Abstract
The process of hepadnavirus reverse transcription involves two template switches during the synthesis of plus-strand DNA. The first involves translocation of the plus-strand primer from its site of generation, the 3′ end of minus-strand DNA, to the complementary sequence DR2, located near the 5′ end of the minus-strand DNA. Plus strands initiated from DR2 are extended to the 5′ end of the minus-strand DNA. At this point, the 3′ end of the minus strand becomes the template via the second template switch, a process called circularization. Elongation of circularized plus-strand DNA generates relaxed circular DNA. Although most virions contain relaxed circular DNA, some contain duplex linear DNA. Duplex linear genomes are synthesized when the plus-strand primer is used at the site of its generation, the 3′ end of the minus-strand template. This type of synthesis is called in situ priming. Although in situ priming is normally low, in some duck hepatitis B virus mutants this type of priming is elevated. For example, mutations within the 3′ end of the minus-strand DNA can lead to increased levels of in situ priming. We report here that these same mutations result in a second defect, a less efficient template switch that circularizes the genome. Although it is not clear how these mutations affect both steps in DNA replication, our findings suggest a commonality in the mechanism of initiation of plus-strand synthesis and the template switch that circularizes the genome.
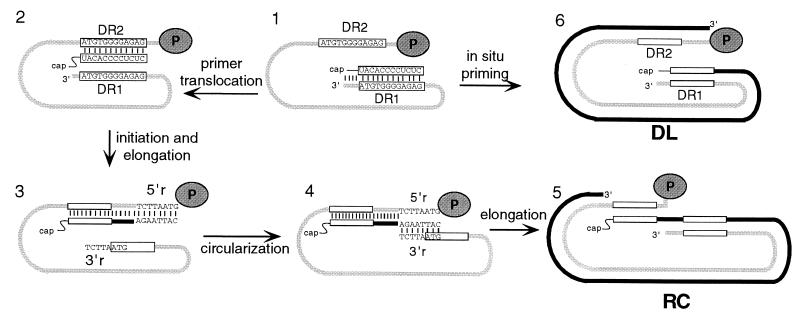
Hepadnaviruses are a family of hepatotropic DNA viruses that carry out genome replication via reverse transcription of an RNA intermediate, the pregenome (for a review, see reference 3). Reverse transcription occurs within nucleocapsids in the cytoplasm of infected liver cells (16). The predominant end product of reverse transcription is relaxed circular (RC) DNA. Its synthesis requires three template switches: one during first-strand, or minus-strand, synthesis (12, 17, 18) and two during second-strand, or plus-strand, synthesis (5, 13, 19). We studied the two template switches during synthesis of the plus strand of the RC form of the duck hepatitis B virus (DHBV) genome. The template for the synthesis of plus-strand DNA is minus-strand DNA, which is copied from the pregenomic RNA. The final stage of minus-strand DNA synthesis involves copying pregenomic RNA to the 5′ end (6). The last RNase H cleavage during minus-strand DNA synthesis generates an oligoribonucleotide that is used as the primer for the initiation of plus-strand DNA synthesis (7) (Fig. 1, part 1). This primer is either 18 or 19 nucleotides (nt) long and contains the DR1 sequence at its 3′ terminus (5). For synthesis of the RC genomic form, the plus-strand primer is translocated to a complementary sequence, DR2, that is near the 5′ end of the minus-strand template (Fig. 1, part 2) (5). Plus-strand DNA primed from DR2 will ultimately yield RC genomes after an additional template switch, called circularization, and elongation (Fig. 1, parts 2 to 5) (5, 10). An 8-nt terminal redundancy on the minus-strand DNA, named r, defines the donor and acceptor sequences for circularization (for an example, see Fig. 1, part 3). All hepadnaviruses described to date support the synthesis of a duplex linear (DL) DNA form (Fig. 1, part 6), which is less abundant than the RC genomic form. For DHBV, the level of DL DNA is typically 10% or less that of RC DNA. The 5′ end of the plus-strand of DL DNA is located at DR1, indicating that the plus-strand primer was used at its site of generation, the 3′ end of minus-strand DNA, rather than being translocated to DR2. This type of initiation of plus-strand synthesis is called in situ priming (15).
FIG. 1.
Synthesis of DHBV plus-strand DNA. Plus-strand DNA synthesis commences upon completion of minus-strand DNA synthesis. (Part 1) The light gray line represents the full-length minus-strand DNA. The dark gray oval labeled “P” represents the P protein covalently attached to the 5′ end of minus-strand DNA. Rectangles with sequences represent DR1 and DR2 on the minus-strand DNA. Final RNase H cleavage generates the plus-strand primer, derived from the 5′ end of the pregenomic RNA and annealed to the 3′ end of minus-strand DNA. The 3′ end of the plus-strand primer contains the DR1 sequence. (Part 2) Primer translocation. For most templates, the plus-strand primer is translocated from DR1 to DR2. Instead of 18 nt of complementarity at DR1, the primer has only 12 nt of complementarity to the DR2 site. DR2 is approximately 50 nt from the 5′ end of the minus-strand DNA template. (Part 3) Initiation and elongation of plus-strand DNA synthesis from DR2 to the 5′ end of the minus-strand DNA template. The black line represents plus-strand DNA. The minus-strand template has an 8-nt terminal redundancy, called r. The sequences of 5′r and 3′r are shown. (Part 4) Circularization. The nascent plus strand, which has the r sequence at its 3′ end, base-pairs with the 3′ end of the minus-strand template via complementarity with 3′r. (Part 5) Resumption of plus-strand DNA synthesis to ultimately generate the RC DNA form. (Part 6) In situ priming generates DL DNA. The plus-strand primer is utilized at DR1. Elongation yields DL DNA.
There are two pathways for the synthesis of plus-strand DNA, but their consequences are not equivalent. RC DNA genomes have a competitive advantage over DL DNA genomes in initiating an infection. In an infection in which the inoculum contains predominantly a variant virus that synthesizes higher-than-wild-type levels of DL DNA, wild-type virus quickly emerges and outcompetes the variant to become the dominant viral species (20–22). Thus, a virus that accurately and efficiently carries out primer translocation and circularization has a competitive advantage over virus that is only modestly defective for one of these template switches.
It has been shown for DHBV that cis-acting mutations can lead to increased synthesis of DL DNA via in situ priming. These mutations were nucleotide substitutions located within and surrounding the DR1 sequence (7, 15). In the present study, we further analyzed several of the mutants that led to the initial description and characterization of the in situ priming phenotype. We found that these mutants were also partially defective for the circularization step.
MATERIALS AND METHODS
Molecular clones.
All molecular clones were derived from DHBV strain 3 (14). All DHBV-containing plasmids contain a head-to-tail dimer of the DHBV strain 3 genome permuted at the single EcoRI site. Details describing the construction of these clones can be found in the work of Staprans et al. (15).
Cell cultures, transfections, and isolation of viral DNA.
The cell line LMH was used in all experiments (2). Culturing of cells and transfection of DHBV-containing plasmid DNA were performed as described previously (8, 9). Viral DNA was isolated from cytoplasmic capsids 3 days after transfection as described before (1). Southern blotting was carried out as previously described (9, 11).
Primer extension analysis.
Primer extension reactions used a thermostable DNA polymerase and 10 reaction cycles as described previously (4). Typically, 500 pg to 1 ng of viral DNA was processed for use in three separate primer extension reactions. For use as an internal standard, each viral DNA was mixed with approximately 500 pg to 1 ng of a DHBV-containing plasmid digested with NcoI (nt 2351) and EcoRV (nt 2650). The end-labeled primer used to measure the level of the plus-strand DNA that initiated from DR2 and elongated to at least the 5′ end of minus-strand DNA has a sequence complementary to that of DHBV at nt 2520 to 2537 (primer B; annealing temperature, 37°C). To measure the level of plus-strand DNA that initiated from DR2 and circularized, we used a primer complementary to nt 2599 to 2622 (primer A; annealing temperature, 55°C). Primer A was also used to measure the level of in situ-primed plus strands from DR1. The level of minus-strand DNA was measured with an end-labeled primer derived from nt 2425 to 2447 (primer M; annealing temperature, 55°C). Quantitation of autoradiographic images was performed with a PhosphorImager from Molecular Dynamics.
Definition and calculation of values. (i) Southern blot analysis.
Southern blotting of wild-type viral DNA isolated from intracellular capsids revealed three major forms. Each of these DNA forms contains a full-length minus strand (4). Two forms, RC and DL, have a full-length plus strand initiating from DR2 and DR1, respectively. The third DNA form, called single-stranded (SS) DNA, is a full-length minus-strand DNA that is primarily, if not completely, single stranded. For wild-type virus, the three forms were found in characteristic proportions (Table 1). These proportions reflect the overall efficiency of the individual steps of plus-strand DNA synthesis. The data in Table 1 were derived by measuring the levels of RC, DL, and SS DNA for a virus and by expressing each of the three DNA forms as a percentage of the total. A deficiency in primer translocation or circularization will lead to a decrease in the proportion of RC DNA and an increase in DL DNA, SS DNA, or both.
TABLE 1.
Proportions of replicative intermediates measured by Southern blotting
| DNA form | % (Mean ± SD)a of DNA form in:
|
||||||
|---|---|---|---|---|---|---|---|
| WT | DR1-13 | DR1-Xho | DR1-Pvu | DR1-12 | DR2-12 | DRs-12 | |
| RC | 70 ± 3 | 32 ± 2 | 34 ± 3 | 6 ± 2 | 8 ± 1 | 60 ± 3 | 15 ± 1 |
| DL | 11 ± 2 | 30 ± 2 | 26 ± 2 | 46 ± 3 | 48 ± 5 | 14 ± 1 | 43 ± 6 |
| SS | 19 ± 1 | 38 ± 2 | 40 ± 5 | 48 ± 4 | 43 ± 5 | 26 ± 5 | 42 ± 6 |
Values are based on three measurements made with intracellular viral DNA isolated from independent transfections, except for the wild-type (WT) and DRs-12 values, which are based on six measurements.
(ii) Primer extension analysis.
Three different primers in three individual primer extension reactions were used to evaluate the efficiency with which minus-strand DNA templates supported the synthesis of plus-strand DNA primed from DR1 or DR2, as well as the circularization of plus-strand DNA primed from DR2. One primer extension reaction (with primer M) allowed measurement of the level of minus-strand DNA, a second reaction (with primer B) allowed measurement of the level of plus-strand DNA primed from DR2, while a third reaction (with primer A) allowed measurement of the level of plus-strand DNA primed from DR2 that had successfully circularized and also allowed the measurement of the level of plus-strand priming from DR1. Because three different primers were used, normalization of the level of viral DNA measured in each primer extension reaction to an internal standard was necessary. Normalized values obtained with primer M represent the level of minus-strand DNA and were termed M. Normalized values obtained with primer B represent the level of plus-strand DNA primed from DR2 and elongated at least to the point of circularization and were termed B(DR2). Two normalized values were obtained with primer A. A(DR2) represents the level of plus-strand DNA that primed from DR2 and that had circularized. A(DR1) represents the level of in situ priming from DR1. Ratios (expressed as percentages) were derived from various pair-wise comparisons of these four measurements (Table 2). The percentages in Table 2 reflect the efficiencies of priming from DR2 [B(DR2)/M], priming from DR1 [A(DR1)/M], and circularization [B(DR2)/A(DR2)]. The efficiency of total plus-strand priming, which is also termed primer utilization, was obtained by the following equation: [B(DR2) + A(DR1)]/M.
TABLE 2.
Efficiency of plus-strand DNA synthesis determined by primer extension
| Plus-strand eventa | % Efficiency (mean ± SD)b for:
|
||||||
|---|---|---|---|---|---|---|---|
| WT | DR1-13 | DR1-Xho | DR1-Pvu | DR1-12 | DR2-12 | DRs-12 | |
| Priming from DR2c | 77 ± 13 | 57 ± 1 | 65 ± 6 | 32 ± 5 | 49 ± 6 | 52 ± 12 | 55 ± 9 |
| Priming from DR2 and circularizationd | 55 ± 5 | 18 ± 4 | 15 ± 5 | 5 ± 2 | 3 ± 1 | 38 ± 9 | 7 ± 2 |
| Circularizatione | 73 ± 14 | 31 ± 7 | 23 ± 5 | 13 ± 5 | 7 ± 3 | 74 ± 6 | 13 ± 4 |
| In situ primingf | 5 ± 1 | 15 ± 4 | 16 ± 4 | 26 ± 3 | 22 ± 3 | 10 ± 3 | 21 ± 4 |
| Primer utilization totalg | 82 | 72 | 81 | 58 | 71 | 62 | 76 |
See Materials and Methods for explanation.
Values are based on three measurements made with intracellular viral DNA isolated from independent transfections, except for the wild-type (WT) and DRs-12 values, which are based on six measurements.
B(DR2)/M value.
A(DR2)/M value.
A(DR2)/B(DR2) value.
A(DR1)/M value.
Priming from DR2 plus in situ priming value.
RESULTS
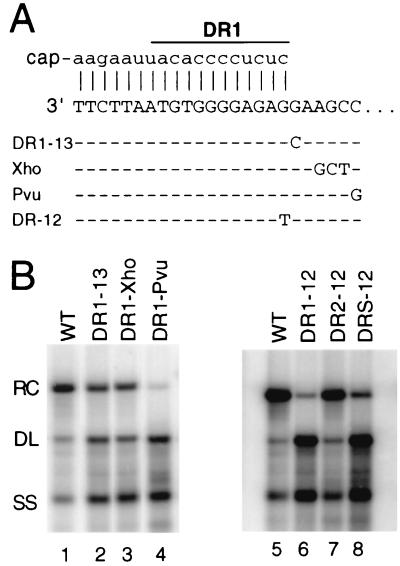
LMH cells were transfected with DNA plasmids to express the in situ priming variant viruses DR1-13, DR1-Xho, and DR1-Pvu (15). These three variants contain base substitutions adjacent to the DR1 sequence at the 3′ end of the minus-strand DNA (Fig. 2A). These base substitutions do not alter the specificity of RNase H cleavage during the generation of the plus-strand primer (15). Southern blotting of viral DNA isolated from intracellular capsids indicated not only the expected increase in the proportion of the DL DNA but also an increase in the proportion of the SS DNA form (Fig. 2B, lanes 1 to 4; Table 1). The increase in the proportion of SS DNA for the variant viruses suggested a defect in the synthesis of plus-strand DNA. Two scenarios seemed likely. The increase in SS DNA could be a consequence of a deficiency in the utilization of the plus-strand primer from either DR2 or DR1 (primer utilization). Alternatively, the accumulation of SS DNA for the mutants could be due to an inhibition of circularization of plus strands that had initiated from DR2. The key difference between these two possibilities would be the presence of a short segment (∼50 nt) of plus-strand DNA primed from DR2 associated with the latter scenario. Unfortunately, our Southern blot analysis did not distinguish between these possibilities.
FIG. 2.
(A) Mutations affecting plus-strand synthesis. The top line represents the mature plus-strand primer base-paired with the 3′ end of minus-strand DNA for wild-type virus. The sequence of DR1, which is 12 nt long, is indicated. Underneath the wild-type sequence are the minus-strand DNA sequences of the variants. Substituted nucleotides are indicated. Dashes represent the wild-type sequence. DR-12 represents base substitution in the DR1-12 (DR1 only), DR2-12 (DR2 only), and DRs-12 (both DR1 and DR2). (B) Southern blotting of in situ priming variant indicates an increased accumulation of DL and SS DNA. Viral DNA was isolated from LMH cells 3 days after the transfection. The positions of RC, DL, and SS DNA are indicated. The blot was hybridized with a genomic-length, minus-strand-specific probe. WT, Wild type.
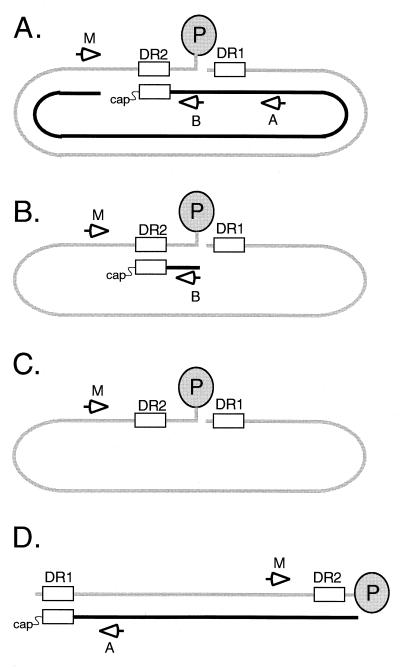
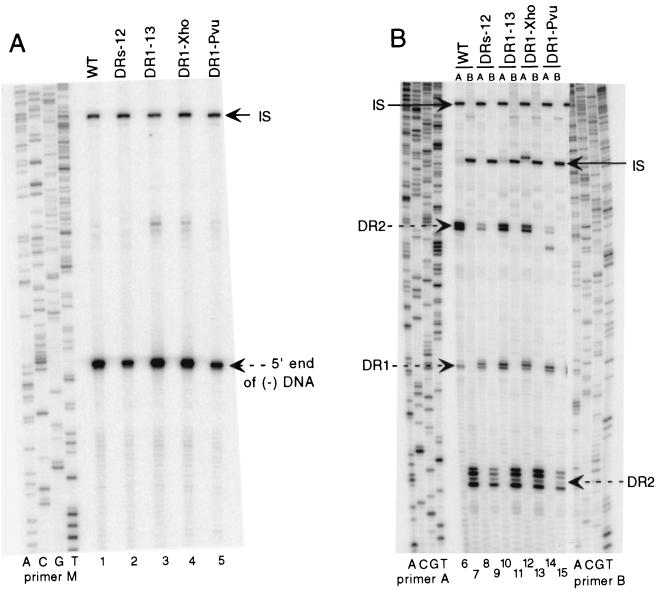
Previously, Havert and Loeb used a method based on primer extension to distinguish a primer utilization defect from a circularization defect (4). This strategy is depicted in Fig. 3. An example of the primer extension analyses for the wild type and variants DR1-13, DR1-Xho, and DR1-Pvu is presented in Fig. 4. First, the level of minus-strand DNA was measured for each sample with primer M (Fig. 4A, lanes 1, 3, 4, and 5). Next, the level of plus-strand DNA primed from DR2 and extending to at least the 5′ end of minus-strand DNA was measured with primer B (Fig. 4B, lanes 7, 11, 13, and 15). This measurement included plus-strand DNA that had successfully circularized and that which had not. In the third primer extension reaction, the levels of plus-strand DNA primed from DR2 that had successfully circularized were measured with primer A (Fig. 4B, lanes 6, 10, 12, and 14). Because primer A anneals 85 nt downstream of the circularization point, in situ-primed plus strands were also detected and measured (Fig. 4B). For each primer extension reaction, the level of viral DNA was normalized to an internal standard. From the normalized primer extension measurements, a series of ratios were calculated for each virus examined (expressed as percentages in Table 2; see also Materials and Methods for descriptions of the calculations). We found for wild-type virus that 82% of minus-strand DNA had plus-strand DNA associated with it, 77% primed from DR2 and 5% primed from DR1 (Table 2). These values were different for each of the variant viruses. As expected, each variant virus had a higher-than-wild-type level of in situ-primed plus-strand DNA (Table 2). The increase measured in the primer extension assay was consistent with the magnitude of the increase in DL DNA measured by Southern blotting (compare Tables 1 and 2). For each of the variant viruses, the level of plus-strand DNA priming from DR2 normalized to the level of minus-strand DNA (Table 2, priming from DR2) was lower than that for wild-type virus. For variants DR1-13 and DR1-Pvu, the decrease in this value was not simply a consequence of increased in situ priming. The sum of plus-strand DNA priming from DR2 and DR1 was lower for DR1-13 and DR1-Pvu than that for the wild type (Table 2, primer utilization total). The magnitude of the defect for DR1-13 and DR1-Pvu was modest, suggesting that the deficiency in primer utilization alone could not account for the increase in SS DNA seen in Southern blotting. In addition, each of the variant viruses was impaired for circularization. For wild-type virus, 73% of the plus strand that initiated from DR2 had successfully carried out circularization (Table 2, circularization efficiency). In contrast, circularization efficiency for the variants was lower. The magnitude of the deficiency in circularization was greater than the defect in primer utilization and would make a greater contribution to the increase in the proportion of SS DNA demonstrated by Southern blotting.
FIG. 3.
Strategy to measure template switches during plus-strand DNA synthesis by using primer extension. Four different replicative intermediates are shown. The gray line represents full-length minus-strand DNA with the P protein (gray oval) attached to the 5′ end. The black line represents plus-strand DNA. Open arrows indicate the positions of annealing of the three end-labeled primers used in the primer extension analyses, i.e., M, B, and A. Primer M measures the level of minus-strand DNA. Primer B measures the level of plus-strand DNA initiating from DR2. Primer A measures the level of plus-strand DNA initiating from DR2 that has circularized and extended. Primer A also detects and measures the level of in situ priming from DR1. (A) RC DNA yields a signal with all three primers. (B) A replicative intermediate that is inhibited for circularization is detected with primers M and B but not primer A. (C) A replicative intermediate in which plus-strand synthesis has not occurred is detected with primer M but not with primers A and B. (D) DL DNA is detected with primers M and A. Primer A yields a signal at DR1 instead of DR2. Primer B does not give rise to a signal because it anneals to the 3′ end of plus-strand DNA.
FIG. 4.
Primer extension analysis of the wild type (WT) and variants DRs-12, DR1-13, DR1-Xho, and DR1-Pvu indicates a circularization defect. Viral DNA was mixed with internal standard DNA, processed (see Materials and Methods), and split into three primer extension reactions. Internal standard DNA is plasmid DNA that contains the 3′ half of the DHBV3 genome and that was cleaved with NcoI and EcoRV. (A) Minus-strand primer extension using primer M. The position of the 5′ end of minus-strand DNA is indicated. IS, EcoRV terminus of the internal standard DNA. The viral signal is normalized to the internal standard signal. The sequence ladder was generated with primer M. (B) Plus-strand primer extension with primers A (lanes 6, 8, 10, 12, and 14) and B (lanes 7, 9, 11, 13, and 15). Primer B anneals to plus-strand DNA before the circularization point and detects plus-strand DNA primed from DR2 (indicated with dashed arrow on right side of panel). Primer A measures the level of plus-strand DNA initiating from DR2 (indicated with dashed arrow on left side of panel) that has successfully circularized. Primer A also detects plus-strand DNA initiated in situ from DR1 (dashed arrow labeled DR1 on left side of panel). IS, NcoI site terminus of the internal standard DNA. Each viral signal is normalized to the IS signal. The sequence ladder on the left was generated with primer A, while the sequence ladder on the right was generated with primer B.
A second set of in situ priming variants also displayed an increase in the proportion of SS DNA when examined by Southern blotting. These viruses were a related series of variants (DR series) that contained a single C-to-A change at the 3′ position of DR1 (DR1-12), DR2 (DR2-12), or both DR1 and DR2 (DRs-12). These base substitutions do not alter the specificity of RNase H cleavage during the generation of the plus-strand primer (15). When the mutation was present at DR1 (Fig. 2B, lanes 6 and 8), an increase in SS DNA was seen (Table 1). When present only at DR2, the mutation led to a smaller increase in SS DNA (Table 1). As previously described (15), the DR1-12 and DRs-12 mutants had four- to fivefold-higher levels of DL DNA than did the wild type, while the DR2-12 mutant had a nearly equivalent level (Table 1). Primer extension analysis was done on these variants (Fig. 4, lanes 2, 8, and 9 for DRs-12; data not shown for DR1-12 and DR2-12). The DR1-12 and DRs-12 variants had a greater-than-fourfold increase in in situ priming of plus-strand synthesis compared to that of the wild type (Table 2, in situ priming). The DR1-12 and DRs-12 variants had lower levels of plus-strand priming from DR2 per minus-strand DNA than that of the wild type (Table 2, priming from DR2 value). More strikingly, DR1-12 and DRs-12 had significant deficiencies in carrying out circularization (Table 2, circularization efficiency). The magnitude of the deficiency in circularization for DR1-12 and DRs-12 could account for the increase in the proportion of SS DNA in Southern blotting. Primer utilization for these mutants was nearly at wild-type levels (Table 2).
DISCUSSION
We present evidence that five variants of DHBV previously shown to have elevated levels of in situ priming also have a second cis-acting defect in the synthesis of plus-strand DNA. We found for these variants that a significant fraction of plus-strand DNA initiating from DR2 was impaired for circularization. This impairment led to an accumulation of an SS DNA intermediate at the expense of mature RC DNA. Our findings emphasize the importance of this region of the genome in the synthesis of RC DNA. A single mutation can increase in situ priming and inhibit circularization, thus reducing the level of RC DNA. In addition, at least three of the variants have a modest defect in primer utilization that made a small contribution to the observed reduction in the level of RC DNA.
Mutations in this region of the genome can reduce the synthesis of RC DNA by simultaneously affecting three aspects of plus-strand DNA synthesis: increasing the level of in situ priming, decreasing the efficiency of circularization of plus-strand DNA primed from DR2, and decreasing utilization of the plus-strand primer. Although all three aspects could be influenced simultaneously, the magnitudes of the effects differed. Since priming from DR2 predominates over priming from DR1, defects in circularization or primer utilization can have a greater influence on the level of RC DNA than an increase in in situ priming. For example, a twofold decrease in the efficiency of circularization will result in a twofold decrease in the level of RC DNA. For increased in situ priming to affect RC DNA levels to a similar extent, a ninefold increase would be necessary. Although we have not attempted to assign a rank order to the three types of defects for the variants, the defects in circularization made substantial contributions to the reduction in RC DNA levels. In addition, this analysis illustrates that describing the phenotype of a mutant virus in terms of the ratio of RC DNA to DL DNA can be inadequate and/or misleading. Our analysis emphasizes the importance of considering the proportion of SS DNA in Southern blot analyses. Also, analysis of DNA extracted from extracellular virions will yield information only about proportions of RC and DL DNA. Circularization and primer utilization defects will not be discerned, and an in situ priming phenotype might be erroneously assigned.
How these mutations affect both the site of initiation of plus-strand DNA synthesis and the efficiency of circularization of plus-strand DNA initiating from DR2 is not clear. Initiation of plus-strand synthesis and circularization are temporally distinct processes, yet our findings suggest that they could be linked mechanistically. In an alternative interpretation, distinct but overlapping cis-acting elements contribute independently to each process. Further genetic analysis of this region of the genome will be necessary to distinguish between these interpretations, which are not necessarily mutually exclusive. The nucleotide substitutions in the DR1-13, DR1-Xho, and DR1-Pvu variants lie within the boundaries of a deletion in a previously described mutant shown to suffer a significant defect in circularization (4). In this mutant, named 2549-2561, the 11 nt between DR1 and epsilon were deleted. From an earlier analysis of 2549-2561, it was proposed that a cis-acting sequence in this region of the genome, which was named 3E, made a positive contribution to the process of circularization (4). It is possible that the DR1-12, DR1-13, DR1-Xho, and DR1-Pvu variants and the 2549-2561 mutant are defective for circularization for the same reason. Interestingly, the 2549-2561 virus did not show increased in situ priming, whereas the DR1-12, DR1-13, DR1-Xho, and DR1-Pvu variants did. In addition, because the DR1-12 and DRs-12 variants have circularization defects, the circularization element apparently extends into the DR1 sequence.
How the sequences affected in the mutants described in this report normally contribute to circularization is not clear at present, but several possibilities seem tenable. The location of cis-acting sequences for circularization at the 3′ end of minus-strand DNA could provide a means to colocalize the 5′end (the donor sequence for circularization) and 3′ end (the acceptor sequence). This colocalization could occur via direct or indirect interactions between the ends of the minus strand. An example of a direct interaction would be an interaction between the P protein that is covalently attached to the 5′ end of the minus strand and the sequences near the 3′ end of minus strand. An example of an indirect interaction would be a protein, other than P, simultaneously binding to the 5′ and 3′ ends of minus-strand DNA to juxtapose the termini. Alternatively, a direct nucleic acid interaction between the ends could be envisaged. Unfortunately, none of these possibilities offers a simple explanation for why these same mutations result in increased levels of in situ priming.
ACKNOWLEDGMENTS
We thank Jeff Habig for critical reading of the manuscript, Ilse Riegel for editorial assistance, and Jesse Summers for thoughtful discussions.
This work was supported by NIH grants GM50263 and CA22443 and ACS grant JFRA-651.
REFERENCES
- 1.Calvert J, Summers J. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J Virol. 1994;68:2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condreay L D, Aldrich C E, Coates L, Mason W S, Wu T T. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem D. Hepadnaviridae and their replication, P. 2703–2737. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 4.Havert M B, Loeb D D. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J Virol. 1997;71:5336–5344. doi: 10.1128/jvi.71.7.5336-5344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lien J-M, Aldrich C A, Mason W S. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol. 1986;57:229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lien J M, Petcu D J, Aldrich C E, Mason W S. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J Virol. 1987;61:3832–3840. doi: 10.1128/jvi.61.12.3832-3840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeb D D, Hirsch R C, Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses; implication for other reverse transcribing elements. EMBOJ. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb D D, Tian R. Transfer of the minus strand of DNA during hepadnavirus replication is not invariable but prefers a specific location. J Virol. 1995;69:6886–6891. doi: 10.1128/jvi.69.11.6886-6891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb D D, Tian R, Gulya K J. Mutations within DR2 independently reduce the amount of both minus- and plus-strand DNA synthesized during hepatitis B virus replication. J Virol. 1996;70:8684–8690. doi: 10.1128/jvi.70.12.8684-8690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb D D, Gulya K J, Tian R. Sequence identity of the terminal redundancies on the minus-strand DNA template are necessary but not sufficient for the template switch during hepadnaviral plus-strand DNA synthesis. J Virol. 1997;71:152–160. doi: 10.1128/jvi.71.1.152-160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller-Hill K, Loeb D D. Previously unsuspected cis-acting sequences for DNA replication revealed by characterization of a chimeric heron/duck hepatitis B virus. J Virol. 1996;70:8310–8317. doi: 10.1128/jvi.70.12.8310-8317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seeger C, Ganem D, Varmus H E. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science. 1986;232:477–484. doi: 10.1126/science.3961490. [DOI] [PubMed] [Google Scholar]
- 14.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 15.Staprans S, Loeb D D, Ganem D. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 17.Tavis J T, Perri S, Ganem D. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G-H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W, Summers J. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W, Summers J. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J Virol. 1998;72:8710–8717. doi: 10.1128/jvi.72.11.8710-8717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y Y, Summers J. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J Virol. 2000;74:5257–5265. doi: 10.1128/jvi.74.11.5257-5265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]