Abstract
The standard method for identifying active Brown Adipose Tissue (BAT) is [18F]-Fluorodeoxyglucose ([18F]-FDG) PET/CT imaging, which is costly and exposes patients to radiation, making it impractical for population studies. These issues can be addressed with computational methods that predict [18F]-FDG uptake by BAT from CT; earlier population studies pave the way for developing such methods by showing some correlation between the Hounsfield Unit (HU) of BAT in CT and the corresponding [18F]-FDG uptake in PET. In this study, we propose training convolutional neural networks (CNNs) to predict [18F]-FDG uptake by BAT from unenhanced CT scans in the restricted regions that are likely to contain BAT. Using the Attention U-Net architecture, we perform experiments on datasets from four different cohorts, the largest study to date. We segment BAT regions using predicted [18F]-FDG uptake values, achieving 23% to 40% better accuracy than conventional CT thresholding. Additionally, BAT volumes computed from the segmentations distinguish the subjects with and without active BAT with an AUC of 0.8, compared to 0.6 for CT thresholding. These findings suggest CNNs can facilitate large-scale imaging studies more efficiently and cost-effectively using only CT.
Subject terms: Obesity, Computer science, Endocrine system and metabolic diseases
The standard method for identifying active brown adipose tissue is costly and exposes patients to radiation. Here, the authors show that convolutional neural networks can predict [18F]-FDG uptake by BAT from unenhanced CT scans and improve the segmentation accuracy compared to conventional CT thresholding.
Introduction
Personalized medicine is commonly associated with the field of oncology. However, it has already become clear from pre-clinical and clinical studies that metabolic diseases are diverse and complex, making them ideal targets for a personalized treatment approach, especially given the fact that obesity and type 2 diabetes have complex genetic backgrounds, which so far are only incompletely understood1. Moreover, the response to modern drugs such as glucagon-like peptide 1 (GLP-1) agonists for the treatment of diabetes2 or to Angiotensin-converting enzyme (ACE) inhibitors for the treatment of hypertension3 is significantly different among different ethnic groups, pointing towards clinically relevant consequences of the genetic background. While the field is evolving rapidly, concepts to target metabolic disease on a personalized level are still uncommon.
Recent research on adipose tissue has yielded promising results toward possible personalized strategies. Adipose tissue can be subdivided into two different types of adipocytes, namely, white and brown adipocytes4. White adipose tissue (WAT) is specialized for storing chemical energy in the form of triglycerides5. In contrast, brown adipose tissue (BAT) dissipates energy in the form of heat in a process called non-shivering thermogenesis through uncoupling protein 1 (Ucp1)6. In the past decade it has become obvious that white adipocytes can transform into another distinct type of energy expending adipocytes which have been called beige adipocytes. Moreover, energy expending adipose tissue depots usually consist of a mixture of brown, beige and white adipocytes7. To improve readability, we will use the term BAT for these depots throughout this paper. Since BAT is an energy-dissipating organ, its activation or deactivation could potentially be used to promote weight loss or gain and improve metabolic control. Over the last decade, researchers have been working on understanding the connection between BAT activity and metabolism, which may contribute to developing personalized treatment strategies for various metabolic diseases8–11.
One central finding, which has been reported numerous times, is the fact that the presence of BAT is extremely heterogeneous in the population, ranging from undetectable levels (because it is absent or inactive) to high levels (active BAT)12. Some studies suggest that active BAT may be partially genetically determined13. Caret et al.14 showed that dysfunction of BAT activity might be a critical factor in the pathogenesis of obesity. A more recent study reported that individuals with active BAT have a lower prevalence of cardio-metabolic diseases15. Furthermore, recent insights in molecular oncology suggest that BAT-deregulation in cancer patients may contribute substantially to cancer-cachexia and its entailed reduction in quality of life and early death16. One example of a personalized strategy is to activate BAT by the use of oral intake of bile acids17 or selective β218 or β3-agonists19 in the context of obesity, or suppress it by propranolol, a non-selective β-blocker20 in the context of cancer-cachexia. By gaining a deeper understanding of BAT through large-scale population studies, it may become possible to create stratified cohorts and develop further personalized treatment strategies targeting BAT.
Despite existing works showing some evidence of the link between BAT activity and metabolic diseases, the studies are limited in terms of the size and genetic diversity of the cohorts. These findings need to be validated on larger cohorts with different genetic backgrounds. However, since BAT activity in humans can currently be non-invasively quantified only by measuring the uptake of [18F]-Fluorodeoxyglucose ([18F]-FDG) by BAT on Positron Emission Tomography (PET) scans, the radiation exposure and the high cost preclude the method’s use for large population analyses. There is a need for alternative non-invasive methods to assess [18F]-FDG uptake by BAT to support the development of personalized treatment strategies in clinical practice. One way towards this end could be through predicting [18F]-FDG uptake by BAT from unenhanced computed tomography (CT) scans. As the cost and the radiation exposure of CT are significantly lower than PET, predicting the uptake by BAT from CT would facilitate imaging larger cohorts, allow patient stratification, and, consequently, accelerate BAT research. Support to this end comes from recent studies that analyzed cohorts with corresponding PET and CT images and showed that BAT had higher tissue density than WAT8,21,22, leading to higher CT intensity as measured by Hounsfield Unit (HU). Furthermore, Baba et al.9 performed cold stimulation studies in rodents and cancer patients and showed a correlation (R = 0.66) between the average HU and [18F]-FDG uptake measured as Standardized Uptake Value (SUV) in the supraclavicular BAT depot.
The existing population studies demonstrate statistical correlations between the voxel-wise HU values to SUVs in the BAT depots and provide motivation for developing more advanced computational methods. Specifically, this direction can be investigated in a predictive framework further to take into account more complex multi-voxel feature dependencies to predict SUV of BAT from CT in the BAT depots. To this end, CNNs have a great potential to predict BAT activity from CT scans thanks to their ability to eliminate the need for feature engineering by automatically learning useful features for a particular task from a training set23–27.
In this study, we propose using CNNs to predict the SUV of BAT from unenhanced CT scans in the restricted regions that are likely to contain BAT. Specifically, we crop the CT scans to cover the supraclavicular region, one of the largest BAT depots in humans28. Then, we train a CNN using a paired [18F]-FDG PET/CT dataset such that it takes these cropped CT scans as input and predicts the [18F]-FDG uptake by BAT measured by SUV. We extensively evaluated the performance of CNNs on datasets from four different cohorts: two of them are interventional (research) cohorts collected after exposing cold on the subjects before acquisition to ensure that BAT is activated if it exists, and the other two are retrospective clinical cohorts with no controlled cold stimulation applied, making 841 [18F]-FDG PET/CT scans from n = 718 subjects in total. The results suggest that CNNs trained on the cold exposure cohorts can be used to classify the subjects into high or low-activity categories using the predicted SUV of BAT only from CT scans. Accurately identifying these classes allows patient stratification by creating cohorts with the desired number of subjects from each category. Obtaining stratified cohorts is extremely useful for researchers to tailor treatments and interventions to specific patient subgroups. We show that CNNs can serve as a useful tool to this end by preselecting cohorts with the desired number of subjects from each BAT activity class, significantly reducing the number of subjects mistakenly included in the selected cohort. The preselected cohort can then undergo actual [18F]-PET/CT scans for more accurate quantification. Thus the actual [18F]-PET/CT scans are only obtained from subjects most relevant to the cohort, substantially lowering the PET acquisition cost for creating such a cohort.
Results
Overview of the experiments
We trained CNNs using the paired [18F]-FDG PET/CT scans from the training set of each cohort such that they take CT scan of the supraclavicular region as input and predict [18F]-FDG uptake by BAT in this region in the corresponding PET scan (see Sec. 4.3 for details). In order to assess the accuracy of the predictions, we first segmented the active BAT regions from the predicted and actual PET scans using SUV thresholding as described in Section 4.4. We then calculated the degree of overlap between these segmented regions using the Dice score29. Dice score measures the degree of overlap between two sets X and Y as 2∣X ∩ Y∣/(∣X∣ + ∣Y∣) and takes a value between 0 and 1, where higher scores indicate better segmentation performance. We conducted a comparative analysis of the performance of CNNs and a HU thresholding-based approach, another method commonly employed for segmenting active BAT from CT scans (see Section 4.5). We used commonly used HU thresholds of -180 and -10 in our experiments following the recent literature30. Subsequently, we investigated if CNNs’ predictions from CT could serve as a basis for classifying patients into two groups, BAT+ and BAT-, based on the presence of active BAT, thus enabling the identification of patients with (BAT+) or without (BAT-) active BAT from a larger population solely from CT without [18F]-FDG PET/CT imaging.
Quantitative results of BAT segmentation
Initially, we assessed the performance of CNNs within the same cohort, which we refer to as intra-cohort experiments, where each CNN was trained and evaluated on the same cohort. We present the results of the intra-cohort experiments quantifying the quality of segmentations using the Dice score in Table 1. We observe that CNNs achieve significantly higher accuracies than HU thresholding-based method for segmenting active BAT on the cold-exposure cohorts. The improvement achieved by CNNs is ∼75% on the Basel cohort and ∼23% on the Granada cohort. We performed permutation test31 and observed that the improvement achieved by CNN is statistically significant with p values << 1%. On the contrary, the Dice score results of the clinical cohorts are very low for both CNNs and the HU thresholding-based method. Additionally, we observed that training CNNs on the Zurich and MSKCC cohorts was not stable. We argue that this is due to the ambiguities when learning a mapping between HU values of BAT and the corresponding SUVs in the cohorts obtained without controlled cold exposure, which we discuss in detail in Sec. 3.
Table 1.
Comparison of intra-cohort performance of CNNs with different baselines in terms of Dice score for detecting active BAT
| Basel | Granada | Zurich | MSKCC | |
|---|---|---|---|---|
| HU thresholding -180, -10 | 0.427 | 0.421 | 0.248† | 0.115 |
| CNN | 0.745† | 0.521† | 0.189 | 0.130 |
† indicates a statistically significant difference between the results of CNN and the HU thresholding method with p < < 1% and bold indicates the best results.
The lack of generalization ability of CNNs beyond the domains they are trained on is a significant limitation in front of deploying them in practical applications32. It has been reported numerous times in medical imaging that a CNN trained on a dataset from one hospital performs poorly on a dataset from another hospital due to changes in imaging parameters, modality, population, and so on33. This motivated us to measure the inter-cohort performance of the CNNs trained on different cohorts to segment active BAT regions. In this experiment, we only used the cold-exposure cohorts, Basel and Granada, since we observed that CNNs trained on Zurich and MSKCC cohorts already performed poorly in the intra-cohort experiments. In particular, we evaluated the performance of the CNN trained on the Basel cohort on the whole Granada cohort and vice versa. The CNN trained on the Basel cohort achieved a Dice score of 0.486 when tested on the whole Granada cohort. This means a performance decrease of ∼7% compared to intra-cohort performance of the Granada model. However, the results are still better than the HU thresholding-based method by ∼15%. The average Dice score achieved by the Granada model on the whole Basel cohort is 0.538, meaning a performance decrease by ∼27% compared to intra-cohort performance of the Basel model. Despite this significant decrease, the results are still higher than thresholding CT scans at HU values -180 and -10 by ∼25%. Additionally, we conducted statistical analysis to measure the generalization performance of the Basel and Granada models. In particular, we applied permutation test31 between the intra and inter-cohort performance of both models. Our analysis revealed no statistically significant difference in the Granada model’s performance across cohorts, with a p-value of 0.77. Conversely, we observed a significant difference in the Basel model’s performance, with a p<< 1%. We discuss the generalization performance of CNNs further in Sec. 3.
Quantitative results on classifying subjects with and without BAT activity for stratification
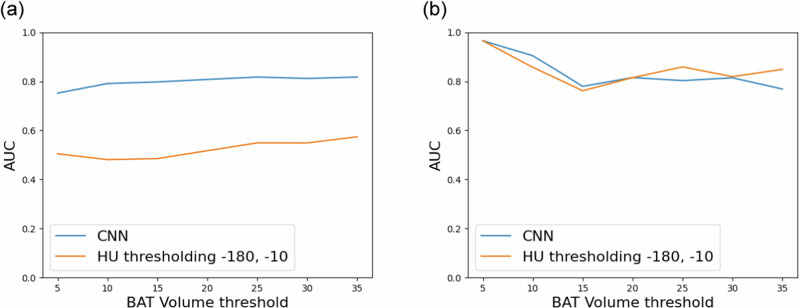
We investigated whether we could use the BAT volumes calculated from the SUV of BAT predicted by CNNs for classifying subjects into two distinct classes based on the presence of active BAT, namely BAT+ and BAT−. The BAT+ class contains subjects with high BAT activity, whereas the BAT− class includes those with low BAT activity. We conducted the same experiment with the BAT volumes computed using the HU thresholding-based method for comparison. To obtain the ground truth labels, we set a threshold for BAT volumes obtained from the actual [18F]-FDG PET scans and labeled the ones above the threshold as BAT+ and the ones below the threshold as BAT−. Then, we computed the area under the receiving operating characteristic curve (AUC)34 using the ground truth samples and predicted BAT volumes to determine the accuracy of classifying subjects as BAT+ and BAT− based on the predicted BAT volumes. Note that the labels and the AUC scores can change depending on the BAT volume thresholds we used when obtaining ground truth labels. Therefore, we plotted the AUC scores as a function of BAT volume threshold for both intra and inter-cohort performance of the Granada model, the CNN trained with the largest cold exposure cohort, in Fig. 1. The results demonstrate that the Granada model can distinguish subjects with and without BAT activity using the predicted BAT volume significantly better than the HU thresholding-based method by improving AUC from ∼0.6 to ∼0.8 in the intra-cohort experiments (Fig. 1a). Additionally, we observed that CNN’s inter-cohort performance is comparable with the HU thresholding-based method; both methods achieve high AUC at almost all BAT volume thresholds (see Fig. 1b).
Fig. 1. AUC scores for classifying subjects as BAT+ and BAT- using the predicted BAT volume at various BAT volume thresholds.
The plots show (a) intra and (b) inter-cohort performance of the Granada model, the CNN trained with the largest cold exposure cohort in our experiments.
Accurately classifying subjects as either BAT+ or BAT− using only CT scans allows for the creation of stratified cohorts of subjects belonging to the class of interest. This approach can be highly beneficial for researchers in several ways, including investigating the impact of treatment strategies on a group, exploring the relationship between BAT activity levels and specific genetic biomarkers, and identifying subgroups of patients who are more likely to respond positively to particular treatments. For instance, researchers interested in understanding the effects of a drug on patients with a specific genetic profile and high BAT activity would typically create a cohort with the relevant genetic profile and then collect [18F]-FDG PET/CT scans from all the patients in the cohort. Unfortunately, the resulting [18F]-FDG PET/CT scans may show that many patients in the cohort have very low BAT activity. This outcome generates a considerable number of [18F]-FDG PET scans that do not serve the intended purpose of the study, are expensive and potentially harmful to patients due to unnecessary radiation exposure. In contrast, researchers can obtain only CT scans from the patients in the cohort and use CNNs to predict which subjects are likely to have high BAT activity. Then, actual [18F]-FDG PET/CT scans are only obtained from those identified by CNNs as BAT+. By doing so, researchers can reduce the number of BAT− subjects mistakenly included in the cohort and save on the associated costs of acquiring [18F]-FDG PET scans that are unnecessary for the targeted study.
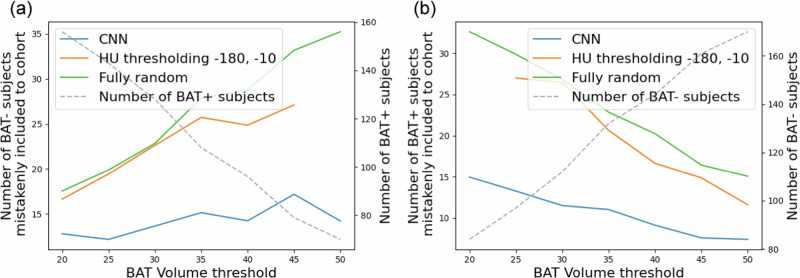
In the following experiment, we quantified the performance of CNNs on creating stratified cohorts (cohorts consisting of only BAT+ or BAT− samples) only using CT scans. To achieve this, initially, we classified subjects as BAT+ and BAT− by thresholding the predicted BAT volumes with a BAT volume threshold as described in the previous experiment. Then, we randomly selected 50 samples among the subjects classified as BAT+ by aiming to obtain a cohort with 50 BAT+ subjects using CT scans. The random selection step was repeated 100 times, and we computed the average number of BAT− samples mistakenly included in the selected cohort. The same experiment was conducted to select 50 subjects among those classified as BAT− to obtain a stratified cohort with BAT− subjects, and the average number of BAT+ samples mistakenly included in the cohort was computed over 100 repeats. We performed these experiments using the BAT volumes predicted by CNN trained on the Granada cohort and the HU thresholding-based method. Additionally, we compared both methods with the conventional, fully random, setting where we randomly chose BAT+ and BAT− subjects from the whole population without using any predicted BAT volume information. In Fig. 2, we present the plots of the number of subjects mistakenly included in a cohort when creating stratified cohorts consisting of 50 BAT+ (Fig. 2a) and 50 BAT− (Fig. 2b) subjects. The study’s results indicate that, compared to both methods, the use of a CNN leads to a substantial reduction in the number of BAT+ subjects mistakenly included in the BAT− cohort and vice versa at every BAT volume threshold. For example, when we selected 50 subjects among the ones classified by the CNN as BAT+, ∼17 BAT− subjects were mistakenly included in this cohort. Whereas, the number of mistakenly included BAT− subjects increases to ∼27 when stratification is done using the BAT volume predictions of the HU thresholding-based method, meaning an improvement ∼37% achieved by the CNN.
Fig. 2. The results demonstrating the patient stratification performance of different methods.
The number of subjects mistakenly included in a cohort when creating stratified cohorts of (a) BAT+ and (b) BAT− subjects. Note that the dashed gray lines in the plots belong to the second y-axes.
Qualitative results
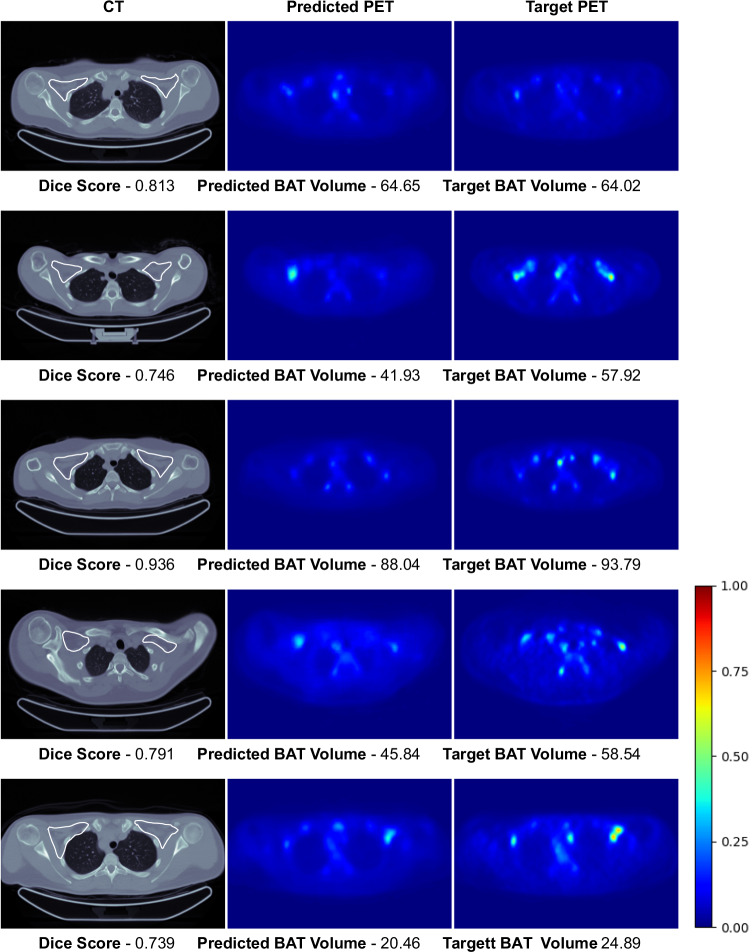
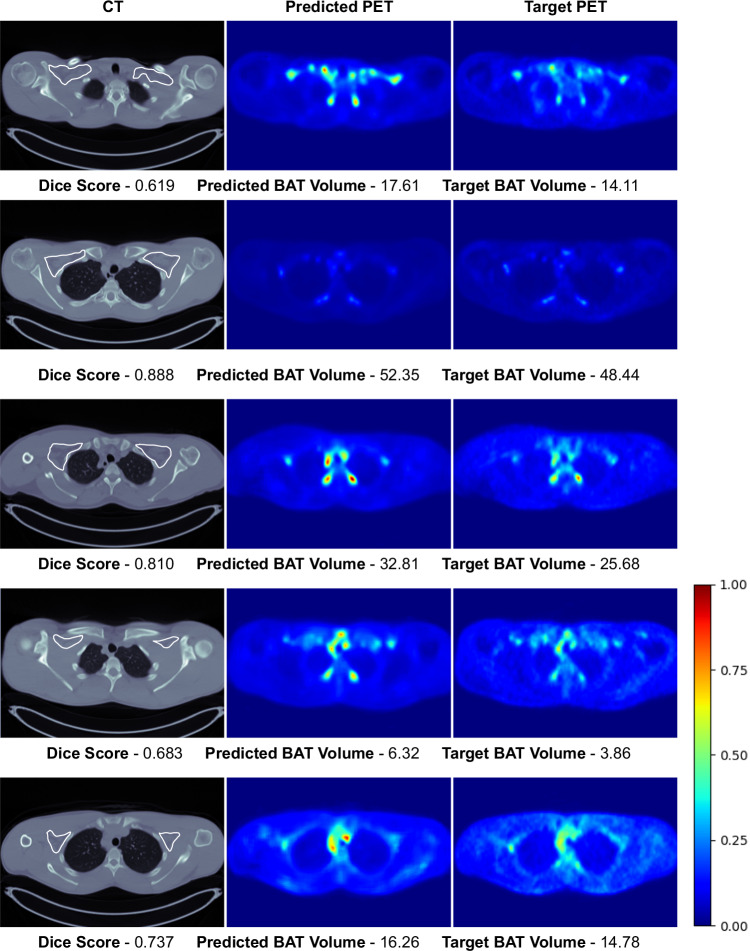
This section presents the qualitative results to visually compare the predicted [18F]-FDG uptake of BAT with the actual [18F]-FDG PET scans. In Figs. 3 and 4, we present the intra-cohort results of the Granada and the Basel models. Here, for the Granada cohort, we only present the results where CNNs achieve high Dice scores since we present some results when discussing the potential reasons for having low scores in Sec. 3. Here, we only present some slices from the scans since we cannot visualize the whole 3D [18F]-FDG PET/CT volumes. Visual results of the whole volumes can be seen via the Gradio User Interface35 in the following link: https://bat.ethz.ch/.
Fig. 3. Visual results of the Granada model’s high-accuracy predictions on the test set of the Granada cohort.
Rough manual segmentation of the supraclavicular region is delineated with white contour on CT scans. Note that each raw corresponds to an axial slice of a different subject and the images are normalized with respect to the max SUV value in the target PET scans for visualization purposes.
Fig. 4. Visual results of the Basel model’s predictions on the test set of the Basel cohort.
Rough manual segmentation of the supraclavicular region is delineated with white contour on CT scans. Note that each raw corresponds to an axial slice of a different subject and the images are normalized with respect to the max SUV value in the target PET scans for visualization purposes.
Discussion
Training issues with the retrospective clinical cohorts
Training CNNs for predicting SUV of BAT from CT aims to find a mapping from HU values of CT to SUV values of PET in a restricted area that corresponds to a BAT depot, the supraclavicular region in our case. There might be ambiguities in learning such a mapping when similar inputs are mapped to different values in the training set, adversely affecting the performance of CNNs. Based on previous studies, we argue that the low performance of the Zurich and MSKCC models may be attributed to such ambiguities that could arise when data is collected without controlled cold stimulation.
Gifford et al.10 conducted an experiment to investigate the impact of cold exposure on the HU and SUVs of supraclavicular BAT. The study involved comparing [18F]-FDG PET/CT scans taken from 17 subjects with BAT, both in thermoneutral conditions and after cold exposure. The results of the study showed that HU values in supraclavicular BAT regions remained relatively stable after cold exposure, whereas SUV values increased significantly. This suggests that there is little difference in HU values between active and inactive BAT, but significant differences in SUV values. As a result, mapping similar HU values to different SUV values can create ambiguities that can lead to unstable training and diminished performance of CNNs.
Such ambiguities are common in cohorts that are not obtained with controlled cold exposure. In the Zurich and MSKCC cohorts, half of the subjects have high PET activity of BAT, while the remaining half shows very low activity. In the second half, some subjects may still have BAT, but it can be inactive. This can create uncertainties in training data because HU levels of active and inactive BAT are similar based on the findings of Gifford et al.10; however, the former is mapped to higher SUV values than the latter. Thus, CNNs trained on Zurich and MSKCC cohorts cannot reliably learn a mapping from HU of BAT to the corresponding SUVs. We do not observe a similar problem in the cold-stimulated cohorts because when there is BAT, it is most likely active, and there is a low chance of having data with inactive BAT. Therefore, HU values of active BAT are consistently mapped to high SUV values, leading to better prediction performance, as observed by the superior results on the cold-stimulated cohorts.
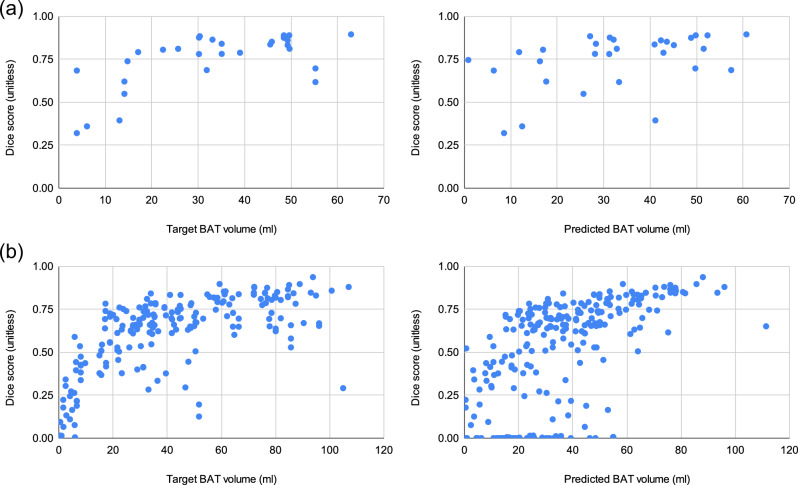
Segmentation accuracy increases in subjects with larger BAT volume
In Fig. 5, we present the scatter plots of predicted BAT volume vs. Dice score for each test subject in the intra-cohort experiments. The plots show that CNNs achieve better BAT segmentation accuracies when the target BAT activity measured by BAT volume is higher, which is especially more evident in the Granada cohort since it is larger. This trend in the scatter plots can be extremely useful when finding an operating point based on the BAT volume predicted by the network. If the researchers are interested in a more accurate delineation of the BAT, they can consider disregarding the predictions when the predicted BAT volume is smaller than a threshold to obtain more reliable predictions. For example, suppose we set such an operating point to 20ml and only consider the samples above this threshold. In that case, the average Dice score increases from 0.521 to 0.598 in the intra-cohort experiments of the Granada cohort. Shifting the operating point to 40ml leads to even more reliable predictions and increases the Dice score to 0.698.
Fig. 5. Scatter plots of Target BAT volume (ml) vs. Dice score (unitless) and Predicted BAT volume (ml) vs. Dice score (unitless) for the intra-cohort experiments.
a CNN trained on Basel cohort. b CNN trained on Granada cohort.
Dataset bias
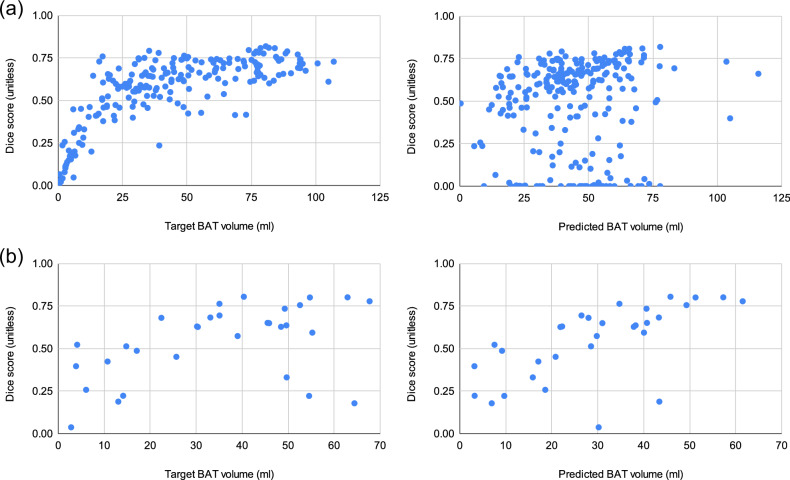
The cold exposure datasets, Basel and Granada, have some bias for active BAT due to both the selection criteria of the subjects in the cohorts (e.g., the Basel cohort is selected by REE, and the Granada cohort is selected from healthy and young adults) and cold exposure. As a result, the majority of the subjects in both cohorts have active BAT, as can be observed from the BAT volume histograms in Supplementary Fig. 1. Therefore, training CNNs on these cohorts leads to models that tend to make over-predictions for the test subjects having small BAT activity, leading to smaller Dice scores for such samples as shown in the scatter plots in Figs. 5 and 6. We present some visual results exemplifying over-predictions by CNNs in Fig. 7.
Fig. 6. Scatter plots of Target BAT volume (ml) vs. Dice score (unitless) and Predicted BAT volume (ml) vs. Dice score (unitless) for the inter-cohort experiments.
a CNN trained on Basel cohort evaluated on Granada cohort. b CNN trained on Granada cohort evaluated on Basel cohort.
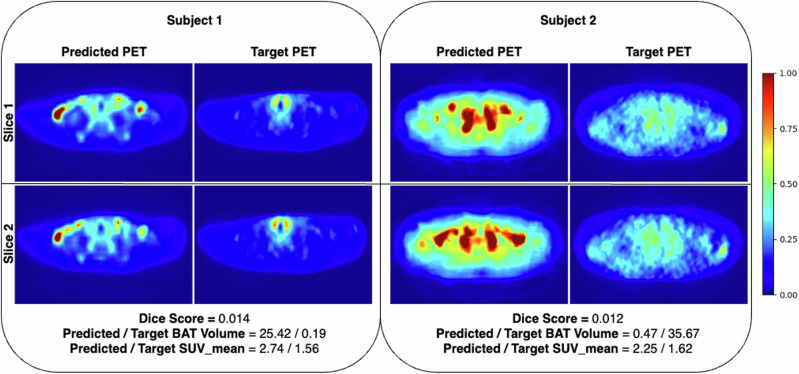
Fig. 7. Visual examples of target and predicted PET scans for the samples with low BAT activity where CNNs make over-predictions in many slices, leading to low quantitative results.
Note that we present examples of two different subjects from the Granada cohort where each row corresponds to a different axial slice of the subjects. In such examples, we observed that CNNs consistently make over-predictions in many slices, possibly due to the bias introduced by the dominance of subjects with high BAT activity in the training set. Note that the images are normalized with respect to the max SUV value in the target PET scans for visualization purposes.
The Granada cohort has a greater diversity in BAT activity levels compared to the Basel cohort, as indicated by Supplementary Fig. 1. Consequently, the Granada model is less vulnerable to over-prediction caused by dataset bias than the Basel model. This is evident from the scatter plots of both models’ predictions on the Granada cohort shown in Figs. 5 and 6. In the Basel model’s predicted BAT volume vs Dice score plot, the samples with the same Dice score are notably shifted towards the right compared to the target BAT volume vs Dice score plot, indicating over-prediction. Conversely, the over-prediction is less pronounced in the Granada model’s predictions.
Generalization
In the inter-cohort experiments, we noted that the Granada model has better generalization performance on the Basel cohort than how Basel model performs on the Granada cohort. This finding was predictable because the Basel cohort is relatively small and predominantly consists of subjects with high BAT activity. Consequently, it is more susceptible to the dataset bias mentioned earlier compared to the Granada cohort. Therefore, to mitigate dataset bias and generalization concerns, we recommend that future studies construct more balanced training cohorts that comprise an equal number of subjects across various BAT activity levels.
Another potential problem that might affect the generalization performance of our models is the variation in some characteristics of the populations in different cohorts. As mentioned before, the Granada cohort consisted of individuals from a specific population with certain self-reported characteristics, such as being sedentary, maintaining a stable body weight, and not being regularly exposed to cold conditions. However, many of these features were not taken into consideration in the Basel cohort. Such discrepancies between cohorts can significantly impact the CNNs’ ability to generalize new data. Conducting generalization experiments without removing these differences does not provide an accurate representation of the generalization performance between cohorts. A more effective method of quantifying the generalization performance between cohorts would be to perform inter-cohort experiments after standardizing these features between the cohorts. We acknowledge that creating such homogenized cohorts can be costly, and this is a limitation of our study. Despite this limitation, we argue that the generalization performance of the Granada model on the Basel cohort would have improved further if these discrepancies between the cohorts had been addressed.
Correlation between Dice score and BMI
In the assessment of the Dice score between the CNN-predicted HU-derived indices (predicted BAT activity) and the actual SUV-derived indices (ground truth BAT activity), an important consideration is the potential influence of confounding factors, such as adiposity, on the predictive accuracy of our model. Adiposity is inversely related to BAT functionality, and increased adipocyte size is associated with higher BMI levels. Thus, we investigated whether the predictive accuracy of our model - as measured by the Dice score - was influenced by the BMI of the subjects in our cohorts. Our analysis revealed a Pearson’s correlation coefficient of -0.131 and -0.076 between the Dice score and BMI for the Granada and Basel cohorts, respectively - suggesting a negligible inverse relationship. This finding is particularly significant as it indicates that the predictive accuracy of our model is not confounded by variations in adiposity levels among subjects. The absence of a significant correlation between BMI and the Dice score substantiates the reliability of the observed agreement between the predicted and actual BAT volumes, affirming that this agreement is not a spurious correlation attributable to adiposity. This result underlines the robustness of our CNN-based predictive model, demonstrating its applicability across a diverse population with varying BMI levels.
Using 2D CNNs
In our experiments, we utilized 2D CNNs and trained them with 2D slices from the axial plane. This approach has been found to produce higher accuracy results compared to 3D CNNs in many medical applications, particularly when the number of 3D volumes is limited. However, a major drawback of 2D CNNs is their inability to utilize 3D information, which can be vital in accurately identifying active BAT regions. In our current setup, each slice is separately predicted by the CNNs without considering any information from neighboring slices. We believe that training 3D CNNs with sufficiently large and well-paired cold-stimulated [18F]-FDG PET/CT datasets could lead to improved predictions through the utilization of 3D information.
Predicting the PET activity of BAT in other BAT depots
In this paper, we primarily focus on predicting the PET activity of BAT from CT scans in the supraclavicular region, as it is one of the largest BAT depots in humans. We then extend our analysis to include multiple BAT regions, aiming to determine whether the capabilities of CNNs can be generalized to predict BAT activity in additional depots, such as the cervical, supraclavicular, and paraspinal areas. The results show that CNNs can predict BAT activity in other BAT depots without loss in accuracy; significantly improving the HU thresholding-based method. The details of this experiment are provided in the Supplementary Material Section A.
Can neural networks predict any functional activity in PET from CT scans?
In this study, we utilize Convolutional Neural Networks (CNNs) to predict PET activity in brown adipose tissue (BAT) from CT scans, based on literature that suggests a correlation between CT’s Hounsfield Units (HU) and PET’s Standardized Uptake Values (SUV) within BAT regions, as previously discussed. The question may arise regarding the broader application of our method for predicting PET activity from CT scans in tissues other than BAT, including tumors. It is important to highlight the intrinsic differences between CT and PET imaging techniques; CT scans offer detailed structural information reflecting stable anatomical features, while PET images reveal dynamic metabolic and biochemical activities that can fluctuate significantly due to a range of factors, including health status and testing conditions. We would like to emphasize that our findings do not imply that our method is universally applicable across different tissues or conditions, given the fundamental differences between the imaging modalities. Our approach is specifically tailored to BAT, relying on distinct CT signatures of BAT activity, which have empirical backing. For the application of neural networks in predicting PET activity for other tissues, like tumors, identifying and thoroughly investigating comparable, specific CT signatures is essential.
Conclusion and future work
In this paper, we proposed using CNNs for predicting the SUV of BAT in the supraclavicular region, one of the largest BAT depots in humans. We used the predicted PET activity to segment the active BAT regions. We performed extensive experiments using four different cohorts, two of which are research cohorts obtained after cold stimulation, and the remaining two are clinical cohorts with no cold stimulation applied. Our results show that active BAT regions can be segmented with high accuracy when the CNNs are trained on cold-stimulated paired [18F]-FDG PET/CT datasets, demonstrating generalization ability to some extent to the other cold exposure cohorts. We also empirically showed that [18F]-FDG PET/CT datasets collected without cold stimulation are unsuitable for training CNNs and discussed why it is the case. One of the most interesting findings of our study is the ability of CNNs on distinguishing subjects with and without active BAT solely from CT scans. Additionally, we demonstrated that this ability of CNNs can be used to create stratified cohorts of BAT+ and/or BAT− subjects, thereby reducing the number of subjects mistakenly assigned to a wrong cohort. This allows researchers to conduct experiments on cohorts with desired characteristics which is extremely useful, e.g., when investigating the effect of a drug on a certain population with large BAT activity. This research represents a significant step towards opportunistic screening for BAT activity, which may be helpful in future research on personalized treatment strategies for metabolism-related diseases by developing predictive models using neural networks with high-quality datasets from sizeable cohorts. By enabling extensive population analyses with reduced radiation exposure and cost, CNNs provide a promising path toward more effective diagnoses and treatments in the future.
In this paper, we employed a CNN-based neural network architecture, specifically the Attention U-Net. Recently, transformer-based models, such as Vision Transformers (ViTs)36, have achieved significant improvements over CNNs in terms of accuracy and generalization across a variety of image analysis tasks37. However, it’s important to note that the superior performance of transformer-based networks largely depends on the availability of extensive labeled datasets. In contexts where only small or medium-sized datasets are available, their performance tends to be less effective compared to that of CNNs36. Given that our study relies on relatively small datasets, we opted for CNNs. Nevertheless, should large cold-exposure [18F]-FDG PET/CT datasets become available for future research, transformer-based architectures have the potential to significantly enhance performance over CNNs.
Methods
Datasets
This section provides information about the datasets from different cohorts used in this study. We mainly group the cohorts used in our experiments into two: 1) interventional (research) cohorts obtained with cold exposure and 2) retrospective (clinical) cohorts obtained without any controlled cold exposure.
The study protocols for each cohort were approved by the Institutional Review Board (IRB) of the centers, as detailed in the corresponding sections for each cohort. These original approvals included permission for the datasets to be used in future research. As a result, specific approval for this study has been waived based on these prior approvals.
Interventional (research) cohorts with cold-exposure
The regional ethics committee at the University of Basel approved the study protocol (approval number EKNZ 2016-01859), and the study was registered at clinicaltrials.gov (NCT03269747) on 2017-09-01. All participants provided written informed consent before being enrolled in the study.
Primary goal of the cohort: The primary goal of this interventional cohort was to see the effect of high-dose glucocorticoid (prednisone) treatment on human brown adipose tissue activity in healthy men. The outcome of the primary study is published in ref. 38.
Participants in the primary study: Healthy male volunteers were recruited between September 2017 and April 2019. The participants underwent a screening visit during which cold-induced thermogenesis (CIT) was measured, and n = 16 participants with a CIT above 5% of resting energy expenditure (REE) were enrolled. [18F]-FDG PET/CT scans were obtained from each participant pre and post-intervention (prednisone or placebo). The subjects in this cohort are aged between 19 and 33 years, with an average age of 24.45 years and a standard deviation of 4.38. Their BMI values range from 18.6 to 27.5, with a mean BMI of 22.5 and a standard deviation of 2.31.
Participants included to this study: We used pre and post-intervention [18F]-FDG PET/CT scans obtained from all n = 16 participants in our experiments. We ensured that pre and post-intervention scans of the same participant were in the same split (one of training, validation, and test splits) during training CNNs.
Mild cold exposure: To activate BAT and assess CIT, participants underwent a controlled mild cold exposure. They were placed on a hospital bed in an air-conditioned study room with a stable ambient temperature of 24∘C in a supine position. Subjects wore shorts and T-shirts and were initially covered with a fleece blanket for measurements under warm conditions. To expose study participants to mild cold, the fleece blanket was removed, and water-perfused cooling mats were placed around the mid-section of the subjects. The cooling mats were perfused with water at a controlled temperature (Hilotherm clinic, Hilotherm GmbH, Argenbühl, Germany). The water temperature was lowered from 25°C to 10°C at a rate of 1°C every 2 minutes.
[18F]-FDG PET/CT Imaging: The imaging procedures for assessment of human supraclavicular BAT activity with a low radiation exposure were developed during a previous trial39. After 120 minutes of cold exposure study, participants received an intravenous bolus of 75 MBq of [18F]-Fluorodeoxyglucose (FDG). Static PET/CT scanning was performed on a Biograph mCT PET/CT scanner (Siemens Healthineers, Erlangen, Germany) after an additional 30 minutes of rest. Low-dose CT scanning was used and confined to the neck and upper thoracic region to reduce exposure to ionizing radiation.
Granada: The study was approved by the Ethics Committee on Human Research of the University of Granada (no. 924) and by the Servicio Andaluz de Salud (Centro de Granada, CEI-Granada, Spain) and was registered at clinicaltrials.gov (NCT02365129) on 2015-02-18. All participants included in the study provided written consent.
Primary goal of the cohort: The interventional cohort was obtained with the primary aim of investigating the supervised exercise training on BAT volume and activity and the outcome of the study is published in40.
Participants in the primary study: All data acquisition was done at the University of Granada (Spain) during the months of October, November, and December in 2015 and 2016. All subjects underwent a comprehensive medical examination and reported themselves to be sedentary ( < 20 min moderate-vigorous physical activity on < 3 days/week), reported a stable body weight over the last 3 months ( < 3 kg change), were not exposed to cold regularly, did not smoke, and did not take any medication. The subjects did not suffer from cardiometabolic disease.
The subjects in this cohort are aged between 18 and 27 years, with an average age of 22.07 years and a standard deviation of 2.23. Their BMI values range from 17.2 to 39.40, with a mean BMI of 24.91 and a standard deviation of 4.62.
In the Granada cohort, 145 participants were initially included in the study. [18F]-FDG PET/CT scans were obtained pre and post-intervention (supervised exercise training) for 107 of them. For the remaining 38 participants, only pre-intervention [18F]-FDG PET/CT scans were obtained since the participants did not continue the study afterward.
Participants included in this study: We used both pre and post-intervention [18F]-FDG PET/CT scans of 107 participants along with the 30 of the 38 pre-intervention scans in our experiments. Our experiments could not include the remaining 8 pre-intervention scans since they were not processed properly in the delivered institution where the experiments in this study were conducted. In total, we used [18F]-FDG PET/CT scans from n = 137 participants in our experiments. We made sure that when training CNNs, the pre and post-intervention scans of each participant were placed together in one of the training, validation, or test sets.
Cold exposure: Cooling protocol was used to activate BAT where the quantified activities were previously reported in41,42. During the cooling period, subjects sat in a cool room (19.5-20°C) wearing a water-perfused cooling vest (Polar Products Inc., Stow, OH, USA). The water temperature was reduced from 16.6°C by ∼2. 2°C per 10 min until the subjects began shivering. After 48-72h of initial cooling, the patients went to the Hospital Virgen de las Nieves where they were again placed in a cool room (19.5-20°C) by wearing the same cooling vest for 2h with the temperature set to ∼4°C above their earlier shivering threshold.
[18F]-FDG PET/CT Imaging: The subjects were given an injection of 18F-FDG (∼185MBq) after first hour of the cooling period. Then, the water temperature was increased by 1°C to avoid visually detectable shivering. After another hour, PET/CT scans of the subjects were obtained using a Siemens Biograph 16 PET/CT scanner (Siemens Healthineers, Erlangen, Germany), scanning two BEDs from the atlas vertebra to thoracic vertebra 6 (approximately).
Retrospective clinical cohorts without any controlled cold-exposure
We used datasets from two retrospective clinical cohorts collected in the University Hospital of Zurich (Zurich cohort) under ethics approval number KEK ZH 2015-0282 and Memorial Sloan Kettering Cancer Center (MSKCC cohort) under IRB approval 19-184. Written consent was waived by the Zurich Cantonal Ethics Committee for the Zurich cohort and by the IRB for the MSKCC cohort.
Primary goal of the cohorts: Both cohorts are retrospective cohorts that were collected from patients at the corresponding hospitals. Some aspects of the Zurich cohort have been investigated in43–46 whereas the MSKCC cohort was not published elsewhere.
Participants: The number of patients included in the Zurich cohort is n = 480 and in the MSKCC cohort is n = 85. These cohorts contain only a single [18F]-FDG PET/CT scan from each patient.
[18F]-FDG PET/CT Imaging: The patient preparation procedure was comparable at both centers: Patients were instructed to arrive fasted (4-6 h), including abstaining from drinking sweetened beverages and chewing gum. Blood glucose was measured prior to the [18F]-FDG injection and had to be below 7 g/l. Patients received ∼4MBq [18F]-FDG per kilogram body weight (Zurich), or ∼400MBq (MSKCC), followed by a 60 ± 5 min. uptake period. Afterward, a low-dose attenuation correction CT scan was acquired (100 − 120kV, ∼80mA), followed by the PET scan from mid-thigh to the vertex of the skull (GE Healthcare ®).
Data pre-processing
We transformed all CT scans with the corresponding rescale and the intercept found in the meta-data in the DICOM files. Voxel values in all raw [18F]-FDG PET scans contain radioactivity amount in terms of MBq/mL. Radioactivity amount in [18F]-FDG PET scans depends on the amount of radioactive material injected into the patient prior to the acquisition, the half-time of the material, and the patient’s weight. We removed the effect of these parameters by converting the voxel values to Standardized Uptake Value (SUV)47.
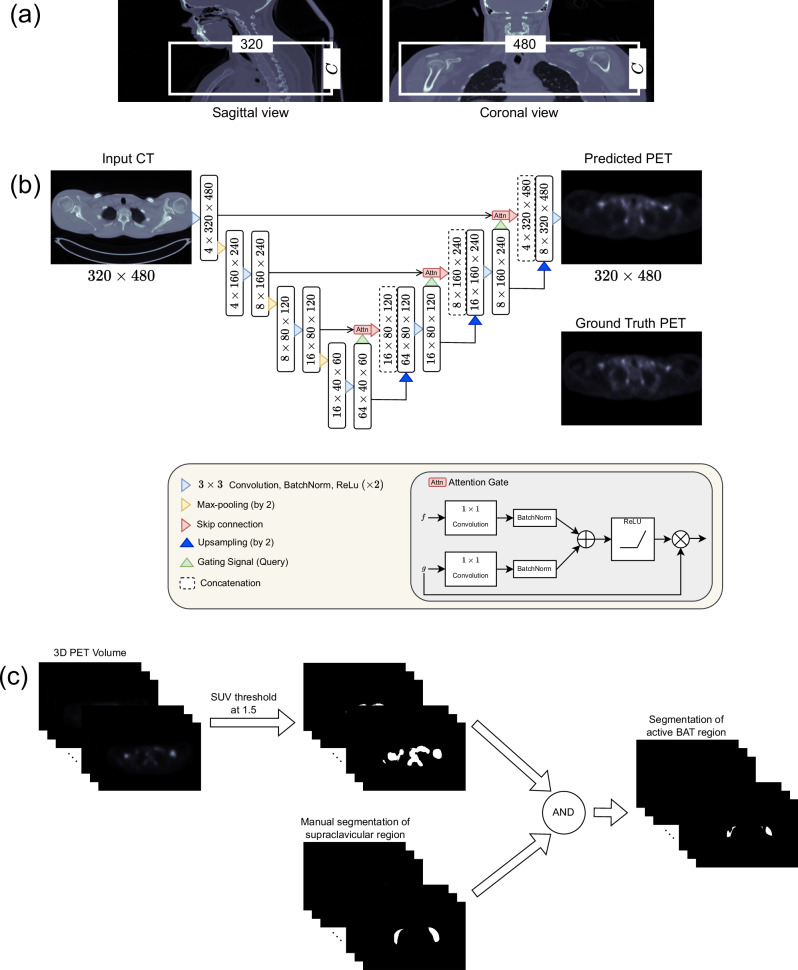
We resampled all volumes, PET and CT, from all cohorts to voxel size of 0.976 × 0.976 × 1.5mm3, the default voxel size of the Granada cohort. We cropped 320 × 480 × C volumes around the supraclavicular region, one of the largest BAT depots in human28, from the original volumes where C denotes the number of slices in the axial plane (see Fig. 8a). Note that C slightly varies from patient to patient which was chosen by an expert by considering the slices where BAT may exist. The crop size of 320 × 480 was determined such that the cropped region encompasses the supraclavicular area and the cropped image’s dimensions are divisible by 23 where 3 is the number of max pooling operations used for downsampling the image size in our network architecture as shown in Fig. 8b.
Fig. 8. Illustration of the flow for predicting PET activity of BAT from CT scans and segmenting the active BAT region.
a Illustration of cropping to obtain a region of interest (ROI) that contains the supraclavicular region. Note that C indicates the number of slices in the axial dimension and can slightly change for different subjects. After cropping, the slices are given as input to the CNN shown in (b). b Schematic of the Attention U-Net architecture. c Detecting active BAT regions from a PET volume. Note that “AND" represents the logical and operator that we used to mask out false positive regions obtained after thresholding.
We applied volume-specific min-max normalization to CT scans, where x is an intensity, and xmin and x99 are the minimum and the 99th percentile values computed over a volume, respectively. Additionally, we normalized the PET scans in a cohort with the 99th percentile SUV value computed over the cohort.
Network architecture and training details
We trained CNNs such that they take pre-processed (as described in the previous section) CT scans of the supraclavicular region as input and predict the corresponding SUV of BAT. We experimented with training CNNs using the paired [18F]-FDG PET/CT datasets introduced in Section 4.1. We used the 2D Attention U-Net48 shown in Fig. 8b as a CNN architecture in our experiments. The architecture has been successfully applied to various medical imaging tasks in the literature49–51. Axial 2D slices were extracted from the 3D volumes for CT and PET and used as inputs and ground truths during training CNNs. We chose a 2D network architecture instead of a 3D one because it has fewer parameters, and has been shown that the former one may lead to better results in various tasks especially when the number of 3D volumes is limited52.
For each dataset, we created 5 different random splits (folds) for training, validation, and testing. In each random split, we split 20% of the 3D volumes for testing and validation and use the remaining volumes for training. We show the number of volumes in training, validation, and test splits for each dataset in Table 2. Note that the scans in the final test, train, and validation splits do not overlap. In our evaluations, we trained 5 different CNNs using each random training and validation split of a dataset and presented the average results.
Table 2.
Number of training, validation, test, and total volumes for each cohort
| Train | Validation | Test | Total | |
|---|---|---|---|---|
| Basel | 20 | 6 | 6 | 32 |
| Granada | 148 | 48 | 48 | 244 |
| Zurich | 282 | 94 | 94 | 480 |
| MSKCC | 51 | 17 | 17 | 85 |
We trained the models for 1000 epochs with a learning rate of 0.003 by minimizing mean square error (MSE) loss between predicted and ground truth PET scans. We used standard data augmentation techniques of translation, rotation, random cropping, scaling, and horizontal/vertical flipping. The models were evaluated during training on the validation set and we picked the model with the lowest MSE loss as the final model. We obtained the predictions for each 3D test volume by giving it as input slice-by-slice to the trained 2D models. Then, we formed the predicted 3D PET volumes by stacking up the 2D predictions.
Segmentation of active BAT regions from PET
The standard procedure for the detection of active BAT regions from PET volumes is applying thresholding to the whole volume and manually eliminating the regions which do not correspond to BAT depots. In our evaluations, we followed the same procedure for detecting active BAT regions from predicted and ground truth BAT volumes as shown in Fig. 8c. We segmented the active BAT regions from PET using the threshold of 1.530. Then, we eliminate the regions that do not correspond to a BAT depot by masking the threshold-based segmentation with the rough manual segmentation of the supraclavicular region delineated from CT by an expert and obtain the final segmentation of active BAT.
HU thresholding-based method
We compared the performance of CNNs with a HU thresholding-based method which enables us to compare the performance of CNNs with a method that can segment active BAT regions without requiring PET scans. We used commonly used HU thresholds of -180 and -10 in the literature30. Note that the thresholding-based approach produces many false positives that do not correspond to a BAT region. Finally, we suppressed the false positives regions detected as active BAT outside of a BAT depot with the ground truth segmentation of the supraclavicular region as shown in Fig. 8c.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was financially supported by 1) Personalized Health and Related Technologies (PHRT), project number 222, ETH domain, 2) The LOOP Zürich - Medical Research Center, Zurich, Switzerland. A.S.B. was supported by a scholarship from the Prof. Dr. Max Cloëtta Foundation. M.J.B. received funding from the Swiss National Science Foundation (PZ00P3_167823), the Bangerter-Rhyner Foundation, Basel, and the Nora van der Meeuwen-Häfliger Foundation, Basel. B.M.T. was funded (Grant RYC2022-036473-I) by MCIN/AEI/ 10.13039/501100011033 and by “ESF Investing in your future". B.D. was funded by Swiss National Science Foundation (SNSF) for the project grant #310030_219550.
Author contributions
E.E. prepared data, implemented CNNs, performed experiments, and wrote the manuscript. E.K. supervised E.E. C.W., N.Z., B.D., and E.K. acquired the funding for this project. A.S.B., M.S., B.M.T., J.R.R., T.S., H.A.V., A.I.B., A.C., D.W., C.I.M., M.J.B., C.W. were involved in data acquisition. A.S.B., M.S., B.M.T., J.R.R., A.I.B., M.J.B., and C.W. provided knowledge about datasets and BAT research. A.S.B., M.S., B.M.T., J.R.R., T.S., H.A.V., A.I.B., A.C., D.W., N.Z., B.D., V.G., C.I.M., M.J.B., C.W., E.K. read the manuscript and provided feedback.
Peer review
Peer review information
Nature Communications thanks Kenji Hirata, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data supporting the findings described in this manuscript are available in the article and in the Supplementary Information and from the corresponding author upon request. The datasets used in the current study are not publicly available due to privacy reasons. Image datasets can be made available after deidentification for any researcher who provides methodologically sound proposals. These proposals should outline the intended use of the data and should be directed to ertunc.erdil@vision.ee.ethz.ch. The proposals must be submitted up to 60 months following the article’s publication. Proposals will be reviewed by the group responsible for each dataset to ensure that they are methodologically sound and ethically appropriate. Once a proposal is accepted, access to the data will be granted after the requestors sign a data access agreement. Approved researchers will receive data via a secure data-sharing platform that will be decided before data sharing, ensuring that all data transfer complies with relevant data protection regulations. The data will be shared for the purposes of replicating the study findings or for conducting additional research related to the study topic and will be available for one year with possible extension. The data will be provided for academic research only, and commercial use is not permitted. Figures 1, 2, 5, 6, and Supplementary Fig. 1 are available as source data. Source data are provided with this paper.
Code availability
The training code is available at https://github.com/eerdil/BAT.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/19/2024
A Correction to this paper has been published: 10.1038/s41467-024-54209-x
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52622-w.
References
- 1.Grarup, N., Sandholt, C. H., Hansen, T. & Pedersen, O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia57, 1528–1541 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Sheu, W. H.-H., Brunell, S. C. & Blase, E. Efficacy and tolerability of exenatide twice daily and exenatide once weekly in asian versus white patients with type 2 diabetes mellitus: a pooled analysis. Diab. Res. Clin. Pract.114, 160–172 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Gupta, A. K. et al. Ethnic differences in blood pressure response to first and second-line antihypertensive therapies in patients randomized in the ascot trial. Am. J. Hypertens.23, 1023–1030 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Dourish, P. What we talk about when we talk about context. Personal. Ubiquitous Comput.8, 19–30 (2004). [Google Scholar]
- 5.Cohen, P. & Spiegelman, B. M. Cell biology of fat storage. Mol. Biol. Cell27, 2523–2527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nedergaard, J. & Cannon, B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab.11, 268–272 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Sun, W., Modica, S., Dong, H. & Wolfrum, C. Plasticity and heterogeneity of thermogenic adipose tissue. Nat. Metab.3, 751–761 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Sampath, S. C., Sampath, S. C., Bredella, M. A., Cypess, A. M. & Torriani, M. Imaging of brown adipose tissue: state of the art. Radiology280, 4–19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba, S., Jacene, H. A., Engles, J. M., Honda, H. & Wahl, R. L. Ct hounsfield units of brown adipose tissue increase with activation: preclinical and clinical studies. J. Nucl. Med.51, 246–250 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Gifford, A., Towse, T. F., Walker, R. C., Avison, M. J. & Welch, E. B. Characterizing active and inactive brown adipose tissue in adult humans using pet-ct and mr imaging. Am. J. Physiol.-Endocrinol. Metab.311, E95–E104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chondronikola, M., Beeman, S. C. & Wahl, R. L. Non-invasive methods for the assessment of brown adipose tissue in humans. J. Physiol.596, 363–378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med.360, 1500–1508 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Vosselman, M. J., Vijgen, G. H., Kingma, B. R., Brans, B. & van Marken Lichtenbelt, W. D. Frequent extreme cold exposure and brown fat and cold-induced thermogenesis: a study in a monozygotic twin. PloS one9, e101653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey, A. L. et al. Ephedrine activates brown adipose tissue in lean but not obese humans. Diabetologia56, 147–155 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Becher, T. et al. Brown adipose tissue is associated with cardiometabolic health. Nat. Med.27, 58–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kir, S. et al. Tumour-derived pth-related protein triggers adipose tissue browning and cancer cachexia. Nature513, 100–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broeders, E. P. et al. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab.22, 418–426 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Straat, M. E. et al. Stimulation of the beta-2-adrenergic receptor with salbutamol activates human brown adipose tissue. Cell Reports Medicine4 (2023). [DOI] [PMC free article] [PubMed]
- 19.Cypess, A. M. et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab.21, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Söderlund, V., Larsson, S. A. & Jacobsson, H. Reduction of fdg uptake in brown adipose tissue in clinical patients by a single dose of propranolol. Eur. J. Nucl. Med. Mol. imaging34, 1018–1022 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Hu, H. H., Chung, S. A., Nayak, K. S., Jackson, H. A. & Gilsanz, V. Differential ct attenuation of metabolically active and inactive adipose tissues-preliminary findings. J. Computer Assist. Tomogr.35, 65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prodhomme, H. et al. Imaging and identification of brown adipose tissue on ct scan. Clin. Physiol. Funct. imaging38, 186–191 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, 234–241 (Springer, 2015).
- 24.Kamnitsas, K. et al. Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med. Image Anal.36, 61–78 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Cheng, S. et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat. Commun.12, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frazer, J. et al. Disease variant prediction with deep generative models of evolutionary data. Nature599, 91–95 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Ziller, A. et al. Medical imaging deep learning with differential privacy. Sci. Rep.11, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundström, E. et al. Automated segmentation of human cervical-supraclavicular adipose tissue in magnetic resonance images. Sci. Rep.7, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dice, L. R. Measures of the amount of ecologic association between species. Ecology26, 297–302 (1945). [Google Scholar]
- 30.Martinez-Tellez, B. et al. The impact of using barcist 1.0 criteria on quantification of bat volume and activity in three independent cohorts of adults. Sci. Rep.8, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmaso, L. & Pesarin, F.Permutation tests for complex data: theory, applications and software (John Wiley & Sons, 2010).
- 32.Ganin, Y. & Lempitsky, V. Unsupervised domain adaptation by backpropagation. In International conference on machine learning, 1180–1189 (PMLR, 2015).
- 33.Karani, N., Erdil, E., Chaitanya, K. & Konukoglu, E. Test-time adaptable neural networks for robust medical image segmentation. Med. Image Anal.68, 101907 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Sangalli, S., Erdil, E., Hötker, A., Donati, O. & Konukoglu, E. Constrained optimization to train neural networks on critical and under-represented classes. Adv. Neural Inf. Process. Syst.34, 25400–25411 (2021). [Google Scholar]
- 35.Abid, A. et al. Gradio: Hassle-free sharing and testing of ml models in the wild. arXiv preprint arXiv:1906.02569 (2019).
- 36.Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. International Conference on Learning Representations (ICLR) (2021).
- 37.Wagner, S. J. et al. Fully transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study. Cancer Cell, 1650–1661 (2023) [DOI] [PMC free article] [PubMed]
- 38.Maushart, C. I. et al. Effect of high-dose glucocorticoid treatment on human brown adipose tissue activity: a randomised, double-blinded, placebo-controlled cross-over trial in healthy men. Ebiomedicine96 (2023). [DOI] [PMC free article] [PubMed]
- 39.Ter Voert, E. E. et al. Low-dose 18 f-fdg tof-pet/mr for accurate quantification of brown adipose tissue in healthy volunteers. EJNMMI Res.10, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Tellez, B. et al. No evidence of brown adipose tissue activation after 24 weeks of supervised exercise training in young sedentary adults in the actibate randomized controlled trial. Nat. Commun.13, 5259 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Tellez, B. et al. A new personalized cooling protocol to activate brown adipose tissue in young adults. Front. Physiol.8, 863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez-Delgado, G. et al. Reliability of resting metabolic rate measurements in young adults: Impact of methods for data analysis. Clin. Nutr.37, 1618–1624 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Sun, W. et al. Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring. Nat. Med.24, 1372–1383 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Becker, A. S., Nagel, H. W., Wolfrum, C. & Burger, I. A. Anatomical grading for metabolic activity of brown adipose tissue. PLoS One11, e0149458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker, A. S. et al. Brown fat does not cause cachexia in cancer patients: A large retrospective longitudinal fdg-pet/ct cohort study. Plos one15, e0239990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker, A. S. et al. In-depth analysis of interreader agreement and accuracy in categorical assessment of brown adipose tissue in (18) fdg-pet/ct. Eur. J. Radiol.91, 41–46 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Lucignani, G., Paganelli, G. & Bombardieri, E. The use of standardized uptake values for assessing fdg uptake with pet in oncology: a clinical perspective. Nucl. Med. Commun.25, 651–656 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Oktay, O. et al. Attention u-net: Learning where to look for the pancreas. Medical Imaging with Deep Learning (MIDL) (2018).
- 49.Islam, M. et al. Brain tumor segmentation and survival prediction using 3d attention unet. In International MICCAI Brainlesion Workshop, 262–272 (Springer, 2019).
- 50.Guo, C. et al. Sa-unet: Spatial attention u-net for retinal vessel segmentation. In 2020 25th International Conference on Pattern Recognition (ICPR), 1236–1242 (IEEE, 2021).
- 51.Lian, S. et al. Attention guided u-net for accurate iris segmentation. J. Vis. Commun. Image Representation56, 296–304 (2018). [Google Scholar]
- 52.Baumgartner, C. F., Koch, L. M., Pollefeys, M. & Konukoglu, E. An exploration of 2d and 3d deep learning techniques for cardiac mr image segmentation. In International Workshop on Statistical Atlases and Computational Models of the Heart, 111–119 (Springer, 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings described in this manuscript are available in the article and in the Supplementary Information and from the corresponding author upon request. The datasets used in the current study are not publicly available due to privacy reasons. Image datasets can be made available after deidentification for any researcher who provides methodologically sound proposals. These proposals should outline the intended use of the data and should be directed to ertunc.erdil@vision.ee.ethz.ch. The proposals must be submitted up to 60 months following the article’s publication. Proposals will be reviewed by the group responsible for each dataset to ensure that they are methodologically sound and ethically appropriate. Once a proposal is accepted, access to the data will be granted after the requestors sign a data access agreement. Approved researchers will receive data via a secure data-sharing platform that will be decided before data sharing, ensuring that all data transfer complies with relevant data protection regulations. The data will be shared for the purposes of replicating the study findings or for conducting additional research related to the study topic and will be available for one year with possible extension. The data will be provided for academic research only, and commercial use is not permitted. Figures 1, 2, 5, 6, and Supplementary Fig. 1 are available as source data. Source data are provided with this paper.
The training code is available at https://github.com/eerdil/BAT.