ctDNA MAF of 1.42% at baseline ([Figure 1]) and, therefore, was eligible to receive an EGFR inhibitor. The patient responded favorably to osimertinib at first restaging, thus confirming the clinical utility and the diagnostic robustness of plasma NGS compared to tissue single-gene testing.
Although ctDNA and radiologic responses were overall concordant, however, the dynamics of cfDNA showed some notable exceptions such as patients LEXO54 and LEXO65 showing radiologic and ctDNA response together with cfDNA increasing levels or patient LEXO33 having radiologic and ctDNA progression with cfDNA decreasing levels ([Table 1], [Fig. 3]).
Discussion
Despite being strongly recommended by scientific agencies19,20, the full implementation of tissue NGS in routine clinical practice remains limited whereas basic single-gene testing is widely available3,21. Further, the use of liquid biopsy to track cancer response remains challenging in the real-world setting with not yet universal reimbursement and uptake by all the healthcare systems22. Here, we described the analytical and clinical performance of a ctDNA multiplex amplicon-based assay that, comparing to the hybrid capture-based technique, features a quicker and less complex workflow while using low quality and quantity of nucleic acid input often present in the real-life clinic. Our case series highlighted the use of ctDNA NGS for confirming the standard tissue findings of conventional single-gene testing while further revealing additional plasma genomic alterations with significant implications in a real-world clinical setting. In this study, we retrospectively evaluated the plasma of patients who were part of a prospective study, showing that the ctDNA evaluation improved the baseline detection of actionable alterations (LEXO14, LEXO16, LEXO33) while enabling the effective tracking of clonal resistance (LEXO33, LEXO37, LEXO44, LEXO51) that would allow prompt patients enrollment in clinical trials.
In this report, the reliable diagnostic accuracy of plasma ctDNA using an amplicon-based NGS assay for DNA-based alterations such as EGFR and BRAF point mutations reaffirmed the performance of this technique on liquid biopsy in such oncogene-driven settings23. The inability to detect the EGFR mutation on plasma in one patient (LEXO51) with pleural effusion echoes findings from the literature, suggesting the notably lower sensitivity of ctDNA in patients with non-shedding intra-thoracic disease compared to those with distant metastases24. Conversely, in line with other recent discouraging results, detecting ALK fusions from plasma using an amplicon-based NGS assay remained challenging, even in high-volume cancers, suggesting the preferred use of hybrid capture-based sequencing in such cases25.
Consistently with literature26,27, compared to tissue single-gene evaluation, NGS applied to ctDNA offered a more nuanced view of the genomic landscape, enhancing our understanding of tumor heterogeneity and pinpointing clinically actionable targets, such as in the seminal case of LEXO16. This patient presented ALK-positive IHC staining on tissue but rapidly progressed on ALK inhibitor, while showing a plasma ctDNA EGFR deletion that was not previously detected by RT-PCR but promptly responded to osimertinib. Thus, ctDNA may play a role in replacing tissue tumor sampling and single-gene testing in some circumstances, as outlined by international recommendations, especially in oncogene-addicted patients28. In this case, since the detection of an impressive EGFR ctDNA MAF of 1.42% in lung cancer patient represented the example of a very unlikely false-positive finding, the liquid biopsy evaluation was valuable to prevent ineffective therapy and avoid unnecessary side effects, suggesting that in the real-world setting monitoring ctDNA molecular status could potentially reflect response before clinical progression or radiologic imaging29.
We then investigated whether ctDNA clearance or a certain degree of ctDNA kinetics reflected by on-treatment variations of MAF values would better correlate with radiologic response. Mostly in the resistance setting, dynamic molecular profiles captured by the serial monitoring of ctDNA using NGS revealed complexities in tumor evolution and therapeutic responses that would not have been identified by conventional single-gene techniques detecting only known hotspot variants on tissue. Here, early ctDNA MAF reduction during first-line standard treatments seemed to predict radiologic responses and longer survival, whereas increasing MAF values with the emergence of co-mutations like BRAFV600E, KRASG12V or TP53M237I seemed to be an early indicator of molecular and radiologic progression, as clinically corroborated by the later aggressive behavior. Notably, concomitant mutations in NSCLC typically portend a poorer prognosis30–32, suggesting the earlier use of ctDNA as a minimally invasive and robust tool for providing crucial insights into potential diagnostic and therapeutic adjustments in the clinic. Notably, considering the negative prognostic impact of co-mutations and the adoption of only single-gene testing on tissue in randomized clinical trials, one could argue about the real-life need for monitoring and adapting cancer treatments using NGS on ctDNA to significantly improve clinical outcomes33. Of note, both LEXO14 and LEXO33 experienced a radiographic progression that matched increasing on- and off-target MAF values at T1, despite showing a relatively long survival that was eventually influenced by second-line treatments. In LEXO54, the sensitivity for the detection of BRAFV600E and monitoring of response to dabrafenib and trametinib reaffirms the diagnostic accuracy of ctDNA for such patients. However, the increase in cfDNA levels, despite a partial radiologic response, further suggests that cfDNA levels might not specifically reflect tumor burden, possibly indicating that other biological processes like apoptosis, necrosis and active secretion are at play, as often described34. Since all the molecular responders showed an ongoing and responding disease whereas molecular non-responders presented with a progressing or high burden disease, these results demonstrated the analytical and clinical validity of an amplicon-based NGS plasma assay in the real-world setting while further confirming the clinical utility of liquid biopsy for the longitudinal monitoring of patients with advanced NSCLC receiving first-line treatments. Hence, this approach can significantly impact the real-world patient management by adding broader molecular profiling and early prognostics for treatment stratification and early access to actively enrolling clinical trials6.
While the exploratory nature of our analyses was hindered by the absence of NGS-based tissue testing, these results underscore the practical challenges and opportunities associated with implementing a liquid biopsy-informed approach for treatment choice and response assessment. While our study emphasizes the potential of liquid biopsy to detect a broader spectrum of genomic variants, it’s important to acknowledge certain limitations that provide direction for future research. First, the retrospective nature and the small sample size of the study necessitate further larger, multi-center validation cohorts. Secondly, the phenomenon of clonal hematopoiesis, which can lead to the presence of non-tumor-related mutations in the bloodstream, poses a challenge to liquid biopsy accuracy, potentially resulting in false-positive results35. In this context, plasma tumor fraction analysis could serve as a potential prognostic and predictive tool to tailor therapy intensity based on individual tumor biology, reducing false-positive ctDNA results while obviating the need for confirmatory tissue testing in selected patients36–38.
Conclusions
These findings accentuate the diagnostic and monitoring prowess of liquid biopsy, which in this instance provided an early indication of on-treatment tumor evolution using an amplicon-based NGS assay, thereby informing potential shifts in therapeutic strategy. This report would add compelling insights into the evolving landscape of advanced NSCLC, underscoring the need for plasma ctDNA analysis and tissue genotyping as complementary tools in the diagnostic and therapeutic decision-making process.
Liquid biopsy can complement existing tissue biomarker testing, particularly for identifying more patients who could benefit from first-line targeted treatment by increasing the number of patients with a proper and well-informed molecular diagnosis. Liquid biopsy may also help identify patients for appropriate second-line targeted therapy, especially through detection of circulating markers of resistance or in patients who did not receive frontline biomarker testing.
This study strengthens the application of ctDNA molecular response assessment as an enrichment strategy. By early identifying patients exhibiting molecular disease progression, this approach has the potential to mitigate the heterogeneity inherent to clinical trials, creating a more homogenous target population and thereby opening a therapeutic window of opportunity. This window would facilitate earlier intervention and potentially overcome primary therapeutic resistance, ultimately leading to improved clinical outcomes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
V.G., A.Go., and TD.BR. contributed to the current work under the Doctoral Programme in Experimental Oncology and Surgery, University of Palermo. The authors thank Mrs Marzia D’alessandro for the English language revision.
Author contributions
Conceptualization: V.G., T.D.B.R., N.B., V.B.; Data curation: T.D.B.R., F.P., G.T.; Formal analysis: V.G., N.B., A.Go., G.R.; Investigation: V.G., N.B., A.Ga.; Methodology: V.G., N.B.; Project administration: V.G., N.B., A.R., V.B.; Resources: V.G., N.B., A.R., V.B.; Software: V.G., A.Ga.; Supervision: L.I., G.B., F.F., G.T., U.M., A.R., V.B., A.Ga.; Validation: V.G., N.B., G.R., F.F., U.M., V.B.; Visualization: V.G., N.B., A.Go.; Writing – original draft: V.G., T.D.B.R., N.B., A.Go.; Writing – review & editing: T.D.B.R., G.R., L.I., G.B., F.F., G.T., U.M., A.R., V.B.; All authors have read and agreed to the published version of the manuscript.
Funding
L.I., G.B., A.R. and A.Ga. were supported by the Piano Nazionale di Ripresa e Resilienza (PNRR) project - Italian Network of excellence for advanced diagnosis (INNOVA) - PNC-E3-2022-23683266 PNC-HLS-DA (C43C22001630001). The remaining authors have not declared a specific grant for this research from any funding agency in the public, commercial, or not-for-profit sectors.
Data availability
Data could be available upon reasonable request to the authors.
Declarations
Competing interests
F.P. received personal fees (as speaker bureau or advisor) from Menarini. G.T. received personal fees (as speaker bureau or advisor) from Roche, MSD, Pfizer, Boehringer Ingelheim, Eli Lilly, BMS, GSK, Menarini, AstraZeneca, Amgen and Bayer, all unrelated to the current work. U.M. received personal fees (as consultant and/or speaker bureau) from Boehringer Ingelheim, Roche, MSD, Amgen, Thermo Fisher Scientific, Eli Lilly, Diaceutics, GSK, Merck and AstraZeneca, Janssen, Diatech, Novartis and Hedera, all unrelated to the current work. The remaining authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Institutional review board
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by Palermo 1 Institutional Ethic Review Board (Statement No. 02/2020, approved on 19 February 2020, AIFA code CE 150109).
Informed consent
Informed consent was obtained from all subjects involved in the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Valerio Gristina, Tancredi Didier Bazan Russo and Nadia Barraco.
These authors jointly supervised this work: Viviana Bazan and Antonio Galvano.
Contributor Information
Giuseppe Badalamenti, Email: giuseppe.badalamenti@unipa.it.
Antonio Russo, Email: antonio.russo@usa.net.
References
- 1.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023, (in eng). CA Cancer J. Clin.73(1), 17–48. 10.3322/caac.21763 (Jan 2023). [DOI] [PubMed]
- 2.Santini, D. et al. Network approach in liquidomics landscape, (in eng). J. Exp. Clin. Cancer Res.42(1), 193. 10.1186/s13046-023-02743-9 (Aug 04 2023). [DOI] [PMC free article] [PubMed]
- 3.Pisapia, P. et al. A narrative review on the implementation of liquid biopsy as a diagnostic tool in thoracic tumors during the COVID-19 pandemic. Mediastinum. 5, 27. 10.21037/med-21-9 (2021). (in eng). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cammarata, G. et al. Extracellular Vesicles-ceRNAs as Ovarian Cancer Biomarkers: Looking into circRNA-miRNA-mRNA Code, (in eng), Cancers (Basel), vol. 14, no. 14, Jul 13 doi: (2022). 10.3390/cancers14143404 [DOI] [PMC free article] [PubMed]
- 5.Gristina, V. et al. Navigating the liquid biopsy minimal residual disease (MRD) in non-small cell lung cancer: making the invisible visible, (in eng). Crit. Rev. Oncol. Hematol.182, 103899. 10.1016/j.critrevonc.2022.103899 (Feb 2023). [DOI] [PubMed]
- 6.Russo, A. et al. Jun., The molecular profiling of solid tumors by liquid biopsy: a position paper of the AIOM-SIAPEC-IAP-SIBioC-SIC-SIF Italian Scientific Societies, (in eng), ESMO Open, vol. 6, no. 3, p. 100164, doi: (2021). 10.1016/j.esmoop.2021.100164 [DOI] [PMC free article] [PubMed]
- 7.Karachaliou, N. et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial, (in eng), JAMA Oncol, vol. 1, no. 2, pp. 149 – 57, May doi: (2015). 10.1001/jamaoncol.2014.257 [DOI] [PubMed]
- 8.Wu, Y. L. et al. Osimertinib in Resected, (in eng). N Engl. J. Med.383, 18, pp. 1711–1723, Oct 29 2020, 10.1056/NEJMoa2027071 [DOI] [PubMed]
- 9.Gregg, J. P., Li, T. & Yoneda, K. Y. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey, (in eng). Transl Lung Cancer Res.8 (3), 286–301. 10.21037/tlcr.2019.04.14 (Jun 2019). [DOI] [PMC free article] [PubMed]
- 10.Malapelle, U. et al. Standardized and simplified reporting of next-generation sequencing results in advanced non-small-cell lung cancer: Practical indications from an Italian multidisciplinary group, (in eng), Crit Rev Oncol Hematol, vol. 193, p. 104217, Jan doi: (2024). 10.1016/j.critrevonc.2023.104217 [DOI] [PubMed]
- 11.Xie, J. et al. Plasma ctDNA increases tissue NGS-based detection of therapeutically targetable mutations in lung cancers, (in eng). BMC Cancer. 23 (1), 294. 10.1186/s12885-023-10674-z (Mar 31 2023). [DOI] [PMC free article] [PubMed]
- 12.Smeltzer, M. P. et al. The International Association for the study of Lung Cancer Global Survey on Molecular Testing in Lung Cancer, (in eng). J. Thorac. Oncol.15(9), 1434–1448. 10.1016/j.jtho.2020.05.002 (Sep 2020). [DOI] [PubMed]
- 13.Vasseur, D. et al. Next-Generation Sequencing on Circulating Tumor DNA in Advanced Solid Cancer: Swiss Army Knife for the Molecular Tumor Board? A Review of the Literature Focused on FDA Approved Test, (in eng), Cells, vol. 11, no. 12, Jun 11 doi: (2022). 10.3390/cells11121901 [DOI] [PMC free article] [PubMed]
- 14.Singh, R. R. Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology, (in eng), Diagnostics (Basel), vol. 12, no. 7, Jun 24 doi: (2022). 10.3390/diagnostics12071539 [DOI] [PMC free article] [PubMed]
- 15.Gristina, V. et al. Clinical Potential of Circulating Cell-Free DNA (cfDNA) for Longitudinally Monitoring Clinical Outcomes in the First-Line Setting of Non-Small-Cell Lung Cancer (NSCLC): A Real-World Prospective Study, (in eng), Cancers (Basel), vol. 14, no. 23, Dec 06 doi: (2022). 10.3390/cancers14236013 [DOI] [PMC free article] [PubMed]
- 16.Venetis, K. et al. Analytical Performance of Next-Generation Sequencing and RT-PCR on Formalin-Fixed Paraffin-Embedded Tumor Tissues for, (in eng), Cells, vol. 11, no. 22, Nov 09 doi: (2022). 10.3390/cells11223545 [DOI] [PMC free article] [PubMed]
- 17.Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1), (in eng), Eur J Cancer, vol. 45, no. 2, pp. 228 – 47, Jan doi: (2009). 10.1016/j.ejca.2008.10.026 [DOI] [PubMed]
- 18.Nguyen, K. T., Sakthivel, G., Milano, M. T., Qiu, H. & Singh, D. P. Oligoprogression in non-small cell lung cancer: a narrative review, (in eng). J. Thorac. Dis.14 (12), 4998–5011. 10.21037/jtd-22-536 (Dec 2022). [DOI] [PMC free article] [PubMed]
- 19.Mosele, F. et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group, (in eng), Ann Oncol, vol. 31, no. 11, pp. 1491–1505, Nov doi: (2020). 10.1016/j.annonc.2020.07.014 [DOI] [PubMed]
- 20.Hendriks, L. E. et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up, (in eng), Ann Oncol, vol. 34, no. 4, pp. 339–357, Apr doi: (2023). 10.1016/j.annonc.2022.12.009 [DOI] [PubMed]
- 21.Bayle, A. et al. ESMO study on the availability and accessibility of biomolecular technologies in oncology in Europe, (in eng), Ann Oncol, vol. 34, no. 10, pp. 934–945, Oct doi: (2023). 10.1016/j.annonc.2023.06.011 [DOI] [PubMed]
- 22.Malapelle, U. Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective., M. Tiseo, A. Vivancos, J. Kapp, M. J. Serrano, and M. Tiemann, Eds., ed: Journal of Molecular Pathology. 2(3):255–273., (2021).
- 23.Mack, P. C. et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: Analysis of over 8000 cases, (in eng), Cancer, vol. 126, no. 14, pp. 3219–3228, Jul 15 2020, 10.1002/cncr.32876 [DOI] [PMC free article] [PubMed]
- 24.Passiglia, F. et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis, (in eng), Sci Rep, vol. 8, no. 1, p. 13379, Sep 06 doi: (2018). 10.1038/s41598-018-30780-4 [DOI] [PMC free article] [PubMed]
- 25.Reclusa, P. et al. Translocation identification in RNA exosomal cargo ((in eng). Transl Cancer Res.8 no. Suppl 1), S76–S78. 10.21037/tcr.2018.11.35 (Jan 2019). [DOI] [PMC free article] [PubMed]
- 26.Malapelle, U. et al. TargetPlex FFPE-Direct DNA Library Preparation kit for SiRe NGS panel: an international performance evaluation study, (in eng). J. Clin. Pathol.75(6), 416–421. 10.1136/jclinpath-2021-207450 (Jun 2022). [DOI] [PubMed]
- 27.Galvano, A. et al. The diagnostic accuracy of, (in eng). Ther. Adv. Med. Oncol.14, 17588359221110162. 10.1177/17588359221110162 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolfo, C. et al. Liquid Biopsy for Advanced NSCLC: a Consensus Statement from the International Association for the study of Lung Cancer, (in eng). J. Thorac. Oncol.16(10), 1647–1662. 10.1016/j.jtho.2021.06.017 (Oct 2021). [DOI] [PubMed]
- 29.Kilgour, E., Rothwell, D. G., Brady, G. & Dive, C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance, (in eng), Cancer Cell, vol. 37, no. 4, pp. 485–495, Apr 13 2020, 10.1016/j.ccell.2020.03.012 [DOI] [PubMed]
- 30.Chang, S. C. et al. Concomitant Genetic Alterations are Associated with Worse Clinical Outcome in EGFR Mutant NSCLC Patients Treated with Tyrosine Kinase Inhibitors, (in eng), Transl Oncol, vol. 12, no. 11, pp. 1425–1431, Nov doi: (2019). 10.1016/j.tranon.2019.07.008 [DOI] [PMC free article] [PubMed]
- 31.Chen, M. et al. Concurrent Driver Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive Non-Small Cell Lung Cancer, (in eng), EBioMedicine, vol. 42, pp. 304–310, Apr doi: (2019). 10.1016/j.ebiom.2019.03.023 [DOI] [PMC free article] [PubMed]
- 32.Gristina, V. et al. Non-small cell lung cancer harboring concurrent egfr genomic alterations: a systematic review and critical appraisal of the double dilemma. 2(2), 173–196. 10.3390/jmp2020016 (2021).
- 33.Gristina, V. & Eze, C. Editorial: real-world data and real-world evidence in lung cancer, (in eng). Front. Oncol.14, 1436077. 10.3389/fonc.2024.1436077 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu, Z., Chen, H., Long, Y., Li, P. & Gu, Y. The main sources of circulating cell-free DNA: apoptosis, necrosis and active secretion, (in eng). Crit. Rev. Oncol. Hematol.157, 103166. 10.1016/j.critrevonc.2020.103166 (Jan 2021). [DOI] [PubMed]
- 35.Hu, Y. et al. False-positive plasma genotyping due to clonal hematopoiesis, (in eng). Clin. Cancer Res.24, 18, pp. 4437–4443, Sep 15 2018, 10.1158/1078-0432.CCR-18-0143 [DOI] [PubMed]
- 36.Reichert, Z. R. et al. Prognostic value of plasma circulating tumor DNA fraction across four common cancer types: a real-world outcomes study, (in eng). Ann. Oncol.34(1), 111–120. 10.1016/j.annonc.2022.09.163 (Jan 2023). [DOI] [PMC free article] [PubMed]
- 37.Russo, A. et al. The challenge of the Molecular Tumor Board empowerment in clinical oncology practice: a position paper on behalf of the AIOM- SIAPEC/IAP-SIBioC-SIC-SIF-SIGU-SIRM Italian Scientific societies, (in eng). Crit. Rev. Oncol. Hematol.169, 103567. 10.1016/j.critrevonc.2021.103567 (Jan 2022). [DOI] [PubMed]
- 38.Russo, A. et al. Sep., The tumor-agnostic treatment for patients with solid tumors: a position paper on behalf of the AIOM- SIAPEC/IAP-SIBioC-SIF Italian Scientific Societies, (in eng), Crit Rev Oncol Hematol, vol. 165, p. 103436, doi: (2021). 10.1016/j.critrevonc.2021.103436 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data could be available upon reasonable request to the authors.