Abstract
We report the identification of a new simian immunodeficiency virus (SIV), designated SIVden, in a naturally infected Dent's Mona monkey (Cercopithecus mona denti), which was kept as pet in Kinshasa, capital of the Democratic Republic of Congo. SIVden is genetically distinct from the previously characterized primate lentiviruses. Analysis of the full-length genomic sequence revealed the presence of a vpu open reading frame. This gene is also found in the virus lineage of human immunodeficiency virus type 1 (HIV-1) and chimpanzee immunodeficiency virus (SIVcpz) and was recently described in viruses isolated from Cercopithecus nictitans, Cercopithecus mona, and Cercopithecus cephus. The SIVden vpu coding region is shorter than the HIV-1/SIVcpz and the SIVgsn, SIVmon, and SIVmus counterparts. Unlike Pan troglodytes schweinfurthii viruses (SIVcpzPts) and Cercopithecus monkey viruses (SIVgsn, SIVmon, and SIVmus), the SIVden Vpu contains the characteristic DSGXES motif which was shown to be involved in Vpu-mediated CD4 and IκBα proteolysis in HIV-1 infected cells. Although it harbors a vpu gene, SIVden is phylogenetically closer to SIVdeb isolated from De Brazza's monkeys (Cercopithecus neglectus), which lacks a vpu gene, than to Cercopithecus monkey viruses, which harbor a vpu sequence.
Simian immunodeficiency viruses (SIV) naturally infect more than 30 species of African nonhuman primates, but current data suggest that the viruses identified to date represent only a fraction of nonhuman primate lentiviruses (6, 13, 24, 54). Most of the SIVs are nonpathogenic in their natural hosts (8, 51), with a few exceptions (52). This is thought to be an example of long-term cospeciation of viruses with their natural hosts (24). This view is supported by the fatal outcome of the zoonotic transmission of primate immunodeficiency viruses in humans and macaques (47). Characterization of newly identified SIV strains is necessary to gain a better understanding of the evolutionary relationships among primate lentiviruses and to identify the potential risk for new zoonotic transmissions into the human population.
Cercopithecidae monkeys are divided into two subfamilies named Colobinae and Cercopithecinae. Cercopithecinae monkeys, from which most nonhuman primate lentiviruses have been isolated, are subdivided into two tribes (Cercopithecini and Papionini) and numerous species and subspecies distributed throughout sub-Saharan Africa (23). Primate lentiviruses for which full-length genome sequences are available have been classified, on the basis of phylogenetic analysis (6, 16), into six approximately equidistant major groups or lineages, as follows: (i) SIVcpz from chimpanzees (Pan troglodytes) including also human immunodeficiency virus type 1 (HIV-1) (21, 31, 45, 56, 69; (ii) SIVsm from sooty mangabey (Cercocebus atys) together with HIV-2 (10, 20, 28); (iii) SIVagm from the four major species of African green monkeys, i.e., grivet (18), sabeus (33), vervet (26), and tantalus (60) (members of the genus Chlorocebus); (iv) SIVsyk from Sykes' monkeys (Cercopithecus mitis albogularis) (27) together with SIVmon from mona monkeys (Cercopithecus mona) (3, 13), SIVgsn from greater spot-nosed guenons (Cercopithecus nictitans) (16), SIVmus from mustached monkeys (Cercopithecus cephus) (13), and SIVdeb from De Brazza's monkeys (Cercopithecus neglectus) (6); (v) the SIVlhoest lineage, composed of SIVlhoest from L'Hoest monkeys (Cercopithecus lhoesti) (25), SIVsun from sun-tailed monkeys (Cercopithecus solatus) (4), and SIVmnd-1 from mandrills (Mandrillus sphinx) (68); and (vi) SIVcol from black and white colobus (Colobus guereza) (15). Several other viruses, notably SIVrcm from red-capped mangabeys (5), SIVmnd-2 from mandrills (61), and SIVdrl from drills (29) showed discordant phylogenetic clustering in different genomic regions, and are therefore considered recombinants (29, 61, 64). Partial sequences are available for SIVtal from talapoin monkeys (Miopithecus talapoin) (50) and for SIVschm from red-tailed guenon (Cercopithecus ascanius schmidti) (70); partial sequences from two species of western colobus are available: SIVwrc from western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus) (14). Serological surveys indicate that other species may also harbor lentiviruses, among them Allen's swamp monkey (Allenopithecus nigrovidis), Diana monkey (Cercopithecus diana), blue monkey (Cercopithecus mitis), and Hamlyn's monkey (Cercopithecus hamlyni) (24, 39, 48). However, such serologic findings need to be confirmed by molecular studies and virus characterization.
For some SIVs, there is good evidence for cospeciation with the host, such as those infecting African green monkeys (1, 33, 44) and the C. lhoesti supergroup (4), whereas some others originated through cross-species transmission from other species. The best known example is that of the human viruses HIV-1 and HIV-2, which originated following cross-species transmission of SIVcpz from chimpanzees and SIVsm from sooty mangabeys, respectively (58). Classification of SIVrcm (5), SIVmnd-2 (61, 64), SIVagm.sab (33), and SIVdrl (29) is complicated because their genomes represent mosaic genomes derived from two or more distinct viral species through prior recombination events. A recent study even suggested that all six phylogenetic lineages might have arisen by recombination (55). All primate lentivirus lineages share the gag, pol, vif, vpr, tat, rev, env, and nef genes but differ by the presence or absence of the vpu/vpx accessory gene in their genome. Viruses of the SIVcpz/HIV-1 lineage, together with SIVgsn, SIVmon, and SIVmus, harbor a vpu gene, while viruses of the SIVsm/HIV-2 lineage, together with SIVrcm, SIVmnd-2, and SIVdrl, harbor a vpx gene; on the other hand, other SIVs harbor neither of the these genes.
In 1999, we launched a lentivirus seroprevalence survey of wild-born monkeys in the Democratic Republic of the Congo (DRC). Here we describe the genetic characterization of a new primate lentivirus (SIVden) harbored by a Cercopithecus denti, a species belonging to the mona subgroup. Six mona species have been described (from west to east: Campbell's monkey, Lowe’s monkey, Mona monkey, crowned monkey, Wolf’s monkey, and Dent’s monkey), together with a few subspecies. Their habitat covers a large region of Africa, from Senegal to Uganda. Two independent sequences from C. mona have been characterized (3, 13), and Wolf's mona was found many years ago to be seropositive for SIV (46). SIVden belongs to the SIVsyk lineage but is genetically distinct from previously characterized primate lentiviruses. Analysis of the entire SIVden genome revealed the presence of a vpu open reading frame. Phylogenetic analyses revealed that the SIVden sequence belongs to the SIVsyk group of viruses and, more specifically, is more closely related to SIVdeb than to viruses belonging to the SIVgsn sublineage, which harbor a vpu gene.
MATERIALS AND METHODS
Animals and serologic testing.
During a serological survey in the DRC, 77 wild-born pet primates were screened for antibodies to human and simian retroviruses. Blood was obtained by venipuncture after the animals were tranquilized with ketamine (10 mg/kg of body weight). Anti-SIV antibodies were tested in Kinshasa with the following commercial HIV antibody tests: ImmunoComb II Bispot HIV 1 and 2 (PBS Orgenics), GENSCREEN HIV 1/2 (Bio-Rad), and Determine (Abbot). Three out of 11 Cercopithecus mona denti were positive for anti-HIV/SIV antibodies.
PCR amplification and cloning.
Viral RNA was recovered from 1 ml of plasma as follows: virions were concentrated by centrifugation for 1 h at 25,000 × g; RNA was extracted with guanidium isothiocyanate (11) and then reverse transcribed into cDNA by using the GeneAmp RNA PCR Core kit (PE Biosystems).
DNA was isolated from whole blood by using a QIAamp blood kit (QIAGEN), according to the manufacturer's instructions. PCR amplification was performed with an automated DNA thermal cycler (GeneAmp PCR system 2400). The pol region was amplified from cDNA using DR1/PolOR for the first round and Polis4/UNIPOL2 for the nested round, as previously described (16). Semi-nested PCR was also performed using the same primer set. PCR fragments (822 and 2,490 bp) were purified by gel extraction (QIAGEN), cloned into the pGEM-T Easy vector (Promega), and sequenced.
The gag-pol region was amplified by semi-nested PCR with specific primers derived from the latter sequence: W5R (5′-AATACTGTGTCTACCGCTACT-3′; nucleotides 2351 to 2331) and W1R (5′-ATCCCTGGGTATGCCTGAAC-3′; nucleotides 2309 to 2290); a primer corresponding to the SIV primer binding site (Kin1;5′-TGGCGCCCGAACAGGGAC-3′; nucleotides 218 to 235) was also used.
The 2,091-bp fragment thus obtained was cloned in the PGEM-T Easy vector (Promega), purified with Mini Prep (QIAGEN), and sequenced. This allowed us to design two sets of primers with which to amplify the complete SIVden genome, by targeting unintegrated circular SIV DNA (6). For the first round, the primers were W20 (5′-CACTAGCAATGGCAACCCTCATTCT-3′; nucleotides 4643 to 4667) and W18R (5′-CACACTACTGCCACTAGTGTGTGC-3′; nucleotides 402 to 379); for the nested round, the primers were W21 (5′-CTAGGGGATATGACAGCAGCAGAAA-3′; nucleotides 4690 to 4714) and W17R (5′-CTTGCTCGGTAAGAGGGTGGTACG-3′; nucleotides 333 to 310). Nested PCRs were performed using the GeneAmp XL PCR kit (PE Biosystems). The 6-kb PCR amplification product was purified and digested with EcoR1, BamH1, and HindIII. The recessed 3′ termini were filled with Taq DNA polymerase to add 3′ A-overhangs, and then DNA fragments were purified and cloned in the PGEM-T Easy vector (Promega).
To check the sequence of the accessory gene region, RNA was reverse transcribed using primer W43R (5′-CAACCTTTTCACAAGCTTGGG; nucleotides 6871 to 6891), and the resulting cDNA was amplified by nested PCR using primer sets located in Pol (W21, 5′-CTAGGGGATATGACAGCAGCAGAAA [nucleotides 4681 to 4705], and W27, 5′-ATCTTCCCGGTACCTAGAAAG [nucleotides 6891 to 6871]) and Env (W42R, 5′-GGTGGATTGATTGCAATACCTC [nucleotides 6866 to 6845], and W30R, 5′-AGAATACTGTGACGTACTGC [nucleotides 6348 to 6329]).
Double-stranded recombinant plasmid DNA and PCR fragments (DNA or cDNA) were sequenced by using cycle sequencing and dye terminator methodologies (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq FS DNA polymerase; PE Biosystems, Warrington, England) on an automated sequencer (ABI 373, Stretch model; Applied Biosystems) and a primer-walking approach based on GENOME express (www.genomex.com).
Sequence comparisons.
Overlapping sequences were joined to reconstitute the full-length genome sequence. The SIVden nucleotide sequence and predicted amino acid sequences were compared to published SIV and HIV sequences. The protein alignment was done with ClustalX (32), and then the multiple alignment was evaluated with T-Coffee (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi). The nucleotides were aligned with MacClade4 (41) to match the protein alignment. The alignments were then verified visually and gap stripped using Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Bayesian scanning analysis.
An exploratory analysis for the presence of any recombination events between the main SIV lineages was performed by Bayesian scanning as implemented in the SlidingBayes program (53) (http://www.kuleuven.ac.be/rega/cev/Software), using the GTR+Gamma model run with a sliding window of 500 nucleotides (nt) moving in steps of 50 nt. The program used for the Bayesian scanning was MrBayes (version 3.0) (30). For every single window, four Markov chains were run for 105 generations with a burn-in of 2 × 104 generations. The following sequences were included as references for the main HIV-1/SIV groups: (i) LAI (HIV-1) and SIVcpzANT; (ii) SIVrcmNG411 and SIVrcmGB1; (iii) SIVsmSL92b and HIV-2/BEN; (iv) SIVagmGRI1 and SIVagmVER3; (v) SIVgsnCM166, SIVgsnCM71, SIVmus, and SIVmon99; (vi) SIVsykKE51 and SIVsyk; (vii) SIVdebCE40 and SIVdebCNE5; and (viii) SIVlhoest524 and SIVlhoest447; moreover, SIVsun and SIVcolCGU1 were included as separate clusters.
Phylogenetic analyses.
In genomic regions with high evidence for discordant evolutionary relationships between the SIV sequences based on the Bayesian approaches, separate phylogenetic analyses were done for the putative nonrecombinant fragments. The borders of these fragments were based on the previous Bayesian analyses. In a previous study, it was clearly shown by testing a similar SIV data set that third codon positions are significantly saturated (16). In contrast, first plus second codon positions still retain enough information to infer reliable phylogenetic relationships. Therefore, we performed phylogenetic analysis using the encoded amino acid sequences to minimize potential saturation error occurring mostly at third codon positions. Phylogenetic trees of amino acid sequences were reconstructed using the Bayesian method as implemented in MrBayes and maximum likelihood under the JTT model of substitution (34) as implemented in the TreePuzzle program (57). Markov chain Monte Carlo analyses were run for 2 × 105 generations with a burn-in of 2 × 104. Alternative topologies were tested whether they were significantly different by using the Shimodaira-Hasegawa test (59) as implemented in TreePuzzle.
Diversity plot.
The sequence similarity between SIVden and other primate lentiviruses was examined on a diversity plot on the concatenated nucleotide alignment of the regions of gag, pol, vif, env, and nef. The fractional nucleotide sequence difference was calculated for a sliding window of 400 nt, and an 80-nt step increment. These analyses were performed with the Simplot program (37).
Nucleotide sequence accession numbers.
The complete sequence of SIVden has been submitted to GenBank database under accession number AJ580407.
RESULTS
In 1999 we launched a serological survey in the DRC, in which we screened wild-born pet primates for antibodies to human and simian retroviruses. Eleven wild-born Dent's Mona monkeys were tested for HIV/SIV-specific antibodies. Three animals were seropositive: a female and one of its offspring and another adult female. The three sera were positive in all three diagnostic tests used on site. In the Immunocomb Bispot HIV 1 and 2 test, only the middle spot, which corresponds to HIV-2 synthetic peptides derived from Env gp36, was reactive with the three sera. The mother and its offspring died of unknown causes before we could take fresh blood samples for virus characterization. A new blood sample was obtained from the remaining female for virus characterization. DNA was purified on the same day and plasma was stored at −80°C. A Congolese family had bought this animal from a hunter at Kinshasa harbor on the Congo River, but the site of capture was unknown. Based on pictures, the animal was classified as a C. mona denti by Colin Groves, after being incorrectly classified as Cercopithecus wolfi (S. Saragosti et al., Molecular characterization of primate lentiviruses from a Cercopithecus wolfi and a Cercopithecus ascanius, Abstr. 8th Annu. HIV Dynam. Evol. Meet., p. 19, 2001).
Genomic organization of SIVden: presence of a vpu gene.
In order to genetically characterize the lentivirus infecting the seropositive C. mona denti monkey, the full-length genome was sequenced by applying the primer-walking approach to an unintegrated circular DNA target (6). The RNA genome sequence (9,679 bp) was compared with that of other primate lentiviruses. Ambiguous sequences and regions of interest were confirmed by independent PCR amplification of DNA and by reverse transcription-PCR on concentrated virions obtained from plasma.
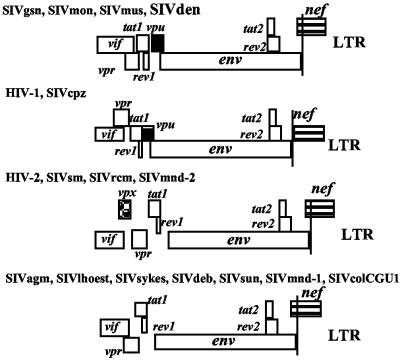
The SIVden genome harbors a vpu gene as do other viruses belonging to the SIVcpz/HIV-1 and SIVgsn lineages (SIVgsn, SIVmon, and SIVmus). However, the SIVden vpu is truncated at its C-terminal end; its length is only 64 residues, whereas the HIV-1/SIVcpz, SIVgsn, SIVmon, and SIVmus vpu genes are, respectively, 74 to 88, 76 to 78, 81, and 76 amino acids long. Moreover, the env and nef ORFs overlap in SIVden, as in other SIVs, whereas they are separate in the HIV-1/SIVcpz lineage (Fig. 1).
FIG. 1.
Genomic organization of SIVden compared to other primate lentiviruses. The gag-pol region was excluded because its structure is identical in all these viruses. The names of the viruses are given at the top of each pattern. LTR, long terminal repeat.
Relationship between SIVden and known SIV/HIV lineages.
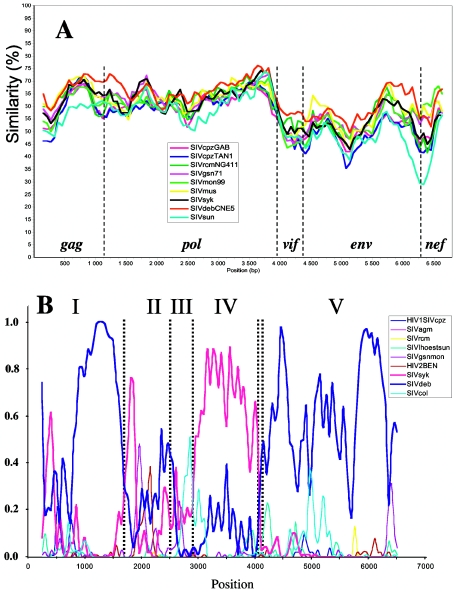
The amino acid sequence identity between SIVden and representatives of other primate lentiviruses was calculated. As shown in Table 1, SIVden was approximately equidistant from all other primate lineages. Gag and Pol were the most closely related proteins, with identities of 67% (SIVdeb) to 45% for Gag and 65% (SIVdeb) to 51% for Pol. Diversity in Env ranged from 53% (SIVdeb) to 30%. The closest Nef sequence was that of SIVdeb (59%), followed by that of SIVagmGRI1 with 52% identity. We also calculated the amino acid identities of SIVmon99 versus SIVgsn, SIVmus, SIVdeb, and SIVden in order to further emphasize that SIVden is not closely related to SIVmon, a virus that belongs to the SIVgsn subcluster, even though both viruses infect members of the C. mona group. To investigate the phylogenetic relationship of SIVden across the genome to representatives of the other primate lentivirus lineage sequences, we performed diversity plot analysis on the gag, pol, vif, env, and nef genes. Vpr, Vpu, Tat, and Rev were not included, because their alignments with the corresponding genes were too ambiguous. Gaps were removed for alignment, and diversity was calculated for a window of 400 nt moved by increments of 80 nt. The diversity plot confirmed the results shown in Table 1, indicating that SIVden is closer to SIVdeb (Fig. 2A). The same results were obtained when diversity plots were generated with protein sequences (data not shown).
TABLE 1.
Protein sequence identities among primate lentiviruses
| Strains compared | % Amino acid identitya
|
|||
|---|---|---|---|---|
| Gag | Pol | Env | Nef | |
| SIVden versus HIV/SIV strains | ||||
| SIVdeb | 67 | 65 | 53 | 59 |
| SIVsyk | 60 | 62 | 45 | 46 |
| SIVmus | 63 | 61 | 47 | 49 |
| SIVmon99 | 60 | 61 | 44 | 48 |
| SIVgsn | 60 | 61 | 44 | 50 |
| SIVsmSL92b | 56 | 60 | 42 | 46 |
| SIVmnd14cg | 58 | 59 | 34 | 50 |
| SIVrcmNG411 | 58 | 59 | 42 | 48 |
| HIV-2 BEN | 56 | 59 | 43 | 48 |
| SIVcpzGab | 58 | 58 | 40 | 46 |
| SIVagmGRIIl | 58 | 58 | 42 | 52 |
| HIV-1_M | 57 | 58 | 40 | 46 |
| HIV-1_N YBF30 | 54 | 58 | 39 | 42 |
| SIVcpzTAN1 | 54 | 58 | 43 | 47 |
| SIVcpzANT | 56 | 57 | 42 | 42 |
| SIVlhoest | 49 | 56 | 35 | 47 |
| SIVmndGB1 | 50 | 56 | 33 | 50 |
| SIVsun | 50 | 55 | 34 | 51 |
| SIVcolGUI1 | 45 | 51 | 30 | 34 |
| SIVmon99 versus SIVsyk lineage strains | ||||
| SIVmus | 75 | 71 | 68 | 62 |
| SIVgsn166 | 75 | 71 | 69 | 72 |
| SIVgsn71 | 75 | 70 | 69 | 72 |
| SIVdebCE40 | 67 | 65 | 53 | 59 |
| SIVdebCNE5 | 67 | 65 | 52 | 55 |
| SIVden | 60 | 61 | 44 | 48 |
SIVden had the highest percent amino acid identity with SIVdeb (boldface).
FIG. 2.
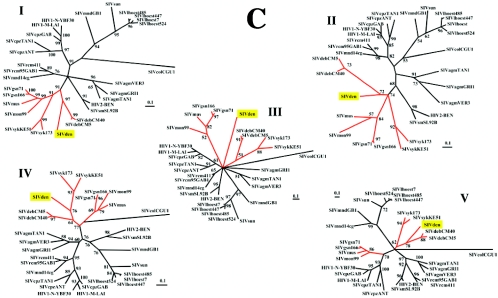
(A) Diversity plot comparing SIVden with SIV and HIV sequences (6,597 nucleotides after degapping). Nucleotide sequence difference is plotted for windows of 400 nucleotides and an 80-nucleotide step increment. (B) Bayesian scanning plot of SIVden against SIV major lineages. The dotted bars indicate the positions of the putative recombination breakpoints in the SIVden genome. (C) Unrooted TreePuzzle trees of fragments I, II, III, IV, and V. The corresponding fragments of the translated sequences were used for the TreePuzzle analysis. The branches of the SIVsyk lineage are shown in red.
Phylogenetic analyses of SIVden sequence.
To investigate for potential recombination between the SIVden and the previously characterized SIV lineages, we performed an exploratory analysis by a full-genomic Bayesian scanning as described previously (53). The results of the Bayesian scanning are shown in Fig. 2B, where the posterior probabilities of different partitions were plotted throughout the alignment. As shown in Fig. 2B, there is evidence that SIVden sequence is more closely related to (i) SIVdeb in regions I and V spanning nucleotides 1 to 1650 and 3900 to 6792, respectively, or (ii) to the SIVsyk lineage in region IV (nt 2901 to 3801), while in regions II and III (nt 1651 to 2502 and 2503 to 2880, respectively) no evidence (high posterior probability support) for close relationship between SIVden and any of the previously characterized SIV sequences was observed.
Previous findings were further confirmed by phylogenetic analyses using maximum-likelihood (ML) and Bayesian methods. ML trees are shown in Fig. 2C. According to the results of phylogenetic analyses in region I, SIVden is more closely related to the SIVdeb sequences, which all cluster together with the SIVgsn, SIVmus, SIVmon, and SIVsyk lineages; Bayesian inference also revealed identical results (data not shown). In region II, SIVden is more closely related to SIVdeb, albeit with lower quartet puzzling support values (72% versus 97% in regions II and I, respectively), while according to Bayesian analysis the SIVden sequence clustered with the SIVgsn, SIVmus, SIVmon, and SIVsyk lineages (posterior probability support, 0.82; data not shown). In region III, SIVden formed a monophyletic clade; Bayesian analysis revealed that SIVden clustered similarly with SIVgsn, SIVmus, SIVmon, SIVsyk, and SIVdeb lineages as in regions I and II (data not shown). In region IV, SIVden is more closely related to the SIVsyk cluster, while it falls within the clade of SIVgsn, SIVmus, SIVmon, SIVsyk, and SIVdeb; similar results were also obtained by Bayesian analysis (data not shown). Finally in region V, SIVden clustered with the SIVdeb lineage and with a deep branch with SIVsyk sequences; interestingly SIVgsn, SIVmus, and SIVmon were not associated with the SIVden, SIVdeb, and SIVsyk clade due to its recombinant nature, as described previously (55). Similar results were also obtained by the Bayesian method (data not shown). Trees in regions I, IV, and V were compared using the Shimodaira-Hasegawa test (59), and in all cases the alternative topologies were significantly different (P < 0.05) from the ML tree.
In conclusion, SIVden was more closely related to the SIVdeb and SIVsyk sequences in regions I, V, and IV, for which phylogenetic results were concordant for both ML and Bayesian methods. In region II, it clustered with the SIVgsn, SIVmus, SIVmon, and SIVsyk lineages, but due to the discrepancy between ML and Bayesian methods, it is not clear whether SIVden is more closely related to either the SIVdeb or SIVsyk, SIVgsn, SIVmus, and SIVmon lineages. Finally, given the very short length and relatively high sequence conservation (3′ terminus of pol) of SIV sequences in region III, we cannot reach conclusions about the phylogenetic relationships of SIVden in this region.
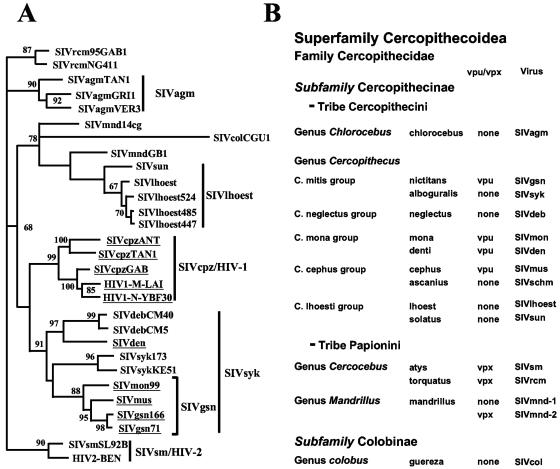
Phylogenetic analysis of the full-length SIVden sequence revealed that it is more closely related to the SIVdeb sequence within the SIVsyk group (Fig. 3A). Indeed, the similarity values, calculated on the pol region (1,877 nt after degapping, corresponding to the available SIVschm pol sequence) between SIVden and SIVdeb, SIVmus, SIVsyk, SIVgsn, SIVmon, and SIVschm were, respectively, 66.5%, 64.4%, 63.5%, 63.1%, 63.0%, and 61.7%. Phylogenetic analysis of the integrase region (54), for which a large number of sequences are available, further confirmed that SIVden is not related to the SIVgsn subcluster; but it clusters with SIVdeb from C. neglectus (65% of bootstraps), while SIVschm from C. ascanius schmidti clustered with the SIVgsn subcluster (97% of bootstraps) and SIVcol clustered with SIVsyk (64%) (data not shown).
FIG. 3.
(A) Phylogenetic relationship of SIVden to other primate lentiviruses. Phylogenetic tree showing the relationship between the SIVden gag, pol, vif, env, and nef regions (2,199 amino acids after degapping) to equivalent sequences from selected HIV/SIV viruses representing the different lineages. Phylogenetic relationships were computed using TreePuzzle. The numbers at the nodes indicate the posterior probability (only values that are >65% are shown). The SIVgsn subcluster is shown with a square bracket. The names of the viral genomes that include a vpu are underlined. (B) Taxonomy of nonhuman primates harboring lentiviruses. Primates harboring lentiviruses are classified according to C. Groves (23). The presence of vpu or vpx is indicated, along with the names of the viruses.
Analysis of the vpu gene.
In HIV-infected cells, Vpu is synthesized from a bicistronic mRNA strand that also encodes the env protein (gp160). The two proteins are synthesized at similar rates. Vpu is a membrane phosphoprotein with an N-terminal hydrophobic membrane-spanning domain and a C-terminal cytoplasmic tail (for a review, see reference 19). Although the Vpu protein shows considerable variability in its sequence and length (42), some invariant amino acids have been identified. Amino acid 23W, which is invariant in HIV-1 group M and group O sequences and in SIVcpz isolated from Pan troglodytes troglodytes (SIVcpzPtt), is present in SIVden and SIVmon99 but not in SIVcpzANT, SIVcpzTAN1, SIVmonNG1, SIVgsn, or SIVmus (Fig. 4). This tryptophan is involved in the structure of the HIV-1 Vpu ion channel (12). The α-helix 1, located in the membrane-proximal cytoplasmic region of Vpu, which is involved in the interaction between Vpu and CD4 and is required for CD4 degradation (65), is predicted to exist in the SIVden Vpu. Vpu promotes CD4 protein degradation, allowing the envelope protein to be transported to the cell surface for virion assembly. This function is regulated by phosphorylation of serine residues 52 and 56 located in the DSGXES motif of the cytoplasmic domain of the Vpu protein. Indeed, this characteristic sequence involved in Vpu-mediated CD4 and IκBα proteolysis is present in SIVden and SIVcpzPtt (SIVcpzGAB, SIVcpzCAM3, SIVcpzCAM5, and SIVcpzUS), whereas the full motif is not found in SIVcpz isolated from Pan troglodytes schweinfurthii (SIVcpzANT and SIVcpzTAN1) or in the SIVgsn subcluster. Only the first serine is present in the Vpu of the SIVcpzPts and SIVgsn groups, while both serines are present in SIVcpzPtt (Fig. 4).
FIG. 4.
Alignment of the vpu sequences of SIVden, the HIV-1/SIVcpz lineage, and SIVgsn. The amino acid sequence of SIVden is shown at the top. The putative transmembrane region (TM) and the α-helix region are indicated. The characteristic DSGXES motif (involved in VPU-mediated CD4 and I-κBα proteolysis) and the highly conserved tryptophan present in the transmembrane region are shaded in black. The dashes indicate gaps introduced to optimize alignment, and dots indicate identical residues.
Analysis of the env gene.
SIVden gp120 harbors the 18 cysteines conserved in most HIV/SIV sequences (6) and two additional cysteines. The first one is located in the N terminus as in the SIVgsn166 and SIVcpzGab sequences, and the second is located in the V2 loop where extra cysteines are frequently found. The major N-linked glycosylation sites are also present with one exception, the site spanning the proximal cysteine of the V2 loop that is present in all the sequences of the SIVsyk lineage (data not shown).
Furthermore, while the V3 loop sequences of viruses from the SIVgsn subcluster, which includes SIVmon, and from SIVcpz viruses are remarkably conserved (13), the SIVden V3 loop sequence differs from all V3 loop sequences hitherto described. In Fig. 5, we chose to align the sequences with SIVmonNG1 in order to highlight the low identity between SIVden and the other sequences, as well as the homology of the SIVgsn subcluster sequences. Moreover, despite the close relationship with SIVdeb, SIVden V3 loop does not resemble SIVdeb V3 loop.
FIG. 5.
Alignment of the V3 loop region. The SIVden sequence is highlighted in gray. The conserved cysteines are indicated by black boxes, the conserved tryptophan is indicated by a light-gray box, and the deletion downstream from the distal cysteine is indicated by a dark-gray box with white dashes. The dashes indicate gaps introduced to optimize alignment, and dots indicate identical residues.
A tryptophan downstream of the V3 loop is highly conserved among primate lentiviruses (Fig. 5). Six amino acids separate the distal cysteine and this tryptophan in the HIV-1/SIVcpz lineage, SIVgsn subcluster, and SIVden, while only five residues are present in the remaining SIVs, including SIVdeb (Fig. 5).
Functional motifs shared by Cercopithecus monkey viruses.
Comparison of Y chromosome-specific (TSPY) sequences of representatives from 10 different Old World monkey genera led Tosi and colleagues to propose that members of the lhoesti group (including L'Hoest's and sun-tailed monkeys) were significantly more closely related to African green monkeys (Chlorocebus aethiops) and patas monkeys (Erythrocebus patas) than they were to other Cercopithecus species including the mona group (66, 67). SIVden as well as SIVsyk, SIVgsn, SIVmus, SIVmon, and SIVdeb derived from species belonging to the Cercopithecus genus.
In light of the new guenon taxonomy, excluding the lhoesti group from the Cercopithecus genus, Bibollet-Ruche et al. (6) inspected sequences from SIV harbored by Cercopithecus in order to highlight shared regulatory and/or protein motifs that would suggest a host-specific function. We checked the SIVden sequence for these signatures.
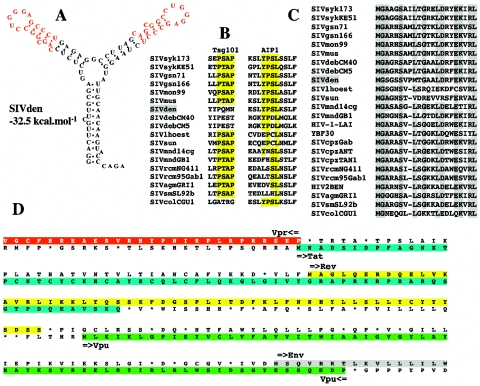
The predicted secondary structure of the Tat-transactivation response (TAR) element for SIVdeb, SIVsyk, SIVgsn, SIVmon, and SIVmus indicated a duplicated stem-loop structure consisting of a one-base bulge (C or U), a 3-bp stem, and a six-base terminal loop with the sequence 5′-CUGGGA-3′ identical to the HIV-1 loop. The predicted secondary structure of SIVden TAR also indicated a duplicated stem-loop with a conserved one-base bulge C more similar to SIVsyk, SIVgsn, SIVmon, and SIVmus than to SIVdeb, despite a closer phylogenetic relationship (Fig. 6A). Moreover, SIVden, SIVdeb, SIVsyk, SIVgsn, SIVmon, and SIVmus share a second bulge two nucleotides upstream from the first one.
FIG. 6.
Features of SIVden. (A) Secondary structure for SIVden TAR with the lowest free energy value (−32.5 kcal/mol). Red letters indicate sequence conserved between the two stems. (B) Alignment of primate lentiviral Gag P6 protein sequences. Only the two putative binding sites for the Tsg101 [P(T/S)AP] and AIP1 [YP(D/S)L] proteins are shown. Conserved amino acids are highlighted in yellow. (C) Alignment of the N terminus of the Gag gene. Dashes indicate space introduced to optimize the alignment. Conserved amino acids are highlighted in gray. Dots indicate amino acid identity at a given residue. (D) Organization of the vpr-env region. The vpr and tat ORFs overlap on a single amino acid residue. The tat ORF is highlighted in blue, while the rev ORF is highlighted in yellow; the vpu ORF is highlighted in green, and the N terminus of the env ORF is highlighted in gray.
We looked also for the two different lentiviral domains, which have been identified in SIV Gag p6 protein sequences. The sequences P(T/S)AP and YPXL interact, respectively, with Tsg101, a cellular protein that facilitates the budding of vesicles into late endosomes, and AIP1, a host protein involved in endosomal sorting and retroviral budding. Inspection of the alignment shows that SIVden, like SIVdeb and SIVcol, lacks a P(T/S)AP site but contains a YPSL site at the C terminus of the p6 protein, present in all other Cercopithecus viruses. This motif is different from the YPDL motif found in SIVdeb (6). Indeed, the YP(D/S)L motif is shared by all SIVs infecting Cercopithecus monkey species, unlike the P(T/S)AP motif, which is not present in SIVdeb and SIVden (Fig. 6B).
Another signature shared by all the viruses belonging to the SIVsyk lineage is an insertion in the N terminus of the Gag protein (Fig. 6C), a region involved in the binding of the precursor Pr55 to membrane (49). However, there is no evidence of a functional significance for this insertion.
SIVden unique features.
Another feature of the SIVden gag gene is located in the nucleocapsid protein Ncp7. The primate lentivirus nucleocapsid protein NCp7 contains two CCHC (Cys-X2-Cys-X4-His-X4-Cys) zinc knuckle domains separated by seven residues. In most SIVs the number of residues is highly conserved, yet 11 residues in SIVden separate the two zinc knuckle domains. The only other SIVs with different numbers of residues are SIVsun (10 residues) and SIVcolCGU1 (28 residues). These changes might modify the RNA-binding properties of these proteins.
SIVden Vif (258 amino acids) is rather longer than the Vif proteins of other SIVs (237 amino acids in SIVgsn and 192 in the HIV-1/SIVcpz lineage). The protein sequences are too dissimilar to allow unambiguous alignment; in particular, the C-terminal region is distinct from that of all known proteins.
Furthermore, the distance between the putative tat/rev splice donor and the N-terminal end of the vpu gene is much longer in SIVden than in other primate lentiviruses, resulting in a greater distance between the tat/rev first exons and the vpu gene and a putative longer open reading frame for Rev (Fig. 6D). This feature is unique to SIVden. Furthermore, it was shown that vpr and tat genes overlap in all SIVs with the exception of SIVdeb (6). SIVden shows an intermediate type with a single amino acid overlap (Fig. 6D).
DISCUSSION
During a seroprevalence survey of wild-caught pet monkeys in Kinshasa, Democratic Republic of Congo, we identified a new primate lentivirus, SIVden. Although the SIVden genome contains a vpu gene, phylogenetic analysis indicated that SIVden was not phylogenetically related to the vpu-harboring viruses of the SIVgsn subcluster.
It was proposed that the vpu gene originated in the common ancestor of SIVgsn, SIVmus, and SIVmon, after the divergence of SIVsyk and SIVdeb (6). However, the fact that SIVden, which carries a Vpu, clusters with SIVdeb, which does not encode a vpu, contradicts this hypothesis. So, three possibilities exist: (i) vpu is a new gene acquired during the evolution of SIV in some Cercopithecus species; (ii) some SIVs have lost the vpu gene during evolution (13); or (iii) the presence of vpu in the SIVden genome results from a recombination event with a virus from a yet unidentified species. The fact that SIVden vpu does not resemble any known vpu excludes the possibility that a recombination event occurred with an already characterized virus. Furthermore, screening of more individuals will be necessary to confirm that Dent's Mona monkey is the natural host of SIVden. Although none of these Vpu proteins has been shown to be functional, it is likely that a gene conserved for so long a period and among different viruses would be essential for replication in vivo.
Primates known to harbor lentiviruses belong to Pan troglodytes and the Cercopithecidae family, the latter being subdivided into two distinct subfamilies, Colobinae and Cercopithecinae. The presence or absence of an extra accessory gene (vpu or vpx) and overlap of the env and nef genes could be used to classify primate lentiviruses into four groups (Fig. 1). Env-nef overlap is shared by all SIVs, with the notable exception of the SIVcpz/HIV-1 lineage, and is unrelated to the presence of a vpu gene. Besides the HIV-1/SIVcpz lineage, the vpu gene is also found in another group comprising SIVgsn, SIVmon, SIVmus, and SIVden. Viruses of the third group have a vpx gene (HIV-2/SIVsm, SIVmnd-2, SIVdrl, and SIVrcm). However, the largest group comprises viruses harboring neither vpx nor vpu (SIVagm, SIVsyk, SIVdeb, SIVlhoest, SIVsun, SIVmnd-1, and SIVcol).
Phylogenetic analysis of the of gag, pol, vif, env, and nef regions or of the gag-pol region (17) confirmed that SIVs isolated from the genus Cercopithecus cluster into two groups. The SIVsyk lineage contains SIVsyk, SIVdeb, SIVden, SIVschm, SIVgsn, SIVmon, and SIVmus; the last three of these SIVs form a separate subcluster that has been named the SIVgsn lineage (13). The L'Hoest lineage includes SIVlhoest, SIVsun, and SIVmnd-1.
The evolutionary history of SIVs is more complex than previously thought. Indeed, the L'Hoest lineage comprises SIVlhoest and SIVsun—a clustering pattern highly suggestive of SIV coevolution with the C. lhoesti superspecies (4)—but also includes SIVmnd-1 (isolated from a Papioni primate) (29, 61, 64). Moreover, SIVsyk, which lacks a vpu gene, and SIVgsn, which carries a vpu gene, were both isolated from C. mitis group members; SIVmus, which carries a vpu gene, and SIVschm, which lacks a vpu gene (unpublished data) were isolated from C. cephus group members. Both SIVmon and SIVden, isolated from two C. mona species, carry a vpu gene, but phylogenetic analysis showed that SIVden is more closely related to SIVdeb than to the SIVgsn subcluster (Fig. 2B and 3A). Thus, the clustering pattern of these viruses does not match their genomic organization. Moreover, comparison of the phylogenetic tree of the gag, pol, vif, env, and nef regions (Fig. 3A) and the taxonomic table (Fig. 3B) shows that although all SIVs derived from Cercopithecus monkeys cluster with high support values, the clustering pattern of these viruses does not match the taxonomic classification of their hosts within the Cercopithecus genus.
The topology of a phylogenetic tree depends on the taxa included in the analysis, and characterization of new full-length genomes should, therefore, provide a more accurate picture of the evolutionary relationships between the different primate lentiviruses.
All these findings imply that host-dependent evolution of viruses within the Cercopithecus genus was not a linear phenomenon but likely involved transmission among closely related nonhuman primates (9), and in some cases, superinfection led to recombination events. Several mosaic genomes have been described (e.g., SIVmnd-2, SIVagm.sab, SIVrcm, and SIVcpz), indicating that cross-species transmission and recombination have long existed (2, 5, 33, 35, 61). The close relationship between SIVmon, SIVmus, and SIVgsn might therefore be explained by cross-species transmissions among mona, mustached, and greater spot-nosed monkeys, as these monkeys share the same habitats and form mixed-species associations (6, 13). SIVden could also have resulted from cross-species transmission. C. mona denti, C. neglectus, and C. ascanius have overlapping habitats, and Dent's Mona monkeys frequently associate with C. ascanius (36). However, SIVden is phylogenetically more closely related to SIVdeb than to SIVschm or to our own full-length SIVschmKin sequence (unpublished data).
Although SIVden has some similarities with SIVcpzANT and carries a vpu gene, it cannot be regarded as a progenitor of the SIVcpz/HIV-1 lineage. Thus, it is conceivable that SIVcpz arose from multiple infections generating a recombinant virus (2) or that an unidentified SIV was transmitted to and successfully spread among chimpanzees, before being transmitted to humans. Alternatively, an unidentified lower primate could be the natural host of both SIVcpz and HIV-1. Six mona monkey species, including C. mona denti, live in a territory extending from west Africa to the Great Lakes region, the area in which the three groups of HIV-1 (M, N, and O) emerged and which corresponds to the geographic biotope of the four chimpanzee subspecies. To date, the viruses harbored by only two of them have been characterized.
Interestingly, both HIV-1 and HIV-2 carry an extra accessory gene (vpu and vpx, respectively), while most SIV genomes do not. The third SIV cross-species transmission event to humans, which was only serologically documented, also involved a vpx-harboring virus, SIVmnd-2 (61). Given the multiple biological functions of Vpu in the HIV-1 replicative cycle, it is intriguing that no functional equivalent of Vpu has yet been found in the other SIV lineages, except for SIVcpz and some recently described SIVs from Cercopithecus monkeys. This is especially paradoxical, as Vpu augments the release of retroviruses that naturally lack a vpu gene, such as HIV-2, visna virus, Moloney murine leukemia virus (22), and simian/human immunodeficiency virus chimeras (SHIV).
SHIV viruses (SHIVKU-1 and SHIV-4) are highly pathogenic, causing AIDS in both rhesus monkeys and pig-tailed macaques (43, 62). Pathogenicity correlates with nef and env expression and is associated with reversion of the vpu start codon (43, 63). Intact vpu and nef genes are essential for acquisition of pathogenicity by nonpathogenic SHIV during serial passage in macaques (40). In viruses carrying neither vpu nor vpx, another protein may be able to positively regulate virion release, as is the case of HIV-2 Env (7).
Characterization of genetic discordances among different lentivirus-host systems may help to identify new candidate viral or host cell targets for the treatment of HIV infection. Because of the critical role of vpu in HIV infection and pathogenesis, both the gene and the protein are good candidate targets.
Further characterization should help to elucidate the origins and evolution of primate lentiviruses and provide insights into the natural history of HIV-1 and HIV-2. Partial or full-length lentivirus sequences have so far been reported in 34 different primate species. Cross-species transmission is an important area of research, as these viruses represent a potential source of new human epidemics (54). Some primate lentivirus strains may not be recognized by commercial HIV-1/HIV-2 screening tests, as it was shown for HIV-1 group O (38), and might thus represent a particular public health threat. It is therefore important to continue to characterize new SIVs from as many different primate species as possible.
Acknowledgments
We thank C. Apetrei, C. Groves, and J. Clewley for their comments and critical reading of the manuscript and Annemie Vandamme for providing the SlidingBayes software.
This work was supported by a grant from the Agence National de Recherche sur le SIDA (ANRS). D.P. was supported by a Marie Curie fellowship from the European Commission (QLK2-CT2001-51062) and by the Hellenic Center for Infectious Disease Control (KEEL).
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, K. L., A. O. Ajao, and J. P. Clewley. 2003. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona). J. Virol. 77:6879-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J. P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. St Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibollet-Ruche, F., E. Bailes, F. Gao, X. Pourrut, K. L. Barlow, J. P. Clewley, J. M. Mwenda, D. K. Langat, G. K. Chege, H. M. McClure, E. Mpoudi-Ngole, E. Delaporte, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2004. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a cercopithecus monkey virus clade. J. Virol. 78:7748-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bour, S., and K. Strebel. 1996. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 70:8285-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broussard, S. R., S. I. Staprans, R. White, E. M. Whitehead, M. B. Feinberg, and J. S. Allan. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charleston, M. A., and D. L. Robertson. 2002. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 51:528-535. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., P. Telfer, P. Reed, L. Zhang, A. Getti, D. D. Ho, and P. A. Marx. 1995. Isolation and characterization of the first simian immunodeficiency virus from a feral sooty mangabey (Cercocebus atys) in West Africa. J. Med. Primatol. 24:108-115. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 12.Cordes, F. S., A. Kukol, L. R. Forrest, I. T. Arkin, M. S. Sansom, and W. B. Fischer. 2001. The structure of the HIV-1 Vpu ion channel: modelling and simulation studies. Biochim. Biophys. Acta 1512:291-298. [DOI] [PubMed] [Google Scholar]
- 13.Courgnaud, V., B. Abela, X. Pourrut, E. Mpoudi-Ngole, S. Loul, E. Delaporte, and M. Peeters. 2003. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 77:12523-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courgnaud, V., P. Formenty, C. Akoua-Koffi, R. Noe, C. Boesch, E. Delaporte, and M. Peeters. 2002. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus). J. Virol. 77:744-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A. M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foley, B. T. 2000. An overview of the molecular phylogeny of lentiviruses, p. 35-43. In C. Kuiken, B. Foley, E. Freed, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinksy (ed.), HIV sequence compendium 2000. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N. Mex.
- 18.Fomsgaard, A., V. M. Hirsch, J. S. Allan, and P. R. Johnson. 1991. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology 182:397-402. [DOI] [PubMed] [Google Scholar]
- 19.Frankel, A. D., and J. A. Young. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 20.Fultz, P. N., H. M. McClure, D. C. Anderson, R. B. Swenson, R. Anand, and A. Srinivasan. 1986. Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys). Proc. Natl. Acad. Sci. USA 83:5286-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 22.Gottlinger, H. G., T. Dorfman, E. A. Cohen, and W. A. Haseltine. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. USA 90:7381-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groves, C. 2001. Primate taxonomy. Smithsonian Institution Press, Washington, D.C.
- 24.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch, V. M., G. Dapolito, P. R. Johnson, W. R. Elkins, W. T. London, R. J. Montali, S. Goldstein, and C. Brown. 1995. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J. Virol. 69:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch, V. M., G. A. Dapolito, S. Goldstein, H. McClure, P. Emau, P. N. Fultz, M. Isahakia, R. Lenroot, G. Myers, and P. R. Johnson. 1993. A distinct African lentivirus from Sykes' monkeys. J. Virol. 67:1517-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 29.Hu, J., W. M. Switzer, B. T. Foley, D. L. Robertson, R. M. Goeken, B. T. Korber, V. M. Hirsch, and B. E. Beer. 2003. Characterization and comparison of recombinant simian immunodeficiency virus from drill (Mandrillus leucophaeus) and mandrill (Mandrillus sphinx) isolates. J. Virol. 77:4867-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 31.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 32.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 33.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 35.Jubier-Maurin, V., P. Sarni-Manchado, F. Veas, N. Vidal, F. Bibollet-Ruche, J. P. Durand, A. Galat-Luong, and G. Cuny. 1995. Regulatory genes of simian immunodeficiency viruses from west African green monkeys (Cercopithecus aethiops sabaeus). J. Virol. 69:7349-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingdon, J. 2001. The Kingdon field guide to African mammals. Academic Press, London, United Kingdom.
- 37.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loussert-Ajaka, I., T. D. Ly, M. L. Chaix, D. Ingrand, S. Saragosti, A. M. Courouce, F. Brun-Vezinet, and F. Simon. 1994. HIV-1/HIV-2 seronegativity in HIV-1 subtype O infected patients. Lancet 343:1393-1394. [DOI] [PubMed] [Google Scholar]
- 39.Lowenstine, L. J., N. C. Pedersen, J. Higgins, K. C. Pallis, A. Uyeda, P. Marx, N. W. Lerche, R. J. Munn, and M. B. Gardner. 1986. Seroepidemiologic survey of captive Old World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys). Int. J. Cancer. 38:563-574. [DOI] [PubMed] [Google Scholar]
- 40.Mackay, G. A., Y. Niu, Z. Q. Liu, S. Mukherjee, Z. Li, I. Adany, S. Buch, W. Zhuge, H. M. McClure, O. Narayan, and M. S. Smith. 2002. Presence of intact vpu and nef genes in nonpathogenic SHIV is essential for acquisition of pathogenicity of this virus by serial passage in macaques. Virology 295:133-146. [DOI] [PubMed] [Google Scholar]
- 41.Maddison, D., and W. Maddison. 2003. MacClade 4: analysis of phylogeny and character evolution, 4.06 ed. Sinauer Assoc., Sunderland, Mass. [DOI] [PubMed]
- 42.McCormick-Davis, C., S. B. Dalton, D. K. Singh, and E. B. Stephens. 2000. Comparison of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids, and a signature sequence motif common to subtype C isolates. AIDS Res. Hum. Retrovir. 16:1089-1095. [DOI] [PubMed] [Google Scholar]
- 43.McCormick-Davis, C., L. J. Zhao, S. Mukherjee, K. Leung, D. Sheffer, S. V. Joag, O. Narayan, and E. B. Stephens. 1998. Chronology of genetic changes in the vpu, env, and nef genes of chimeric simian-human immunodeficiency virus (strain HXB2) during acquisition of virulence for pig-tailed macaques. Virology 248:275-283. [DOI] [PubMed] [Google Scholar]
- 44.Muller, M. C., N. K. Saksena, E. Nerrienet, C. Chappey, V. M. Herve, J. P. Durand, P. Legal-Campodonico, M. C. Lang, J. P. Digoutte, and A. J. Georges. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 67:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller-Trutwin, M. C., S. Corbet, S. Souquiere, P. Roques, P. Versmisse, A. Ayouba, S. Delarue, E. Nerrienet, J. Lewis, P. Martin, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. SIVcpz from a naturally infected Cameroonian chimpanzee: biological and genetic comparison with HIV-1 N. J. Med. Primatol. 29:166-172. [DOI] [PubMed] [Google Scholar]
- 46.Nicol, I., D. Messinger, P. Dubouch, J. Bernard, I. Desportes, R. Jouffre, R. Snart, P. Nara, R. C. Gallo, and D. Zagury. 1989. Use of Old World monkeys for acquired immunodeficiency syndrome research. J. Med. Primatol. 18:227-236. [PubMed] [Google Scholar]
- 47.Novembre, F. J., J. De Rosayro, S. P. O'Neil, D. C. Anderson, S. A. Klumpp, and H. M. McClure. 1998. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J. Virol. 72:8841-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohta, Y., T. Masuda, H. Tsujimoto, K. Ishikawa, T. Kodama, S. Morikawa, M. Nakai, S. Honjo, and M. Hayami. 1988. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int. J. Cancer 41:115-122. [DOI] [PubMed] [Google Scholar]
- 49.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osterhaus, A. D., N. Pedersen, G. van Amerongen, M. T. Frankenhuis, M. Marthas, E. Reay, T. M. Rose, J. Pamungkas, and M. L. Bosch. 1999. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin). Virology 260:116-124. [DOI] [PubMed] [Google Scholar]
- 51.Pandrea, I., R. Onanga, C. Kornfeld, P. Rouquet, O. Bourry, S. Clifford, P. T. Telfer, K. Abernethy, L. T. White, P. Ngari, M. Muller-Trutwin, P. Roques, P. A. Marx, F. Simon, and C. Apetrei. 2003. High levels of SIVmnd-1 replication in chronically infected Mandrillus sphinx. Virology 317:119-127. [DOI] [PubMed] [Google Scholar]
- 52.Pandrea, I., R. Onanga, P. Rouquet, O. Bourry, P. Ngari, E. J. Wickings, P. Roques, and C. Apetrei. 2001. Chronic SIV infection ultimately causes immunodeficiency in African non-human primates. AIDS 15:2461-2462. [DOI] [PubMed] [Google Scholar]
- 53.Paraskevis, D., P. Lemey, M. Salemi, M. Suchard, Y. Van De Peer, and A. M. Vandamme. 2003. Analysis of the evolutionary relationships of HIV-1 and SIVcpz sequences using Bayesian inference: implications for the origin of HIV-1. Mol. Biol. Evol. 20:1986-1996. [DOI] [PubMed] [Google Scholar]
- 54.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salemi, M., T. De Oliveira, V. Courgnaud, V. Moulton, B. Holland, S. Cassol, W. M. Switzer, and A. M. Vandamme. 2003. Mosaic genomes of the six major primate lentivirus lineages revealed by phylogenetic analyses. J. Virol. 77:7202-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santiago, M. L., F. Bibollet-Ruche, E. Bailes, S. Kamenya, M. N. Muller, M. Lukasik, A. E. Pusey, D. A. Collins, R. W. Wranghm, J. Goodall, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2003. Amplification of a complete simian virus genome from a fecal RNA of a wild chimpanzee. J. Virol. 77:2233-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 58.Sharp, P. M., E. Bailes, R. R. Chaudhuri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of acquired immune deficiency syndrome viruses: where and when? Philos. Trans. R. Soc. Lond. B 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 60.Soares, M. A., D. L. Robertson, H. Hui, J. S. Allan, G. M. Shaw, and B. H. Hahn. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394-399. [DOI] [PubMed] [Google Scholar]
- 61.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stephens, E. B., C. McCormick, E. Pacyniak, D. Griffin, D. M. Pinson, F. Sun, W. Nothnick, S. W. Wong, R. Gunderson, N. E. Berman, and D. K. Singh. 2002. Deletion of the vpu sequences prior to the env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology 293:252-261. [DOI] [PubMed] [Google Scholar]
- 63.Stephens, E. B., S. Mukherjee, M. Sahni, W. Zhuge, R. Raghavan, D. K. Singh, K. Leung, B. Atkinson, Z. Li, S. V. Joag, Z. Q. Liu, and O. Narayan. 1997. A cell-free stock of simian-human immunodeficiency virus that causes AIDS in pig-tailed macaques has a limited number of amino acid substitutions in both SIVmac and HIV-1 regions of the genome and has offered cytotropism. Virology 231:313-321. [DOI] [PubMed] [Google Scholar]
- 64.Takehisa, J., Y. Harada, N. Ndembi, I. Mboudjeka, Y. Taniguchi, C. Ngansop, S. Kuate, L. Zekeng, K. Ibuki, T. Shimada, B. Bikandou, Y. Yamaguchi-Kabata, T. Miura, M. Ikeda, H. Ichimura, L. Kaptue, and M. Hayami. 2001. Natural infection of wild-born mandrills (Mandrillus sphinx) with two different types of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 17:1143-1154. [DOI] [PubMed] [Google Scholar]
- 65.Tiganos, E., J. Friborg, B. Allain, N. G. Daniel, X. J. Yao, and E. A. Cohen. 1998. Structural and functional analysis of the membrane-spanning domain of the human immunodeficiency virus type 1 Vpu protein. Virology 251:96-107. [DOI] [PubMed] [Google Scholar]
- 66.Tosi, A. J., T. R. Disotell, J. C. Morales, and D. J. Melnick. 2003. Cercopithecine Y-chromosome data provide a test of competing morphological evolutionary hypotheses. Mol. Phylogenet. Evol. 27:510-521. [DOI] [PubMed] [Google Scholar]
- 67.Tosi, A. J., D. J. Melnick, and T. R. Disotell. 2004. Sex chromosome phylogenetics indicate a single transition to terrestriality in the guenons (tribe Cercopithecini). J. Hum. Evol. 46:223-237. [DOI] [PubMed] [Google Scholar]
- 68.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]
- 69.Vanden Haesevelde, M. M., M. Peeters, G. Jannes, W. Janssens, G. van der Groen, P. M. Sharp, and E. Saman. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346-350. [DOI] [PubMed] [Google Scholar]
- 70.Verschoor, E. J., Z. Fagrouch, I. Bontjer, H. Niphuis, and J. L. Heeney. 2004. A novel simian immunodeficiency virus isolated from a Schmidts' guenon (Cercopithecus ascanius schmidti). J. Gen. Virol. 85:21-24. [DOI] [PubMed] [Google Scholar]