Abstract
Viral initiator proteins are polypeptides that form oligomeric complexes on the origin of DNA replication (ori). These complexes carry out a multitude of functions related to initiation of DNA replication, and although many of these functions have been characterized biochemically, little is understood about how the complexes are assembled. Here we demonstrate that loss of one particular interaction, the dimerization between E1 DNA binding domains, has a severe effect on DNA replication in vivo but has surprisingly modest effects on most individual biochemical activities in vitro. We conclude that the dimer interaction is primarily required for initial recognition of ori.
Viral replication initiator proteins, such as the E1 protein from papillomaviruses and T antigen from simian virus 40, have been studied extensively, and many of the activities that are associated with these proteins have been characterized at the biochemical level (1, 8, 15). However, little information exists about which oligomeric forms are responsible for particular biochemical activities and how the initiator binding sites in ori direct complex assembly. A recently characterized interaction is the dimer interactions between the DNA binding domains (DBDs) of bovine papillomavirus E1, when the protein is bound to the origin of DNA replication (ori) (2, 5). We find that, surprisingly, mutations in the dimer interface that prevent dimerization have very modest effects on most biochemical activities of the E1 protein with the exception of ori-specific DNA binding. We know that the dimerization is important for sequence-specific DNA binding in two ways: the binding affinity is reduced for mutants that cannot bind as dimers, and the nature of binding is different because the mutation physically prevents E1 from binding as a dimer (5, 17). All the defects that we observe for the E1 A206R mutant can be explained in this context, and although the dimer interaction may be directly involved in other activities, we have failed to uncover any evidence for such an involvement. We conclude that the severe in vivo phenotype of these mutations is related to ori-specific DNA binding and that ATPase, DNA helicase, and melting and unwinding activities of E1 are not directly affected by the failure to dimerize.
In the context of the E1 DBD, mutations in the dimer interface, such as V202R and A206R, preclude binding of the DBD as a dimer to the paired sites in ori (3, 5). To determine the effects of these mutations on DNA replication in vivo, we generated the V202R and A206R mutations in the E1 expression vector pCGE1 (18). After transfection of CHO cells with an ori plasmid, an E2 expression vector (pCGE2) and expression vectors encoding either the wild-type (wt) or mutant E1, we harvested low-molecular-weight DNA on days 2, 3, and 4 as described previously (18). After linearization with the enzyme HindIII and digestion with DpnI, which cleaves unreplicated (methylated) DNA, we analyzed the resulting DNA samples using Southern blotting. wt E1 supported DNA replication at a robust level (Fig. 1A, lanes 1 to 3), while the mutant E1 V202R showed a trace amount of replicated plasmid DNA (lanes 4 to 6). With the E1 A206R mutant, no plasmid replication was detected (lanes 7 to 9), indicating that, as has been observed for human papillomavirus 11, dimerization is essential for DNA replication in vivo (17).
FIG. 1.

(A) E1 dimerization mutants are defective for DNA replication in vivo. Two point mutations (V202R and A206R) were generated in the dimerization surface in the context of the full-length E1 protein in the expression vector pCGE1 and tested for activity in a transient DNA replication assay. An ori plasmid, expression vectors for the wt or mutant E1 proteins, and an E2 expression vector were transfected into CHO cells. Low-molecular-weight DNA was harvested at 2, 3, and 4 days after transfection (indicated above the lanes), cleaved with DpnI, linearized, and analyzed by Southern blotting. The migration of replicated, DpnI-resistant plasmid DNA is indicated by an arrow. (B) E1 A206R is partially defective for in vitro DNA replication. The ability of wt E1 and E1 A206R proteins to support DNA replication in a cell-free replication system were compared. Four quantities (50, 100, 200, and 400 ng) of wt E1 (lanes 3 to 6) and E1 A206R (lanes 7 to 10) were compared in an in vitro DNA replication assay as described previously (12), and the products were analyzed by agarose gel electrophoresis. In lane 1, no E1 was added; in lane 2, no template DNA was added. The position of replication intermediates (RI) is shown. The levels of incorporation in lanes 3 to 6 were >1, 5, 59, and 32 pmol, respectively, and for lanes 7 to 10 >1, >1, 2, and 21 pmol, respectively.
We expressed and purified the E1 A206R mutant from Escherichia coli using our previously described procedures (13). The A206R mutation had no apparent effect on the expression or stability of the protein. The most comprehensive in vitro assay of replication-related activities is cell-free DNA replication, which tests all the known aspects of initiator function (16). We compared the wt and mutant E1 proteins for in vitro DNA replication activity using a mixture of S100 extract and high-salt nuclear extract from 293 cells (12) (Fig. 1B). Although the E1 A206R mutant failed to support DNA synthesis at the levels wt E1 was capable of (compare lanes 3 to 6 and 7 to 10), higher levels of the mutant E1 protein could restore DNA synthesis, and similar levels of synthesis were observed at approximately four- to eightfold-higher levels (compare lanes 4 and 5 and lanes 9 and 10). The magnitude of this effect is similar to that observed for a plasmid lacking ori where 5- to 10-fold increases in E1 levels is sufficient to restore DNA synthesis to the same levels as an ori+ plasmid (19). These results indicate that dimerization of the initiator is not essential under the conditions used for cell-free DNA replication.
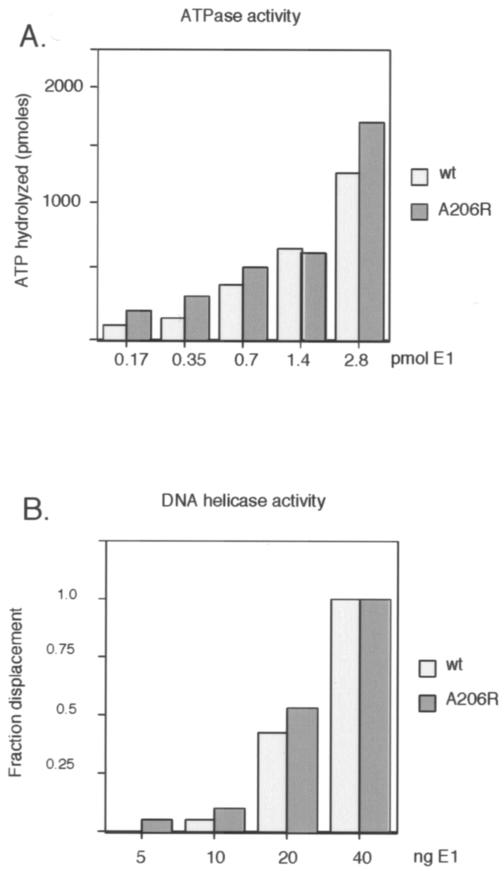
To define further the steps that may be affected by the failure of E1 to dimerize, we tested the E1 A206R mutant for ATPase activity (Fig. 2A) and nonspecific DNA helicase activity (Fig. 2B). As shown in Fig. 2A, ATPase activities at four different concentrations of E1, in the presence of single-stranded DNA (ssDNA), differed very little between the wt E1 and E1 A206R proteins. This indicates that the failure to dimerize does not affect this activity and provides assurance that the A206R mutation does not disrupt the overall folding of the protein. Similarly, in the DNA helicase assay using a partially double-stranded M13 template, the activities of the wt and mutant proteins were indistinguishable, indicating that under these conditions the dimerization defect had no impact on the ability of E1 to form the hexameric helicase.
FIG. 2.
The dimerization defect of E1 A206R does not affect the ATPase and DNA helicase activities of the E1 protein. (A) Five quantities of wt and mutant E1 (0.17, 0.35, 0.7, 1.4, and 2.8 pmol) were compared for the ability to hydrolyze ATP in the presence of ssDNA essentially as described previously (4). The reaction products were analyzed by thin-layer chromatography, and the quantity of hydrolyzed ATP was determined. (B) E1 A206R and wt E1 proteins have similar DNA helicase activities. The DNA helicase activity of E1 A206R and wt E1 were compared using a partially double-stranded M13 template as a substrate, essentially as described previously (14). A 50-mer oligonucleotide with 28 bp of complementarity to M13 was end labeled and annealed to M13 DNA, generating a substrate with a 22-nucleotide 3′ tail. Four concentrations of E1 (5, 10, 20, and 40 ng) were incubated with the substrate, and the reaction was terminated by the addition of sodium dodecyl sulfate to 0.1%. The samples were analyzed on agarose gels, and the fraction of displaced oligonucleotide was determined.
We next measured the ability of the mutant protein to melt ori DNA using a permanganate reactivity assay (11) (Fig. 3A) and the ability to unwind a double-stranded ori fragment (Fig. 3B). wt E1 gives rise to a characteristic pattern of permanganate reactivity on both flanks of the E1 binding sites (lanes 2 to 4), consistent with the patterns that have been observed previously (6, 9). The E1 A206R mutant gave rise to the same overall pattern; however, the level of reactivity was significantly lower (ca. fourfold [lanes 5 to 7]).
FIG. 3.
(A) E1 A206R shows a slight defect for ori melting. An ori probe labeled on the top strand was incubated with three concentrations (200, 400, and 800 ng) of wt E1 (lanes 2 to 4, respectively) or E1 A206R (lanes 5 to 7, respectively) in the presence of ATP and then treated with 6 mM potassium permanganate as described previously (10). (B) A206R has wt activity for unwinding in the absence of competitor DNA (−comp) but is defective in the presence of competitor DNA (+comp). wt E1 and E1 A206R were used in an ori fragment unwinding assay essentially as described previously (7). In lane 1, the probe was denatured by boiling; in lane 2, no E1 was added. In lanes 1 to 8, no competitor was present; in lanes 9 to 14, 10 ng of poly(dI-dC) (∼100-fold excess over probe) was added prior to the addition of protein. The samples were separated on 11% acrylamide gels (acrylamide:bis ratio of 29:1). The positions of ssDNA (ss) and double-stranded DNA (ds) are shown to the right.
We also tested the E1 A206R mutant protein for its ability to unwind a DNA fragment containing the origin of DNA replication. This unwinding assay requires sequence-specific DNA binding. We observed a very slight difference (less than twofold) between the wt protein and the mutant protein consistent with the results from the permanganate reactivity assay (Fig. 3B, compare lanes 3 to 5 and 6 to 8). We also performed unwinding assays under conditions where we introduced nonspecific competitor DNA together with the probe. Addition of 2 ng of competitor DNA [poly(dI-dC)] affected unwinding by wt E1 to a minimal extent (50% unwinding in lane 5 compared to 48% unwinding in lane 11) but affected unwinding by the E1 A206R mutant to a greater extent (30% unwinding in lane 8 compared to 16% unwinding in lane 14), indicating that dimerization contributes to DNA binding specificity. A distinction between these assays and the helicase assays is that the melting and unwinding processes are dependent on sequence-specific DNA binding, while for the helicase assay no such requirement exists.
These results indicated that the defect of the E1 A206R mutant might be related to ori-specific DNA binding. We therefore compared ori binding of wt E1 and E1 A206R proteins in a gel shift assay in the absence and presence of nonspecific competitor DNA (Fig. 4A to C). In the absence of competitor DNA, the wt and mutant E1 proteins had similar DNA binding activities (compare lanes 2 to 6 in Fig. 4A with lanes 2 to 6 in Fig. 4B). The presence of competitor DNA reduced binding for both wt and mutant E1; however, binding of the mutant E1 showed a greater reduction (compare lanes 7 to 11 in Fig. 4A to lanes 7 to 11 in Fig. 4B) especially at low concentrations of protein, indicating that E1 A206R binds ori with lower specificity than wt E1.
FIG. 4.
E1 A206R binds ori with lower specificity than wt E1. (A) Five concentrations of E1 (1, 2, 4, 8, and 16 ng) were used in a DNA binding assay (13)with an ori fragment as the probe in the absence (lanes 1 to 6) or presence (lanes 7 to 11) of 2 ng of nonspecific competitor DNA [poly(dI-dC)] (+ Comp). In lane 1, no E1 was added. (B) Five concentrations of E1 A206R was used in the absence (lanes 2 to 6) or presence (lanes 7 to 11) of nonspecific competitor DNA. In lane 1, no E1 was added. (C) The results from the electrophoretic mobility shift assay were quantitated, and the level of binding was plotted as a function of the concentration of E1 for wt E1 and for the E1 A206R mutant protein.
Here we demonstrate that a severe defect in DNA replication results from the disruption of the DBD dimer interface. Interestingly, in the biochemical assays, we observe only modest defects for the dimerization mutant. The most severe defect is observed in in vitro DNA replication assays, where the effect is ∼10-fold. In the other assays, only under conditions where sequence-specific DNA binding is challenged, can we observe a significant difference between wt E1 and E1 A206R proteins. A simple explanation for this observation is that under the in vitro conditions, DNA binding specificity is not challenged. In contrast, under in vivo conditions, the presence of host DNA requires a higher degree of sequence-specific DNA binding, which in turn requires dimerization, consistent with the observations of Titolo et al. (17). These results indicate that the severe defect that we observe for DNA replication in vivo most likely corresponds to a failure of the E1 A206R mutant protein to carry out the first step in initiation, i.e., recognition of ori.
Acknowledgments
This work was supported by a grant from NIH (CA 13106) to A.S.
REFERENCES
- 1.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 2.Chen, G., and A. Stenlund. 1998. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J. Virol. 72:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, G., and A. Stenlund. 2001. The E1 initiator recognizes multiple overlapping sites in the papillomavirus origin of DNA replication. J. Virol. 75:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellison, V., and B. Stillman. 1998. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J. Biol. Chem. 273:5979-5987. [DOI] [PubMed] [Google Scholar]
- 5.Enemark, E. J., G. Chen, D. E. Vaughn, A. Stenlund, and L. Joshua-Tor. 2000. Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol. Cell 6:149-158. [PubMed] [Google Scholar]
- 6.Gillette, T. G., M. Lusky, and J. A. Borowiec. 1994. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc. Natl. Acad. Sci. USA 91:8846-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 8.Lin, B. Y., A. M. Makhov, J. D. Griffith, T. R. Broker, and L. T. Chow. 2002. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol. Cell. Biol. 22:6592-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders, C. M., and A. Stenlund. 1998. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 17:7044-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders, C. M., and A. Stenlund. 2000. Transcription factor-dependent loading of the E1 initiator reveals modular assembly of the papillomavirus origin melting complex. J. Biol. Chem. 275:3522-3534. [DOI] [PubMed] [Google Scholar]
- 11.Sasse-Dwight, S., and J. D. Gralla. 1989. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 264:8074-8081. [PubMed] [Google Scholar]
- 12.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedman, T., J. Sedman, and A. Stenlund. 1997. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J. Virol. 71:2887-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seo, Y. S., and J. Hurwitz. 1993. Isolation of helicase alpha, a DNA helicase from HeLa cells stimulated by a fork structure and signal-stranded DNA-binding proteins. J. Biol. Chem. 268:10282-10295. [PubMed] [Google Scholar]
- 15.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 16.Stillman, B. 1989. Initiation of eukaryotic DNA replication in vitro. Annu. Rev. Cell Biol. 5:197-245. [DOI] [PubMed] [Google Scholar]
- 17.Titolo, S., K. Brault, J. Majewski, P. W. White, and J. Archambault. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus E1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]