Abstract
The cytosolic matrix domain (MD) located between amino acids (aa) 103 and 124 of the large hepatitis B virus envelope protein L is essential for virion formation. We reduced the distance between MD and the transmembrane domain (TD; aa 254 to 272) by deletions starting at aa 132. Six mutants with deletions of up to aa 234 were wild type, and four mutants with slightly larger deletions were blocked with respect to virion morphogenesis. Thus, the minimal distance between MD and TD was around 26 aa. This spacer might be required by MD to reach contact sites on the capsid.
Hepatitis B virus (HBV) particles are formed by budding of preformed cytoplasmic icosahedral nucleocapsids through intracellular membranes containing three viral envelope proteins: the long L, middle-sized M, and short S proteins (1, 14). These proteins are expressed from a single open reading frame by the usage of three different start codons and a common stop codon without the involvement of RNA splicing and are therefore coterminal. Only the L and S proteins are required for the envelopment of capsids (3). The transmembrane topology of the S protein is determined by an uncleaved N-terminal type I signal (sig I) and a central type II signal/anchor domain (sig II) (7). In the L protein (total length: 400 amino acids [aa] for genotype A) the type I signal in its C-terminal S domain is not functional. Therefore, the N-terminal domain of L (preS) and the N-terminal part of the S domain up to aa 253 (start of the transmembrane domain [TD] of sig II) initially remain at the cytosolic side of the endoplasmic reticulum membrane (Fig. 1E) (4, 8, 15, 19, 20). By a poorly understood mechanism, the topology of approximately half of the L chains changes posttranslationally, resulting in a luminal disposition of preS. The L isomer with cytoplasmic preS is essential for virion formation (6, 8, 20).
FIG. 1.
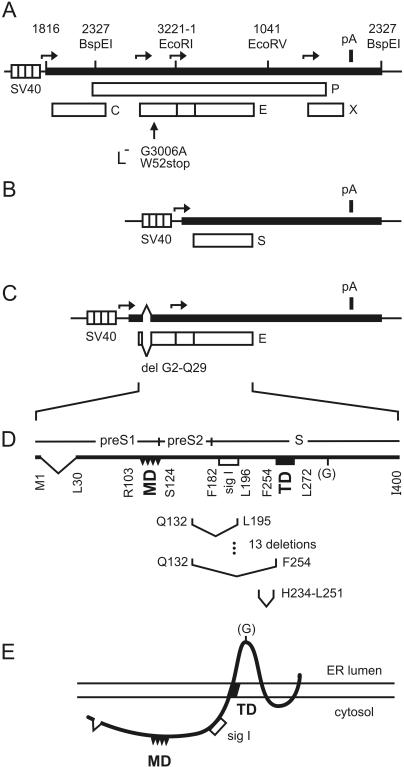
Maps of plasmids and proteins. (A) Expression vector for the L-negative HBV genotype A genome. Expression of the overlength HBV genome (black bar) was driven by the SV40 early promoter (striped box). Restriction enzyme sites are indicated. Numbering starts with the dC of the unique EcoRI site. Transcription start sites of the SV40 promoter and of HBV promoters are indicated by horizontal arrows. Open reading frames for the core (C), polymerase (P), envelope (E), and X (X) proteins are shown as open boxes. The L− mutation (vertical arrow) changes nucleotide 3006 to dA and preS1 aa 52 to a stop codon. pA, polyadenylation signal. (B) Expression vector for the S protein. (C) Expression vector for the L protein. Mutations constructed in this work were introduced into an SV40 expression vector carrying the E open reading frame with an N-terminal deletion (del) of aa 2 to 29 of preS1 to release the secretion block of L. (D) Linear map of L protein. The preS1-preS2-S domain structure of the L protein and the boundaries of the N-terminal deletion, of MD, of sig I, and of TD are shown. The regions spanned by 13 increasing deletions and 1 additional deletion constructed in this work are shown below. G in parentheses, facultative N-glycosylation site. (E) Two-dimensional model of the transmembrane L protein. The distance between MD exposed at the cytosolic side and the endoplasmic reticulum (ER) membrane was shortened by increasing deletions.
A small domain between aa 103 and 124 in the cytosolic portion of L termed the matrix domain (MD) has been identified by genetic approaches to play an essential role in HBV nucleocapsid envelopment (Fig. 1D and E) (2, 9, 12). Subtle mutations in this region like the exchange of 2 aa blocked virion formation without affecting subviral particle formation. Neither N-terminal deletions in preS up to aa 103 (5, 11) nor substitutions downstream of MD between aa 125 and 171 (2, 12) inhibited virion formation. Small deletions between aa 209 and 229 (13) also blocked virion morphogenesis; however, the effect of these changes was mediated by the S protein and not by the L protein. Less deleterious mutations in this region of S like substitutions of 2 adjacent aa did not affect virus formation (V. Bruss, unpublished data).
These observations suggest that the 22-aa-long MD is the only region in the 253-aa-long N-terminal cytoplasmic portion of L where the primary amino acid sequence is essential for capsid envelopment. The exact function of MD is unknown. One plausible model is that MD interacts with the capsid to initiate or drive budding (10). This model is supported by in vitro binding assays using HBV core particles and synthetic peptides derived from HBV envelope proteins or in vitro-expressed truncated L proteins (16, 21). Candidate binding sites on the capsid have been mapped to the base of the 2.5-nm-long spikes protruding from the capsid surface and to a rim around pores in the capsid shell (17).
To gain further insights into the functions of L in virion morphogenesis, we introduced 13 increasing deletions in L, all beginning at aa 132 immediately downstream of MD in the background of a simian virus 40 (SV40) expression vector for the L protein (Fig. 1D and Table 1). Mutants are designated according to the number of the C-terminal amino acid of the deletion. We tested the L derivatives for stable expression, secretion as components of subviral lipoprotein particles (Fig. 2), and support of virion morphogenesis (Fig. 3). The L background carried an additional constant deletion from aa 2 to 29 (Fig. 1C) not influencing virion formation but releasing the secretion block mediated by the wild-type L protein (5, 18) and therefore facilitating the interpretation of the phenotype of L derivatives.
TABLE 1.
Geno- and phenotypes of the analyzed L mutantsa
| Deletion | Expression | Secretion | Virion formation |
|---|---|---|---|
| Q132-L195 | + | + | − |
| Q132-D207 | + | + | − |
| Q132-S212 | + | − | NDb |
| Q132-L216 | + | + | + |
| Q132-V221 | + | + | + |
| Q132-G224 | + | + | + |
| Q132-S227 | + | + | + |
| Q132-S232 | + | + | + |
| Q132-H234 | + | + | + |
| Q132-C239 | + | + | − |
| Q132-C243 | + | + | − |
| Q132-L251 | + | + | − |
| Q132-F254 | + | + | − |
| H234-L251 | ND | ND | + |
Expression: detection of L derivatives in transfected cells after 1-h pulse-labeling and 24-h chase (Fig. 2). Secretion: secretion of L derivatives from cells also expressing the wild-type S protein (Fig. 2). Virion formation: complementation of an L-negative HBV genome with respect to virion secretion (Fig. 3).
ND, not determined.
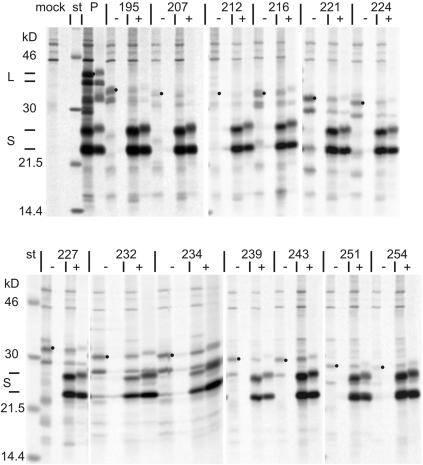
FIG. 2.
Stable expression and secretion of L derivatives. Numbers correspond to the C-terminal ends of the deletions. Lanes: −, transfection with derivatives of the L expression plasmid shown in Fig. 1C; +, cotransfection with the S expression plasmid shown in Fig. 1B. Materials from cell lysates and media were loaded onto the left and right lanes of each panel, respectively. P, parental construct. Dots mark the position of the N-glycosylated L derivative in the intracellular material. Molecular masses of the standard proteins (st) are indicated on the left. The positions of the parental L and S proteins are marked by short bars on the left.
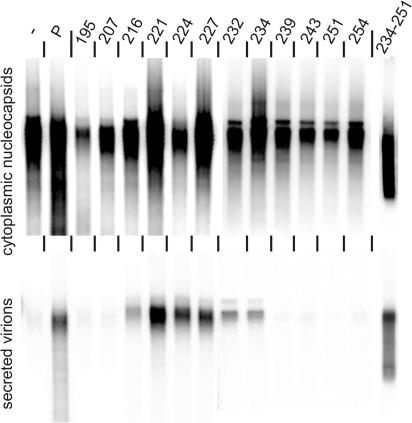
FIG. 3.
Complementation of the L-negative HBV genome by L derivatives. Cytoplasmic (upper panel) or secreted, virion-derived (lower panel) nucleocapsids were detected by the endogenous polymerase reaction. Single numbers correspond to the C-terminal ends of the deletions in L. 234-251, derivative of parental L expression plasmid (Fig. 1C) lacking codons 234 to 251 of the L open reading frame; −, transfection without L expression vector; P, complementation with the parental L expression vector.
Stable expression of the L variants was tested in the absence and presence of additional S protein by transfection of COS7 cells with the plasmids shown in Fig. 1B and C, pulse-chase labeling with radioactive methionine, and immunoprecipitation from cell lysates and media with a polyclonal antiserum against hepatitis B surface antigen (for technical details, see reference 5). The materials from cells and media were loaded adjacent to each other onto a polyacrylamide gel and are depicted after electrophoresis by fluorography in Fig. 2. The left (right) lane of each pair shows the intracellular (secreted) material. The S and M proteins were coexpressed from the parental plasmid (lanes P) due to the internal HBV promoter in the preS domain. The L, S, and glycosylated M proteins were released from the cells as expected. All 13 L mutants were detectable in cell lysates after the chase period (Fig. 2, left lane of each pair of lines designated with a minus sign) as a double band with a 3-kDa molecular mass difference due to a facultative N-glycosylation in the S domain. Mutants 207, 212, and 254 were only weakly expressed but still detectable. The deletions removed the start codon for S expression. In the absence of S, no L derivative was secreted from the cells, as expected (Fig. 2, right lane of panels marked with a minus sign). However, coexpression of wild-type S mobilized most of the L mutants and they appeared probably as components of mixed subviral particles in the culture medium (Fig. 2, right lanes of panels labeled with a plus sign). Mutants 195, 207, and 212 could not or could only barely be detected in the medium after S coexpression. Possibly the interaction between L and S required to form the macromolecular complex of mixed subviral lipoprotein particles (22) was blocked in these three mutants.
To test whether the L variants can achieve the function of the wild-type L protein in virion formation, they were coexpressed in Huh7 cells together with a genomic HBV construct carrying a nonsense mutation at codon 52 of preS1 blocking wild-type L expression (Fig. 1A). Five days posttransfection, nucleocapsids from cells and virions from media were immunoprecipitated with specific antisera and the presence of nucleocapsids and virions was measured by a radioactive endogenous polymerase reaction (for technical details, see reference 3). The isolated labeled genomes were visualized by agarose gel electrophoresis and fluorography. Intracellular nucleocapsids could be easily detected in all cases (Fig. 3, upper panel). Secretion of virions was only supported by mutants 216 to 234 (Fig. 3, lower panel).
The reason for the failure of mutants 195 and 207 to support virion morphogenesis was unclear (Table 1). Possibly, aberrant folding of the cytosolic preS domain of these mutants prevented a functional interaction of MD with the capsid. Mutants 216 to 234 supported virion formation, clearly demonstrating that the region from aa 132 to 234 of L was dispensable for morphogenesis. This area encloses the stretch from aa 209 to 229 of L shown by Loffler-Mary and colleagues to be dispensable for nucleocapsid envelopment (13). L mutants 239 to 254 failed to complement the L-negative HBV genome. However, the amino acid sequence between residues 234 and 251 was not important as demonstrated by the functional L mutant 234-251 (Fig. 3, right lane) carrying a deletion with these boundaries (Fig. 1D). The expression plasmid for this mutant carried a missense mutation of the M and S start codons because coexpression of an S protein with the 234-251 deletion had a transdominant negative effect on virion formation (data not shown).
These results demonstrate that a minimal distance rather than a specific sequence is required between MD and TD. This minimal distance was around 26 aa since mutant 239 lacking only 5 additional aa between MD and TD relative to the functional mutant 234 was strongly blocked in virion morphogenesis. A candidate binding site for factors interacting with the capsid during envelopment was mapped to the base of the protruding spike and the rim of pores on the capsid (17). The spike has a length of 24 aa in alpha-helical conformation (23). This corresponds quite well to the minimal distance between MD and TD of L determined in this work and fits the model that MD binds to one or both of the sites mapped on the capsid surface.
Acknowledgments
We acknowledge Otto Götze for support.
This work was supported by the Deutsche Forschungsgemeinschaft, Graduiertenkolleg 521 and Sonderforschungsbereich 402, project C2.
REFERENCES
- 1.Bruss, V. 2004. Envelopment of the hepatitis B virus nucleocapsid. Virus Res. 106:199-209. [DOI] [PubMed] [Google Scholar]
- 2.Bruss, V. 1997. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 71:9350-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., and R. Thomssen. 1994. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J. Virol. 68:1643-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss, V., and K. Vieluf. 1995. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J. Virol. 69:6652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eble, B. E., D. R. MacRae, V. R. Lingappa, and D. Ganem. 1987. Multiple topogenic sequences determine the transmembrane orientation of the hepatitis B surface antigen. Mol. Cell. Biol. 7:3591-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo, J. T., and J. C. Pugh. 1997. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J. Virol. 71:1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenhoff, R. J., and J. Summers. 1994. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J. Virol. 68:4565-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Pogam, S., and C. Shih. 2002. Influence of a putative intermolecular interaction between core and the pre-S1 domain of the large envelope protein on hepatitis B virus secretion. J. Virol. 76:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1998. Role of the pre-S2 domain of the large envelope protein in hepatitis B virus assembly and infectivity. J. Virol. 72:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loffler-Mary, H., J. Dumortier, C. Klentsch-Zimmer, and R. Prange. 2000. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270:358-367. [DOI] [PubMed] [Google Scholar]
- 14.Nassal, M. 1996. Hepatitis B virus morphogenesis. Curr. Top. Microbiol. Immunol. 214:297-337. [DOI] [PubMed] [Google Scholar]
- 15.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poisson, F., A. Severac, C. Hourioux, A. Goudeau, and P. Roingeard. 1997. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology 228:115-120. [DOI] [PubMed] [Google Scholar]
- 17.Ponsel, D., and V. Bruss. 2003. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J. Virol. 77:416-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prange, R., A. Clemen, and R. E. Streeck. 1991. Myristylation is involved in intracellular retention of hepatitis B virus envelope proteins. J. Virol. 65:3919-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swameye, I., and H. Schaller. 1997. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J. Virol. 71:9434-9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan, W. S., M. R. Dyson, and K. Murray. 1999. Two distinct segments of the hepatitis B virus surface antigen contribute synergistically to its association with the viral core particles. J. Mol. Biol. 286:797-808. [DOI] [PubMed] [Google Scholar]
- 22.Wunderlich, G., and V. Bruss. 1996. Characterization of early hepatitis B virus surface protein oligomers. Arch. Virol. 141:1191-1205. [DOI] [PubMed] [Google Scholar]
- 23.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]