Abstract
Biodiversity data, particularly species occurrence and abundance, are indispensable for testing empirical hypothesis in natural sciences. However, datasets built for research programmes do not often meet FAIR (findable, accessible, interoperable and reusable) principles, which raises questions about data quality, accuracy and availability. The 21st century has markedly been a new era for data science and analytics and every effort to aggregate, standardise, filter and share biodiversity data from multiple sources have become increasingly necessary. In this study, we propose a framework for refining and conforming secondary biodiversity data to FAIR standards to make them available for use such as macroecological modelling and other studies. We relied on a Darwin Core base model to standardise and further facilitate the curation and validation of data related including the occurrence and abundance of multiple taxa of a region that encompasses estuarine ecosystems in an ecotonal area bordering the easternmost Amazonia. We further discuss the significance of feeding standardised public data repositories to advance scientific progress and highlight their role in contributing to the biodiversity management and conservation.
Keywords: Darwin Core standard, FAIR data, Golfão Maranhense, secondary data
Introduction
High-quality, openly available biodiversity datasets (e.g. species occurrence, abundance, traits) are indispensable for the monitoring of species and ecosystems and to improve the development of conservation and management policies (Wetzel et al. 2015, Wetzel et al. 2018). Biodiversity data under FAIR (findable, accessible, interoperable and reusable) principles also help optimising editorial processes for academic publications accelerating peer review, increasing the visibility of scientific papers and improving citation rates (Costello et al. 2013, Piwowar and Vision 2013). Efforts to build and maintain repositories of FAIR data principles have been undertaken by biodiversity data collectors and curators (Hackett et al. 2019), who strive to organise, standardise and share data from a variety of primary (e.g. fieldwork) and secondary (e.g. literature) sources, for example, global initiatives, such as the "Global Biodiversity Information Facility - GBIF" (https://www.gbif.org/), which provide access to comprehensive biodiversity datasets and facilitate their reuse. This is particularly important in the modern era of big data science, which challenges our ability to organise, filter and analyse large and complex datasets (Cao 2016).
The availability of biodiversity data is influenced by several factors, including geographic region, scientific interest and resource availability (e.g. financial and infrastructure constraints), which can affect the quality and type of data produced (Amano et al. 2016). In addition, biodiversity data are not always publicly available, requiring users to conduct extensive searches in scientific publications, technical reports or by directly contacting the researcher who collected the data (Costello et al. 2013). Therefore, conducting systematic literature reviews is often necessary to compile comprehensive databases focusing on biodiversity research programmes. This method is recommended as it provides a rigorous way to search for relevant literature, allowing for peer replication and ensuring data validity and reliability (Xiao and Watson 2019). However, the retrieved data may be in inconsistent and sometimes confusing formats, requiring additional effort from users, including finding separate metadata files, reorganising and renaming fields, integrating aggregated data and discarding poorly documented or questionable data (Parr et al. 2012, Hunt et al. 2015, Culley 2017). The fundamental goal of the structure and standardisation of biodiversity data is to enable them to be understood and used by anyone and to be continuously updated and integrated with other datasets (Borregaard and Hart 2016). There are several guidelines and standards for biodiversity data, including the Darwin Core (DwC; Wieczorek et al. (2012); Darwin Core Maintenance Group (2014) and the DMPTool (https://dmptool.org/).
Finally, data must be shared and archived to ensure findability, accessibility and reusability. Data sharing can occur in a variety of ways, ranging from private sharing on request to depositing data on a public platform. Often, authors make their data available as supplementary material in scientific publications and post datasets on public websites. Sharing biodiversity data is essential for ecological research, conservation and management, education and policy decision-making (Ganzevoort et al. 2017). In addition, ecologists often use shared data for comparative studies, syntheses (e.g. meta-analyses), model parameterisation and reproducibility testing (Michener 2015). Data archiving is a critical final step that allows data to be reused for further analyses and syntheses to address new questions. Whitlock (2011) outlines optimal procedures for archiving ecological and evolutionary data, including selecting the appropriate repository and ensuring the accuracy of data and metadata. Good archiving practices facilitate long-term preservation of data, making them accessible for future research and applications.
Research programmes from megadiverse regions, such as the Neotropics, face difficulties in retrieving, organising and providing quality data due to their intrinsic complexity, which includes unrecognised species and unresolved taxa complexes and taxonomy. To address these challenges and to improve the reusability of secondary biodiversity data, our study had three main objectives:
to create a scheme or "pipeline" to improve the usability of secondary data by locating the data, performing quality control, standardising the data and archiving and sharing it;
to test our pipeline through a case study, demonstrating the step-by-step management of secondary data according to the FAIR principles; and
to evaluate if and how our approach can improve our understanding of the dynamics of regional biodiversity distribution and conservation, promote new scientific studies and knowledge and enhance our ability to generate hypotheses.
Retrieving biodiversity data is not an easy task, as the use of systematic literature searches alone does not guarantee the quality of the data. Additionally, commonly used pipelines for retrieving, standardising and making secondary data available typically overlook grey literature, despite its potential for biodiversity studies. The proposed pipeline is a combination of data retrieval and data management tools that are typically used separately, such as systematic review and the Darwin Core Standard. Following this Biodiversity Data Retrieval Pipeline ensures that secondary data are cleaned, normalised, shared and archived according to the FAIR Data Principles. Additionally, we discuss the challenges of efficient data retrieval, the potential reuse of secondary data in future studies and its limitations. Finally, we predict that initiatives to collect biodiversity data and make it available for reuse can improve knowledge and advance conservation efforts to protect the species, communities and ecosystems of these regions.
Material and methods
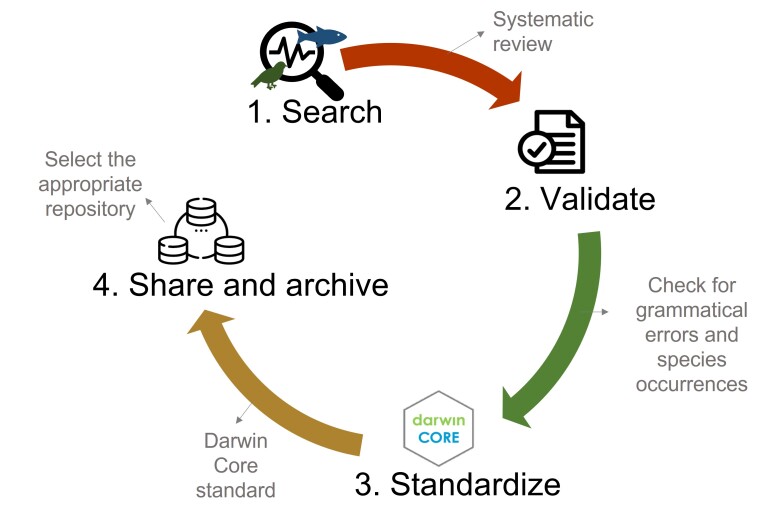
The Biodiversity Data Retrieval Pipeline was built following four stages (Fig. 1): (1) Search; (2) Validate; (3) Standardise; and (4) Share and archive. All the steps are described below.
Figure 1.
Step-by-step guide for the proposed Biodiversity Data Retrieval Pipeline to retrieve secondary biodiversity data from various sources (e.g. scientific articles, technical reports, theses, dissertations, databases) according to the FAIR principles (findable, accessible, interoperable and reusable).
To test the pipeline, we selected the Golfão Maranhense, a region located in the extreme north of the Amazon (Brazil), due to its richness, ecological diversity and importance as an ecotonal mosaic between the Amazon Forest and the dry ecosystems of eastern South America and because of the scarcity of knowledge about the biodiversity in the region. Although reports on biodiversity in the region exist, they are presented in heterogeneous forms, including scientific articles and non-peer-reviewed technical reports, making it difficult to understand the true distribution of biodiversity richness in the region.
Study area
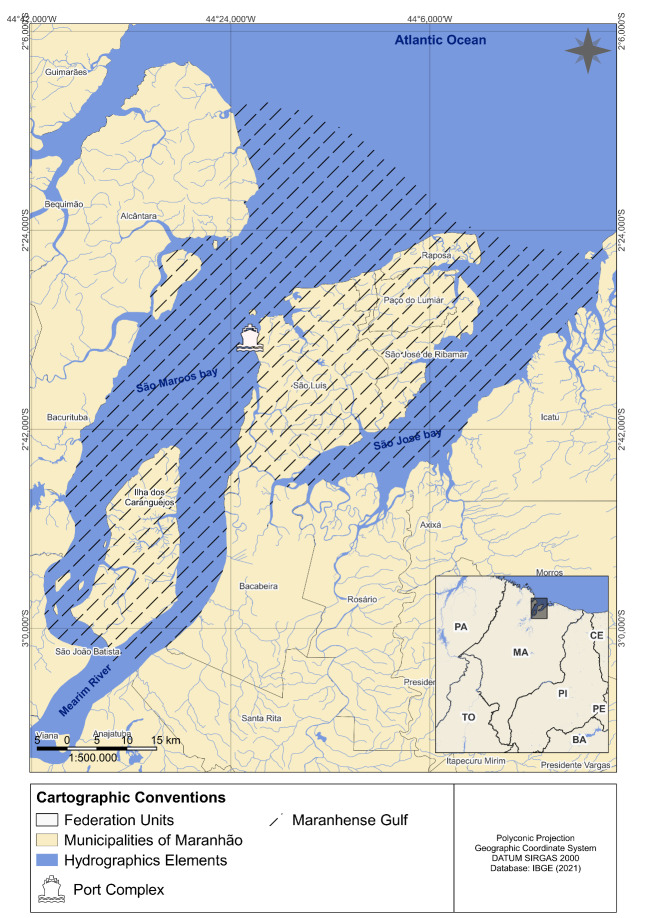
The study was conducted in the Golfão Maranhense (Maranhão State, Brazil) including 13 municipalities in the surroundings (Fig. 2). The Golfão Maranhense is a vast estuarine complex located in eastern Amazonia (Brazil) and is formed by the São Marcos and São José bays separated by the island of São Luís. This region is an area of high ecological relevance known as the “Macromaré” Mangrove Coast of Amazonia where lies the largest continuous mangrove system in the world, with about 5,414 km of mangroves in north-western Maranhão and 2,177 km in north-eastern Pará (Souza Filho 2005). The climate is tropical humid, with well-defined dry (July to December) and rainy (January to June) seasons and average temperatures around 26oC. The area is characterised by semi-diurnal macrotidal with average variations of 4 m and maximum of 7 m, with maximum tidal currents exceeding 4 m/s (Rebelo-Mochel 1997). São Marcos and São José Bays are port areas that hold significant importance for maritime activities, trade and transportation within the region.
Figure 2.
Map showing the Golfão Maranhense region, an estuary in the eastern Amazon (Brazil). This is where the secondary biodiversity data was retrieved.
Step 1: Search- systematic review
The first step in retrieving secondary data is to find the data. To do this, a systematic review of the literature is recommended. We conducted a systematic review performing searches in the platforms Science Direct and Google Scholar and public data repositories such as GBIF, VertNet, Wikiaves and SpeciesLink. During the search on the platforms, we included all the works found, such as scientific articles, books, theses and dissertations. Our search across platforms covered both published (i.e. papers and books) and unpublished literature (i.e. theses, dissertations and environmental consultancy reports focused on licensing). The searches were carried out over two months (June and July 2021) in Brazil. To ensure transparency, completeness and consistency, we followed the "Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)" guidelines. The PRISMA framework, with its checklist and flow diagram, facilitates reader comprehension and allows for the assessment of the reliability and validity of the findings.
We followed four steps:
Biotic group selection - We have chosen eight biotic groups that represent a substantial proportion of the terrestrial and aquatic biodiversity: mammals (Mammalia), reptiles including turtles, lizards and snakes (Testudines, Squamata), birds (Aves), amphibians (Amphibia), plants (Magnoliophyta), fishes, phytoplankton and benthos.
Keywords - keywords were defined considering each biotic group (Suppl. material 1). The keywords must include the name of the biotic group (e.g. Amphibians) and the study area (e.g. Maranhão) and may vary according to the needs of each biotic group.
Inclusion criteria - Our inclusion criteria were twofold: (a) studies that were conducted in Golfão Maranhense and; (b) studies that included both the geographic coordinates and the finest possible taxonomic level.
Data selection - We selected 76 variables to be extracted from the selected studies. These variables were classified into three main categories: (a) General Information - data about the published work, such as title, year, keywords and objectives of the study; (b) Sampling Events - information about when and where the sampling of target taxa occurred, such as date, sampling method, location and geographic coordinates; (c) Occurrences - description of the collected individual, such as epithet, life stage and conservation status. The species' conservation status was sourced from the International Union for Conservation of Nature (IUCN) and the Brazilian Ministry of the Environment (MMA) and applies at the species level rather than the individual level. All variables are described in Suppl. material 2.
Step 2: Validate- data quality and control
To ensure that the data have the lowest possible error rate, they need to go through a validation process. This process reduces the chances that the final data will contain grammatical errors, which can make it difficult to understand and species that have been incorrectly identified.
We conducted a manual validation process for the Golfão Maranhanse data that we optimised in two steps:
1. Identifying and fixing errors - We conducted a thorough examination of the data to identify and correct any errors or inaccuracies.
These errors included:
(a) Scientific names (e.g. “Boanaranipcs” [wrong] vs. “Boanaraniceps” [correct]);
(b) Geographic coordinates (e.g. “44,321605 [typo without negative sign]” vs. “-44,321605 [correct]”);
(c) Date: we standardised the sampling dates to "Start Month" "Start Year" "End Month" and "End Year" (the original data column, verbatim date, contains the day of sampling, if available).
Additionally, our data cleaning process involved removing duplicates and standardising entries to ensure consistency. By meticulously correcting these issues, we ensured the data's integrity and reliability, making it suitable for further analysis and interpretation.
2. Checking the records - Species occurrence data are susceptible to misidentification and taxonomic inconsistencies, making this a challenging and dynamic task. To ensure the reliability and validity of the species occurrence data, we reviewed the relevant literature for known geographic species distributions and compared them with the collected points. Our team of taxa specialists meticulously checked each entry for inconsistencies and up-to-date taxonomy, according to the most recent accepted taxonomy of each group. Any mismatches between known and collected geographic distributions served as a first alert, indicating the need for further investigation. Additionally, we reviewed the literature for changes in synonymy and updated the occurrence records accordingly.
Step 3: Standardise
To standardise data from the Golfão Maranhense region, we used the Darwin Core standard (DwC) (Wieczorek et al. 2012). In addition, we manually added columns for data that are not covered by the DwC (e.g. conservation status). DwC is one of the most widely used standards for biodiversity data used as a language for sharing biodiversity data that can be understood by human users and interpreted by computational systems. The DwC provides a straightforward, stable standard that simplifies the process of publishing biodiversity data, promoting the sharing, use and reuse of openly accessible biodiversity data (Wieczorek et al. 2012). Additionally, DwC allows users to adapt terms that name the columns for various applications, including the checklists of species in an area (DoNascimiento et al. 2017).
Step 4: Share and archive
The last step is to choose the right repository to store the data. For species occurrence data, GBIF (GBIF 2024) may be the best option, as it is a specific repository for biodiversity data that guarantees data quality and open access. Other repositories that are popular and should be considered:
- Integrated Digitized Biocollections (iDigBio 2024);
- Dryad (2024);
- Open Science Framework (OSF 2024);
Statistical analysis
To test whether the number of occurrences depended on the number of taxa in each group, a simple linear regression was performed using R software.
Results
Step 1: Search- Systematic review
Considering all biotic groups, a total of 161 bibliographical references, including papers and technical reports were included in the systematic review of the literature (Fig. 3). In addition, we included species occurrence records from four public repositories (GBIF, VertNet, Wikiaves, SpeciesLink). Considering only published papers, the group included in the largest number of published papers and reports was plants (n = 59) and the group with the least data sources was benthos (n = 11) (Suppl. material 3 “Preferred Reporting Items for Systematic Reviews and Meta-Analyses – PRISMA”, separated by groups).
Figure 3.

Flowchart of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) for all groups showing the process of selecting studies throughout systematic review. The selection process includes three stages: (1) identifying the database and choosing the papers; (2) scanning the references and selecting the papers to be included; (3) including the selected papers.
Step 2: Validate- data quality and control
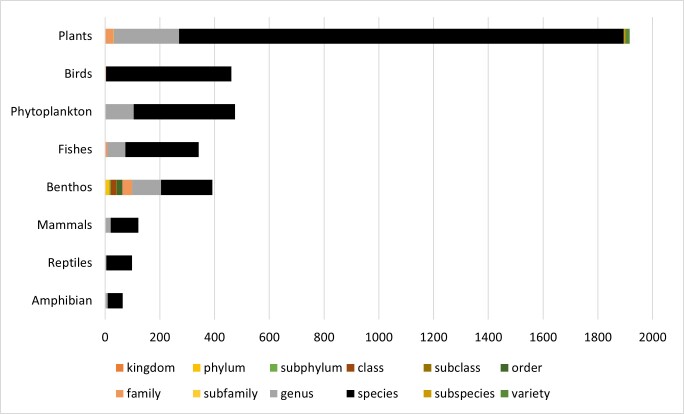
A total of 2,070 occurrence events were obtained from bibliographic references and 43,947 were obtained by public repositories (n = 46,017) from 3,871 taxa. These include birds (Aves, 458 species; three other taxonomic level), amphibians (Amphibia, 55 species; nine to the genus level), reptiles (two Crocodylia; 86 Squamata; 11 Testudines); mammals (Class Mammalia; 101 species; 21 to the genus level), fish (268 species, 74 other taxonomic levels), phytoplankton (370 species; 105 other taxonomic levels), benthos (188 species; 204 other taxonomic levels) and plants (1,624 species; 292 other taxonomic levels) (Suppl. material 4). Most of the taxa were identified to species (81%) and genus (14%) level (Fig. 4). Benthos accounted for the highest number of occurrence events, with 12,510 records and reptiles had the lowest number of occurrence events recorded (570).
Figure 4.
Proportion of each taxonomic level identified for each biological group in the secondary data recovered from the Golfão Maranhense area.
Data were carefully analysed by specialists in each group to check for inconsistencies in identification, spelling and, as much as possible, potentiality of identification correctness (e.g. check if the geographic locations were within expected known geographic distribution for each taxon, checking vouchers when possible). A total of 93 occurrence events were deleted, including 92 from taxa that were not correctly identified (76 birds and 16 mammals) and one bird specimen that was a victim of animal trafficking.
Step 3: Standardise
Of the 76 variables extracted from the studies, 59 were standardised using DwC terms and 17 were adapted due to the lack of appropriate terms for these variables within the current DwC models (Suppl. material 2).
Step 4: Share and archive
We decided not to publish the database in a specialised database such as GBIF because it contains secondary data that includes information extracted from other databases, including GBIF itself. This would result in duplication of information. We decided to store the data in the Open Science Framework (OSF). OSF is an open access repository that maintains version control, allowing them to track changes to their projects over time. The OSF also assigns Digital Object Identifiers (DOIs), making the data citable and ensuring its long-term preservation. The database is publicly available on the link (Marques 2024). In addition, the database will be accessible through an online platform developed using PowerBI software (Microsoft corporation 2024). This platform will be developed and will be freely available, promoting the dissemination of knowledge related to the biodiversity of the Golfão Maranhense region.
Statistical analysis
The number of occurrences was dependent on the number of taxa in each sampled group (R² = 0.47, p = 0.03). While amphibians and non-bird reptiles were represented by low numbers of both taxa and occurrences, plants, birds and phytoplankton were highly represented for both occurrences and richness. On the other hand, the group “benthos” had a high number of occurrences and a low number of taxa (Fig. 5).
Figure 5.
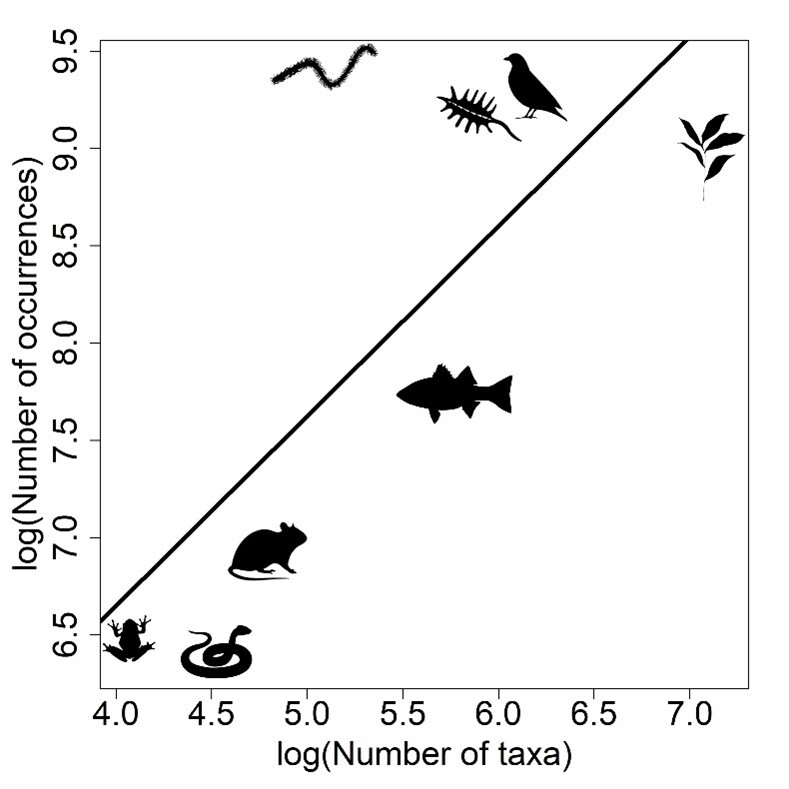
Relationships between numbers of taxa and occurrences gathered through an extensive review of secondary biodiversity data from the Golfão Maranhense area, in the estuarine regions of eastern Amazonia.
Discussion
We proposed a workflow to improve our ability to recover higher quality biodiversity data using secondary data sources. We were able to extract a large amount of information about the biodiversity of the Golfão Maranhense and transform this unrelated data into organised and re-usable data. This systematic approach ensured data accuracy and reliability, facilitating the potential reuse of information in future studies. A further step that we have begun to take for some groups is the systematic survey of museum collections and analyses, focusing on relevant questions that we have identified along the way (e.g. general patterns of occurrence of migratory birds, sampling biases and gaps for many groups etc.).
Researchers can use existing datasets, such as those obtained through our biodiversity data retrieval method, to conduct a wide range of studies to advance scientific research (Pernat et al. 2024). For example, secondary data can be used to conduct meta-analyses (e.g. Biggs et al. (2020)) for comparative studies across different geographic regions and time, to support ecological modelling of species distributions (e.g. Fletcher et al. (2019)), habitat preferences and potential impacts of environmental change (Bayraktarov et al. 2019) as long as they are used judiciously. However, finding high-quality secondary data can be challenging, as evidenced by a recent survey in which most researchers reported that data finding can be arduous (73%) or difficult (19%) (Gregory et al. 2020). Several initiatives have been launched to collect, standardise, store and make biodiversity data openly available (Costello et al. 2013). For example, international repositories, such as “GBIF” (GBIF 2024) and “Freshwater Biodiversity Data Portal- BioFresh” (http://data.freshwaterbiodiversity.eu/) (for other repositories, see Culina et al. (2018)).
Long-term monitoring datasets can help to understand patterns and changes in ecological variables over time (Costa and Magnusson 2010, Magnusson et al. 2021). This can help identify ecological shifts and potential drivers of biodiversity change (e.g. Marques et al. (2022)). We are currently using our database to increase our knowledge of mammal, bird, fish, amphibian, reptile, marine phytoplankton and benthic species in the Golfão Maranhense. We are also studying the patterns and determinants of floristic variation in the region and the temporal variation of migratory birds in the São Marcos Bay region. While data collected in standardised monitoring programmes, such as LTER (Vanderbilt et al. 2015), can be directly linked to FAIR's standardised data repositories, other secondary data that are not standardised may be important to rescue, treat and use, as they can be thoroughly revised and curated beforehand and stored with some rule of error estimation to build robust hypotheses to investigate and understand biodiversity patterns.
While secondary data can be a valuable resource for scientific research, it is crucial to recognise and address its limitations and ideally estimate the errors within. Common challenges include species identification accuracy, geographic coordinate precision and data entry errors. In addition, datasets from different studies may differ in their sampling methods, data structure and definitions of key variables, making direct comparisons difficult. Finally, some datasets may not be openly accessible, which has implications for data availability and complicates data access and sharing policies.
Other limitations are the sampling and temporal biases, which can arise when working with secondary data, making data interpretation more challenging. Sampling bias occurs when the data sampling disproportionately favours certain species or areas over others, for example, the concentration of specimen records in more easily accessible sites, such as major cities, roads and navigable rivers (Boakes et al. 2010). In addition, logistics and human interference are factors that can explain research probability (e.g. 64% of research probability in Amazon; Carvalho et al. (2023)). Temporal bias, on the other hand, refers to the uneven distribution of data across time periods. Secondary data sources may include data collected over different time spans, reflecting historical variations in research focus, funding availability or changes in data-recording practices. Consequently, certain time periods may be over-represented, while others may be sparsely covered or entirely absent. Additionally, the difficulty of conducting research in regions with limited accessibility introduces challenges that restrict the ability to gather data from remote areas. Thus, remote regions potentially hosting unique biodiversity hotspots are often under-represented or completely absent from the dataset.
In our study, sampling bias is evident in the São Marcos Bay area, where an industrial ship port is located. Thus, most of the data were obtained from environmental monitoring reports in the region linked to the environmental licensing process. These reports conducted in a port area inherently prioritise certain species and ecological aspects more relevant to the licensing process, overlooking other important components of biodiversity. Within our database, it becomes apparent that some species records originate from technical reports that are not easily available. For example, we found 365 species and varieties of phytoplankton in technical reports, but 101 were not previously catalogued on the Brazilian Biodiversity Platform REFLORA (Jardim Botânico do Rio de Janeiro 2024) for the Maranhão region. This underscores the fact that the retrieval of biodiversity data can yield enhancements in the comprehension of species composition existing within the defined geographical area.
Conclusions
The workflow that we employed has facilitated the retrieval of biodiversity data from the ecologically rich and megadiverse Golfão Maranhense region in Maranhão, Brazil. By combining a systematic review approach with standardised worksheets with a Darwin Core base, we were able to effectively search and explore a wide range of scientific articles, technical reports and specialised public repositories. The potential use of secondary data for the advancement of scientific research is significant although it must be taken with care and analysed with precautions observing all bias limitation and filters involved. Many technical survey reports were produced in the Golfão Maranhense linked to environmental licensing process for the port and surrounding activities. By using existing datasets, researchers can carry out a wide range of activities which include meta-analyses, comparative studies, ecological modelling and, most of all, building hypotheses and producing experiment designs to monitor diversity in a standardised base. Our study highlights the value of systematic review methods and the need for an approach to address data limitations and biases. Likewise, this method can facilitate collaboration amongst researchers, enable comparative analyses across different datasets and support evidence-based conservation strategies and policy-making.
Supplementary Material
Keywords
Nubia Marques
Data type
table
Brief description
Keywords used in the systematic review of each biotic group.
File: oo_1105742.docx
Table Darwin Core (DwC)
Nubia Marques
Data type
table
Brief description
Table containing the Darwin Core (DwC) standard terms that were used to make the table and extract the information from the bibliographic references previously selected in the systematic review. Label = name of the column in the DwC standard; Definition = Brief definition of what each column means.
File: oo_1105743.docx
Flowchart of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)
Nubia Marques
Data type
images
Brief description
Flowchart of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) separated by groups showing the process of selecting studies throughout the systematic review. The selection process includes three stages: (1) identifying the database and choosing the papers; (2) scanning the references and selecting the papers to be included; (3) including the selected papers.
File: oo_1105747.docx
List of species from the Golfão Maranhense (Maranhão State, Brazil)
Nubia Marques
Data type
Table
Brief description
List of species from the Golfão Maranhense (Maranhão State, Brazil) that were retrieved through the systematic literature review.
File: oo_1105748.docx
Acknowledgements
We would like to thank the Environmental Management of the Ponta da Madeira Maritime Terminal for their support in developing the project. We are grateful to the reviewer Pedro Cardoso for his suggestions for improving the manuscript and to the collectors of the data we retrieved from the literature. Daniel M. Casali is currently being funded by the grant #2022/00044-7, São Paulo Research Foundation (FAPESP)
Hosting institution
Vale Institute of Technology
Conflicts of interest
No conflict of interest to declare
Disclaimer: This article is (co-)authored by any of the Editors-in-Chief, Managing Editors or their deputies in this journal.
References
- Amano Tatsuya, Lamming James D. L., Sutherland William J. Spatial gaps in global biodiversity information and the role of citizen science. BioScience. 2016;66(5):393–400. doi: 10.1093/biosci/biw022. [DOI] [Google Scholar]
- Bayraktarov Elisa, Ehmke Glenn, O'Connor James, Burns Emma L., Nguyen Hoang A., McRae Louise, Possingham Hugh P., Lindenmayer David B. Do Big Unstructured Biodiversity Data Mean More Knowledge? https://www.frontiersin.org/articles/10.3389/fevo.2018.00239. Frontiers in Ecology and Evolution. 2019;6 doi: 10.3389/fevo.2018.00239. [DOI] [Google Scholar]
- Biggs Christopher R., Yeager Lauren A., Bolser Derek G., Bonsell Christina, Dichiera Angelina M., Hou Zhenxin, Keyser Spencer R., Khursigara Alexis J., Lu Kaijun, Muth Arley F., Negrete Jr. Benjamin, Erisman Brad E. Does functional redundancy affect ecological stability and resilience? A review and meta-analysis. https://onlinelibrary.wiley.com/doi/abs/10.1002/ecs2.3184. Ecosphere. 2020;11(7) doi: 10.1002/ecs2.3184. [DOI] [Google Scholar]
- Boakes Elizabeth H., McGowan Philip J. K., Fuller Richard A., Chang-qing Ding, Clark Natalie E., O'Connor Kim, Mace Georgina M. Distorted Views of Biodiversity: Spatial and Temporal Bias in Species Occurrence Data. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1000385. PLOS Biology. 2010;8(6) doi: 10.1371/journal.pbio.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard Michael Krabbe, Hart Edmund M. Towards a more reproducible ecology. https://onlinelibrary.wiley.com/doi/10.1111/ecog.02493. Ecography. 2016;39(4):349–353. doi: 10.1111/ecog.02493. [DOI] [Google Scholar]
- Cao Longbing. Data science and analytics: a new era. International Journal of Data Science and Analytics. 2016;1(1):1–2. doi: 10.1007/s41060-016-0006-1. [DOI] [Google Scholar]
- Carvalho Raquel L., Resende Angelica F., Barlow Jos, França Filipe M., Moura Mario R., Maciel Rafaella, Alves-Martins Fernanda, Shutt Jack, Nunes Cassio A., Elias Fernando, Silveira Juliana M., Stegmann Lis, Baccaro Fabricio B., Juen Leandro, Schietti Juliana, Aragão Luiz, Berenguer Erika, Castello Leandro, Costa Flavia R. C., Guedes Matheus L., Leal Cecilia G., Lees Alexander C., Isaac Victoria, Nascimento Rodrigo O., Phillips Oliver L., Schmidt Fernando Augusto, Ter Steege Hans, Vaz-de-Mello Fernando, Venticinque Eduardo M., Vieira Ima Célia Guimarães, Zuanon Jansen, Ferreira Joice, Carvalho Raquel L., Resende Angelica F., Barlow Jos, França Filipe, Moura Mario R., Maciel Rafaella, Alves-Martins Fernanda, Shutt Jack, Nunes Cassio A., Elias Fernando, Silveira Juliana M., Stegmann Lis, Baccaro Fabricio B., Juen Leandro, Schietti Juliana, Aragão Luiz, Berenguer Erika, Castello Leandro, Costa Flavia R. C., Guedes Matheus L., Leal Cecilia G., Lees Alexander C., Isaac Victoria, Nascimento Rodrigo O., Phillips Oliver L., Schmidt Fernando Augusto, Ter Steege Hans, Vaz-de-Mello Fernando, Venticinque Eduardo M., Vieira Ima Célia Guimarães, Zuanon Jansen, Ferreira Joice, Geber Filho Adem Nagibe Dos Santos, Ruschel Ademir, Calor Adolfo Ricardo, De Lima Alves Adriana, Muelbert Adriane Esquivel, Quaresma Adriano, Vicentini Alberto, Piedade Alexandra Rocha Da, Oliveira Alexandre Adalardo De, Aleixo Alexandre, Casadei-Ferreira Alexandre, Gontijo Alexandre, Hercos Alexandre, Andriolo Aline, Lopes Aline, Pontes-Lopes Aline, Santos Allan Paulo Moreira Dos, Oliveira Amanda Batista Da Silva De, Mortati Amanda Frederico, Salcedo Ana Karina Moreyra, Albernaz Ana Luisa, Fares Ana Luisa, Andrade Ana Luiza, Oliveira Pes Ana Maria, Faria Ana Paula Justino, Batista Anderson Pedro Bernadina, Puker Anderson, Bueno Anderson S., Junqueira André Braga, Holanda De Andrade André Luiz Ramos, Ghidini André Ricardo, Galuch André V., Menezes Andressa Silvana Oliveira De, Manzatto Angelo Gilberto, Correa Anne Sthephane A. S., Queiroz Antonio C. M., Zanzini Antonio Carlos Da Silva, Olivo Neto Antonio Miguel, Melo Antonio Willian Flores De, Guimaraes Aretha Franklin, Castro Arlison Bezerra, Borges Augusto, Ferreira Aurélia Bentes, Marimon Beatriz S., Marimon-Junior Ben Hur, Flores Bernardo M., De Resende Bethânia Oliveira, Albuquerque Bianca Weiss, Villa Boris, Davis Bradley, Nelson Bruce, Williamson Bruce, Melo Bruna Santos Bitencourt De, Cintra Bruno B. L., Santos Bruno Borges, Prudente Bruno Da Silveira, Luize Bruno Garcia, Godoy Bruno Spacek, Rutt Cameron L., Duarte Ritter Camila, Silva Camila V. J., Ribas Carla Rodrigues, Peres Carlos A., Azevêdo Carlos Augusto Silva De, Freitas Carlos, Cordeiro Carlos Leandro, Brocardo Carlos Rodrigo, Castilho Carolina, Levis Carolina, Doria Carolina Rodrigues Da Costa, Arantes Caroline C., Santos Cássia Anicá Dos, Jakovac Catarina C., Silva Celice Alexandre, Benetti Cesar João, Lasmar Chaim, Marsh Charles J., Andretti Christian Borges, Oliveira Cinthia Pereira De, Cornelius Cintia, Alves Da Rosa Clarissa, Baider Cláudia, Gualberto Cláudia G., Deus Claudia Pereira De, Monteiro Jr. Cláudio Da Silva, Santos Neto Cláudio Rabelo Dos, Lobato Cleonice Maria Cardoso, Santos Cleverson Rannieri Meira Dos, Penagos Cristian Camilo Mendoza, Costa Daniel Da Silva, Vieira Daniel Luis Mascia, Aguiar Daniel Praia Portela De, Veras Daniel Silas, Pauletto Daniela, Braga Danielle De Lima, Storck-Tonon Danielle, Almeida Daniely Da Frota, Douglas Danyhelton, Amaral Dário Dantas Do, Gris Darlene, Luther David, Edwards David P., Guimarães David Pedroza, Santos Deane Cabral Dos, Campana Débora Rodrigues De Souza, Nogueira Denis Silva, Silva Dennis Rodrigues Da, Dutra Dhâmyla Bruna De Souza, Rosa Dian Carlos Pinheiro, Silva Diego Armando Silva Da, Pedroza Diego, Anjos Diego V., Melo Lima Diego Viana, Silvério Divino V., Rodrigues Domingos De Jesus, Bastos Douglas, Daly Douglas, Barbosa Edelcilio Marques, Arenas Edith Rosario Clemente, Oliveira Edmar Almeida De, Santos Ednaira Alencar Dos, Santana Edrielly Carolinne Carvalho De, Guilherme Edson, Vidal Edson, Campos-Filho Eduardo Malta, Van Den Berg Eduardo, Morato Elder Ferreira, Da Silva Elidiomar R., Marques Elineide E., Pringle Elizabeth G., Nichols Elizabeth, Andresen Ellen, Farias Emanuelle De Sousa, Siqueira Emely Laiara Silva De, De Albuquerque Emília Zoppas, Görgens Eric Bastos, Cunha Erlane José Rodrigues Da, Householder Ethan, Novo Evlyn Márcia Moraes De Leão, Oliveira Fabiana Ferreira De, Roque Fabio De Oliveira, Coletti Fabrício, Reis Fagno, Moreira Felipe F. F., Todeschini Felipe, Carvalho Fernanda Antunes, Coelho De Souza Fernanda, Silva Fernando Augusto Barbosa, Carvalho Fernando Geraldo, Cabeceira Fernando Gonçalves, d’Horta Fernando Mendonça, Mendonça Fernando P., Florêncio Fernando Prado, Carvalho Fernando Rogério De, Arruda Filipe Viegas De, Nonato Flávia Alessandra Da Silva, Santana Flávia Delgado, Durgante Flavia, Souza Flávia Kelly Siqueira De, Obermuller Flávio Amorim, Castro Flávio Siqueira De, Wittmann Florian, Sales Francisco Matheus Da Silva, Neto Francisco Valente-, Salles Frederico Falcão, Borba Gabriel Costa, Damasco Gabriel, Barros Gabriel Gazzana, Brejão Gabriel Lourenço, Jardim Gabriela Abrantes, Prance Ghillean T., Lima Gisiane Rodrigues, Desidério Gleison Robson, Melo Gracilene Da Costa De, Carmo Guilherme Henrique Pompiano Do, Cabral Guilherme Sampaio, Rousseau Guillaume Xavier, Da Silva Gustavo Cardoso, Schwartz Gustavo, Griffiths Hannah, Queiroz Helder Lima De, Espírito-Santo Helder M. V., Cabette Helena Soares Ramos, Nascimento Henrique Eduardo Mendonça, Vasconcelos Heraldo L., Medeiros Herison, Aguiar Hilton Jeferson Alves Cardoso De, Leão Híngara, Wilker Icaro, Gonçalves Inês Correa, De Sousa Gorayeb Inocêncio, Miranda Ires Paula De Andrade, Brown Irving Foster, Santos Isis Caroline Siqueira, Fernandes Itanna Oliveira, Fernandes Izaias, Delabie Jacques Hubert Charles, De Abreu Jadson Coelho, Gama Neto Jaime De Liege, Costa Janaina Barbosa Pedrosa, Noronha Janaína Costa, De Brito Janaina Gomes, Wolfe Jared, Santos Jean Carlos, Ferreira-Ferreira Jefferson, E Gomes Jerrian Oliveira, Lasky Jesse R., De Faria Falcão Jéssica Caroline, Costa Jessica Gomes, Cravo Jessica Soares, Guerrero Jesús Enrique Burgos, Muñoz Gutiérrez Jhonatan Andrés, Carreiras João, Lanna João, Silva Brito Joás, Schöngart Jochen, Mendes Aguiar Jonas José, Lima Jônatas, Barroso Jorcely G., Noriega Jorge Ari, Pereira Jorge Luiz Da Silva, Nessimian Jorge Luiz, Souza Jorge Luiz Pereira De, De Toledo José Julio, Magalhães José Leonardo Lima, Camargo José Luís, Oliveira José Max B., Ribeiro José Moacir Ferreira, Silva José Orlando De Almeida, Da Silva Guimarães José Renan, Hawes Joseph E., Andrade-Silva Joudellys, Revilla Juan David Cardenas, Da Silva Júlia Santana, Da Silva Menger Juliana, Rechetelo Juliana, Stropp Juliana, Barbosa Julianna Freires, Do Vale Julio Daniel, Louzada Julio, Cerqueira Silva Július César, Da Silva Karina Dias, Melgaço Karina, Carvalho Karine Santana, Yamamoto Kedma Cristine, Mendes Keila Rêgo, Vulinec Kevina, Maia Laís Ferreira, Cavalheiro Larissa, Vedovato Laura Barbosa, Demarchi Layon Oreste, Giacomin Leandro, Dumas Leandro Lourenço, Maracahipes Leandro, Brasil Leandro Schlemmer, Ferreira Leandro Valle, Calvão Lenize Batista, Maracahipes-Santos Leonardo, Reis Leonardo Pequeno, Da Silva Letícia Fernandes, De Oliveira Melo Lia, Carvalho Lidiany Camila Da Silva, Casatti Lílian, Amado Lílian Lund, De Matos Liliane Stedile, Vieira Lisandro, Prado Livia Pires Do, Alencar Luana, Fontenele Luane, Mazzei Lucas, Navarro Paolucci Lucas, Zanzini Lucas Pereira, Carvalho Lucélia Nobre, Crema Luciana Carvalho, Brulinger Luciane Ferreira Barbosa, Montag Luciano Fogaça De Assis, Naka Luciano Nicolas, Azara Ludson, Silveira Luis Fábio, Nunes Luis Gabriel De Oliveira, Rosalino Luís Miguel Do Carmo, Mestre Luiz A. M., Bonates Luiz Carlos De Matos, Coelho Luiz De Souza, Borges Luiz Henrique Medeiros, Lourenço Luzia Da Silva, Freitas Madson Antonio Benjamin, Brito Maiara Tábatha Da Silva, Pombo Maihyra Marina, Da Rocha Maíra, Cardoso Maira Rodrigues, Guedes Marcelino Carneiro, Raseira Marcelo Bassols, Medeiros Marcelo Brilhante De, Carim Marcelo De Jesus Veiga, Simon Marcelo Fragomeni, Pansonato Marcelo Petratti, Dos Anjos Marcelo Rodrigues, Nascimento Marcelo Trindade, Souza Márcia Regina De, Monteiro Marcília Gabriella Tavares, Da Silva Márcio Joaquim, Uehara-Prado Marcio, Oliveira Marco Antonio De, Callisto Marcos, Vital Marcos José Salgado, O Santos Marcos Pérsi Dantas, Silveira Marcos, Oliveira Marcus Vinicio Neves D., Pérez-Mayorga María Angélica, Carniello Maria Antonia, Lopes Maria Aparecida, Silveira Maria Aurea Pinheiro De Almeida, Esposito Maria Cristina, Maldaner Maria Eduarda, Passos Maria Inês S., Anacléto Maria José Pinheiro, Costa Maria Katiane Sousa, Martins Maria Pires, Piedade Maria Teresa Fernandez, Irume Mariana Victória, Costa Marília Maria Silva Da, Maximiano Marina Franco De Almeida, Freitas Marina Guimarães, Cochrane Mark A., Gastauer Markus, Almeida Marllus Rafael Negreiros, Souza Mateus Fernando De, Catarino Michel, Costa Batista Michela, Massam Mike R., Martins Mila Ferraz De Oliveira, Holmgren Milena, Almeida Morgana, Dias Murilo S., Espírito Santo Nádia Barbosa, Benone Naraiana Loureiro, Ivanauskas Natalia Macedo, Medeiros Natália, Targhetta Natalia, Félix Nathalia Silva, Ferreira Nelson, Hamada Neusa, Campos Nubia, Giehl Nubia França Da Silva, Metcalf Oliver Charles, Silva Otávio Guilherme Morais Da, Cerqueira Pablo Vieira, Moser Pamela, Miranda Patrícia Nakayama, Peruquetti Patricia Santos Ferreira, Alverga Paula Palhares De Polari, Prist Paula, Souto Paula, Brando Paulo, Pompeu Paulo Dos Santos, Barni Paulo Eduardo, Graça Paulo Mauricio De Alencastro, Morandi Paulo S., Cruz Paulo Vilela, Da Silva Pedro Giovâni, Bispo Pitágoras C., Camargo Plínio Barbosa De, Sarmento Priscila S. De M., Souza Priscila, Andrade Rafael Barreto De, Braga Rafael Benzi, Boldrini Rafael, Bastos Rafael Costa, Assis Rafael Leandro De, Salomão Rafael P., Leitão Rafael Pereira, Mendes Raimundo N. G., Carpanedo Rainiellen De Sá, Melinski Ramiro Dário, Ligeiro Raphael, E Pérez Raúl Enriqu Pirela, Barbosa Reinaldo Imbrozio, Cajaiba Reinaldo Lucas, Silvano Renato Azevedo Matias, Salomão Renato Portela, Hilário Renato Richard, Martins Renato Tavares, Perdiz Ricardo De Oliveira, Vicente Ricardo Eduardo, Silva Ricardo José Da, Koroiva Ricardo, Solar Ricardo, Silva Richarlly Da Costa, S De Lima Robson Borge, Silva Robson Dos Santos Alves Da, Mariano Rodolfo, Ribeiro Rodrigo Arison Barbosa, Fadini Rodrigo Ferreira, Oliveira Rodrigo Leonardo Costa De, Feitosa Rodrigo Machado, Matavelli Rodrigo, Mormul Roger Paulo, Da Silva Rogério Rosa, Zanetti Ronald, Barthem Ronaldo, Almeida Rony Peterson Santos, Ribeiro Sabina Cerruto, R Costa Neto Salustiano Vila Da, Nienow Samuel, Oliveira Sérgio Augusto Vidal De, Borges Sérgio Henrique, Milheiras Sérgio, Ribeiro Sérvio Pontes, Couceiro Sheyla Regina Marques, Sousa Sidney Araújo De, Rodrigues Silvia Barbosa, Dutra Silvia Leitão, Mahood Simon, Vieira Simone Aparecida, Arrolho Solange, Silva Sonaira Souza Da, Triana Stefania Pinzón, Laurance Susan, Kunz Sustanis Horn, Alvarado Swanni T., Rodrigues Taís Helena Araujo, Santos Talitha Ferreira Dos, Machado Tatiana Lemos Da Silva, Feldpausch Ted R., Sousa Thaiane, Michelan Thaisa Sala, Emilio Thaise, Brito Thaline De Freitas, André Thiago, Barbosa Thiago Augusto Pedroso, Miguel Thiago Barros, Izzo Thiago Junqueira, Laranjeiras Thiago Orsi, Mendes Thiago Pereira, Silva Thiago Sanna Freire, Krolow Tiago Kütter, Begot Tiago Octavio, Baker Timothy R., Domingues Tomas F., Giarrizzo Tommaso, Bentos Tony Vizcarra, Haugaasen Torbjørn, Peixoto Ualerson, Pozzobom Ully Mattilde, Korasaki Vanesca, Ribeiro Vanessa Soares, Scudeller Veridiana Vizoni, Oliveira Victor Hugo Fonseca, Landeiro Victor Lemes, Santos Ferreira Victor Rennan, Silva Victória De Nazaré Gama, Gomes Vitor Hugo Freitas, Oliveira Vívian Campos De, Firmino Viviane, Santiago Wagner Tadeu Vieira, Beiroz Wallace, Almeida Wanessa Rejane De, Oliveira Washington Luis De, Silva Wegliane Campelo Da, Castro Wendeson, Dáttilo Wesley, Cruz Wesley Jonatar Alves Da, Silva Wheriton Fernando Moreira Da, Magnusson William E., Laurance William, Milliken William, Paula William Sousa De, Malhi Yadvinder, Shimabukuro Yosio Edemir, Lima Ysadhora Gomes De, Shimano Yulie, Feitosa Yuri. Pervasive gaps in Amazonian ecological research. https://linkinghub.elsevier.com/retrieve/pii/S0960982223008631. Current Biology. 2023;33(16) doi: 10.1016/j.cub.2023.06.077. [DOI] [PubMed] [Google Scholar]
- Costa Flávia Regina Capellotto, Magnusson William Ernest. The Need for Large-Scale, Integrated Studies of Biodiversity - the Experience of the Program for Biodiversity Research in Brazilian Amazonia. http://doi.editoracubo.com.br/10.4322/natcon.00801001. Natureza & Conservação. 2010;08(01):3–12. doi: 10.4322/natcon.00801001. [DOI] [Google Scholar]
- Costello Mark, Michener William, Gahegan Mark, Zhang Zhi-Qiang, Bourne Philip. Biodiversity data should be published, cited, and peer reviewed. Trends in Ecology & Evolution. 2013;28 doi: 10.1016/j.tree.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Culina Antica, Baglioni Miriam, Crowther Tom W., Visser Marcel E., Woutersen-Windhouwer Saskia, Manghi Paolo. Navigating the unfolding open data landscape in ecology and evolution. https://www.nature.com/articles/s41559-017-0458-2. Nature Ecology & Evolution. 2018;2(3):420–426. doi: 10.1038/s41559-017-0458-2. [DOI] [PubMed] [Google Scholar]
- Culley Theresa M. The frontier of data discoverability: Why we need to share our data. Applications in Plant Sciences. 2017;5(10) doi: 10.3732/apps.1700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DoNascimiento Carlos, Herrera-Collazos Edgar Esteban, Herrera-R. Guido A., Ortega-Lara Armando, Villa-Navarro Francisco A., Oviedo José Saulo Usma, Maldonado-Ocampo Javier A. Checklist of the freshwater fishes of Colombia: a Darwin Core alternative to the updating problem. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5674168/ ZooKeys. 2017:25–138. doi: 10.3897/zookeys.708.13897. [DOI] [PMC free article] [PubMed]
- Dryad Data Dryad. https://datadryad.org/ https://datadryad.org/
- Fletcher Robert J., Hefley Trevor J., Robertson Ellen P., Zuckerberg Benjamin, McCleery Robert A., Dorazio Robert M. A practical guide for combining data to model species distributions. Ecology. 2019;100(6) doi: 10.1002/ecy.2710. [DOI] [PubMed] [Google Scholar]
- Ganzevoort Wessel, van den Born Riyan J. G., Halffman Willem, Turnhout Sander. Sharing biodiversity data: citizen scientists’ concerns and motivations. Biodiversity and Conservation. 2017;26(12):2821–2837. doi: 10.1007/s10531-017-1391-z. [DOI] [Google Scholar]
- GBIF What is GBIF? 2024. https://www.gbif.org/what-is-gbif https://www.gbif.org/what-is-gbif
- Gregory Kathleen, Groth Paul, Scharnhorst Andrea, Wyatt Sally. Lost or found? discovering data needed for research: Supplementary materials. https://assets.pubpub.org/ml515gnb/28646d0a-bc50-42c9-a185-db71e447877c.pdf. Harvard Data Science Review. 2020 doi: 10.1162/99608f92.e38165eb. [DOI]
- Hackett Rachel A., Belitz Michael W., Gilbert Edward E., Monfils Anna K. A data management workflow of biodiversity data from the field to data users. https://onlinelibrary.wiley.com/doi/abs/10.1002/aps3.11310. Applications in Plant Sciences. 2019;7(12) doi: 10.1002/aps3.11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt Victoria M., Jacobi Sarah K., Knutson Melinda G., Lonsdorf Eric V., Papon Shawn, Zorn Jennifer. A data management system for long‐term natural resource monitoring and management projects with multiple cooperators. Wildlife Society Bulletin. 2015;39(3):464–471. doi: 10.1002/wsb.547. [DOI] [Google Scholar]
- iDigBio Integrated Digitized Biocollections. https://www.idigbio.org/ https://www.idigbio.org/
- Janeiro Jardim Botânico do Rio de. Flora e Funga do Brasil. http://floradobrasil.jbrj.gov.br/ [2024-07-31T00:35:05+00:00]. http://floradobrasil.jbrj.gov.br/
- Magnusson William Ernest, Lima Albertina Pimental, Aragón Susan, Rosa Clarissa Alves da, Brocardo Carlos Rodrigo, Fadini Rodrigo F. Long-term standardized ecological research in an amazonian savanna: A laboratory under threat. https://repositorio.inpa.gov.br/handle/1/38327. Volume 93, Número e20210879. 2021 doi: 10.1590/0001-3765202120210879. [DOI] [PubMed]
- Marques, et al. https://osf.io/gh9ym/ https://osf.io/gh9ym/
- Marques Nubia CS, Machado Ricardo B., Aguiar Ludmilla MS, Mendonca-Galvão Luciana, Tidon Rosana, Vieira Emerson M., Marini-Filho Onildo J., Bustamante Mercedes. Drivers of change in tropical protected areas: Long-term monitoring of a Brazilian biodiversity hotspot. https://www.sciencedirect.com/science/article/pii/S2530064422000116. Perspectives in Ecology and Conservation. 2022;20(2):69–78. doi: 10.1016/j.pecon.2022.02.001. [DOI] [Google Scholar]
- Michener William K. Ecological data sharing. https://www.sciencedirect.com/science/article/pii/S1574954115001004. Ecological informatics. 2015;29:33–44. doi: 10.1016/j.ecoinf.2015.06.010. [DOI] [Google Scholar]
- corporation Microsoft. Power BI. [2024-06-01T00:17:34+00:00];https://powerbi.microsoft.com 2024
- OSF Open Science Framewor. https://osf.io/ https://osf.io/
- Parr Cynthia S., Guralnick Robert, Cellinese Nico, Page Roderic D. M. Evolutionary informatics: unifying knowledge about the diversity of life. Trends in Ecology & Evolution. 2012;27(2):94–103. doi: 10.1016/j.tree.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Pernat Nadja, Canavan Susan, Golivets Marina, Hillaert Jasmijn, Itescu Yuval, Jarić Ivan, Mann Hjalte M. R., Pipek Pavel, Preda Cristina, Richardson David M., Teixeira Heliana, Vaz Ana Sofia, Groom Quentin. Overcoming biodiversity blindness: Secondary data in primary citizen science observations. Ecological Solutions and Evidence. 2024;5(1) doi: 10.1002/2688-8319.12295. [DOI] [Google Scholar]
- Piwowar Heather A., Vision Todd J. Data reuse and the open data citation advantage. https://peerj.com/articles/175/?report=reader. PeerJ. 2013;1 doi: 10.7717/peerj.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo-Mochel F. In: Mangrove ecosystem studies in Latin America and Africa. Kjerfve B, Lacerda LD, Diop EHS, editors. UNESCO; Paris: 1997. Mangroves on Sao Luís Island, Maranhão.145-154 [Google Scholar]
- Souza Filho Pedro Walfir Martins. Costa de manguezais de macromaré da Amazônia: cenários morfológicos, mapeamento e quantificação de áreas usando dados de sensores remotos. https://www.scielo.br/j/rbg/a/548fTgMXRHTmSTYBXNhfxbc/?lang=pt. Revista Brasileira de Geofísica. 2005;23:427–435. doi: 10.1590/S0102-261X2005000400006. [DOI] [Google Scholar]
- Vanderbilt Kristin L., Lin Chau-Chin, Lu Sheng-Shan, Kassim Abd Rahman, He Honglin, Guo Xuebing, Gil Inigo San, Blankman David, Porter John H. Fostering ecological data sharing: collaborations in the International Long Term Ecological Research Network. https://esajournals.onlinelibrary.wiley.com/doi/10.1890/ES14-00281.1. Ecosphere. 2015;6(10):1–18. doi: 10.1890/ES14-00281.1. [DOI] [Google Scholar]
- Wetzel Florian T., Saarenmaa Hannu, Regan Eugenie, Martin Corinne S., Mergen Patricia, Smirnova Larissa, Tuama Éamonn Ó, García Camacho Francisco A., Hoffmann Anke, Vohland Katrin, Häuser Christoph L. The roles and contributions of Biodiversity Observation Networks (BONs) in better tracking progress to 2020 biodiversity targets: a European case study. http://www.tandfonline.com/doi/full/10.1080/14888386.2015.1075902. Biodiversity. 2015;16(2-3):137–149. doi: 10.1080/14888386.2015.1075902. [DOI] [Google Scholar]
- Wetzel Florian T., Bingham Heather C., Groom Quentin, Haase Peter, Kõljalg Urmas, Kuhlmann Michael, Martin Corinne S., Penev Lyubomir, Robertson Tim, Saarenmaa Hannu. Unlocking biodiversity data: Prioritization and filling the gaps in biodiversity observation data in Europe. https://www.sciencedirect.com/science/article/pii/S0006320717300629. Biological conservation. 2018;221:78–85. doi: 10.1016/j.biocon.2017.12.024. [DOI] [Google Scholar]
- Whitlock Michael C. Data archiving in ecology and evolution: best practices. https://www.cell.com/trends/ecology-evolution/fulltext/S0169-5347(10)00269-7. Trends in Ecology & Evolution. 2011;26(2):61–65. doi: 10.1016/j.tree.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Wieczorek John, Bloom David, Guralnick Robert, Blum Stan, Döring Markus, Giovanni Renato, Robertson Tim, Vieglais David. Darwin Core: an evolving community-developed biodiversity data standard. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0029715. PLOS One. 2012;7(1) doi: 10.1371/journal.pone.0029715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Yu, Watson Maria. Guidance on conducting a systematic literature review. http://journals.sagepub.com/doi/10.1177/0739456X17723971. Journal of Planning Education and Research. 2019;39(1):93–112. doi: 10.1177/0739456X17723971. [DOI] [Google Scholar]
- Zenodo https://zenodo.org/ https://zenodo.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Keywords
Nubia Marques
Data type
table
Brief description
Keywords used in the systematic review of each biotic group.
File: oo_1105742.docx
Table Darwin Core (DwC)
Nubia Marques
Data type
table
Brief description
Table containing the Darwin Core (DwC) standard terms that were used to make the table and extract the information from the bibliographic references previously selected in the systematic review. Label = name of the column in the DwC standard; Definition = Brief definition of what each column means.
File: oo_1105743.docx
Flowchart of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)
Nubia Marques
Data type
images
Brief description
Flowchart of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) separated by groups showing the process of selecting studies throughout the systematic review. The selection process includes three stages: (1) identifying the database and choosing the papers; (2) scanning the references and selecting the papers to be included; (3) including the selected papers.
File: oo_1105747.docx
List of species from the Golfão Maranhense (Maranhão State, Brazil)
Nubia Marques
Data type
Table
Brief description
List of species from the Golfão Maranhense (Maranhão State, Brazil) that were retrieved through the systematic literature review.
File: oo_1105748.docx