Abstract
Antiretroviral drug-resistant human immunodeficiency virus type 1 (HIV-1) is a major, growing, public health problem. Immune responses targeting epitopes spanning drug resistance sites could ameliorate development of drug resistance. We studied 25 individuals harboring multidrug-resistant HIV-1 for T-cell immunity to HIV-1 proteins and peptides spanning all common drug resistance mutations. CD8 T cells targeting epitopes spanning drug-induced mutations were detected but only in the 3 individuals with robust HIV-specific T-cell activity. Novel CD8 T-cell responses were detected against the common L63P and L10I protease inhibitor fitness mutations. Induction of T-cell immunity to drug-resistant variants was demonstrated in simian human immunodeficiency virus-infected macaques, where both CD8 and CD4 T-cell immune responses to reverse transcriptase and protease antiretroviral mutations were elicited using a novel peptide-based immunotherapy. T-cell responses to antiretroviral resistance mutations were strongest in the most immunocompetent animals. This study suggests feasible strategies to further evaluate the potential of limiting antiretroviral drug resistance through induction of T-cell immunity.
Antiretroviral (ARV) therapies have dramatically reduced the mortality rate from human immunodeficiency virus (HIV) in the developed world (22). Unfortunately, current ARV therapies are not curative, and many treated patients develop resistance to one or more drugs (11), which is costly and may lead to complete treatment failure and death. As ARV therapy becomes increasingly accessible, the global burden of ARV resistance will likely increase dramatically. Newer, more potent and less complicated treatment regimens and efforts to maximize patient compliance should help limit this, but additional strategies are needed. Characterizing immune defenses against ARV drug-resistant strains could initiate novel strategies to reduce rates of ARV drug resistance.
ARV drug resistance is associated with specific mutations in the viral genome. For example, lamivudine usage is commonly associated with the amino acid substitution methionine (M) to valine (V) at position 184 of the HIV type 1 (HIV-1) reverse transcriptase (RT) enzyme (M184V), rendering the virus resistant to this drug (29, 34). Similar mutations have been described for all inhibitors of RT and protease enzymes currently in clinical use. The development of resistance is frequently associated with a reduction in viral replicative capacity, and a series of compensatory “fitness” mutations have also been observed (16).
T-cell immune responses are important in obtaining partial control of HIV replication. Vaccines based on inducing cell-mediated immunity have shown promise in simian models and are progressing to clinical trials (1, 4, 24, 30). However, mutational escape from CD8 T cells has also been observed at the individual and population levels (3, 17). It may be beneficial if the new protein sequences generated following the development of ARV mutations were recognized as novel T-cell epitopes, potentially providing an immune barrier against the development of resistance. Prior studies have examined the interaction between CD8 T-cell responses and drug resistance in selected patient groups (12, 27, 28). Three of 52 (primarily HLA A2 positive) individuals from these studies had detectable CD8 T-cell responses to ARV drug-resistant forms of HIV-1 but not against the wild type. Only responses to 5 ARV-induced mutations were examined. The frequency of T-cell responses to epitopes spanning the more than 30 relatively common drug resistance mutations, in an unselected cohort of ARV-treated subjects, is unknown. We examined T-cell responses directed to the wild type and drug-induced mutations in patients harboring multidrug-resistant HIV-1 and assessed whether T-cell responses against epitopes spanning sites of ARV drug-resistant mutations could be induced in simian human immunodeficiency virus (SHIV)-infected macaques.
MATERIALS AND METHODS
Patient cohort.
Human Research Ethics approval was granted to conduct this study. Subjects with ARV drug-resistant HIV-1 likely to be capable of generating T-cell responses to HIV were studied. Patients who met these inclusion criteria were recruited: HIV-positive adults attending the Melbourne Sexual Health Clinic with a current CD4 count of >50, at least one detectable plasma viral RNA measurement in the last 12 months, and viral genotyping within 24 months demonstrating 3 or more drug-induced mutations in RT (n = 21) (M41L, E44D, K65R, D67N, T69D, K70R, L74V, V75T, A98G, K103N, V118I, Q151 M, Y181C, M184V, M184I, Y188L, G190A, L210W, T215F, T215Y, K219Q) or protease (n = 13) (L10I, K20R, D30N, M46I, G48V, I50V, F53L, I54V, L63P, V82A, V82T, I84V, L90M) (Table 1). Genotyping of the RT and protease genes of the predominant HIV-1 species in plasma was kindly performed by Chris Birch and Tracey Middleton at the Victorian Infectious Diseases Reference Laboratory using an ABI sequencing method as previously described (5).
TABLE 1.
Antiretroviral drug resistance mutations, peptides, and frequencies in various cohorts
| Mutation | Sequence of 17-mer used to analyze T-cell responses | Prevalence in cohort (%)
|
||
|---|---|---|---|---|
| VIDRL | Stanforda | This report | ||
| Protease | ||||
| L10I | QITLWQRPIVTIKIGGQ | 28 | 64 | 40 |
| K20R | TIKIGGQLREALLDTGA | 6 | 15 | 12 |
| D30N | ALLDTGADNTVLEEMNL | 3 | 5 | 0 |
| M46I | LPGRWKPKIIGGIGGFI | 10 | 30 | 16 |
| G48V | GRWKPKMIVGIGGFIKV | 4 | 11 | 8 |
| I50V | WKPKMIGGVGGFIKVRQ | 1 | 0 | 0 |
| F53L | KMIGGIGGLIKVRQYDQ | 1 | 12 | 4 |
| I54V | MIGGIGGFVKVRQYDQI | 10 | 38 | 20 |
| L63P | KVRQYDQIPIEICGHKA | 59 | 85 | 84 |
| V82A | TVLVGPTPANIIGRNLL | 10 | 39 | 28 |
| V82T | TVLVGPTPTNIIGRNLL | 1 | 4 | 4 |
| I84V | LVGPTPVNVIGRNLLTQ | 7 | 30 | 8 |
| L90M | VNIIGRNLMTQIGCTLN | 22 | 61 | 36 |
| RT | ||||
| M41L | ALVEICTELEKEGKISK | 32 | 47 | 60 |
| E44D | EICTEMEKDGKISKIGP | 8 | 12 | 12 |
| K65R | NTPVFAIKRKDSTKWRK | 8 | 2 | 0 |
| D67N | PVFAIKKKNSTKWRKLV | 15 | 35 | 48 |
| T69D | FAIKKKDSDKWRKLVDF | 6 | 12 | 16 |
| K70R | AIKKKDSTRWRKLVDFR | 6 | 25 | 40 |
| L74V | KDSTKWRKVVDFRELNK | 9 | 12 | 12 |
| V75T | DSTKWRKLTDFRELNKR | 1 | 0 | 0 |
| A98G | VQLGIPHPGGLKKKKSV | 2 | 5 | 12 |
| K103N | PHPAGLKKNKSVTVLDV | 20 | 61 | 16 |
| V118I | DVGDAYFSIPLDKDFRK | 7 | 18 | 36 |
| Q151M | RYQYNVLPMGWKGSPAI | 1 | 2 | 4 |
| Y181C | KQNPDIVICQYMDDLYV | 13 | 28 | 28 |
| M184V | PDIVIYQYVDDLYVGSD | 35 | 41 | 68 |
| G190A | QYMDDLYVASDLEIGQH | 9 | 14 | 28 |
| L210W | IEELRQHLWRWGFFTPD | 8 | 29 | 36 |
| T215F | QHLLRWGFFTPDKKHQK | 3 | 11 | 24 |
| T215Y | QHLLRWGFYTPDKKHQK | 12 | 42 | 44 |
| K219Q | RWGFTTPDQKHQKEPPF | 3 | 15 | 36 |
The Stanford HIV drug resistance database can be found at http://hivdb.stanford.edu. Data shown refer to patients treated with ≥3 protease inhibitors (protease data) and nonnucleoside reverse transcriptase inhibitors (reverse transcriptase data).
Peptides.
A panel of 34 17-mer peptides were synthesized (Mimotopes, Croydon, Australia) corresponding to common ARV resistance mutations, as listed above. These 17-mer peptides contained the mutant amino acid substitution as the middle (ninth) amino acid such that any cleaved 9-mer would contain the mutant amino acid (Table 1). A control set of 17-mer peptides, containing the wild-type amino acid sequence, was also synthesized. Lyophilized peptides were reconstituted in 100% dimethyl sulfoxide (DMSO; Sigma, St. Louis, Mo.). Four pools of peptides, containing either mutant or wild-type RT or protease peptides, were subsequently made. Six pools of 15-mer peptides overlapping by 11 amino acids comprising all 9 HIV-1 proteins (HIV-1 subtype B Env MN and consensus subtype B Gag, Pol1, Pol2, Rev/Tat/Vpu [RTV], and Vif/Nef/Vpr [VNV]; kindly supplied by the NIH AIDS Research and Reference Reagent Program) were used to detect HIV-1-specific immune responses. The large peptide pool spanning Pol was split into two pools, termed Pol1 (peptides 1 to 125) and Pol2 (peptides 126 to 249). Smaller 9-mer peptides were also made to define the minimal epitope (GL Biochem, Shanghai, China, or kindly made by David Jackson, University of Melbourne, Australia).
T-cell assays.
Intracellular cytokine staining (ICS) for gamma interferon (IFN-γ) was used to measure CD4 and CD8 T-cell responses as previously described (9, 15). Briefly, 200 μl of whole blood was incubated at 37°C with peptides at a final concentration of 1 μg/ml/peptide in the presence of costimulatory antibodies to CD28 and CD49d. Brefeldin A (Sigma) was added after 2 h (10 μg/ml), and the incubation was terminated after 7 h. Negative-control (DMSO alone) and positive-control (combined Staphylococcus enterotoxin B and pokeweed mitogen; Sigma) wells were included. Cells were surface stained with CD4-fluorescein isothiocyanate, CD3-phycoerythrin, and CD8-peridinin chlorophyll protein complex, lysed, permeabilized, and stained intracellularly with IFN-γ-allophycocyanin. Analysis was performed on a FACSort and with Cell Quest software. A result was considered positive if there were at least 0.05% CD3+ CD8+ (or CD3+ CD4+) T lymphocytes expressing IFN-γ and the number of antigen-stimulated cells expressing IFN-γ was three times above the number of control-stimulated cells. All antibodies and flow cytometry instruments and software were obtained from Becton Dickinson, San Diego, California.
HLA typing and restriction of T-cell responses.
HLA class I typing of individuals was kindly performed by the Victorian Transplant and Immunogenetics Service (Parkville, Australia) using sequence-based typing of exons 2 and 3 (10).
The following HLA class I homozygote and transfected cell lines (kindly supplied by Nicole Mifsud and Simon Knowles, Australian Red Cross, South Melbourne, Australia) were used to restrict T cell epitopes: 9010 (A*6802, B*5301, Cw*0401), 9007 (A*0201, B*5701, Cw*0602), 92CLC14 (A*11, B*35, Cw*0401), 721.221.B*3501 (B*3501), 721.221 parental (no HLA class I expression), 9088 (A*0101, B*0801, C*0701), and T265 (A*0101, B*0702, C*0702). To major histocompatibility complex (MHC) restrict T-cell responses, 1 million cells were pulsed with 1 μg/ml of peptide for 30 min at 37°C. Excess peptide was washed off, and cells were incubated with whole blood for 7 h (with the last 5 h in the presence of brefeldin A) as per ICS protocol (see above).
Alternatively, a peptide-specific cell line was derived from peripheral blood mononuclear cells (PMBC). Autologous PBMC (3 × 106) were pulsed with target peptide and cultured in RPMI with 10% fetal calf serum and interleukin-2 (20 μg/ml). At 7 days, cells were fed with autologous phytohemagglutinin blasts (at a ratio of 5:1), which had been previously been pulsed with the target peptide for 1 h at 37°C and then gamma-irradiated. At 13 days, cells were incubated with a matrix of cell lines (which had been pulsed with the target peptide) in a standard ICS assay as described above.
Vaccination of Macaca nemestrina with ARV drug-resistant peptides.
Ethics approval was obtained to vaccinate 8 juvenile outbred M. nemestrina (previously infected with SHIVmn229) (9) by using a novel technique involving three doses of peptide-pulsed whole blood (5 μg/ml/peptide pulsed onto 9 ml of whole blood) over 8 weeks, at 4-week intervals (8). The pools of 34 ARV drug-resistant 17-mer peptides (Table 1) were solubilized in DMSO. Nine milliliters of acid citrate dextrose-anticoagulated blood was centrifuged to remove 3 ml of plasma and then pulsed with 5 μg/ml/peptide of the resistant peptide pools for 1 h at 37°C. The peptide-pulsed autologous blood sample was then immediately reinfused intravenously. Sequential blood samples were assessed for CD4 and CD8 T-cell responses to the ARV drug-resistant peptides by ICS as described above.
RESULTS
Patient cohort.
Patients attending a large urban HIV clinic were studied. Since February 1998, 254 samples had been submitted to the Victorian Infectious Disease Reference Laboratory (VIDRL) for HIV-1 genotypic analysis of 173 patients (Fig. 1). Of these, 72 had been performed within the last 2 years and contained at least 1 common ARV drug resistance mutation; the 34 amino acid substitutions selected for analysis were those common to both the VIDRL and Stanford databases of genotypic HIV resistance mutations (Table 1). Twenty-five (35%) of the 72 people had at least 3 mutations, a CD4 count of >50/μl, and detectable HIV-1 RNA in plasma in the preceding year and were, therefore, likely to be generating T-cell immune responses to HIV-1 (19). Clinician interviews revealed three additional subjects who were managed at the clinic (but had had genotype testing performed through other institutions) and fit the inclusion criteria. From this initial cohort of 28 subjects, 3 were lost to follow-up prior to enrollment. All 25 remaining eligible subjects participated in the study.
FIG. 1.
Flowchart of subject enrollment in the study of T-cell responses in ARV drug-resistant HIV-1. All eligible patients were recruited from a single site, the Melbourne Sexual Health Centre. All patients had HIV viral load (VL) measurements, CD4 counts, and HIV genotyping performed at the Victorian Infectious Diseases Reference Laboratory, except 3 additional subjects who were monitored at the clinic but had resistance testing performed at another institution.
Consistent with recruiting subjects harboring multidrug-resistant HIV-1, the participants had been treated previously with a mean of 6.4 ARV drugs (range, 2 to 14) and were currently taking, on average, 3.2 ARV drugs (range, 2 to 6) (Table 2). The cohort had HIV-1 strains with an average of 7.9 (range, 3 to 16) drug resistance mutations. The average numbers of protease and RT mutations were 2.6 and 5.2, respectively. The relative frequency of mutations in our cohort was comparable to those of the Stanford and VIDRL databases (Table 1). The subjects had modest levels of immunodeficiency: mean current and nadir peripheral CD4 T-cell counts were 407/μl and 190/μl, respectively (Table 2).
TABLE 2.
| Subject no. | Current ARV therapy | Previous ARV therapy | ARV drug resistance mutations (protease; RT) | HIV RNA count in plasma (copies/ml plasma) | Current CD4 count (per μl blood) | Nadir CD4 count (per μl blood) |
|---|---|---|---|---|---|---|
| 1 | LPV/r, TNF, 3TC | AZT, SQV, ddC, ddI, ABC, NVP, INV, HU | L10I, L63P, M41L, L74V, Y181C, M184V, G190A, T215Y | <50 (19,100)b | 356 | 223 |
| 2 | LPV/r, TNF, ddI | AZT, 3TC, SQV, ddC, d4T, NVP | D67N, K70R, K103N, G190A, T215F, K219Q | <400 (1,390) | 290 | 173 |
| 3 | RTV, ATV, 3TC, TNF | AZT, SQV, ddI, d4T, INV, ddC | L63P, M41L, D67N, K70R, V118I, Q151M, Y181C, T215F, K219Q | 38,900 | 110 | 41 |
| 4 | LPV/r, NVP, ddI | AZT, 3TC, SQV, ddC, d4T, INV | L63P, L90M, D67N, T69D, K70R, M184V, L210W, T215F, K219Q | 87 | 318 | 182 |
| 5 | LPV/r, ddI, 3TC | AZT, ddC, ABC, NVP, TNF | M46I, I54V, L63P, V82T, L90M, D67N, K70R, M184V, K219Q | 139,000 | 519 | 389 |
| 6 | ABC, NVP, ddI | AZT, 3TC, RTV, INV, TNF | L63P, M41L, M184V, T215Y | 2,930 | 637 | 430 |
| 7 | LPV/r, EFV | AZT, 3TC, ddC | L63P, M41L, D67N, T69D, K70R, A98G, M184V, L210W, T215F, K219Q | <50 (7,050) | 308 | 190 |
| 8 | LPV/r, TNF, 3TC, AZT | SQV, ddI, d4T, INV, RTV, NVP, NFV, ABC, AMP | L10I, M46I, L63P, I84V, L90M, M41L, V118I, Y181C, M184V, G190A, L210W, T215Y | 100,000 | 110 | 120 |
| 9 | LPV/r, TNF, SQV | AZT, 3TC, ddC, ddI, ABC | L10I, L63P, M184V | <50 (2,160) | 414 | 210 |
| 10 | LPV/r, INV, ABC | AZT, 3TC, ddI, d4T, NVP, TNF | Y181C, M184V, T215Y | 3,140 | 333 | 40 |
| 11 | nil | AZT, 3TC, d4T INV | L10I, M46I, I54V, L63P, V82A, L90M, M41L, D67N, V118I, M184V, T215Y | 115,000 | 518 | 428 |
| 12 | ABC, TNF, EFV, LPV, ATV | AZT, 3TC, SQV, ddC, ddI, d4T, NFV, INV | L10I, M46I, L63P, I84V, L90M, M41L, T69D, V118I, T215F, K219Q | <400 (500) | 271 | 132 |
| 13 | d4T, NVP, 3TC | AZT, ddC | L63P, K70R, M184V | 1,830 | 370 | 300 |
| 14 | d4T, NVP, 3TC | AZT, ddC, INV, NFV, ABC, EFV | M41L, M184V, Y188L, L210W, T215Y | 14,100 | 1144 | 301 |
| 15 | LPV/r, NVP, TNF | AZT, 3TC, ABC, ddC, ddI, d4T, INV, NFV, HU, EFV | L63P, M41L, K103N | <50 (369,000) | 357 | 112 |
| 16 | ABC, AZT, 3TC | d4T, NVP, EFV | L63P, A98G, M184V | 2,970 | 821 | 290 |
| 17 | nil | AZT, 3TC, ddC, d4T, NVP, INV, HU, EFV, LPV/r, ABC, TNF | D67N, K70R, G190A, K219Q | 830 | 176 | 15 |
| 18 | LPV/r, TNF, ddI | AZT, 3TC, SQV, ddC, d4T, INV, RTV | L10I, I54V, L63P, V82A, L90M, M41L, V118I, M184V, L210W, T215Y | 2,540 | 426 | 119 |
| 19 | TNF, NVP, ddI | AZT, 3TC, SQV, ddC | K20R, L90M, K70R, M184V, K219Q | 970 | 776 | 436 |
| 20 | LPV/r, ABC, ATV | AZT, 3TC, SQV, ddC, ddI, d4T, NVP, DEL, HU, AMP | L10I, K20R, F53L, I54V, L63P, V82A, L90M, M41L, E44D, D67N, K70R, A98G, V118I, G190A, L210W, T215Y | 5,780 | 394 | 127 |
| 21 | AZT, 3TC, ABC, TNF, RTV, ATZ | SQV, ddI, d4T, INV, HU, EFV, LPV/r, AMP | L10I, L63P, V82A, Y181C, M184V | 100,000 | 55 | 24 |
| 22 | ABC, TNF, NVP, 3TC | AZT, d4T, SQV | L10I, G48V, I54V, L63P, V82A, M41L, D67N, K70R, K103N, Y181C, G190A, T215F, K219Q | <400 (5,370) | 317 | 137 |
| 23 | ABC, AZT, 3TC, NVP | SQV, ddI, d4T, INV, NFV | L10I, L63P, V82A, M41L, D67N, L74V, V118I, M184V, L210W, T215Y | 2,330 | 381 | 178 |
| 24 | INV, RTV, TNF, ddI | AZT, 3TC, d4T, NVP, ABC, LPV | L63P, M41L, E44D, D67N, T69D, L74V, V118I, Y181C, M184V, G190A, L210W, T215Y | <400 (55,400) | 289 | 130 |
| 25 | RTV, EFV, ATV, T20 | AZT, SQV, ddC, ddI, d4T, NVP, INV, HU, DEL, LPV/r, TNF, RTV, ABC, NFV | L10I, K20R, G48V, L63P, V82A, L90M, M41L, E44D, D67N, V118I, M184V, L210W, T215Y, K219Q | 150 | 476 | 28 |
LPV/r, lopinavir/ritonavir; TNF, tenofovir; 3TC, lamivudine; AZT, zidovudine; SQV, saquinavir; ddC, dalcytabine; ddI, didanosine; ABC, abacavir; NVP, nevirapine; INV, indinavir; HU, hydroxyurea; d4T, stavudine; RTV, ritnoavir; NFV, nelfinavir; AMP, amprenavir; EFV, efavirenz; ATV, atazanavir; DEL, delavirdine; T20, enfurvitide; nil, no ARV therapy.
For subjects with undetectable HIV-1 RNA in plasma, the last detectable plasma RNA level in the preceding 12 months is noted in parentheses.
Means ± standard deviations for each category are as follows: number of drugs in current ARV therapy, 3.2 ± 1.2, number of drugs in previous ARV therapy, 6.4 ± 2.8; number of ARV drug resistance mutations, 7.8 ± 3.9; HIV RNA count in plasma, 21,294 ± 42,328 copies/ml plasma; current CD4 count, 407 ± 239/μl blood; nadir CD4 count, 190 ± 131/μl blood.
HIV-1 specific cellular immunity in multidrug-resistant subjects.
General analysis of T-cell immune responses to HIV revealed that 18 (72%) of our cohort had a detectable CD8 HIV-specific immune response to ≥1 HIV-1 protein (Table 3). The strongest and most common responses were directed against the Gag protein, followed by the pool containing peptides from the Nef, Vif, and Vpr proteins. Responses to the Pol1 peptide pool, which includes peptides from the wild-type RT and protease enzymes, were seen in 44% of the subjects and averaged 0.8%. HIV-specific CD4 T-cell responses were only detected in 12 (48%) of the subjects and were much weaker than the CD8 T-cell responses (Table 3), mostly ranging from 0.1% to 0.5%.
TABLE 3.
HIV-1-specific T-cell responses in subjects with ARV drug-resistant HIV-1a
| Subject no. | % CD8 T-cell response to protein
|
% CD4 T-cell response to protein
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag | Pol1 | Pol2 | Env | RTV | VNV | Total | Gag | Pol1 | Pol2 | Env | RTV | VNV | Total | |
| 1 | 2.6 | —b | — | — | — | 0.5 | 3.1 | 0.2 | — | — | — | — | — | 0.2 |
| 2 | 0.1 | 0.2 | 0.3 | — | — | — | 0.6 | 0.1 | — | — | — | — | — | 0.1 |
| 3 | 0.8 | 0.1 | 0.1 | 0.2 | — | 0.1 | 1.3 | 0.1 | — | 0.1 | — | — | 0.1 | 0.3 |
| 4 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 5 | — | 0.1 | — | — | — | 0.1 | 0.2 | — | — | — | — | — | — | — |
| 6 | 0.1 | 0.1 | 0.1 | — | — | 0.2 | 0.5 | — | — | — | — | — | 0.1 | 0.1 |
| 7 | 0.5 | 0.3 | — | — | — | 0.5 | 1.3 | 0.1 | 0.1 | 0.1 | — | — | 0.1 | 0.4 |
| 8 | 2.4 | 0.6 | 0.5 | 1.5 | 0.5 | 2.2 | 7.7 | 0.1 | 0.3 | 0.1 | 0.1 | 0.2 | 0.2 | 1.0 |
| 9 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 10 | 0.1 | — | — | — | — | — | 0.1 | — | — | — | — | — | — | — |
| 11 | 0.2 | — | — | — | — | — | 0.2 | — | — | — | — | — | — | — |
| 12 | 0.9 | — | — | — | — | — | 0.9 | — | — | — | — | — | — | — |
| 13 | 1.1 | 0.2 | 0.2 | 0.1 | — | 0.4 | 1.9 | 0.1 | 0.1 | — | — | — | 0.1 | 0.3 |
| 14 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 15 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 16 | 2.0 | — | 1.2 | 0.3 | — | 1.0 | 4.5 | 0.1 | — | — | — | — | — | 0.1 |
| 17 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 18 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 19 | 3.0 | 0.5 | — | 0.4 | — | 0.8 | 4.7 | — | — | — | — | — | 0.3 | 0.3 |
| 20 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 21 | 0.4 | — | — | 0.4 | — | — | 0.8 | — | — | — | — | — | — | — |
| 22 | 0.9 | 3.8 | 2.6 | 1.2 | — | 1.1 | 9.6 | 0.3 | 0.3 | 0.3 | 0.2 | — | 0.2 | 1.3 |
| 23 | 12.7 | 2.8 | 0.6 | 0.6 | — | 3.6 | 20.3 | 0.6 | 0.3 | — | — | — | — | 0.9 |
| 24 | 3.1 | 0.6 | 1.2 | 1.1 | — | 0.3 | 6.3 | 0.4 | — | 0.1 | 0.1 | — | — | 0.6 |
| 25 | 0.1 | — | 0.1 | 0.1 | 0.1 | 0.2 | 0.6 | 0.1 | — | 0.1 | 0.1 | 0.1 | 0.1 | 0.5 |
| Total (%) (% mean ± SD) | 68 (1.8 ± 3.0) | 44 (0.8 ± 1.3) | 40 (0.7 ± 0.8) | 40 (0.6 ± 0.5) | 8 (0.3 ± 0.3) | 52 (0.8 ± 1.0) | 72 (2.7 ± 4.6) | 40 (0.3 ± 0.5) | 16 (0.4 ± 0.4) | 20 (0.1 ± 0.1) | 12 (0.1 ± 0.1) | 4 (0.1) | 28 (0.1 ± 0.1) | 48 (0.2 ± 0.3) |
RTV, pool containing peptides from HIV-1 proteins rev, tat, and vpu; VNV, pool containing peptides from HIV-1 proteins vif, nef, and vpr. Subjects 8, 22, and 23 (in boldface type) had CD8 T-cell responses to pools containing drug-resistant peptides.
Obtained from repeat testing. Negative responses are indicated with a dash (<0.05% or <3-fold above background response).
T-cell responses against peptides spanning resistance mutations.
We then analyzed T-cell immune responses against a pool of 17-mer peptides spanning common RT and protease resistance mutations (Table 1). Two pools containing wild-type protease and RT 17-mer peptides, without the resistance amino acid changes at position 9, were used as controls. If responses were detected, additional samples were obtained to map the relevant minimal epitope.
In 3 (12%) of the 25 subjects (subjects 8, 22, and 23), CD8 T-cell immune responses were detected against ARV mutant peptides; no ARV mutant peptide-specific CD4 T-cell responses were detected. Interestingly, the 3 subjects mounting CD8 responses to ARV mutant peptides had the strongest overall CD8 and CD4 responses to HIV-1 proteins (Table 3).
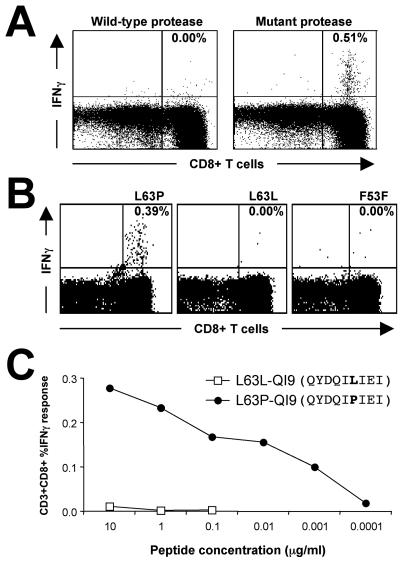
In subject 22, we detected a CD8 T-cell immune response to the resistant protease pool but not to the wild-type protease pool (0.47% cf. 0.00%) (Fig. 2A). This response was directed exclusively against the L63P peptide and failed to recognize the wild-type sequence (Fig. 2B); this subject harbored the L63P mutation. The minimal epitope was QYDQIPIEI (L63P-QI9, L-to-P mutation underlined) (Table 4). A titration curve to L63P-QI9 showed responses down to a concentration of only 1 ng/ml (Fig. 2C). Using a matrix of homozygous and transfected cell lines, we found that HLA-Cw*0401 presented L63P-QI9 (Table 5). To further confirm that HLA-Cw*0401 and not A*1101 restricts this epitope, we tested L63P-specific T cells against the cell line T265, expressing A1101 but not Cw*0401 (HLA type A*1101/3001, B*1302/1502, Cw*0602/0801) and no significant T cell response was detected.
FIG. 2.
T-cell recognition of ARV resistance mutation L63P in subject 22. (A) Recognition of mutant protease peptides. Whole blood was stimulated with a pool of 13 ARV drug-resistant protease peptides (right panel) (Table 1) or a corresponding pool of wild-type protease peptides. Expression of CD8 and intracellular IFN-γ on gated CD3+ lymphocytes is shown. (B) CD8 T cells from subject 22 specifically respond to an epitope spanning the L63P protease fitness mutation. ICS results using 17-mer peptides with the ninth amino acid (the 63rd of the protease protein) either being the wild-type L (L63L) or mutant P (L63P) are shown. An irrelevant adjacent protease 17-mer peptide (F53F) (Table 1) is also shown for comparison. Percent specific CD8 T-cell responses are noted in each panel. (C) A titration panel of the minimal 9-mer epitope (identified in Table 4) of either the wild-type protease containing L at position 63 or mutant protease containing P at position 63.
TABLE 4.
Fine mapping of CD8 T-cell response spanning L63P and L10I mutations
| Peptide sequence (peptide)a | CD8+ IFN-γ response (%) at peptide concn (μg/ml)b:
|
|
|---|---|---|
| 1 | 10 | |
| KVRQYDQIPIEICGHKA(L63P) | 0.14 | |
| KVRQYDQIP | 0.00 | |
| VRQYDQIPI | 0.00 | 0.00 |
| RQYDQIPIE | 0.01 | 0.00 |
| QYDQIPIEI(QI9) | 0.07 | 0.12 |
| YDQIPIEIC | 0.00 | 0.00 |
| DQIPIEICG | 0.00 | 0.00 |
| QIPIEICGH | 0.00 | |
| IPIEICGHK | 0.00 | |
| PIEICGHKA | 0.00 | |
| QITLWQRPIVTIKIGGQ(L10I) | 0.07 | |
| QITLWQRPI | 0.02 | |
| ITLWQRPIV | 0.03 | |
| TLWQRPIVT | 0.03 | |
| LWQRPIVTI | 0.02 | |
| WQRPIVTIK | 0.04 | |
| QRPIVTIKI(QI9) | 0.17 | |
| RPIVTIKIG(RG9) | 0.14 | |
| PIVTIKIGG | 0.01 | |
| IVTIKIGGQ | 0.04 | |
Boldface type indicates the amino acid mutation.
Boldface numbers indicate positive responses.
TABLE 5.
HLA restriction of L63P and L10I CD8 T-cell responses
| Epitope restricted | Cell line | HLA-A type | HLA-B type | HLA-C type | CD8 T-cell response (%) |
|---|---|---|---|---|---|
| L63P | 9007 | A*0201 | B*5701 | Cw*0602 | 0.10 |
| 9010 | A*6802 | B*5301 | Cw*0401 | 0.41a | |
| 92CLC14 | A*11 | B*35 | Cw*0401 | 0.54a | |
| 721.221 parent | 0.08 | ||||
| 721.221 B*3501 | B*3501 | 0.10 | |||
| Subject 22 PBMC | A*0201 | B*1518 | Cw*0401 | 0.26a | |
| A*1101 | B*3501 | Cw*0704 | |||
| L10I | 9012 | A*0201, | B*0701 | Cw*0701 | 0.00 |
| 9088 | A*0101 | B*0801 | Cw*0701 | 1.90a | |
| T230 | A*0205 | B*1510 | Cw*0202 | 0.00 | |
| A*3201 | B*4002 | Cw*0304 | |||
| T240 | A*0201 | B*1501 | Cw*0702 | 0.03 | |
| A*0301 | B*6701 | ||||
| T265 | A*0101 | B*0702 | Cw*0303 | 0.31a | |
| B*1507 | Cw*0702 | ||||
| Subject 23 PBMC | A*0101 | B*0702 | Cw*0701 | NDb | |
| A*0205 | B*0801 | Cw*0702 |
Positive responses in antigen-presenting cells expressing the proposed restricting allele are indicated by boldface type.
ND, not determined in concurrent experiment; previous experiments demonstrated responses in autologous blood (Fig 4).
In subject 8, 0.30% of CD8 T cells were expressing IFN-γ in response to the resistant RT pool and 0.37% expressed IFN-γ in response to the wild-type RT pool. This response in the ARV drug-resistant peptide pool was directed solely at the K65R 17-mer (Fig. 3A) and against the K65K 17-mer within the wild-type pool. Subsequent testing of the 9-mers NTPVFAIKK (NK9) and NTPVFAIKR (NR9) at the N terminus of our original 17-mer confirmed them as the minimal epitopes, respectively. Titration of the CD8 T-cell responses to the 8-mer (NK8) peptide lacking the C-terminal K or R amino acid residue showed substantially weaker responses, demonstrating that the C terminal peptide (position 65) is required to elicit this response (Fig. 3C).
FIG. 3.
T-cell recognition of ARV resistance mutation K65R response in subject 8. (A) CD8 T-cell responses to an epitope spanning the K65R RT mutation also recognize the wild-type RT sequence. ICS results using 17-mer peptides with the ninth amino acid (the 65th of the RT protein) either being the wild-type K (K65K) or mutant R (K65R) are shown. An irrelevant adjacent RT 17-mer peptide (D67D) (Table 1) is also shown for comparison. (B) A titration panel of the minimal 9-mer epitope of either the wild-type RT (NK9) containing K at position 9 (amino acid 65 of RT) or mutant containing R at position 9 (NR9). (C) Titration panel of 9-mer NR9 peptide or 8-mer peptide lacking the C-terminal amino acid.
The wild-type NK9 epitope, with K at residue 65 of RT, is a well-described CD8 T-cell epitope known to be restricted by the HLA A3 supertype (21). HLA class I typing of this patient identified HLA A*6801, part of the A3 superfamily. Previous reports demonstrate that epitopes restricted by the A3 superfamily are capable of binding peptides with either K or R at their C termini (7, 33). Titration of the 9-mer peptides derived from the wild-type (NK9) and ARV drug-resistant (NR9) epitopes showed that they stimulate CD8 T-cell responses with equal efficiency down to a concentration of 1 ng/ml (Fig. 3B). Virus from subject 8 did not harbor the K65R mutation (Table 2), suggesting that this response was generated against wild-type virus and that the K65R mutation would not result in T-cell escape.
In subject 23 (HLA class I type A*0101/0205, B*0702/0801, Cw*0701/0702), a CD8 T-cell response was repeatedly detected against the pool containing resistant protease peptides. The strength of this response (compared to the wild-type protease pool) fluctuated between 0.08% and 0.21% when measured serially over 6 months. Subsequent testing revealed modest CD8 T-cell responses against both the V82A and the L10I 17-mer peptides but not their wild-type counterparts (Fig. 4); subject 23 harbored both the L10I and V82A mutations. It has been documented that the wild-type 9-mer peptide LVGPTPVNI (which is within our wild-type V82V 17-mer peptide) is restricted by HLA A2 (21). HLA A2 individuals also recognize the mutant counterpart LVGPTPANI (12) within the V82A mutant 17-mer. The V82A response we detected in subject 23 did not, however, recognize the V82V wild-type peptide nor a different resistance variant, V82T, nor the adjacent mutation I84V (Fig. 4A). Unfortunately, despite detecting this low-level V82A-specific CD8 T-cell response from multiple separate blood samples, we were not able to conclusively fine map or restrict this response prior to the subject moving overseas.
FIG. 4.
T-cell recognition of ARV resistance mutations V82A and L10I in subject 23. (A) CD8 T-cell responses to the V82A 17-mer peptide but no T-cell recognition of the wild-type V82V peptide or ARV resistance mutation V82T or I84V peptides. (B) CD8 T-cell response to L10I 17-mer peptide but not to L10L wild-type peptide. (C) The response to L10I was mapped to the adjacent 9-mers QI9 and RG9 at 1 μg/ml (Table 4), but a titration study at lower peptide concentrations showed the response was strongest to the L10I-QI9 epitope.
Although CD8 T-cell responses against epitopes spanning the 10th amino acid in protease (wild-type L10L) have been described in HLA A2-positive persons (14), specific recognition of the L10I fitness mutation has not been described. The presence of L10I-specific CD8 T cells was confirmed on subsequent blood samples and the minimal epitope mapped to the 9-mer QRPIVTIKI (L10I-QI9) containing the L-to-I change (underlined) at position 4 of this 9-mer (Table 4; Fig. 4B). This suggests that the response is not a variation of the wild-type L10L response, as this recognizes the 9-mer VTLWQRPLV with the relevant amino acid at position 8 (underlined). The L10I-QI9-specific T cells were HLA restricted using a panel of cell lines and found to be restricted to HLA A*0101 (Table 5).
Immune responses induced to resistant HIV-1 peptides in macaques.
Having detected CD8 T-cell immune responses to ARV drug-resistant peptides in some of our subjects, we examined whether we could induce such responses in nonhuman primates. Such a vaccine strategy could ultimately be studied to prevent or delay drug resistance by providing a T-cell immune barrier against mutations. Eight pigtail macaques, previously infected with SHIVmn229 (9) were vaccinated with a combined pool of 34 ARV drug-resistant mutant protease and RT peptides (Table 1) pulsed onto autologous blood. This technique induced CD4 and CD8 T-cell immune responses to SHIV and hepatitis C virus peptides in macaques in previous studies (8).
Both CD4 and CD8 T-cell immune responses to peptides spanning ARV resistance mutations were readily detected after 2 or 3 immunizations with autologous blood pulsed with the ARV drug-resistant peptides in most of the immunized macaques (Fig. 3; Table 6). No responses were detected against the wild-type RT and protease peptides prior to vaccinations. The one macaque with undetectable levels of SHIV viremia and normal CD4 T-cell counts (macaque 4295) had detectable CD4 and CD8 T cells on multiple occasions that were boosted with multiple immunizations (Fig. 5A). In another macaque, 4277, which had low SHIV RNA levels and a high CD4 T-cell count, 2.31% of all CD4 T cells were responding to the pool of ARV drug-resistant peptides at week 8. Antiretroviral drug-resistant peptide-specific CD8 T-cell responses of 0.17%, 1.25%, and 0.35% were also observed at weeks 5, 8, and 13, respectively, in animal 4277. In the third immunocompetent animal with high total CD4 T-cell counts (monkey 4296), CD4 and CD8 T cells specific for the ARV drug-resistant peptides were also detected. Thawed PBMC from animal 4296 from week 10 (2 weeks after the third vaccination) were also available to confirm this response and assess T-cell immune responses to either the RT or protease resistant pool and compare them to the wild-type peptide pools. CD4 and CD8 T-cell responses were directed against at least one of the peptides in both the RT and protease pools but not against the comparator wild-type peptides (Fig. 5B), demonstrating the immune responses induced in this animal specifically recognized the mutant amino acids conferring drug resistance.
TABLE 6.
Induction of CD8 and CD4 T-cell responses to ARV drug-resistant peptides in macaques
| Animal no. | Total CD4 T-cell count (% of total lymphocytes) | Baseline SHIV viral load (log10 RNA copies/ml plasma) | Baseline T-cell response to pool of ARV drug-resistant peptides (%)a
|
Maximum T-cell response detected to ARV drug-resistant peptides postvaccination (%)a
|
||
|---|---|---|---|---|---|---|
| CD8 | CD4 | CD8 | CD4 | |||
| 4295 | 30.03 | <3.20 | <0.10 | <0.10 | 0.18 | 0.18 |
| 4296 | 28.76 | 4.19 | <0.10 | <0.10 | 0.23 | 0.31 |
| 4277 | 26.16 | 4.70 | <0.10 | <0.10 | 1.25 | 2.31 |
| 4386 | 18.61 | 4.30 | <0.10 | <0.10 | 0.10 | 0.10 |
| H8 | 16.96 | 4.92 | <0.10 | <0.10 | 0.18 | 0.20 |
| 4194 | 5.98 | 5.87 | <0.10 | 0.11 | 0.10 | 0.41 |
| 4241 | 1.52 | 6.16 | <0.10 | <0.10 | <0.10 | 0.46 |
| 1.1705 | 0.08 | 5.35 | <0.10 | <0.10 | <0.10 | <0.10 |
T-cell responses of ≥0.10% are shown in boldface type.
FIG. 5.
Induction of T-cell immunity to ARV drug-resistant peptides in macaques. Macaques were immunized with 3 monthly doses of autologous blood pulsed for 1 h with a pool of 34 17-mer peptides spanning common ARV drug resistance mutations. (A) Kinetic analysis of CD4 and CD8 T-cell responses specific for the combined pool of RT and protease ARV drug-resistant peptides (n = 34) before and after each immunization (arrows) in SHIV-infected macaque 4295. (B) CD8 and CD4 T-cell responses to individual peptide pools of either resistant RT or resistant protease, in comparison with a combined comparator pool of wild-type protease and RT peptides, were analyzed in macaque 4296 after 3 immunizations. Percent CD8 (left hand panels) or CD4 (right hand panels) T-cell responses to the peptides are shown in the upper right quadrants of the panels.
Of the 8 macaques studied (Table 6), 5 had progressive SHIV infection with significant CD4 depletion and higher plasma SHIV viral loads, and immune responses were less efficiently induced in these animals. The four animals with intermediate levels of immunodeficiency (H8, 4194, 4241, and 4386; CD4 T-cell counts, 1.5 to 18.6%; SHIV viral loads, 4.3 to 6.2 log10 copies/ml) had sporadically detected CD4 T-cell immune responses to the ARV drug-resistant peptide pool (range, 0.10 to 0.41%) to the set of drug-resistant peptides (Table 6). Low-level CD8 T-cell immune responses to the resistant peptides were generated in 2 of these 4 animals. The most immunodeficient macaque (1.7105; CD4 T-cell count, 0.08%; SHIV viral load, 5.4 log10 copies/ml) failed to mount CD4 or CD8 T-cell immune responses to ARV drug-resistant epitopes at any time point.
DISCUSSION
This study has two important findings: first, the identification of novel CD8 T-cell immune responses to drug resistance and fitness mutations in individuals with infection with drug-resistant HIV-1; second, immune responses targeting ARV-induced mutations in protease and RT can be induced, in the setting of limited immunodeficiency, by a novel vaccine technique.
We recruited all 25 available subjects infected with strains of HIV-1 encoding multiple ARV drug resistance mutations. Three (12%) subjects generated CD8 T-cell responses to peptides spanning ARV resistance mutations, including one subject with 2 responses. Importantly, these 3 subjects had the highest levels in our cohort of CD8 and CD4 T-cell responses to other regions of HIV-1. These observations suggest that, although ARV resistance mutations were recognized in a minority of our cohort, recognition may be improved in subjects with relative immunocompetence.
Previous studies have mapped immune responses to drug-resistant peptides already described for their wild-type counterparts (e.g., M41 M/L, M184 M/V, and V82V/A) in HLA A2-positive individuals (12, 27, 28). The defining of the HLA Cw*0401-restricted L63P-QI9 epitope, spanning the L63P mutation in HIV-1 protease, is unique in that the wild-type peptide L63L has not been previously described as a class I cytotoxic T lymphocyte (CTL) epitope and we did not detect any responses to this wild-type peptide. We also detected and mapped another novel response in another subject to the L10I mutation, restricted by the allele HLA A*0101, which did not recognize the L10L wild-type variant.
Both the L63P and L10I mutations in protease are located at sites distant to its active binding site and primarily act to restore HIV-1 replicative fitness (20, 32), although they may be the primary source of resistance in some instances (18). Preventing the development of the L63P or L10I mutations could potentially have a broader benefit than preventing any other single mutation, given that they are the most common mutations in our cohort and the Stanford and VIDRL databases (2, 6, 36).
The V82A mutation is associated with resistance to indinavir (23) and, to a lesser extent, lopinavir (13). Only subject 23 had a detectable immune response directed against this epitope, which has previously been described in HLA A2-positive individuals (12). Subject 8 recognized the NR9 epitope spanning the K65R RT mutation, which occurs in some patients exposed to RT inhibitors, particularly didanosine, abacavir, and tenofovir (25, 35). There was equivalent recognition of both the wild-type (NK9) and mutant (NR9) epitopes down to a peptide concentration of 1 ng/ml. This is consistent with the conserved nature of the K→R substitution at residue 9, which preserves binding to the HLA-A3 superfamily of MHC class I molecules (31). Interestingly, subject 8 did not have virus containing the K65R mutation despite having 12 other ARV resistance mutations and exposure to multiple ARV drugs that induce K65R. The potential preventive role of CD8 T-cell responses directed against this mutation could be studied if sufficient numbers of HIV-1-infected HLA A3-positive subjects were studied for the presence or absence of the K65R mutation (17).
Interestingly, although only 3 subjects had detectable responses to ARV mutations, other subjects had both the appropriate amino acid sequences and HLA phenotype yet did not have detectable T-cell responses to ARV resistance mutations. Three of the four HLA Cw*0401-positive subjects in our cohort (subjects 5, 13, and 22) had the L63P mutation and had detectable CD8 T-cell responses to some part of HIV-1, but only subject 22 had a response to the L63P peptide. A primary factor in the recognition of multiple CD8 T-cell epitopes in HIV-infected subjects is the degree of CD4 T-cell help available to augment CD8 T cells. Remarkably, the 3 patients responding to ARV drug-resistant peptides had the highest total levels of HIV-specific CD4 T cells (0.9 to 1.3%), as measured by ICS, at the time of enrollment. This high level of cognate CD4 T-cell help likely played a role in not only the recognition of the ARV resistance mutations but also in the dramatically high levels of CD8 T-cell responses to other parts of HIV-1 (7.7 to 20.3% of all CD8 T cells). Generating high-level HIV-specific CD4 T-cell help is clearly a critical goal of HIV immunotherapy (26).
To model the ability to induce T-cell responses against new epitopes generated by ARV resistance mutations in HIV-infected subjects, we administered peptide-pulsed blood to SHIV-infected macaques, a technique that can induce high levels of specific CD4 and CD8 T-cell responses in macaques (8). Three of the 8 macaques administered 3 doses of peptide-pulsed blood developed CD4 and CD8 T-cell responses to the combined pool containing ARV drug resistance mutations. Not surprisingly, the most immunocompetent SHIV-infected macaques generated the strongest and most consistently detectable T-cell immunity, suggesting parameters for future clinical studies. It is important to stress that although encouraging immune responses were detected in this nonhuman primate study, the ability to induce such responses in humans and the utility of inducing T-cell responses targeting ARV resistance mutations in preventing or delaying the emergence of ARV drug resistance requires further evaluation.
Importantly, however, both CD4 and CD8 T-cell responses to separate pools of RT and protease ARV drug-resistant peptide pools were identified in one macaque, suggesting at least a reasonably broad response across multiple determinants. The breadth of T-cell immunity will likely be of considerable importance in preventing ARV resistance, since several mutational pathways exist for the development of resistance against some ARV drugs. The breadth of responses will be dependent in part on the various MHC alleles of individual hosts capable of presenting the new potential epitopes generated by ARV resistance mutations. Future studies will map responses to individual ARV drug-resistant epitopes to more precisely determine the breadth of immunity induced.
The CD4 T-cell responses induced to epitopes spanning ARV resistance mutations using peptide-pulsed blood vaccine technology are particularly exciting. Such CD4 responses were not observed in our cohort nor in other smaller, previously described cohorts (12, 27, 28). Our data, and other reports, strongly suggest HIV-specific CD4 T-cell responses are linked to effective long-term immunity (26).
In summary, this study demonstrates that individuals infected with drug-resistant HIV-1 can develop CD8 T-cell immune responses specifically targeting ARV drug mutations. The identification of the very common fitness mutations (L63P and L10I) as CD8 T-cell epitopes restricted by relatively common HLA alleles suggests the potential for a broad application of these findings. Further, our macaque study provides a first step forward for subsequent testing of whether T-cell immunity to ARV drug-resistant HIV can be induced in HIV-infected humans, with the ultimate aim of assessing these for protective efficacy against developing ARV drug-resistant HIV.
Acknowledgments
We thank Robert De Rose, Caroline Fernandez, Miranda Smith, Richard Sydenham, and Nicole Mifsud for advice and expert technical assistance; Chris Birch and Tracey Middleton for helpful discussions and kindly providing the genotyping; Kit Fairley, Anne Mijch, Norm Roth, Tina Schmidt, Tim Read, Kerri Boyd, and other staff at the Melbourne Sexual Health Centre for their kind help; Simon Mallal for helpful discussions; and the subjects who very kindly participated in this study.
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Ammaranond, P., P. Cunningham, R. Oelrichs, K. Suzuki, C. Harris, L. Leas, A. Grulich, D. A. Cooper, and A. D. Kelleher. 2003. Rates of transmission of antiretroviral drug resistant strains of HIV-1. J. Clin. Virol. 26:153-161. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 5.Birch, C., T. Middleton, G. Hales, D. Cooper, M. Law, S. Crowe, J. Hoy, and S. Emery. 2003. Limited evolution of HIV antiretroviral drug resistance-associated mutations during the performance of drug resistance testing. J. Acquir. Immune Defic. Syndr. 32:57-61. [DOI] [PubMed] [Google Scholar]
- 6.Brindeiro, R. M., R. S. Diaz, E. C. Sabino, M. G. Morgado, I. L. Pires, L. Brigido, M. C. Dantas, D. Barreira, P. R. Teixeira, and A. Tanuri. 2003. Brazilian Network for HIV Drug Resistance Surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS 17:1063-1069. [DOI] [PubMed] [Google Scholar]
- 7.Chang, K. M., N. H. Gruener, S. Southwood, J. Sidney, G. R. Pape, F. V. Chisari, and A. Sette. 1999. Identification of HLA-A3- and -B7-restricted CTL response to hepatitis C virus in patients with acute and chronic hepatitis C. J. Immunol. 162:1156-1164. [PubMed] [Google Scholar]
- 8.Chea, S., C. J. Dale, R. De Rose, I. A. Ramshaw, and S. J. Kent. 2005. Enhanced cellular immunity in macaques following a novel peptide immunotherapy. J. Virol. 79:3748-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dale, C. J., R. De Rose, I. Stratov, S. Chea, D. C. Montefiori, S. Thomson, I. A. Ramshaw, B. E. Coupar, D. B. Boyle, M. Law, and S. J. Kent. 2004. Efficacy of DNA and fowlpox virus priming/boosting vaccines for simian/human immunodeficiency virus. J. Virol. 78:13819-13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen, J. A., and B. Dupont. 2004. HLA 2004: immunobiology of the human MHC. In The proceedings of the 13th International Histocompatibility Workshop and Congress, vol. I and II. IHWG Press, Seattle, Wash.
- 11.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson, A. C., S. G. Deeks, J. D. Barbour, B. D. Heiken, S. R. Younger, R. Hoh, M. Lane, M. Sallberg, G. M. Ortiz, J. F. Demarest, T. Liegler, R. M. Grant, J. N. Martin, and D. F. Nixon. 2003. Dual pressure from antiretroviral therapy and cell-mediated immune response on the human immunodeficiency virus type 1 protease gene. J. Virol. 77:6743-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korber, B. T. M., C. Brander, B. F. Haynes, R. Koup, C. Kuiken, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.). 2002. HIV molecular immunology. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, New Mexico. [Online.]
- 15.Maecker, H. T., H. S. Dunn, M. A. Suni, E. Khatamzas, C. J. Pitcher, T. Bunde, N. Persaud, W. Trigona, T. M. Fu, E. Sinclair, B. M. Bredt, J. M. McCune, V. C. Maino, F. Kern, and L. J. Picker. 2001. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J. Immunol. Methods 255:27-40. [DOI] [PubMed] [Google Scholar]
- 16.Menendez-Arias, L., M. A. Martinez, M. E. Quinones-Mateu, and J. Martinez-Picado. 2003. Fitness variations and their impact on the evolution of antiretroviral drug resistance. Curr. Drug Targets Infect. Disord. 3:355-371. [DOI] [PubMed] [Google Scholar]
- 17.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 18.Muzammil, S., P. Ross, and E. Freire. 2003. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry 42:631-638. [DOI] [PubMed] [Google Scholar]
- 19.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 20.Ohtaka, H., A. Schon, and E. Freire. 2003. Multidrug resistance to HIV-1 protease inhibition requires cooperative coupling between distal mutations. Biochemistry 42:13659-13666. [DOI] [PubMed] [Google Scholar]
- 21.Propato, A., E. Schiaffella, E. Vicenzi, V. Francavilla, L. Baloni, M. Paroli, L. Finocchi, N. Tanigaki, S. Ghezzi, R. Ferrara, R. Chesnut, B. Livingston, A. Sette, R. Paganelli, F. Aiuti, G. Poli, and V. Barnaba. 2001. Spreading of HIV-specific CD8+ T-cell repertoire in long-term nonprogressors and its role in the control of viral load and disease activity. Hum. Immunol. 62:561-576. [DOI] [PubMed] [Google Scholar]
- 22.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 23.Richman, D. D., and S. Staszewski. 2000. HIV drug resistance and its implications for antiretroviral treatment and strategies, 2nd ed. International Medical Press Ltd., London, United Kingdom.
- 24.Robinson, H. L., D. C. Montefiori, R. P. Johnson, K. H. Manson, M. L. Kalish, J. D. Lifson, T. A. Rizvi, S. Lu, S. L. Hu, G. P. Mazzara, D. L. Panicali, J. G. Herndon, R. Glickman, M. A. Candido, S. L. Lydy, M. S. Wyand, and H. M. McClure. 1999. Neutralizing antibody-independent containment of immunodeficiency virus challenges by DNA priming and recombinant pox virus booster immunizations. Nat. Med. 5:526-534. [DOI] [PubMed] [Google Scholar]
- 25.Roge, B. T., T. L. Katzenstein, N. Obel, H. Nielsen, O. Kirk, C. Pedersen, L. Mathiesen, J. Lundgren, and J. Gerstoft. 2003. K65R with and without S68: a new resistance profile in vivo detected in most patients failing abacavir, didanosine and stavudine. Antivir. Ther. 8:173-182. [PubMed] [Google Scholar]
- 26.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 27.Samri, A., G. Haas, J. Duntze, J. M. Bouley, V. Calvez, C. Katlama, and B. Autran. 2000. Immunogenicity of mutations induced by nucleoside reverse transcriptase inhibitors for human immunodeficiency virus type 1-specific cytotoxic T cells. J. Virol. 74:9306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitt, M., E. Harrer, A. Goldwich, M. Bauerle, I. Graedner, J. R. Kalden, and T. Harrer. 2000. Specific recognition of lamivudine-resistant HIV-1 by cytotoxic T lymphocytes. AIDS 14:653-658. [DOI] [PubMed] [Google Scholar]
- 29.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Schipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, C. Christopherson, et al. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411-1419. [DOI] [PubMed] [Google Scholar]
- 30.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 31.Sidney, J., H. M. Grey, S. Southwood, E. Celis, P. A. Wentworth, M. F. del Guercio, R. T. Kubo, R. W. Chesnut, and A. Sette. 1996. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum. Immunol. 45:79-93. [DOI] [PubMed] [Google Scholar]
- 32.Sune, C., L. Brennan, D. R. Stover, and T. Klimkait. 2004. Effect of polymorphisms on the replicative capacity of protease inhibitor-resistant HIV-1 variants under drug pressure. Clin. Microbiol. Infect. 10:119-126. [DOI] [PubMed] [Google Scholar]
- 33.Threlkeld, S. C., P. A. Wentworth, S. A. Kalams, B. M. Wilkes, D. J. Ruhl, E. Keogh, J. Sidney, S. Southwood, B. D. Walker, and A. Sette. 1997. Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J. Immunol. 159:1648-1657. [PubMed] [Google Scholar]
- 34.Wainberg, M. A., H. Salomon, Z. Gu, J. S. Montaner, T. P. Cooley, R. McCaffrey, J. Ruedy, H. M. Hirst, N. Cammack, J. Cameron, et al. 1995. Development of HIV-1 resistance to (-)2′-deoxy-3′-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS 9:351-357. [PubMed] [Google Scholar]
- 35.White, K. L., N. A. Margot, T. Wrin, C. J. Petropoulos, M. D. Miller, and L. K. Naeger. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 46:3437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong, P., L. Kang, Q. Pan, F. Konings, S. Burda, L. Ma, Y. Xue, X. Zheng, Z. Jin, and P. Nyambi. 2003. Identification and distribution of HIV type 1 genetic diversity and protease inhibitor resistance-associated mutations in Shanghai, P. R. China. J. Acquir. Immune Defic. Syndr. 34:91-101. [DOI] [PubMed] [Google Scholar]