Abstract
A sequence-independent PCR amplification method was used to identify viral nucleic acids in the plasma samples of 25 individuals presenting with symptoms of acute viral infection following high-risk behavior for human immunodeficiency virus type 1 transmission. GB virus C/hepatitis G virus was identified in three individuals and hepatitis B virus in one individual. Three previously undescribed DNA viruses were also detected, a parvovirus and two viruses related to TT virus (TTV). Nucleic acids in human plasma that were distantly related to bacterial sequences or with no detectable similarities to known sequences were also found. Nearly complete viral genome sequencing and phylogenetic analysis confirmed the presence of a new parvovirus distinct from known human and animal parvoviruses and of two related TTV-like viruses highly divergent from both the TTV and TTV-like minivirus groups. The detection of two previously undescribed viral species in a small group of individuals presenting acute viral syndrome with unknown etiology indicates that a rich yield of new human viruses may be readily identifiable using simple methods of sequence-independent nucleic acid amplification and limited sequencing.
The identification of previously uncharacterized viruses infecting humans and animals remains a constant task. The development of molecular tools for that purpose, including immunoreactive cDNA expression library screening, representational difference analysis, DNA microarrays, and use of degenerate PCR primers, has resulted in the identification of hepatitis C virus (HCV) (8), human herpesvirus 8 (7), the severe acute respiratory syndrome (SARS) coronavirus (39, 40), GB virus C (GBV-C)/hepatitis G virus (HGV) (22, 34), hantavirus (26), TT virus (TTV) (24, 27), and herpesviruses in monkeys (12, 32).
Allander et al. reported on a method for the DNase sequence-independent single primer amplification (DNase-SISPA) of nucleic acids in serum (1). A related method was recently used to identify a novel human coronavirus in a cell culture supernatant (37). Amplification products are subcloned and sequenced, and their similarities to known viruses were tested using BLASTn (for nucleic acid similarity) and tBLASTx (for protein similarity) (1). Conceptually, DNase-SISPA is most related to shotgun library sequencing using the nonspecific linker amplification method recently used to identify viruses in seawater and human feces (3, 4).
Evolutionary relationships remain detectable at the amino acid level for longer time periods than they do at the DNA sequence level (18). The ability to perform amino acid-based similarity searches using the translated products of sequenced amplification products therefore allows the identification of viral sequences more divergent from already known viruses than do methods relying on nucleic acid hybridization.
DNase-SISPA was used here to determine whether known and previously uncharacterized viruses could be identified in the plasma samples of 25 patients suffering from acute viral infection syndrome.
MATERIALS AND METHODS
Study subjects.
Specimens for analysis were plasma samples from subjects screened for acute human immunodeficiency virus (HIV) infection in the University of California, San Francisco (UCSF) Options Project but found to be HIV negative. The screening process has been described previously (14). In brief, participants with recent possible exposure to HIV and with two or more symptoms compatible with acute retroviral syndrome were screened for anti-HIV antibodies and plasma HIV type 1 (HIV-1) RNA (bDNA; Bayer Diagnostics, Emeryville, California). Study staff performed a structured interview in which participants were asked whether they had any of 21 symptoms compatible with acute HIV infection or other viral illnesses, including fever, rash, fatigue, malaise, pharyngitis, nausea, diarrhea, headache, myalgias, and arthralgias. All subjects consented to participate in a protocol approved by the UCSF Institutional Review Board before specimens were collected. This study was approved by the UCSF Committee on Human Research.
DNase-SISPA.
Plasma samples were treated by the method of Allander et al. (1). Briefly, 100 μl of plasma was filtered through a 0.22-μm filter (Ultrafree MC; Millipore, Bedford, MA) and treated with 250 U of DNase I (Roche Diagnostics, Mannheim, Germany) to remove contaminating human DNA. DNase I-resistant nucleic acids were purified using either the QIAamp Viral RNA Mini kit or the QIAamp DNA Blood Mini kit (QIAGEN, Valencia, CA). To detect viral RNA, first-strand cDNA synthesis was performed using 200 U of SuperScript II RNase H− reverse transcriptase (Invitrogen, Carlsbad, CA) primed with 5 pmol random hexamers (GIBCO, Gaithersburg, MD) at 42°C for 1 h. Thirteen microliters of a second-strand cDNA synthesis mix, including 1 U of RNase H and 4 U of DNA polymerase I (Invitrogen, Carlsbad, CA), was added to the cDNA mix and incubated at 12°C for 1 h followed by 1 h at 22°C. To detect viral DNA, a complementary strand was generated by incubating 30 of the 60 μl of extracted DNA, 5 units of Klenow fragment (exo-) (New England Biolabs, Beverly, MA), and 5 pmol of random hexamers at 37°C for 1 h. Double-stranded DNA samples were then digested with the restriction enzyme Csp6.I (Fermentas, Hanover, MD). Adaptors composed of hybridized oligonucleotides NBam24 (5′AGGCAACTGTGCTATCCGAGGGAG3′) and NCsp11 (5′TACTCCCTCGG3′) (80 pmol) were then ligated to the restricted DNA using T4 DNA ligase (Invitrogen, Carlsbad, CA) (1). Two microliters of the ligation reaction was used to prime PCR containing 50 pmol of NBam24 and 2.5 units of Taq polymerase (Promega, Madison, WI). Cycling conditions were as follows: 40 cycles, with 1 cycle consisting of 94°C for 1 min and 72°C for 3 min.
Analysis of DNase-SISPA-amplified DNA.
Amplified PCR products were analyzed by polyacrylamide gel electrophoresis (PAGE). The distinct DNA bands seen by PAGE were excised and pooled. The DNA of the pooled PAGE bands from each plasma sample was purified and subcloned into pGEM-T Easy. Because excised DNA bands were pooled prior to subcloning, we were not able to relate each sequence to a particular PAGE band. Insert-containing plasmids were purified and sequenced using a flanking PCR primer.
Amplification of full viral genomes.
Subcloned and sequenced fragments of parvovirus 4 (PARV4) (located at the extreme 5′ and 3′ positions) were used to design PCR primers (pr-3B-04303-46F [5′TGCCTTACCATTCACTGACGC3′] and pr-7R-04303-174R [5′TTGGCAAGGGTAAAAGGCAT3′]) to amplify the intervening 4.3-kb region. The fragment was then sequenced using primer walking. The 5′ end of the PARV4 genome was amplified and sequenced using the 5′ rapid amplification of cDNA end (RACE) kit (Invitrogen, Carlsbad, CA). The PARV4 genome was first linearly amplified (60 cycles, with 1 cycle consisting of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min) using Taq polymerase and PARV4-specific primer pr4303-377R (5′ACTCCTTCTGCAGCTGGTGTC3′). Amplification products were purified using QIAquick columns (QIAGEN) and a poly(C) tail added to the 3′ end using deoxycytidine and terminal deoxynucleotidyl transferase (Invitrogen, Carlsbad, CA). The 5′ region was then amplified with 2.5 units of Taq polymerase using an anchor abridged primer (5′GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGTTG3′) (Invitrogen, Carlsbad, CA), pr4303-377R, and Taq polymerase (2.5 U) for 60 cycles (1 cycle consisting of 94°C for 30s, 50°C for 30s, and 72°C for 2 min).
Complete circular genomes for both small anellovirus 1 (SAV-1) and SAV-2 were generated by PCR using abutting primers of opposite polarity designed within a subcloned region. Primers used for amplification of SAV-1 were prSAV-1-IF-784F (5′GTGGCGAATGGCTGAGTTTACC3′) and prSAV-1-IR-988R (5′GTGTTGGTGTCTGTAAAAGGTCATAACAC3′), and primers for SAV-2 were pr5412-2201 (5′GTGGCGAATGGCTGAGTTTAC3′) and pr5412-97 (5′TCTTCTTACCTTGTACCGGCG3′). Amplification reactions contained 2.5 U Taq polymerase, and cycling conditions were as follows: 40 cycles, with 1 cycle consisting of 94°C for 18 s, 54°C for 21 s, and 72°C for 1 min 30 s. The fragments were then sequenced using primer walking.
Phylogenetic analysis of PARV4.
Phylogenetic analysis was performed using sequences representing full-length genomes from all species from the Parvovirinae subfamily (23). In addition, sequences of recently identified parvoviruses, including two bovine parvoviruses (bovine parvovirus 2 [BPV-2] [GenBank accession no. AF406966] and BPV-3 [AF406967]) (1), an A6 human virus (AY064476) (25), and the minute virus of canines (AF495467) (33) were included. Sequences were aligned using CLUSTAL X (36), and a neighbor-joining tree (nucleotide distance with Jukes-Cantor correction, pair-wise gap deletion), with bootstrap resampling (100 replicates) was constructed using MEGA software (20).
Phylogenetic analysis of TTV-like viruses.
TTV-like sequences amplified from two of the study subjects were aligned to a data set comprised of previously described complete genome sequences of TTV genotypes (GenBank accession no. AB008394 [TA278, genotype 1], AF122916 [JA1, genotype 2], AF247138 [T3PB, genotype 3], AB017613 [TUS01, genotype 11], AB028668 [TJN01, genotype 12], and AB028669 [TJN02, genotype 13]) and human TTV-like minivirus sequences (GenBank accession no. AB041962 [TGP96], AB038628 [CLC205], AB038626 [CLC138], AB026931 [CBD279], AB026930 [CBD231], AB038629 [NLC023], AB038630 [NLC026], AB038631 [NLC030], AB026929 [CBD203], AB038627 [CLC156], and AF291073 [PB4TL]). The data set also included complete genome sequences from nonhuman primates (GenBank sequence accession numbers are indicated in brackets), such as chimpanzees (Pt-TTV6 [AB041957] and Pt-TTV8-II [AB041963]), macaques (Macaca fascicularis) (Mf-TTV3 [AB041958] and Mf-TTV9 [AB041959]), tamarin (Sanguinis oedipus) (So-TTV2 [AB041960]), and owl monkeys (Aotus trivirgatus) (At-TTV3 [AB041961]), tree shrews (Tupaia belangeri chinensis) (Tbc-TTV14 [AB057358]), and other mammals, such as dogs (Cf-TTV10 [AB076002]), pigs (Sd-TTV31 [AB076001]), and cats (Fc-TTV4 [AB076003]) (17, 28-30). Noncoding regions of the genomes were aligned using CLUSTAL W with default settings and edited by eye to maximize alignment of regions of homology. The large open reading frame (ORF) was aligned using the inferred amino acid sequence of the encoded protein in CLUSTAL W. The introduction of increasing gap penalties identified several regions of sequence homology in the coding sequence. Phylogenetic comparison of sequences was carried out using maximum likelihood (HKY85 model with gamma distribution for estimation of likelihoods), maximum parsimony, and neighbor-joining methods (Jukes-Cantor and Timura-Nei distances) using both PAUP and MEGA software packages (19).
Nucleotide sequence accession number.
Sequence reported herein have been assigned GenBank accession numbers AY622908 to AY622915, AY622918 to AY622920, AY622922 to AY622960, AY728236 to AY728255, AY787829 to AY787831, and AY819629 to AY819640 and AY816323.
RESULTS
Detection of HCV sequences in seropositive samples.
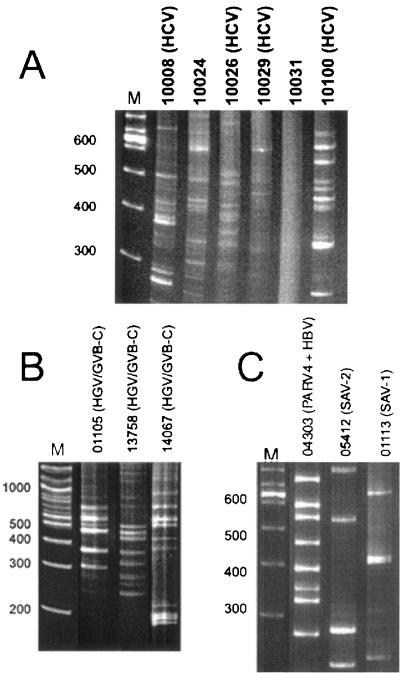
The efficiency of DNase-SISPA was initially tested using six HCV-seropositive plasma samples. Human DNA contaminating the plasma samples was first removed by DNase digestion. Viral nucleic acids protected from DNase digestion by their viral coats were then extracted and purified before reverse transcription to cDNA using random hexamer primers. Second-strand DNA synthesis was performed using RNase H and DNA polymerase I. The double-stranded DNA was then digested by a 4-base-pair-specific restriction endonuclease (Csp6.I), resulting in two overhanging bases to which a complementary oligonucleotide linker is ligated. A single PCR primer complementary to the ligated linker was then used to PCR amplify the sequences between the Csp6.I restriction sites. The PCR products were then analyzed by PAGE (Materials and Methods) (1). The sequence-independent amplification of viral genomes is thought to result in amplicons of limited genetic complexity visible as PAGE DNA bands, while amplification of the much larger human or bacterial DNA genomes is thought to result in a more complex mixture of amplicons seen as a PAGE DNA smear (1).
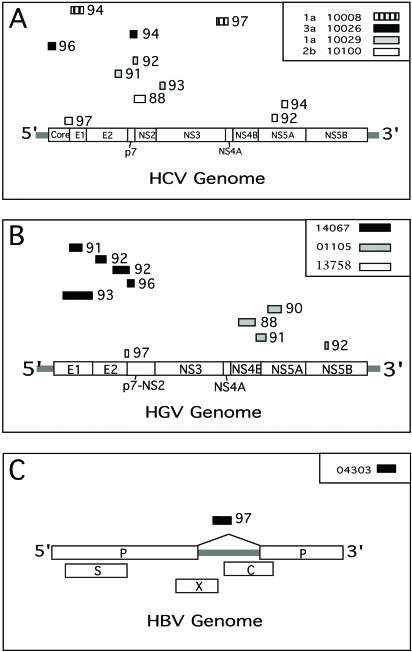
One sample showed only a DNA smear, while different band patterns were observed for the other five samples (Fig. 1A). The DNA PAGE bands were excised and pooled separately from each of these five samples before DNA purification and subcloning. From a total of 31 plasmid inserts sequenced, 11 were HCV sequences. Using BLASTn similarity searches, fragments belonging to HCV genotypes 1a, 2b, and 3a were identified in four plasma samples (Fig. 2A).
FIG. 1.
DNase-SISPA amplification products from (A) six HCV-positive plasma samples used to test methodology, (B) three RNA-extracted plasma samples, and (C) three DNA-extracted plasma samples. PCR products were analyzed on a 6.5% polyacrylamide gel. Patient identification numbers are shown over the lanes. Viral sequences identified are shown in parentheses. Lanes M contain molecular weight markers in base pairs.
FIG. 2.
Genomic locations of subcloned viral sequences homologous to HCV (A), GBV-C/HGV (B), and HBV (C). Patient identification numbers (and HCV genotypes in panel A) are indicated in the symbol key, and viral subtypes are indicated for HCV sequences. Nucleotide percent similarity values are indicated adjacent to the subcloned fragments.
Selection of patients with viral infection syndrome.
From 261 individuals presenting with acute viral syndrome, who were screened for HIV-1 infection between June 1996 and June 2002, the 25 subjects with symptoms most likely to be virus infection related were selected for this study. These 25 subjects had a range of 11 to 17 of the 21 potential virus infection-related symptoms assessed at screening. The most common reported symptoms (in order) were fatigue, malaise, night sweats, and headache. Twenty-three of the 25 subjects were male, and all reported potential sexual exposure to HIV in the prior 6 months; two also reported using injection drugs in the prior 6 months.
Detection of DNA and RNA viruses.
All samples were processed by DNase-SISPA for the presence of both RNA and DNA viruses (see Materials and Methods). Samples from three individuals processed for RNA viruses yielded distinct bands (Fig. 1B), while the other samples yielded only DNA smears. Following extraction of the amplified material from the gel, subcloning, and sequencing, followed by BLASTn analyses, a fraction of the subclones from these three subjects were shown to be highly homologous to the flavivirus GBV-C/HGV with E scores of 0 to 10−28 (Fig. 2B). The number of GBV-C/HGV subclones relative to the total number of subclones sequenced was one of nine for patient 13758, four of six for patient 01105, and five of seven for patient 14067.
An additional three samples screened for DNA viruses also yielded distinct bands (Fig. 1C), while the other samples yielded only DNA smears. One out of 20 subcloned sequences from subject 04303 was highly homologous to HBV (BLASTn E score of 10−39) (Fig. 2C). All other sequences from these three patients resulted in very weak BLASTn E scores of >0.002.
In order to search for similarity at the amino acid level, the sequences were then analyzed using tBLASTx, which translates a sequence into its six possible reading frames and searches for amino acid similarity against the entire sequence database translated in the same fashion. Thirteen out of 20 subclones from patient 04303 yielded tBLASTx E scores of 0.035 to 10−42 against parvoviruses. The regions of homology ranged in size from 120 to 450 bp. Four out of 15 sequences from patient 01113 (E scores of 10−4 to 10−23) and 2 out of 10 sequences from patient 05412 (E scores of 10−7 to 10−11) ranging in size from 120 to 350 bp also showed similarity to the anelloviruses TorqueTenoVirus (TTV) (formerly called TT virus) and TorqueTenoMiniVirus (TTMV) (16). To further characterize these viral sequences, viral genomes were PCR amplified and sequenced.
Sequencing of new human parvovirus.
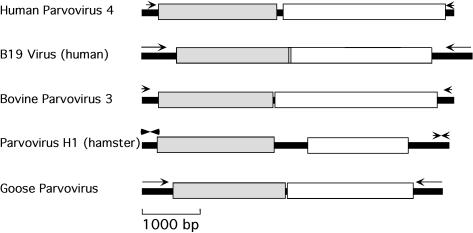
Two methods were used to amplify the full genome of the virus with homology to parvoviruses. PCR primers were first designed on the basis of DNase-SISPA-generated fragment sequences expected by tBLASTx to be located near the 5′ and 3′ ends of a parvovirus genome. Long-range PCR yielded the intervening portions of the parvovirus genome. The 5′ end of the linear genome was then acquired using the 5′ RACE method. The resulting, almost full-length genome sequence was 5,268 bp in length and contained two ORFs (Fig. 3) (GenBank accession number AY622943). tBLASTx searches showed that ORF1- and ORF2-encoded proteins showed significant homology to the nonstructural and capsid proteins of other parvoviruses, respectively. On the basis of its genome size, ORF structure, and homology to parvoviruses and its human host, this linear DNA virus was tentatively named parvovirus 4 (PARV4) after the three closely related replication-competent human parvoviruses B19, V9, and A6. The infected patient was a daily injection drug user homeless at the time of evaluation. He complained of fatigue, night sweats, pharyngitis, neck stiffness, vomiting, diarrhea, arthralgias, and confusion.
FIG. 3.
Genetic organization of PARV4 compared to those of B19, BPV-3, parvovirus H1, and goose parvoviruses. The gray and white boxes represent the genes encoding for nonstructural and structural proteins, respectively. The arrows indicate the positions of the terminal repeat sequences. The arrows at the extremities of parvovirus H1 denote that the terminal repeat sequences are dissimilar.
Phylogenetic analysis of new parvovirus PARV4.
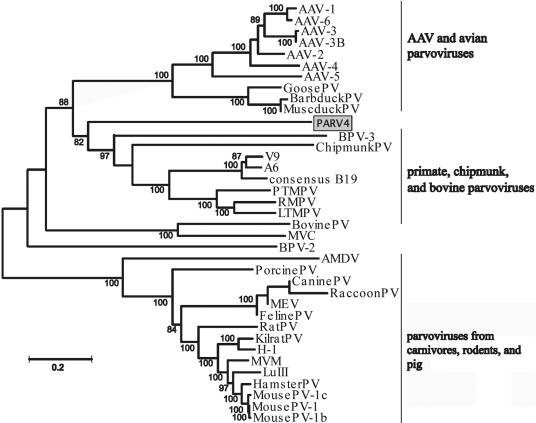
Three major evolutionary groups of viruses have recently been identified within the Parvovirinae subfamily (23). To establish the evolutionary relationship of PARV4 to other parvoviruses, phylogenetic analysis of the genomes from all known members of the Parvovirinae subfamily was performed (Fig. 4).
FIG. 4.
Phylogenetic analysis of the PARV4 genome and other members of the Parvoviridae subfamily. Abbreviations: Barbduck PV, Barbary duck parvovirus; MuscduckPV, Muscovy duck parvovirus; PTMPV, pig-tailed macaque parvovirus; RMPV, rhesus macaque parvovirus; LTMPV, long-tailed macaque parvovirus; MVC, minute virus of canines; AMDV, Aleutian mink disease virus; MEV, mink enteritis virus; Kilrat, Kilham rat parvovirus; MVM, minute virus of mice.
Our analysis indicated that PARV4 was not closely related to any known parvoviruses and represent a deeply rooted lineage between two parvovirus groups: (i) adeno-associated viruses (AAV) and avian parvoviruses and (ii) primate, chipmunk, and BPV-3 (Fig. 4). Phylogenetic analyses were also performed separately for ORF1 and ORF2. The topologies of these trees were similar to that of the full-genome tree (data not shown). The amino acid similarity between PARV4 and other parvoviruses was below 30%. PARV4 has the largest amino acid similarity in ORF1 with the AAV/avian parvoviruses (23.9 to 28.6% for AAV-4) and in ORF2 with the primate parvoviruses (23.0 to 29.5% for B19 consensus). Both PARV4 ORFs have the least amino acid similarity with the group of parvoviruses found in carnivores/rodents and pigs (12.1 to 17.2%). Recombination analysis using the bootscan method was performed on PARV4 to determine whether it was a recombinant of other known parvoviruses. Short genetic regions within the PARV4 genome that were more similar to the chipmunk parvovirus and BPV-3 were identified. However, the short length of these regions of homology and the low bootstrap support for these associations suggested that PARV4 was not a recombinant (data not shown). Our results therefore indicated that PARV4 is a unique member of the Parvoviridae family that is not closely related to any other known human or animal parvovirus.
Sequencing of new anelloviruses SAV-1 and SAV-2.
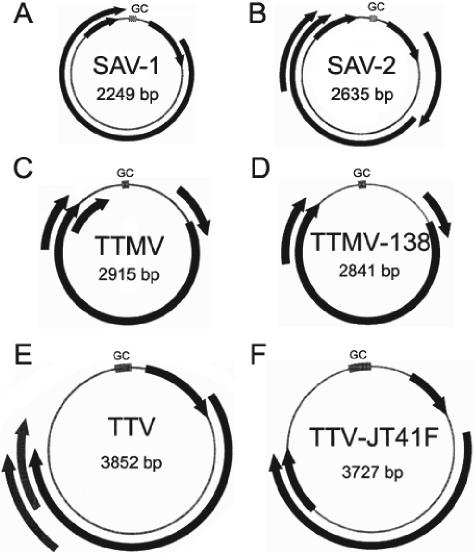
To acquire the remainder of the anellovirus-like genomes, sequence gaps were also filled by PCR and sequenced using primer walking. Anelloviruses have circular genomes (16), and their sequences were therefore assembled using long-range PCR with amplification primers facing opposite directions. The viral sequence from patient 01113 was 2,249 bp in length and contained at least three potential ORFs (Fig. 5A) (GenBank accession number AY622908), and the virus from sample 05412 was 2,635 bp in length and contained at least five potential ORFs (Fig. 5B) (GenBank accession number AY622909). Such ORF structures differed from those observed for TTV and TTMV, which have three or four ORFs and larger genomes (Fig. 5C and D) (16). These two viral sequences were classified as anelloviruses on the basis of their circular DNA nature and the presence of regions of homology to TTV and TTMV in the large ORF and untranslated region. The provisional names assigned to the viruses from samples 01113 and 05412 are small anellovirus 1 (SAV-1) and small anellovirus 2 (SAV-2), respectively. The patient infected with SAV-1 was a homosexual male with one recent sexual partner and a history of injection drug use. He developed symptoms, including fatigue, headaches, fever, night sweats, nausea, diarrhea, genital ulcers, and a rash, that lasted 1 to 2 weeks. The male patient infected with SAV-2 had multiple male sexual partners prior to developing symptoms and never used injection drugs. He developed symptoms, including fatigue, headaches, fevers, night sweats, oral ulcers, diarrhea, 4-kg weight loss, myalgias, and a rash, that lasted 1 to 2 weeks.
FIG. 5.
Genetic organization of (A) SAV-1, (B) SAV-2, (C) TTMV, (D) TTMV-238, (E) TTV, and (F) TTV-JT41F. Arrows represent ORFs detected in each virus. The GC-rich region (GC) has a GC content greater than 72%. ORFinder (National Center for Biotechnology Information) was used to determine the ORFs for each virus as described in Materials and Methods.
Phylogenetic analysis of new anelloviruses SAV-1 and SAV-2.
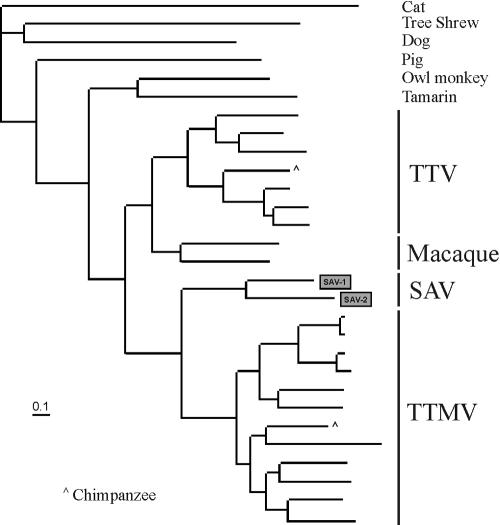
To better understand the relationship of SAV-1 and SAV-2 to other anelloviruses, phylogenetic analyses were performed with the long ORF regions (Fig. 6). The high degree of divergence within the anellovirus group and the smaller size of the new genomes prevented the generation of whole-genome alignments. This analysis showed that SAV-1 and SAV-2 were related but clustered independently from other known human anellovirus groups (TTV and TTMV-like viruses). SAV sequences were also distinct from TTV-related viruses obtained from chimpanzees and all other nonhuman primates (Fig. 6). Similar results were obtained with a phylogenetic analysis of the untranslated region alone (data not shown). Because of the low degree of homology, only the first 783 amino acids of the longest ORF could be reliably aligned. The percent similarity between the two SAVs was 54, and the SAVs were approximately equidistant to human TTV and TTMV (32 to 35% similarity). SAV similarity to monkey TTVs ranged from 28 to 33%. Because of their distinct genomic organization, length, and distant phylogenetic relationship to other anelloviruses, we postulate that SAV-1 and SAV-2 may be members of a third group of anelloviruses.
FIG. 6.
Phylogenetic analysis of the large ORFs of SAV, TTV, and TTMV found in a range of mammalian species. All viral sequences originate from humans unless indicated otherwise.
Cloning of nonviral sequences.
In addition to the viral sequences described above, we identified five human genome sequences from a total of 67 subclones sequenced from the six samples from patients with acute infection symptoms. The amplification of human DNA presumably reflects residual human DNA in plasma following DNase treatment. Sequences with similarities (tBLASTx E scores of <2 × 10−3) to bacterial sequences were also detected (10 sequences). Similarities to a Rhodobacter capsulatus (an anaerobic purple nonsulfur soil bacteria) sequence (E score of 4 × 10−7) and to Brucella melitensis/suis (E score of 10−4) were seen in two subclones from patient 04303. A subclone from patient 01113 showed similarities to Pseudomonas putida (E score of 10−9), and a single subclone from patient 05412 yielded E scores of 2.5 × 10−5 to 10−4 to Pseudomonas fluorescens, Neisseria meningitidis, Pseudomonas putida, and Haemophilus influenzae. The wide range of E scores to bacterial sequences in the database indicated that these subcloned sequences were not likely to reflect the presence of these exact bacterial species but of related species. A large fraction of subclones (21/67) showed no detectable similarity to any deposited sequences using either BLASTn or tBLASTx (E score of >2 × 10−3). The origin of these sequences warrants further investigations.
DISCUSSION
DNase-SISPA was used to analyze 25 plasma samples from individuals presenting with acute viral infection syndrome. GBV-C was detected in three individuals. A previously undescribed parvovirus was detected in another individual who was also coinfected with HBV, and two new anelloviruses were identified in two other individuals. The detection of three previously undescribed viruses in a limited number of individuals using only very limited sequencing reflects the general utility of sequence-independent molecular methods for virus discovery.
GBV-C RNA or antibodies to its E2 protein have been found in 5.5% of blood donors and 89% of intravenous drug users in the United States (10). GBV-C viremia has been shown to last at least 3 years (13). Since the pathogenicity of GBV-C remains uncertain (6), detection of GBV-C RNA in 3/25 patients may simply reflect its high prevalence, rather than a causative role in the symptoms of these individuals.
A recent phylogenetic analysis demonstrated that the Parvovirinae subfamily could be organized into three main groups: (i) primate and chipmunk parvoviruses; (ii) rodent, pig, and carnivore parvoviruses; and (iii) AAV and avian parvoviruses (23). Here we report the finding of a new human parvovirus that clustered independently of these three groups of vertebrate parvoviruses (Fig. 4). The detection of a parvovirus highly distinct from the B19 group (B19, V9, and A6 viruses) or AAV group (AAV1 through AAV6) suggests that the number of parvoviruses able to replicate in humans may be larger than currently appreciated. The PCR primers currently used to test for B19 viremia would not be expected to detect PARV4 (11, 31) and parvoviruses are of particular concern in the manufacture of plasma-derived products due to their resistance to viral inactivation methods (2, 5). Transmission of B19 parvovirus occurs via the respiratory route, through blood-derived products transfusion and from mother to fetus (15). The codetection of HBV infection in this patient indicated that this other virus may have also played a role in his symptoms. Since parvovirus B19 infection can be pathogenic, further investigations of the prevalence of PARV4 and its association with pathogenic conditions are under way.
Anelloviruses infecting humans include TTV and its smaller relative TTMV (24). Both TTV and TTMV are genetically highly diverse groups and are classified into multiple genotypes (35). Both viruses are detected at very high frequencies in healthy blood donors and primates (9, 21, 35). These viruses have no currently recognized clinical consequences and may therefore be part of normal human viral flora (16, 38). Phylogenetic analysis at two loci indicated that SAV-1 and SAV-2 were phylogenetically related but distinct from both the TTV and TTMV groups and may therefore represent two strains of a third group of human anelloviruses. Preliminary studies indicate that the prevalence of SAV may be as high as that of TTV/TTMV (data not shown). The discovery of an additional group of TTV-related viruses in humans further highlights the extreme sequence diversity of this viral family.
As in recent nonspecific approaches to identifying new viral sequences, over half the sequences identified were unrelated to any known sequences in GenBank (3, 4). The possible origin of these sequences is being further investigated using enhanced sequence similarity searches.
We identified three new viruses, PARV4, SAV-1, and SAV-2, in plasma samples from patients presenting with multiple symptoms of acute viral infection. PARV4 represents a new member of the Parvoviridae family, and SAV-1 and SAV-2 are potential founding members of a third group of the Anellovirus genus. The pathogenicity of these viruses remains to be determined. The detection of two new viral species in the plasma samples of 25 subjects with symptoms of viral infection, only 6 of which were analyzed by DNA sequencing, does suggest that other human viruses may be readily identifiable using simple molecular methods requiring only limited sequencing.
Acknowledgments
This work was supported by a gift from the Blood Systems Foundation to E.D.
REFERENCES
- 1.Allander, T., S. U. Emerson, R. E. Engle, R. H. Purcell, and J. Bukh. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA 98:11609-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylis, S. A., N. Shah, and P. D. Minor. 2004. Evaluation of different assays for the detection of parvovirus B19 DNA in human plasma. J. Virol. Methods 121:7-16. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnouf, T., and M. Radosevich. 2000. Reducing the risk of infection from plasma products: specific preventative strategies. Blood Rev. 14:94-110. [DOI] [PubMed] [Google Scholar]
- 6.Chams, V., C. Fournier-Wirth, A. Chabanel, P. Herve, and C. Trepo. 2003. Is GB virus C alias “hepatitis” G virus involved in human pathology? Transfus. Clin. Biol. 10:292-306. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood borne non-A, non-B viral hepatitis genome. Science 244:359-361. [DOI] [PubMed] [Google Scholar]
- 9.Desai, S. M., A. S. Muerhoff, T. P. Leary, J. C. Erker, J. N. Simons, M. L. Chalmers, L. G. Birkenmeyer, T. J. Pilot-Matias, and I. K. Mushahwar. 1999. Prevalence of TT virus infection in US blood donors and populations at risk for acquiring parenterally transmitted viruses. J. Infect. Dis. 179:1242-1244. [DOI] [PubMed] [Google Scholar]
- 10.Dille, B. J., T. K. Surowy, R. A. Gutierrez, P. F. Coleman, M. F. Knigge, R. J. Carrick, R. D. Aach, F. B. Hollinger, C. E. Stevens, L. H. Barbosa, G. J. Nemo, J. W. Mosley, G. J. Dawson, and I. K. Mushahwar. 1997. An ELISA for detection of antibodies to the E2 protein of GB virus C. J. Infect. Dis. 175:458-461. [DOI] [PubMed] [Google Scholar]
- 11.Durigon, E. L., D. D. Erdman, G. W. Gary, M. A. Pallansch, T. J. Torok, and L. J. Anderson. 1993. Multiple primer pairs for polymerase chain reaction (PCR) amplification of human parvovirus B19 DNA. J. Virol. Methods 44:155-165. [DOI] [PubMed] [Google Scholar]
- 12.Greensill, J., J. A. Sheldon, N. M. Renwick, B. E. Beer, S. Norley, J. Goudsmit, and T. F. Schulz. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J. Virol. 74:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez, R. A., G. J. Dawson, M. F. Knigge, S. L. Melvin, C. A. Heynen, C. R. Kyrk, C. E. Young, R. J. Carrick, G. G. Schlauder, T. K. Surowy, B. J. Dille, P. F. Coleman, D. L. Thiele, J. R. Lentino, C. Pachucki, and I. K. Mushahwar. 1997. Seroprevalence of GB virus C and persistence of RNA and antibody. J. Med. Virol. 53:167-173. [DOI] [PubMed] [Google Scholar]
- 14.Hecht, F. M., M. P. Busch, B. Rawal, M. Webb, E. Rosenberg, M. Swanson, M. Chesney, J. Anderson, J. Levy, and J. O. Kahn. 2002. Use of laboratory tests and clinical symptoms for identification of primary HIV infection. AIDS 16:1119-1129. [DOI] [PubMed] [Google Scholar]
- 15.Heegaard, E. D., B. L. Petersen, C. J. Heilmann, and A. Hornsleth. 2002. Prevalence of parvovirus B19 and parvovirus V9 DNA and antibodies in paired bone marrow and serum samples from healthy individuals. J. Clin. Microbiol. 40:933-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hino, S. 2002. TTV, a new human virus with single stranded circular DNA genome. Rev. Med. Virol. 12:151-158. [DOI] [PubMed] [Google Scholar]
- 17.Inami, T., T. Obara, M. Moriyama, Y. Arakawa, and K. Abe. 2000. Full-length nucleotide sequence of a simian TT virus isolate obtained from a chimpanzee: evidence for a new TT virus-like species. Virology 277:330-335. [DOI] [PubMed] [Google Scholar]
- 18.Koonin, E. V., and M. Y. Galperin. 2003. Sequence—evolution—function: computational approaches in comparative genomics. Kluwer Academic, Boston, Mass. [PubMed]
- 19.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 21.Leary, T. P., J. C. Erker, M. L. Chalmers, S. M. Desai, and I. K. Mushahwar. 1999. Improved detection systems for TT virus reveal high prevalence in humans, non-human primates and farm animals. J. Gen. Virol. 80:2115-2120. [DOI] [PubMed] [Google Scholar]
- 22.Linnen, J., J. Wages, Jr., Z. Y. Zhang-Keck, K. E. Fry, K. Z. Krawczynski, H. Alter, E. Koonin, M. Gallagher, M. Alter, S. Hadziyannis, P. Karayiannis, K. Fung, Y. Nakatsuji, J. W. Shih, L. Young, M. Piatak, Jr., C. Hoover, J. Fernandez, S. Chen, J. C. Zou, T. Morris, K. C. Hyams, S. Ismay, J. D. Lifson, J. P. Kim, et al. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271:505-508. [DOI] [PubMed] [Google Scholar]
- 23.Lukashov, V. V., and J. Goudsmit. 2001. Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J. Virol. 75:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mushahwar, I. K., J. C. Erker, A. S. Muerhoff, T. P. Leary, J. N. Simons, L. G. Birkenmeyer, M. L. Chalmers, T. J. Pilot-Matias, and S. M. Dexai. 1999. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc. Natl. Acad. Sci. USA 96:3177-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen, Q. T., S. Wong, E. D. Heegaard, and K. E. Brown. 2002. Identification and characterization of a second novel human erythrovirus variant, A6. Virology 301:374-380. [DOI] [PubMed] [Google Scholar]
- 26.Nichol, S. T., C. F. Spiropoulou, S. Morzunov, P. E. Rollin, T. G. Ksiazek, H. Feldmann, A. Sanchez, J. Childs, S. Zaki, and C. J. Peters. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914-917. [DOI] [PubMed] [Google Scholar]
- 27.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto, H., T. Nishizawa, M. Takahashi, A. Tawara, Y. Peng, J. Kishimoto, and Y. Wang. 2001. Genomic and evolutionary characterization of TT virus (TTV) in tupaias and comparison with species-specific TTVs in humans and non-human primates. J. Gen. Virol. 82:2041-2050. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, H., T. Nishizawa, A. Tawara, Y. Peng, M. Takahashi, J. Kishimoto, T. Tanaka, Y. Miyakawa, and M. Mayumi. 2000. Species-specific TT viruses in humans and nonhuman primates and their phylogenetic relatedness. Virology 277:368-378. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto, H., M. Takahashi, T. Nishizawa, A. Tawara, K. Fukai, U. Muramatsu, Y. Naito, and A. Yoshikawa. 2002. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J. Gen. Virol. 83:1291-1297. [DOI] [PubMed] [Google Scholar]
- 31.Patou, G., D. Pillay, S. Myint, and J. Pattison. 1993. Characterization of a nested polymerase chain reaction assay for detection of parvovirus B19. J. Clin. Microbiol. 31:540-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose, T. M., K. B. Strand, E. R. Schultz, G. Schaefer, G. W. Rankin, Jr., M. E. Thouless, C. C. Tsai, and M. L. Bosch. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 71:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz, D., B. Green, L. E. Carmichael, and C. R. Parrish. 2002. The canine minute virus (minute virus of canines) is a distinct parvovirus that is most similar to bovine parvovirus. Virology 302:219-223. [DOI] [PubMed] [Google Scholar]
- 34.Simons, J. N., T. J. Pilot-Matias, T. P. Leary, G. J. Dawson, S. M. Desai, G. G. Schlauder, A. S. Muerhoff, J. C. Erker, S. L. Buijk, M. L. Chalmers, et al. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. USA 92:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thom, K., C. Morrison, J. C. Lewis, and P. Simmonds. 2003. Distribution of TT virus (TTV), TTV-like minivirus, and related viruses in humans and nonhuman primates. Virology 306:324-333. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-Van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viazov, S., R. S. Ross, C. Varenholz, R. Lange, M. Holtmann, C. Niel, and M. Roggendorf. 1998. Lack of evidence for an association between TTV infection and severe liver disease. J. Clin. Virol. 11:183-187. [DOI] [PubMed] [Google Scholar]
- 39.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, D., A. Urisman, Y. T. Liu, M. Springer, T. G. Ksiazek, D. D. Erdman, E. R. Mardis, M. Hickenbotham, V. Magrini, J. Eldred, J. P. Latreille, R. K. Wilson, D. Ganem, and J. L. DeRisi. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]