Abstract
Sperm typing is an effective way to study recombination rate on a fine scale in regions of interest. There are two strategies for the amplification of single meiotic recombinants: repulsion-phase allele-specific PCR and whole genome amplification (WGA). The former can selectively amplify single recombinant molecules from a batch of sperm but is not scalable for high-throughput operation. Currently, primer extension pre-amplification is the only method used in WGA of single sperm, whereas it has limited capacity to produce high-coverage products enough for the analysis of local recombination rate in multiple large regions. Here, we applied for the first time a recently developed WGA method, multiple displacement amplification (MDA), to amplify single sperm DNA, and demonstrated its great potential for producing high-yield and high-coverage products. In a 50 μl reaction, 76 or 93% of loci can be amplified at least 2500- or 250-fold, respectively, from single sperm DNA, and second-round MDA can further offer >200-fold amplification. The MDA products are usable for a variety of genetic applications, including sequencing and microsatellite marker and single nucleotide polymorphism (SNP) analysis. The use of MDA in single sperm amplification may open a new era for studies on local recombination rates.
INTRODUCTION
A detailed knowledge of linkage disequilibrium (LD) patterns across the human genome was widely considered a prerequisite for comprehensive association testing (1). Recent data have shown that LD in human populations is highly structured into discrete blocks with limited haplotype diversity (2–5). This LD structure was believed to result from the interplay between recombination hotspots (3,5,6) and the demographic history of human populations (7,8). Little is known about the role of recombination in shaping LD patterns in populations, although statistical approaches may provide some clues (9–11). The answer to this question may lie in comparison of population LD structure with the distribution of meiotic crossovers. Sperm typing can identify the distribution of male local meiotic recombination rate, which can at least partially explain the LD pattern, as exemplified by Jeffreys et al. (3).
Two strategies have been taken for detecting highly localized meiotic recombination hotspots in sperm. One is to amplify single recombinant molecules using repulsion-phase allele-specific PCR from a large batch of sperm DNA, followed by localization of crossover sites (3,12–14). The other is to scan thousands of single sperm cells to identify and localize meiotic recombinants (15–17). The former is efficient for fine mapping of crossover sites in a defined region of several kilobases without laborious work on screening recombinants as required in the latter. However, it can only be used in studies on small regions of <10 kb and the workers have to be very careful of possible contamination from artificial recombinants resulting from template switching during PCR amplification (18,19). For the second method, single sperm cells were first pre-amplified by either multiple PCR or whole genome amplification (WGA) to produce sufficient DNA for further multiple genotyping reactions to identify and localize meiotic recombinants. Although multiple PCR has been more widely used for single sperm analysis (20–22), WGA seems to be more promising and preferred for fine mapping of meiotic recombination sites because it can amplify many more marker loci than multiple PCR. Primer extension pre-amplification (PEP) was developed to amplify single sperm DNA on the whole genome level in 1992 (23) and first applied for localization of recombination sites in 1994 (15). Recently, Cullen et al. (16) used PEP to pre-amplify 20 031 single sperm cells, followed by genotyping using 48 short tandem repeat (STR) markers in a 3.3 Mb interval encompassing the major histocompatibility complex (MHC). Unfortunately, their study did not achieve high enough density to allow comparison with the LD map, owing to insufficient PEP products and low marker density.
Recently, multiple displacement amplification (MDA), using Φ29 DNA polymerase and random exonuclease-resistant hexamer primers, has been demonstrated to be very efficient for balanced amplification and generation of long DNA products (>10 kb) from a small amount of DNA, or directly from whole blood (24). The high processivity and fidelity of Φ29 DNA polymerase provides an advantage to MDA in terms of yield, accuracy and coverage over the other PCR-based WGA methods (24,25). Amplification of single lymphocytes or blastomeres using MDA has been successfully carried out (26,27). Up to 92% coverage was estimated from 100 PCRs by detecting 20 different loci in five single lymphocytes, although at heterozygous loci, a high rate of allele dropout (31%) was observed (26).
In this paper, we demonstrate that MDA can be utilized to produce high-coverage and high-yield WGA products from single sperm cells, and the resulting MDA products can be used for a variety of genetic applications, including sequencing and microsatellite marker (STR) and single nucleotide polymorphism (SNP) analysis. It was estimated that 76% of the genomic sequence can be amplified at least 2500-fold, or 93% at least 250-fold. We also showed that the second-round MDA can further give an averaged 236-fold amplification. MDA can therefore be quite useful for amplifying single sperm DNA for a large amount of genotyping reactions and this may open a new avenue for single sperm analysis.
MATERIALS AND METHODS
Sperm sample and genomic DNA preparation
Fertile men considering vasectomy were approached for consent to enter the study. Once informed consent was obtained, a blood sample and semen sample (by masturbation) were collected. The blood sample (10 ml) was collected into tubes with acid citrate dextrose (ACD) and centrifuged at 3300 g at room temperature for 10 min. The intermediate layer where white blood cells were concentrated was collected and resuspended in phosphate-buffered saline (PBS) for further processing for DNA analysis. Genomic DNA was extracted from white blood cells using the standard phenol–chloroform method. DNA concentration was determined using a Hoefer DyNA Quant 200 Fluorometer.
Sperm lysis
Sperm cells were counted with a hemacytometer, diluted to a concentration of either 0.8 or 3 cells/3 μl with PBS and 16 aliquots were prepared of each dilution. Three microliter of diluted sperm cells were dispensed into 200 μl PCR tubes and frozen at −80°C overnight. An aliquot of 3.5 μl of freshly prepared lysis solution (0.1 M DTT, 0.4 M KOH and 10 mM EDTA) was then added, mixed well by gentle vortex and incubated for 10 min on ice for eight aliquots of the dilution of 3 cells/3 μl, or at 65°C for the other aliquots. Lysis was stopped by adding 3.5 μl of neutralizing buffer (buffer B in REPLI-g kit, Qiagen Inc.). The dilution of 3 cells/3 μl was picked to test whether 65°C incubation could lyse sperm cells better or not, and the dilution of 0.8 cells/3 μl was used to obtain aliquots containing single sperm cells. Aliquots named after S01, S02…S16 below were prepared from the dilution of 0.8 cells/3 μl
Multiple displacement amplification
WGA was achieved using REPLI-g™ kit according to the manufacturer's manual (Qiagen Inc.). All samples were pre-amplified by MDA. A PBS blank was included as a negative control. A reaction in a total volume of 50 μl was performed at 30°C overnight and then terminated at 65°C for 10 min. Amplified DNA products were then stored at −20°C. Dilutions of 5- or 50-fold (referred as 1/5C0 and 1/50C0, respectively, below) were used for further sequencing, the coverage test and microsatellite and SNP genotyping analysis. One microliter of a 10-fold diluted S16 MDA product was used as template for the second-round MDA.
PCR and sequencing analysis
In order to determine the aliquots that were successfully pre-amplified by MDA, three genes—TOP1, P53 and CYP1A2—were selected for PCR testing using 1 μl of 1/5C0 MDA product. Primers used are listed in set A of Table 1. A 20 μl mixture was prepared for each reaction and included 1× HotStarTaq buffer, 2.5 mM Mg2+, 0.2 mM dNTP, 0.3 μM of each primer, 1 U HotStarTaq polymerase (Qiagen Inc.) and 1 μl template DNA. The cycling program was 95°C for 15 min; 40 cycles of 94°C for 15 s, 56°C for 30 s, 72°C for 1 min; 72°C for 2 min. Amplified fragments representative of the three genes (TOP1, P53 and CYP1A2) were 1080, 643 and 550 bp in length, respectively. PCR products were checked on 1.5% agarose gels. For the aliquots of the 0.8 cells/3 μl dilution, those MDA products in which at least one of the three genes got amplified were selected for further analyses.
Table 1.
Primers for PCR in this study
| Fragments | Forward primers | Reverse primers | |
|---|---|---|---|
| Set A | TOP1 | 5′-CCCCTAGATTGATGCCAAGA-3′ | 5′-ACTTTAGCCCGGACTAGCAA-3′ |
| P53 | 5′-CTCATCTTGGGCCTGTGTTA-3′ | 5′-ACCGCTTCTTGTCCTGCTT-3′ | |
| CYP1A2 | 5′-CAGAATGCCCTCAACACCTT-3′ | 5′-TCACACAGCTGGTCTGAGGT-3′ | |
| Set B | TOP1 | 5′-TGACTCATCTCTTATGGTTGCAG-3′ | 5′-CAACCCACTGCTGAATGATG-3′ |
| P53 | 5′-GCCTCTTGCTTCTCTTTTCC-3′ | 5′-ACCGCTTCTTGTCCTGCTT-3′ | |
| CYP1A1 | 5′-ATTTCCAGCTGCTGTCACAT-3′ | 5′-CTGGTCTCCAGCTGCCTTTA-3′ | |
| PIK3CA | 5′-TTTGCTCCAAACTGACCAAAC-3′ | 5′-ACTCCAAAGCCTCTTGCTCA-3′ | |
| C6orf195 | 5′-TGATAGGCATTTCTTTTGAAACTGG-3′ | 5′-ACAGGGAAGCAAAGCACTTC-3′ | |
| DKKL1 | 5′-CATCTGCAAAGTGGGCTGAATAGG-3′ | 5′-CCGTGGCAGCTTAATGATCC-3′ | |
| SHH | 5′-TCTCGGAACTCAATGCCCTGTC-3′ | 5′-GGTCCAGGAAAGTGAGGAAG-3′ | |
| ADCYAP1 | 5′-CAAGTGCTGTTCAACTCAGGGA-3′ | 5′-GGCGATGCTAGTAGTCTGGACC-3′ | |
| MSH2 | 5′-TCCATTGGTGTTGTGGGTGT-3′ | 5′-TCAGTTTCCCCATGTCTCCA-3′ | |
| PTEN | 5′-TGAAGACCATAACCCACCACAG-3′ | 5′-CTAGGGCCTCTTGTGCCTTT-3′ | |
| PMS2 | 5′-CTGGATGCTGGTGCCACTAA-3′ | 5′-GCCTGGCACACCGTAAGAA-3′ | |
| CAT | 5′-TACCAGCTCCAGTGGTCAGG-3′ | 5′-CCCATCCTGTCAGATTTTAGTACTTT-3′ | |
| Set C | TOP1 (1) | 5′-GCATGAATCTCAGCTCTTTCCA-3′ | 5′-GATGGGTTAGGCCCTCTTAAA-3′ |
| TOP1 (2) | 5′-GATGACTTGGGCTCTCCCTTT-3′ | 5′-GGGGAAACAGCATTGATCCTA-3′ | |
| CYP1A1 (1) | 5′-CTGCAGCCAGATCAGTGTCTA-3′ | 5′-GACACAGTGATTGGCAGGTC-3′ | |
| CYP1A1 (2) | 5′-TTTTGCGTATTTATGTTGCAGA-3′ | 5′-AGGCTCTGCTTTGATTGAGG-3′ |
A total of 12 genes, including TOP1, P53, CYP1A1, PIK3CA, C6orf195, DKKL1, SHH, ADCYAP1, MSH2, PTEN, PMS2 and CAT, were examined to estimate amplification coverage. Almost all genes are located on different chromosomes, except for SHH and PMS2, which are both on chromosome 7 (PMS2 on the p arm and SHH on the q arm). The standard PCR method described above was used to amply 12 sequences of length 162–351 bp, one for each gene, from 1 μl of 1/50C0 MDA product. Primers used to amplify these fragments are listed in set B of Table 1. Five microliter PCR products were then loaded on 1.5% agarose gels and checked for the presence of target fragments. For those samples in which some primers did not work, the 1/5C0 MDA products were submitted to PCR again with the failed primers. To assess the fidelity of WGA from single cells, we used another four pairs of primers (set C in Table 1) to amplify four DNA fragments (two in TOP1 and two in CYP1A1) with a total length of ∼3.3 kb, and these fragments were then sequenced using ABI Big Dye Terminator V3 kit and ABI3100 capillary sequencer (AppliedBiosystems, Foster City, CA). PCRs were prepared and performed as described above. These four fragments were selected because they include five SNPs: rs6129731, rs2235362, rs6102287, rs1048943 and rs17861120, which are heterozygous in the studied sample, so the sequencing data can also provide haplotype information and help check whether the MDA products does come from single sperm cells or not.
Quantification of gene copy number
We quantified the copy number for nine genes with the absolute quantification program of SDS2.0 software using an ABI7900 machine. These nine genes were same as those used in MDA coverage analysis, except for three genes, DKKL1, SHH and MSH2, which were excluded owing to bad standard curves. Primers for quantification were same as those used for coverage analysis. Human genomic DNA from Promega (Catalog no. G1521) was serially diluted as input DNA for the construction of standard curves. Each reaction was duplicated three times. A 20 μl mixture was prepared for each reaction and included 0.05× SYBR (Molecular Probes, Eugene, OR), 1× HotStarTaq buffer, 2.5 mM Mg2+, 0.2 mM dNTP, 0.3 μM of each primer, 1 U HotStarTaq polymerase (Qiagen Inc.) and 1 μl of 1/50C0 MDA products. The cycling program was 95°C for 15 min; 40 cycles of 94°C for 15 s, 56°C for 30 s, 72°C for 30 s. We determined the total number of gene copies amplified and their standard deviations in a 50 μl reaction by multiplying those in 1 μl of 1/50C0 MDA products by 2500. For those samples in which real-time PCR did not work, 0, 250 and 2500 were assigned to those samples with no amplification or amplified in 1/5C0 or 1/50C0 MDA products in the coverage test, respectively.
STR and SNP genotyping
In order to test whether the WGA products from single cells were useful for STR and SNP genotyping analysis, five STR markers (D11S902, D11S937, D11S987, D7S486 and D10S547) were typed using the ABI 3100 sequencer and seven SNPs (rs4575595, rs11868324, rs4267385, rs5369, rs1800541, rs1535721 and rs1323690) were examined with Taqman Assay-on-demand kits (AppliedBiosystems). The S01 sample was not included owing to accidental loss. All these markers were heterozygous in the sampled individual. One microliter of 1/5C0 MDA products were added as templates in the reactions. Five microsatellite sites were separately amplified in a 6 μl reaction containing 1× HotStarTaq buffer, 2.5 mM Mg2+, 0.2 mM dNTP, 0.3 U HotStarTaq polymerase (Qiagen Inc.), 0.06 μl of fluorescence-labeled primer mixture from ABI PRISM® Linkage Mapping Sets V2.5 and 1 μl of template DNA. The cycling program was 95°C for 15 min; 10 cycles of 94°C for 15 s, 55°C for 15 s, 72°C for 30 s; 28 cycles of 89°C for 15 s, 55°C for 15 s, 72°C for 30 s; 72°C for 1 min. The PCR products for D11S987, D11S937 and D7S486 or for D10S547 and D11S902 were mixed for subsequent gel loading. Data for STR markers were collected with the 3100 Data Collection V1.1 software and analyzed with GeneMapper™ V3.0. PCR for Taqman SNP typing was performed in a 5 μl reaction containing 2.5 μl 2× Taqman universal PCR master mixture, 0.125 μl 40× assay mixture and 1 μl template DNA. The cycling program was 95°C for 10 min; 45 cycles of 92°C for 15 s, 60°C for 1 min. Genotypes were determined by allelic discrimination program in SDS 2.1 software.
RESULTS
Successful amplification of whole genomic DNA in cells by MDA depends on efficient release of DNA from chromosomes. DNA molecules are packed very densely in the spermatozoon nucleus and a much tougher method is needed to isolate DNA from sperm cells than from other tissue cells (28). The manual for the REPLI-g kit suggests incubation on ice during alkaline lysis of blood or tissue culture cells. Here, we showed that incubation at 65°C could provide much better genome amplification from minute sperm cells than incubation on ice (Figure 1). Eight aliquots from the dilution of 3 cells/3 μl were subject for alkaline lysis on ice and another eight aliquots were lysed at 65°C. All three genes tested (TOP1, P53 and CYP1A2) were shown to undergo amplification successfully in seven out of eight aliquots if incubated at 65°C during alkaline lysis. In contrast, of the eight aliquots lysed on ice, only two have all three genes amplified, one aliquot has two and three have just one amplified. Moreover, many fragments showed much fainter bands than those amplified from MDA products of aliquots lysed at 65°C, indicating a lower efficiency of MDA in those samples lysed on ice. We thus selected incubation at 65°C for alkaline lysis of 16 aliquots (S01–S16) from the dilution of 0.8 cells/3 μl, which were selected for detailed analysis of efficiency of genome amplification of single sperm cell by MDA.
Figure 1.
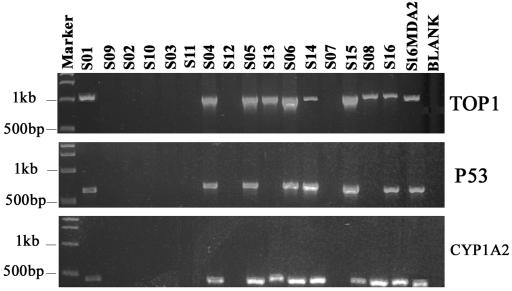
PCR amplification of the three fragments of the TOP1, P53 and CYP1A2 genes in MDA products of 16 sperm aliquots treated using two different lysis methods. The fragments are 1080 bp in length for TOP1, 643 bp for P53 and 550 bp for CYP1A2. Eight aliquots from the dilution of 3 cells/3 μl were lysed on ice and the other eight were lysed at 65°C.
All of the three genes tested (TOP1, P53 and CYP1A2) were successfully amplified in 7 of 16 sperm aliquots at the 0.8 cells/3 μl dilution (S01, S04, S05, S06, S14, S15 and S16). For S08 and S13, only TOP1 and CYP1A2 genes were amplified (Figure 2). For the others, no amplification was observed. A total of 9 out of 16, or ∼56%, sperm aliquots were therefore selected for further studies. This observation was consistent with the expectation of cumulative Poisson distribution (55%) with a mean of 0.8 for aliquots with at least one sperm, so most aliquots containing sperm cell(s) should be amplified, although we could not estimate the success rate. Two of them (S06 and S14) may have contained more than one sperm based on further sequencing and STR genotyping below.
Figure 2.
PCR amplification of the three fragments of the TOP1, P53 and CYP1A2 genes in MDA products of S01–S16 samples. The fragments are 1080 bp in length for TOP1, 643 bp for P53 and 550 bp for CYP1A2. S01–S16 indicate MDA products from 16 sperm aliquots. S16MDA2 is the second-round MDA product using 1 μl of 1/10C0 S16 MDA product as resource DNA. BLANK here is the MDA product of the negative control, in which 3 μl PBS buffer was added instead of sperm aliquot.
Coverage
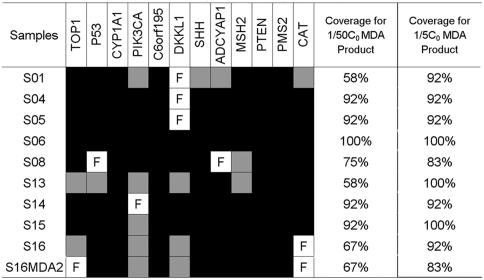
The coverage of the amplification can be estimated based on the success rate of amplification of the 12 fragments (see Materials and Methods) tested. Most of the 12 fragments were successfully amplified in all nine sperm aliquots (Figure 3). For those seven aliquots containing single sperm cells (S01, S04, S05, S08, S13, S15 and S16), the success rate ranges from 58 to 92% for 1/50C0 with an average of 76%. If 1/5C0 MDA products were used, the coverage increased to 93%. About 76% of the genes can therefore be amplified at least 2500-fold or 93% of the genes can be amplified at least 250-fold in single sperm cell suspensions. The second-round MDA product, S16MDA2, from sample S16, had almost the same amplification pattern.
Figure 3.
PCR amplification of 12 fragments using 1 μl of 1/5C0 or 1/50C0 MDA products as templates. The amplification result is indicated with a black square for successful amplification in 1/50C0, a gray square for successful amplification in 1/5C0 and a framed F for no amplification in both.
Yield
MDA yields for the nine aliquots containing sperm and the S16MDA2 product were 13–35 μg (Table 2), and the yields for other aliquots without sperm were 15–35 μg. It was clear that background amplification in MDA was quite high, so total DNA yield did not provide much information on MDA efficiency, and thus a detailed quantitative analysis became necessary. Here, we quantitated nine genes in MDA products of nine sperm aliquots and the S16MDA2 (Table 2). Although the specific fragment was amplified in S16 and S16MDA2 for P53, S15 for CYP1A1, S13 for C6orf195 and S13 for ADCYAP1, another unspecific fragment was also observed in each of these reactions. The unspecific amplification makes it impossible to determine the accurate copy number. We, therefore, considered these reactions as failed and estimated the gene copy number based on PCR results in Figure 3. The PIK3CA gene, which was not amplified from 1 μl of 1/50C0 S13 or S15 MDA product in the coverage test, was estimated to have two or four copies. This inconsistency occurred because the PCR condition for amplifying the PIK3CA fragment in the coverage test was possibly not optimized for several template molecules. Gene copy number in these MDA products varied greatly across samples and gene loci. In the 50 μl MDA products for seven single sperm cells (S01, S04, S05, S08, S13, S15 and S16), ∼51% of fragments could be amplified at least 250 000-fold. The human DNA representative rate (R.R. in Table 2) in MDA products was estimated as observed average copy number per nanogram DNA (A.C.N./ng in Table 2) divided by 280. Here, 280 is considered as the copy number of unique genes per nanogram human genomic DNA (Qiagen Genomic DNA Handbook). Some genes were over-amplified in some sperm samples, which caused the human DNA representative rate to be >100% in the amplified sperm samples, such as S04, S06 and S16. The representative rate for second-round MDA product was reduced from 105 to 15% for S16MDA2. Even though the representative rate for second-round MDA samples was dramatically decreased, the second-round MDA can give an amplification of from 0- to 500-fold for eight genes, with an average of 236-fold (Table 3).
Table 2.
Total copy number for nine genes (TOP1, P53, CYP1A1, C6orf195, ADCYAP1, PTEN, PMS2 and CAT) and the human DNA representative rate in each 50 μl MDA product
| S01 | S04 | S05 | S06 | S08 | S13 | S14 | S15 | S16 | S16 MDA2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| TOP1 | 2.2E+6 ± 0.4E+6 | 2.1E+7 ± 0.1E+7 | 23.0E+6 ± 0.5E+6 | 1.6E+8 ± 0.2E+8 | 4.1E+3 ± 0.5E+3 | 250* | 2.6E+7 ± 0.2E+7 | 4.1E+3 ± 0.5E+3 | 250* | 0* |
| P53 | 2.4E+5 ± 0.5E+5 | 3.6E+5 ± 0.2E+5 | 6.1E+4 ± 0.5E+4 | 2.1E+6 ± 0.1E+6 | 0* | 250* | 2.1E+6 ± 0.1E+6 | 1.2E+7 ± 0.1E+7 | 2500* | 2500* |
| CYP1A1 | 5.5E+5 ± 1.1E+5 | 3.9E+7 ± 0.1E+7 | 3.9E+6 ± 0.7E+6 | 33.5E+4 ± 0.4E+4 | 2.1E+6 ± 0.2E+6 | 15.3E+4 ± 0.5E+4 | 7.1E+3 ± 2.5E+3 | 2500* | 3.4E+7 ± 0.1E+7 | 1.0E+7 ± 0.2E+7 |
| PIK3CA | 250* | 3.8E+5 ± 0.4E+5 | 4.1E+5 ± 0.6E+5 | 1.3E+7 ± 0.2E+7 | 2.9E+6 ± 0.6E+6 | 5.0E+3 ± 0.1E+3 | 0* | 1.0E+4 ± 1.0E+4 | 250* | 250* |
| C6orf195 | 5.8E+6 ± 0.3E+6 | 3.1E+5 ± 0.1E+5 | 8.9E+3 ± 0.3E+3 | 1.3E+6 ± 0.1E+6 | 6.1E+4 ± 1.7E+4 | 2500* | 3.2E+6 ± 0.5E+6 | 5.4E+5 ± 0.4E+5 | 4.2E+5 ± 0.5E+5 | 1.0E+5 ± 0.1E+5 |
| ADCYAP1 | 250* | 3.6E+6 ± 0.2E+6 | 8.8E+3 ± 3.6E+3 | 18.7E+5 ± 0.3E+5 | 0* | 2500* | 7.0E+5 ± 1.1E+5 | 2.4E+5 ± 0.1E+5 | 6.1E+5 ± 0.2E+5 | 2.0E+5 ± 0.2E+5 |
| PTEN | 13.7E+4 ± 0.6E+4 | 23.3E+4 ± 0.2E+4 | 5.6E+3 ± 4.5E+3 | 3.2E+6 ± 0.2E+6 | 4.8E+6 ± 0.6E+6 | 3.6E+6 ± 0.3E+6 | 4.4E+3 ± 0.7E+3 | 16.7E+6 ± 0.2E+6 | 5.8E+5 ± 0.2E+5 | 2.9E+5 ± 0.2E+5 |
| PMS2 | 2.6E+5 ± 0.2E+5 | 1.4E+6 ± 0.1E+6 | 2.1E+6 ± 0.1E+6 | 9.6E+5 ± 0.6E+5 | 1.6E+6 ± 0.4E+6 | 12.4E+5 ± 0.2E+5 | 19.7E+5 ± 0.2E+5 | 9.7E+5 ± 0.7E+5 | 4.5E+5 ± 0.1E+5 | 1.8E+5 ± 0.3E+5 |
| CAT | 250* | 1.6E+6 ± 0.1E+6 | 9.9E+3 ± 1.4E+3 | 5.0E+6 ± 0.4E+6 | 2.3E+4 ± 0.7E+4 | 2.8E+6 ± 0.4E+6 | 3.7E+6 ± 1.9E+6 | 5.1E+3 ± 0.6E+3 | 0* | 0* |
| Yield (μg) | 13 | 20 | 29 | 26 | 32 | 29 | 26 | 35 | 14 | 30 |
| A.C.N./ng | 80 | 379 | 114 | 774 | 40 | 30 | 158 | 98 | 295 | 41 |
| R.R. (%) | 29 | 135 | 41 | 276 | 14 | 11 | 56 | 35 | 105 | 15 |
Total copy number was displayed in mean ± SD format and the numbers are written in scientific format. A.C.N./ng is for average copy number per ng DNA and R.R. for human DNA representative rate. For those samples on which real-time PCR did not work (labeled with asterisk), 0, 250 and 2500 were separately assigned in accordance with no amplification, successful amplification in 1/5C0 and 1/50C0 MDA products displayed in Figure 3.
Table 3.
Amplification rate for nine genes after second-round MDA treatment
| Genes used for quantification analysis | TOP1 | P53 | CYP1A1 | PIK3CA | C6orf195 | ADCYAP1 | PTEN | PMS2 | CAT |
|---|---|---|---|---|---|---|---|---|---|
| Total copy number | |||||||||
| S16MDAa | 0.5 | 5 | 68 995 | 0.5 | 840 | 1225 | 1155 | 905 | 0 |
| S16MDA2b | 0 | 2500 | 10 320 000 | 250 | 100 000 | 202 500 | 295 000 | 182 500 | 0 |
| Amplification rate | 0 | 500 | 150 | 500 | 119 | 165 | 255 | 202 | /c |
aThe input template DNA for the second-round MDA reaction, which is 1 μl of 1/10C0 MDA product of a single sperm aliquot S16. The copy number for each gene in the input DNA was determined by the total copy number of these genes in MDA products of S16 divided by 500.
bThe total 50 μl second-round MDA product of S16.
cUnable to calculate.
Sequencing
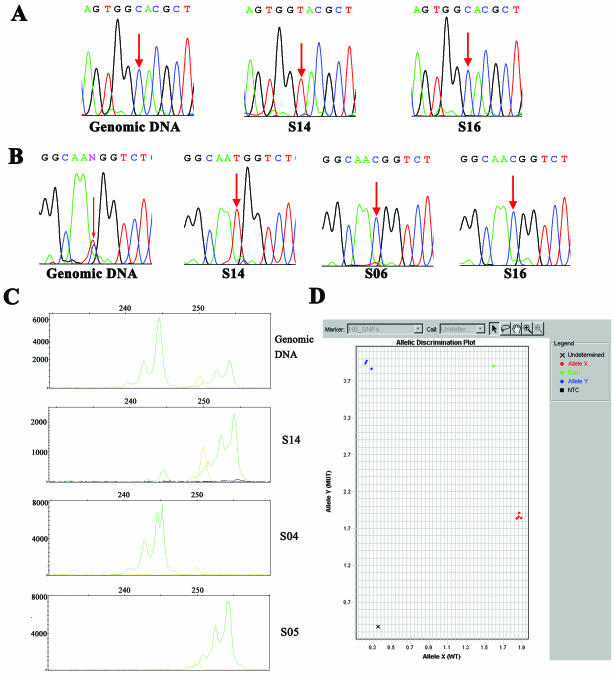
We sequenced four fragments (overall ∼3.3 kb) in all 10 MDA products (nine sperm aliquots and one second-round MDA product) and the leukocyte genomic DNA. Only one artificial mutation was observed, in sample S14 (Figure 4A). The error rate of MDA amplification in a single or a few sperm cells was therefore estimated to be 3.65 × 10−5 in an overall 27.4 kb sequenced. All MDA products from sperm aliquots, except S06 showed single alleles at five SNP sites in the four fragments (Table 4). Rs1048943 and rs17861120 were observed to be heterozygous in S06, although the peak height of one allele was much lower that that of the other (Figure 4B, rs1048943). Together with the STR data (see below), S06 and S14 were deduced to contain more than one sperm, whereas the other seven aliquots were considered to have just one sperm each. Rs6129731, rs2235362 and rs6102287 are in the same Contig NT_011362.9. Rs1048943 and rs17861120 are in another Contig NT_010194.16. From Table 4, it is obvious that haplotypes for the first three SNPs are AAA and CCG, and haplotypes for the latter two SNPs are CG and TA.
Figure 4.
MDA products were useful for a variety of genetic applications, including sequencing and SNP and STR analysis. (A) A mutation at 45803241 of NT_010194.16 is marked by red arrows in the S14 MDA product. (B) Sequencing traces for rs1048943 (marked by red arrows) and its flanking 10 nt in genomic DNA, S14, S06 and S16 MDA products. S06 has two alleles, although the C allele was much preferred for amplification. (C) The electropherogram for the D10S547 locus in genomic DNA, S14, S04 and S05 samples displayed by GeneMapper V3.0. Their genotypes were displayed in Table 5. S14 was observed to have two alleles (244 and 254), although allele 254 showed to be preferentially amplified. The orange peak is a 250 bp marker. (D) The interface for the identification of genotypes at rs1535721 locus by Taqman assay using the ABI7900. The green one is for genomic DNA sample, blue for S05, S13 and S15, and red for S06, S08, S14, S16 and S16MDA2. × is for S04, for which the amplification failed.
Table 4.
Genotypes of genomic DNA and nine sperm aliquots at five SNP loci determined by sequencing
| SNP ID | Contigs | Position in contigs | Genomic DNA | S01 | S04 | S05 | S06 | S08 | S13 | S14 | S15 | S16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6129731 | NT_011362.9 | 4713602 | A/C | ? | A | A | A | C | A | C | A | C |
| rs2235362 | NT_011362.9 | 4797334 | A/C | C | ? | A | A | C | A | C | A | C |
| rs6102287 | NT_011362.9 | 4797565 | A/G | G | ? | A | A | G | A | G | A | G |
| rs1048943 | NT_010194.16 | 45803303 | T/C | C | C | T | C/t | T | C | T | ? | C |
| rs17861120 | NT_010194.16 | 45817497 | A/G | G | G | A | A/g | A | G | A | G | G |
? indicates no PCR amplification, and the italic small letter indicates the allele with amplification of much lower efficiency.
Genotyping
Five STRs were successfully typed with the ABI 3100 sequencer and GeneMapper software in 59 out of 65 reactions. The genotypes for the nine sperm aliquots and S16MDA2 are described in Table 5. S06 and S14 were shown to be heterozygous at D11S937 and D10S547, respectively (Figure 4C, D10S547). The allele 168 for D11S937 and 254 for D10S547 were preferentially amplified. S06 was also observed to be heterozygous at two SNP sites as described in Table 4. This indicated that the two aliquots contained at least two sperm cells. Seven SNPs located in three genes, ACE, EDN1 and ELF5, were successfully typed on the ABI7900 machine with Assay-on-demand kits in 84/91 reactions (Figure 4D, rs1535721). Genotypes for each sperm aliquot are described in Table 6. Haplotypes for ACE, EDN1 and ELF5 were easily determined to be GAT/AGC, GT/AG and AG/GA, respectively. S16MDA2 had exactly the same genotypes with S16 sample for all STRs and SNPs examined.
Table 5.
Genotypes of genomic DNA, eight sperm aliquots and second-round MDA product of S16 at five microsatellite (STR) loci
| Samples | D11S902 | D11S937 | D11S987 | D7S486 | D10S547 |
|---|---|---|---|---|---|
| Genomic DNA | 155/161 | 156/168 | 94/116 | 224/236 | 244/254 |
| S04 | 161 | 156 | 94 | 224 | 244 |
| S05 | 161 | 168 | ? | 236 | 254 |
| S06 | 155 | 156/168 | 94 | 236 | 254 |
| S08 | 155 | 168 | 116 | ? | 254 |
| S13 | 161 | 168 | ? | 236 | 254 |
| S14 | 155 | 156 | 94 | ? | 244/254 |
| S15 | 161 | 168 | 116 | 224 | 254 |
| S16 | ? | 156 | 94 | 236 | 244 |
| S16MDA2 | ? | 156 | 94 | 236 | 244 |
? indicates no amplification and the italic number in this table indicates the allele with amplification of much lower efficiency.
Table 6.
Genotypes of seven SNPs in genomic DNA, eight sperm aliquots and the second-round MDA product of S16 determined by Taqman technology
| Samples | ACE | EDN1 | ELF5 | ||||
|---|---|---|---|---|---|---|---|
| rs4575595 | rs11868324 | rs4267385 | rs5369 | rs1800541 | rs1535721 | rs1323690 | |
| Genomic DNA | G/A | G/A | T/C | G/A | T/G | G/A | G/A |
| S04 | G | A | T | G | T | ? | ? |
| S05 | ? | ? | C | A | G | A | G |
| S06 | G | A | T | G | T | G | A |
| S08 | G | A | T | A | G | G | A |
| S13 | A | G | C | A | G | A | G |
| S14 | G | A | T | A | G | G | A |
| S15 | ? | ? | ? | G | T | A | G |
| S16 | G | A | T | A | G | G | A |
| S16MDA2 | G | A | T | A | G | G | A |
? indicates no amplification.
DISCUSSION
Four methods for WGA: PEP (23), degenerate oligonucleotide primed-PCR (DOP-PCR) (29), ligation-mediated PCR (LM-PCR) (30) and MDA (24), were developed and used for studies on single or mutiple cells. (16,25,27,30–32). Only a single primer was used in LM-PCR in contrast to the DOP and PEP, in which millions of different primers were included, LM-PCR can thus amplify whole genomic sequences more evenly and produce less bias than DOP and PEP (33), although it requires more steps, including restriction endonuclease digestion and ligation of digested DNA with primers. PEP was first reported by Zhang et al. (23) to amplify single sperm DNA and then utilized by other researchers for single sperm analysis (15,16). Recently developed MDA was demonstrated to have a high amplification potential and great loci representation with less than threefold bias (24). It has been successfully applied for WGA from laser capture-microdissected fresh tissue as well as single leukocytes and blastomere cells (25,26,34), although it was reported to fail in amplifying highly degraded samples, such as formalin-fixed, paraffin-embedded tissues (35,36).
The MDA method was used here to amplify whole genome DNA from single sperm cells and achieved high efficiency. In this study, when 1 μl of 1/50C0 MDA products were used as PCR templates, 76% reactions succeeded, and when 1 μl of 1/5C0 was used, the rate increased to 93%. This means that in a 50 μl reaction, 76 or 93% of loci could be amplified at least 2500- or 250-fold, respectively, from single sperm DNA, in contrast to only 78% of loci amplified 30-fold by the PEP method (23). Second-round MDA product analysis showed that a further ∼236-fold amplification could be obtained. Considering MDA is isothermally performed and can be done in an incubator, the MDA reaction volume may be extended to 100 μl or more. Pre-amplification of the single sperm genome can therefore provide sufficient DNA for thousands of genotyping reactions.
Our study used real-time PCR to quantitate the copy number for nine unique genes in 10 MDA products. Variable amplification was observed. This variation seemed to result from random template selection, not from gene difference. For eight genes, copy number varied from zero to several thousand or more in 1 μl of 1/50C0 MDA products from different aliquots. Only one gene (PMS2) showed similar amplification efficiencies across all eight samples. Preferred amplification and allele dropout were observed to be quite common in MDA products from single leukocyte cells (data not shown). These were consistent with observations reported from other studies on single or a small number of cells (25,26). Many researchers have used WGA products from a limited number of cells (<5) for CGH or LOH analysis (26,30,31). Based on data from this study, we suggest much care should be taken in explaining these CGH or LOH data.
Our sequencing data showed that MDA products were appropriate for sequencing projects. Here, a total of 27.4 kb nucleotides of MDA products from single sperm cells were sequenced and only one mutation was identified, although we do not know whether this mutation came from the sperm itself or MDA treatment. The error rate of MDA was estimated to be 3.65 × 10−5. In fact, during the amplification of single DNA molecules, many mutations in the first two strands replicated, which created a 1:3 ratio of mutated to normal allele, can be detected by sequencing, so the incorporation error rate for Φ29 DNA polymerase should be <3.65 × 10−5.
Microsatellite slippage is a big problem in WGA from single cells using PCR-based methods, especially those in which thermophilic polymerases without strand displacement activity, such as Taq or Pfu, were used (26,37,38). The MDA method utilizes the Φ29 mesophilic polymerase for primer extension. The Φ29 polymerase has strong strand displacement activity and is able to extend >70 kb on DNA templates. Even stable secondary structure in the template does not affect the processivity and strand displacement ability of the enzyme (39). Polymerase slippage was demonstrated to include four major steps: (i) arrest of DNA synthesis within direct repeats, (ii) dissociation of the polymerase from the template, (iii) unpairing of the 3′ end of the newly synthesized strand and its reannealing to the neighboring repeat and (iv) resumption of DNA synthesis (37). The aforementioned features of Φ29 polymerase, which prevent the dissociation of the enzyme from the template, are therefore theoretically able to drastically reduce the rate of slippage at microsatellite sites when Φ29 polymerase is used, in contrast to other polymerases without strand displacement activity. Hellani et al. (26) has observed no microsatellite slippage at 16 loci in MDA products of 40 single leukocytes. Our data also showed that all alleles at five STR sites detected in MDA products of eight sperm aliquots were identical to parental alleles in genomic DNA, indicating no microsatellite slippage occurred. It was, therefore, demonstrated that MDA products from single sperm cells were useful for further microsatellite analysis.
From the genotyping data of single sperm cells, we can easily determine the two haplotypes in the sample studied. For example, haplotypes composed of rs6129731, rs2235362 and rs6102287 should be AAA and CCG (Table 4), and for the ACE gene they are GAT and AGC (Table 6). Currently, almost all available haplotypes with a high density of SNPs in human population have been inferred by statistical methods from phase-unknown genotypes in random samples or trio families (see data from PGA, EGP and HapMap projects). Further LD analysis was performed based on the inferred haplotype data. Little is known about how well these inferred haplotypes match the true ones, and how much inference error can affect the LD pattern. It is necessary to investigate some representative regions by comparing experimentally determined true haplotypes with those inferred statistically. Here, we demonstrated that MDA in single sperm cells can produce sufficient DNA for thousands of genotyping reactions. By genotyping DNA from leukocytes and MDA products from one or two single sperm cells, the true haplotypes in a large chromosome region can therefore be determined.
In this study, we found that sperm lysis at 65°C instead of on ice, as described in the manual supplied, could provide much better amplification. It is possible that warm incubation is more efficient in releasing DNA from the chromosomes. Except for alkaline lysis, some other studies also used proteinase K to treat cells before subjecting them to MDA (26,34). Rook et al. (26) tried alkaline lysis on ice and proteinase K digestion of laser capture-microdissected tissues, and decreased allele imbalance was observed for proteinase K treatment. Using proteinase K for lysis of sperm cells may, therefore, also produce good amplification.
We have demonstrated that MDA is an effective way for global genome amplification in single sperm cells and can provide sufficient single sperm DNA for genotyping in large quantity. It also makes possible fine mapping of male meiotic recombination sites on a <10 kb level, and experimental determination of haplotypes in a large chromosome region, or even a whole chromosome. These maps can unambiguously help better understand LD patterns in human population, thus further facilitating association studies. It can also improve our knowledge about the mechanism of recombination, especially how recombination is initiated.
Acknowledgments
The authors thank Dr C. Stuart Baxter, Ming Dong, Ge Zhang, Huifeng Xi and all anonymous reviewers for helpful comments and Joanna Watson for help in STR and SNP genotyping. The authors also thank Dr Marian Miller for her help on figure edition. This work was supported by grants from National Institutes of Health (NIH), USA. Funding to pay the Open Access publication charges for this article was provided by NIH, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goldstein D.B. Islands of linkage disequilibrium. Nature Genet. 2001;29:109–111. doi: 10.1038/ng1001-109. [DOI] [PubMed] [Google Scholar]
- 2.Daly M.J., Rioux J.D., Schaffner S.F., Hudson T.J., Lander E.S. High-resolution haplotype structure in the human genome. Nature Genet. 2001;29:229–232.. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 3.Jeffreys A.J., Kauppi L., Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 4.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 5.Nejentsev S., Godfrey L., Snook H., Rance H., Nutland S., Walker N.M., Lam A.C., Guja C., Ionescu-Tirgoviste C., Undlien D.E., et al. Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum. Mol. Genet. 2004;13:1633–1639. doi: 10.1093/hmg/ddh169. [DOI] [PubMed] [Google Scholar]
- 6.Kauppi L., Sajantila A., Jeffreys A.J. Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum. Mol. Genet. 2003;12:33–40. doi: 10.1093/hmg/ddg008. [DOI] [PubMed] [Google Scholar]
- 7.Wang N., Josh A.M., Zhang K., Chakraborty R., Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K., Akey J.M., Wang N., Xiong M., Chakraborty R., Jin L. Randomly distributed crossovers may generate block-like patterns of linkage disequilibrium: an act of genetic drift. Hum. Genet. 2003;113:51–59. doi: 10.1007/s00439-003-0941-5. [DOI] [PubMed] [Google Scholar]
- 9.Wiehe T., Mountain J., Parham P., Slatkin M. Distinguishing recombination and intragenic gene conversion by linkage disequilibrium patterns. Genet. Res. 2000;75:61–73. doi: 10.1017/s0016672399004036. [DOI] [PubMed] [Google Scholar]
- 10.Reich D.E., Schaffner S.F., Daly M.J., McVean G., Mullikin J.C., Higgins J.M., Richter D.J., Lander E.S., Altshuler D. Human genome sequence variation and the influence of gene history, mutation and recombination. Nature Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 11.Phillips M.S., Lawrence R., Sachidanandam R., Morris A.P., Balding D.J., Donaldson M.A., Studebaker J.F., Ankener W.M., Alfisi S.V., Kuo F.S., et al. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nature Genet. 2003;33:382–387. doi: 10.1038/ng1100. [DOI] [PubMed] [Google Scholar]
- 12.Jeffreys A.J., Murray J., Neumann R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell. 1998;2:267–273. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 13.Jeffreys A.J., Ritchie A., Neumann R. High resolution analysis of haplotype diversity and meiotic crossover in human TAP2 recombination hotspot. Hum. Mol. Genet. 2000;9:725–733. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 14.May C.A., Shone A.C., Kalaydjieva L., Sajantila A., Jeffreys A.J. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nature Genet. 2002;31:272–275. doi: 10.1038/ng918. [DOI] [PubMed] [Google Scholar]
- 15.Hubert R., MacDonald M., Gusella J., Arnheim N. High resolution localization of recombination hot spots using sperm typing. Nature Genet. 1994;7:420–424. doi: 10.1038/ng0794-420. [DOI] [PubMed] [Google Scholar]
- 16.Cullen M., Perfetto S.P., Klitz W., Nelson G., Carrington M. High-resolution patterns of meiotic recombination across the human major histocompatibility complex. Am. J. Hum. Genet. 2002;71:759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider J.A., Peto T.E., Boone R.A., Boyce A.J., Clegg J.B. Direct measurement of the male recombination fraction in the human beta-globin hot spot. Hum. Mol. Genet. 2002;11:207–215. doi: 10.1093/hmg/11.3.207. [DOI] [PubMed] [Google Scholar]
- 18.Pääbo S., Irwin D.M., Wilson A.C. DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 19.Zangenberg G., Huang M.M., Arnheim N., Erlich H. New HLA-DPB1 alleles generated by interallelic gene conversion detected by analysis of sperm. Nature Genet. 1995;10:407–414. doi: 10.1038/ng0895-407. [DOI] [PubMed] [Google Scholar]
- 20.Furlong R.A., Goudie D.R., Carter N.P., Lyall J.E., Affara N.A., Ferguson-Smith M.A. Analysis of four microsatellite markers on the long arm of chromosome 9 by meiotic recombination in flow-sorted single sperm. Am. J. Hum. Genet. 1993;52:1191–1199. [PMC free article] [PubMed] [Google Scholar]
- 21.Girardet A., Lien S., Leeflang E.P., Beaufrere L., Tuffery S., Munier F., Arnheim N., Claustres M., Pellestor F. Direct estimation of the recombination frequency between the RB1 gene and two closely linked microsatellites using sperm typing. Eur. J. Hum. Genet. 1999;7:239–242. doi: 10.1038/sj.ejhg.5200250. [DOI] [PubMed] [Google Scholar]
- 22.Park C., Frank M.T., Lewin H.A. Fine-mapping of a region of variation in recombination rate on BTA23 to the D23S22-D23S23 interval using sperm typing and meiotic breakpoint analysis. Genomics. 1999;59:143–149. doi: 10.1006/geno.1999.5869. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasken R.S., Egholm M. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 2003;21:531–535. doi: 10.1016/j.tibtech.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Handyside A.H., Robinson M.D., Simpson R.J., Omar M.B., Shaw M.A., Grudzinskas J.G., Rutherford A. Isothermal whole genome amplification from single and small numbers of cells: a new era for preimplantation genetic diagnosis of inherited disease. Mol. Hum. Reprod. 2004;10:767–772. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- 27.Hellani A., Coskun S., Benkhalifa M., Tbakhi A., Sakati N., Al-Odaib A., Ozand P. Multiple displacement amplification on single cell and possible PGD applications. Mol. Hum. Reprod. 2004;10:847–852. doi: 10.1093/molehr/gah114. [DOI] [PubMed] [Google Scholar]
- 28.Carter K.L., Robertson B.C., Kempenaers B. A differential DNA extraction method for sperm on the perivitelline membrane of avian eggs. Mol. Ecol. 2000;9:2149–2150. doi: 10.1046/j.1365-294x.2000.01114.x. [DOI] [PubMed] [Google Scholar]
- 29.Cheung V.G., Nelson S.F. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl Acad. Sci. USA. 1996;93:14676–14679. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein C.A., Schmidt-Kittler O., Schardt J.A., Pantel K., Speicher M.R., Riethmuller G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl Acad. Sci. USA. 1999;96:4494–4499. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells D., Sherlock J.K., Handyside A.H., Delhanty J.D. Detailed chromosomal and molecular genetic analysis of single cells by whole genome amplification and comparative genomic hybridisation. Nucleic Acids Res. 1999;27:1214–1218. doi: 10.1093/nar/27.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D., Liu C., DeVries S., Waldman F., Cote R.J., Datar R.H. LM-PCR permits highly representative whole genome amplification of DNA isolated from small number of cells and paraffin-embedded tumor tissue sections. Diagn. Mol. Pathol. 2004;13:105–115. doi: 10.1097/00019606-200406000-00007. [DOI] [PubMed] [Google Scholar]
- 33.Stoecklein N.H., Erbersdobler A., Schmidt-Kittler O., Diebold J., Schardt J.A., Izbicki J.R., Klein C.A. SCOMP is superior to degenerated oligonucleotide primed-polymerase chain reaction for global amplification of minute amounts of DNA from microdissected archival tissue samples. Am. J. Pathol. 2002;161:43–51. doi: 10.1016/S0002-9440(10)64155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rook M.S., Delach S.M., Deyneko G., Worlock A., Wolfe J.L. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am. J. Pathol. 2004;164:23–33. doi: 10.1016/S0002-9440(10)63092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G., Brennan C., Rook M., Wolfe J.L., Leo C., Chin L., Pan H., Liu W.H., Price B., Makrigiorgos G.M. Balanced-PCR amplification allows unbiased identification of genomic copy changes in minute cell and tissue samples. Nucleic Acids Res. 2004;32:e76. doi: 10.1093/nar/gnh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G., Maher E., Brennan C., Chin L., Leo C., Kaur M., Zhu P., Rook M., Wolfe J.L., Makrigiorgos G.M. DNA amplification method tolerant to sample degradation. Genome Res. 2004;14:2357–2366. doi: 10.1101/gr.2813404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canceill D., Viguera E., Ehrlich S.D. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 1999;274:27481–27490. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 38.Viguera E., Canceill D., Ehrlich S.D. In vitro replication slippage by DNA polymerases from thermophilic organisms. J. Mol. Biol. 2001;312:323–333. doi: 10.1006/jmbi.2001.4943. [DOI] [PubMed] [Google Scholar]
- 39.Blanco L., Bernad A., Lazaro J.M., Martin G., Garmendia C., Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]