RNA-dependent RNA replication is a special process reserved exclusively for RNA viruses but not cellular RNAs. Almost all RNA viruses (except retroviruses) undergo RNA-dependent RNA replication by a virus-encoded RNA-dependent RNA polymerase (RdRP), which specifically replicates the viral RNA genome. Satellite viral RNAs do not encode their own polymerase but rely on the RdRP from the coexisting helper virus for their replication. Hepatitis delta virus (HDV) and plant viroids present an exception which still confounds the conventional thinking. None of them encode an RdRP, and yet they can undergo robust RNA replication autonomously once inside the cells. It is intuitive that they have to replicate their RNA genome using a cellular enzyme. However, unlike plants and lower animal species, which encode RdRPs that are putatively involved in gene silencing (11, 13, 57, 58), no mammalian cells have been shown to encode any RdRP or its equivalent. Nevertheless, the RNA interference phenomena suggest the possibility that RNA-templated RNA amplification may also take place in the mammalian cells (61). Thus, viroids and HDV present both a challenge and an opportunity for understanding a potentially physiological flow of genetic information from RNA to RNA in the mammalian cells. Viroids and HDV differ in that viroids do not encode any protein, whereas HDV encodes a protein (hepatitis delta antigen [HDAg]) which is intimately involved in RNA replication. In addition, HDV RNA not only has to replicate itself but also needs to transcribe a subgenomic mRNA species that encodes HDAg. The transcription of the HDAg-encoding mRNA has all of the hallmarks of the conventional mRNA transcription in the cells except for the nature of the template (DNA versus RNA). Therefore, HDV represents a hybrid of the conventional DNA-dependent transcription and the unique RNA-dependent RNA synthesis in the absence of an RdRP.
HDV causes chronic, and occasionally fulminant, hepatitis (20). It is always associated with hepatitis B virus (HBV) infection because HDV cannot form virus particles on its own (54). It was prevalent worldwide, with a particularly high prevalence rate in the Mediterranean countries. In recent years, the incidence of new HDV infection has precipitously declined, partially due to HBV vaccination. Nevertheless, the scientific mystery surrounding HDV replication thickens.
STRUCTURE OF HDV RNA AND HDAg
HDV contains a circular RNA genome of 1.7 kilobases which can replicate on its own but requires HBV as the helper virus to supply HBV surface antigen for the production of virus particles. The genome sequence is nearly self-complementary such that the viral RNA forms a rodlike double-stranded RNA structure under the native conditions (29, 70). As such, HDV RNA is very similar to viroid RNAs; indeed, part of the HDV RNA sequence is evolutionarily related to viroids (15). However, HDV RNA is 3 to 4 times longer than the typical viroid, with the additional sequences being attributed to a coding sequence for HDAg on the complementary (antigenomic sense) strand. HDV RNA contains ribozyme activities on both the genomic and antigenomic strands, which cleave the respective RNA strand in cis (73). This is another feature shared between HDV RNA and viroids. Although HDV RNA assumes a rodlike structure under the native conditions, it undergoes dynamic conformational changes under different conditions. For example, the RNA sequence around the ribozyme domain forms several helices to assume the ribozyme-active conformation (16). Presumably, HDV RNA structure also changes to other conformations during RNA replication and editing (see below). The regulatory mechanisms for controlling RNA conformation are likely crucial for the biology of HDV replication.
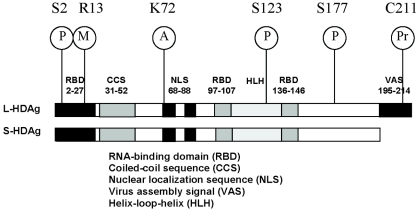
The production of HDAg is a unique feature that distinguishes HDV from viroids. HDAg consists of two distinct species, the small form (S-HDAg; 195 amino acids, 24 kDa) and the large form (L-HDAg; 214 amino acids, 27 kDa) (31). The difference between the two species resides in the extra 19 amino acids at the C terminus of the large form. These two HDAg forms share several functional domains, including RNA-binding motifs, a nuclear localization signal, a coiled-coil domain, a helix-loop-helix motif, and a C-terminal stretch of proline- and glycine-rich sequence (Fig. 1). L-HDAg contains an additional 19 amino acids at the very end, which consists of a stretch of variable but genotype-specific, membrane-attaching sequence and serves as the virion assembly signal (33). S-HDAg is required for HDV RNA replication (30), whereas L-HDAg is required for virion assembly (7).
FIG. 1.
The functional domains and sites of posttranslational modifications of HDAg. P, phosphorylation; M, methylation; A, acetylation; Pr, prenylation.
HDAg is modified posttranslationally by phosphorylation (8, 51), acetylation (50), and methylation (34) and, in the case of L-HDAg, isoprenylation (19). Methylation of Arg-13, acetylation of Lys-72, and phosphorylation of Ser-177 and Ser-123 have been reported to affect the subcellular localization of HDAg and RNA replication (26, 34, 50, 65). Most of these modifications are important for the functions of S-HDAg in HDV RNA replication. Isoprenylation of L-HDAg is required for virus assembly (19).
AN OVERVIEW OF THE HDV REPLICATION CYCLE
In natural infection, HDV enters cells through a cellular receptor shared with HBV, since HDV incorporates HBV surface antigen as its envelope protein. The identity of this receptor is still not known. The genomic RNA (considered a negative-strand RNA by virtue of the fact that the HDAg-coding sequence is present on the antigenomic strand) is then replicated into the antigenomic strand. Because the cell culture and animal systems for HDV infection (primary hepatocytes and woodchucks, chimpanzees, or mice) are inefficient or cumbersome, most studies of the HDV replication cycle have been carried out by transfecting cultured cells with HDV cDNA of more than the length of a single genome (a dimer, a trimer, or a minimum of 1.2 times [1.2×] the genomic length). A functional S-HDAg, which is generated from the mRNA transcript of the transfected HDV cDNA or, alternatively, provided in trans from a plasmid, is required for the initiation of HDV RNA replication. It should be noted that this experimental approach introduces an artificial requirement of a DNA-dependent transcription step in order to generate a precursor HDV RNA, which, in turn, leads to subsequent RNA replication. An alternative approach involves the transfection of the 1.2×-genomic-length HDV RNA together with an mRNA encoding HDAg (46); this approach circumvents the need for the cDNA step and thus avoids the artificial requirement of the DNA-dependent transcription. The development of this RNA transfection method has led to very significant revisions of the previous concepts of HDV RNA replication, which were based largely on the cDNA transfection methods.
In the cells undergoing HDV RNA replication, both the genomic and antigenomic strands of HDV RNA are detected. Both RNA strands exist mainly as monomers, but dimers, trimers, and higher oligomers are also detected. The detection of these RNA species, similar to the corresponding RNA species of viroids, led to the proposal that HDV RNA replication occurs via a rolling-circle mechanism (4). But the definitive proof of this mechanism was not available until recently, when the metabolic labeling and kinetic tracing studies of HDV RNA intermediates finally proved that the dimer and higher oligomer species are precursors to the monomers (40, 43). The cleavage of the RNA products is carried out by the ribozyme inherent in these RNA molecules (28). Site-specific mutagenesis studies of the cleavage sites led to the conclusion that both the genomic and the antigenomic ribozyme activities are required for successful HDV RNA replication (44). Therefore, HDV RNA replication likely occurs by a double-rolling-circle mechanism involving both genomic and antigenomic strands (Fig. 2). This model implies that, once RNA replication is initiated, it can theoretically continue to elongate without the need to be reinitiated. The monomer RNA species consists of both linear and circular forms. The circular form is the functional RNA template, but the linear form can also be used for replication (21). The circularization of HDV RNA was initially thought to be carried out by a ribozyme (a reverse form of RNA cleavage) (59), but recent studies have suggested that it may be mediated by cellular ligases (53).
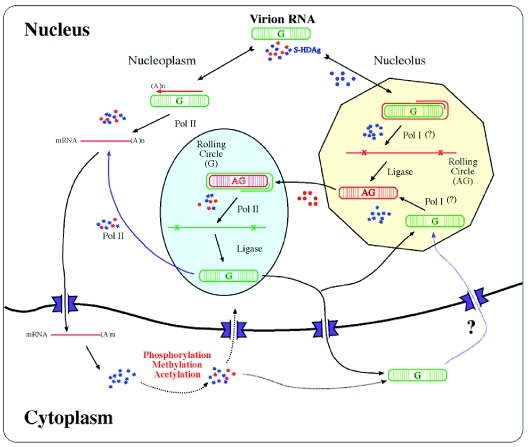
FIG. 2.
A hypothetical model of the HDV RNA replication cycle. The viral genome is first transported via the unmodified HDAg into the nucleolus, where the antigenomic (AG) RNA is synthesized by a rolling-circle mechanism. This step does not require HDAg modification (blue stars represent unmodified HDAg, and red stars represent modified HDAg). The responsible enzyme is postulated to be pol I. The AG RNA is transported via the modified HDAg to a nucleoplasmic organelle for the synthesis of genomic (G) RNA. This transport and the subsequent rolling-circle replication of the G RNA require the modified HDAg. Some viral RNA is transported to an unidentified site for transcription of the mRNA, which is then exported to the cytoplasm for translating HDAg. The HDAg requirement of mRNA transcription is still not clear. The newly synthesized HDAg is modified in the cytoplasm. The G RNA is transported back to the nucleolus, and some of it is transported to the cytoplasm. The fate of the G RNA in the cytoplasm is still not known; some may be transported back to the nucleus. The natures of the subnuclear compartments housing the synthesis of the various RNA species are not certain, although it is hypothesized that antigenomic RNA synthesis occurs in the nucleolus.
In addition to the monomer and multimer RNA species, there is an mRNA species of 0.8 kb which represents the HDAg-coding sequence on the antigenomic strand (10). This is the RNA species that serves as the functional mRNA for translating HDAg (35). This RNA has all the hallmarks of mRNA (e.g., it is capped and polyadenylated) and is associated with polysomes. Thus, distinct from plant viroids, HDV must carry out mRNA transcription in concert with the replication of the genomic and antigenomic RNA. A significant new understanding in this regard is that this mRNA is transcribed continuously throughout the replication cycle (46), in contrast to the previous proposal that it is transcribed only at the beginning of RNA replication (23, 67).
During the late stage of HDV RNA replication, an editing event takes place on the antigenomic strand (6, 37), and this event converts the amber termination codon of the S-HDAg reading frame to a trp-coding sequence, thus extending the reading frame for an additional 19 amino acids to encode L-HDAg. Since L-HDAg is required for virus assembly, the editing enables the production of virus particles. The editing is carried out by a cellular double-stranded RNA-adenosine deaminase (72).
TRANSCRIPTION VERSUS REPLICATION
The first event in the HDV replication cycle is probably the transcription of the 0.8-kb mRNA species, since this mRNA is necessary for encoding S-HDAg, which, in turn, is required for the initiation of HDV RNA replication. How does the mRNA transcription fit into the replication strategy of HDV RNA, which uses a rolling-circle mechanism? The previous proposal from Taylor's laboratory was that a poly(A) addition signal present in the HDV antigenomic RNA causes the interruption of rolling-circle replication, generating a poly(A)-tailed mRNA. As the replication proceeds, the poly(A) addition signal is suppressed by the very protein product (HDAg) of this mRNA species, thus allowing the continuation of rolling-circle RNA replication beyond the poly(A) addition signal, generating multimer RNA species (23, 67). According to this model, the mRNA species would be synthesized only early in the RNA replication cycle. However, this hypothesis has two problems. First, the suppression of poly(A) addition by HDAg was demonstrated only in DNA-dependent transcription; it has not been proven that this will also occur in RNA-templated transcription. Second, the mRNA would be produced only at the beginning of RNA replication; this is conceptually problematic because HDAg needs to be continually synthesized throughout the replication cycle. In particular, after the RNA editing has occurred, an mRNA encoding L-HDAg has to be transcribed anew. This would not be possible with this hypothesis. Instead, a more recent study showed that the amount of the 0.8-kb mRNA was maintained, after the initial period of steady increase, at the same level throughout the HDV replication cycle (46). These results suggest that the transcription of the mRNA and the replication of the HDV genome are independent processes and occur concurrently. However, there is also a conceptual difficulty with this new model; namely, how are mRNA transcription and RNA replication coordinated on the same RNA template (genomic strand) inasmuch as the replication process has to go past the poly(A) addition signal? This dilemma can be resolved by a recent finding; namely, the transcription of the mRNA and replication of the genomic RNA into antigenomic RNA may be carried out by different polymerases and probably in different subnuclear compartments (see below), thus physically and biochemically separating these two functions. From the limited amount of available information, it appears that the metabolic requirements of mRNA transcription and those of antigenomic RNA synthesis are significantly different (Table 1).
TABLE 1.
Comparison of the genomic and antigenomic RNA syntheses and mRNA transcriptiona
| Characteristic | Genomic synthesis (AG to G) | Antigenomic synthesis (G to AG) | mRNA transcription |
|---|---|---|---|
| Sensitivity to α-amanitin | Sensitive (inhibited at 1-5 μg/ml) | Resistant (inhibited at >100 μg/ml) | Sensitive (inhibited at 1-5 μg/ml) |
| S-HDAg requirement | |||
| S-177 phosphorylation | Yes | No | ? |
| R-13 methylation | Yes | No | ? |
| K-72 acetylation | Yes | ? | ? |
| Assisted by recombinant S-HDAg from E. coli | No | Yes | ? |
| Cytoplasmic transport after synthesis | Yes | No | Yes |
| Inhibition by L-HDAg | Yes | No | ? |
| Polymerases | pol II | pol I (?) | pol II |
| Site of synthesis | Nucleoplasm | Nucleolus | Nucleoplasm |
AG, antigenomic RNA; G, genomic RNA.
The initiation site of the antigenomic RNA synthesis has been determined to be near one end of the rod structure of HDV RNA (22, 46), where the putative promoter for HDV RNA synthesis is located (1). However, whether the initiation site of antigenomic RNA synthesis is identical to that of mRNA transcription is not known. Also, the initiation point of the genomic RNA synthesis is not yet known.
REPLICATION OF GENOMIC VERSUS ANTIGENOMIC RNA
There is an overabundance of the genomic RNA strand compared to the antigenomic strand in the cells replicating HDV RNA (the genomic RNA strand is approximately 30-fold more abundant) (10), indicating that the genomic RNA is more robustly synthesized. Thus, the majority of HDV RNA synthesis detected in the cells under the steady-state condition represents the synthesis of the genomic RNA strand. There must be regulatory mechanisms to differentially enable the synthesis of the two RNA strands. Both genomic and antigenomic RNAs consist of similar RNA intermediates (dimers and longer multimers) and similar ratios of circular and linear RNA molecules, indicating that both strands are made by a similar rolling-circle mechanism. However, the metabolic requirements for the synthesis of the two strands are different (Table 1). Genomic RNA synthesis (from the antigenomic template) is sensitive to a low concentration of α-amanitin (1 μg/ml), whereas the antigenomic RNA synthesis (from the genomic template) is surprisingly resistant to a very high concentration (100 μg/ml) of the drug (by comparison, the transcription of the 0.8-kb mRNA is also sensitive to the low concentration of α-amanitin) (40, 43, 47). The genomic RNA synthesis requires an S-HDAg that is phosphorylated and methylated, whereas the antigenomic RNA synthesis can be mediated by an unmethylated S-HDAg (34, 49, 60). Certain phosphorylation-defective mutants (S177A) and methylation-defective mutants (R13A) of HDAg can mediate the antigenomic RNA synthesis but not the genomic RNA synthesis. Correspondingly, antigenomic RNA, but not genomic RNA, synthesis can be mediated by the recombinant S-HDAg derived from Escherichia coli, which is most likely not modified (60). The genomic RNA synthesis is inhibited by L-HDAg when the latter is expressed at the beginning of the replication cycle, whereas the antigenomic RNA synthesis is not inhibited (45). Furthermore, the genomic RNA is exported to the cytoplasm immediately after its synthesis, whereas the antigenomic RNA is not (40), suggesting that these two RNA strands are associated with different transcription machineries which have different RNA-exporting capabilities. Also, certain site-specific mutations of HDV RNA sequence affect the synthesis of one RNA strand but not the other (69), indicating that the syntheses of these two strands are differentially regulated.
It should be noted that both genomic and antigenomic RNA strands appear to be synthesized continuously during the entire replication cycle. Unlike the case for the replication of most other RNA viruses, there is no temporal switch between the syntheses of the two RNA strands in the HDV replication cycle. Therefore, their syntheses likely occur concurrently but separately.
THE ROLE OF HDAg IN HDV RNA REPLICATION
S-HDAg is absolutely required for HDV RNA replication. Site-specific mutations on most sites in the HDAg-coding sequences inhibit the ability of HDV RNA to replicate; the defects can be complemented in trans by a wild-type S-HDAg. S-HDAg is required throughout the replication cycle, being involved in both the initiation and the elongation (maintenance) of HDV RNA replication (unpublished observations). This fact suggests that S-HDAg is likely a component of the RNA synthetic machinery, in addition to its role as a transporter of HDV RNA to the site of RNA replication. All of the known functional domains of HDAg, including phosphorylation, acetylation, and methylation sites, the coiled-coil domain, RNA-binding motifs, and the C-terminal domain, are important for HDV RNA replication. Certain modifications of HDAg affect genomic or antigenomic RNA syntheses differentially.
HDAg has many features reminiscent of a transcription factor: it is a nuclear protein and contains coiled-coil and helix-loop-helix domains and a stretch of proline- and glycine-rich sequence at the C terminus. It is both acetylated and methylated, very much like many DNA-dependent transcription factors. It shares some sequence similarity with transcription elongation factor NELF-A (the subunit A of negative elongation factor) (76). Furthermore, HDAg binds to DNA-dependent RNA polymerase II (pol II). HDAg has also been shown to bind several other cellular factors, including some pol II transcription factors, such as YY1 (Y. H. Wu-Lee, personal communication), and nucleolar proteins, such as B23, and to colocalize with nucleolin (32). But HDAg has also been shown to localize in other parts of the nucleus (3, 74). During active HDV RNA replication, it tends to form speckle structures in the nucleus (3). In a cell-free transcription system, in which pol II mediates transcription initiation from an HDV RNA fragment, S-HDAg promotes RNA elongation (76). In this regard, HDAg serves as a transcription elongation factor, a function very similar to that of Tat of human immunodeficiency virus (75). It also has similar activities in DNA-templated, pol II-mediated transcription in vitro. However, the role of HDAg in the initiation of HDV RNA replication has not been directly demonstrated. HDAg has also been shown to have a very limited sequence similarity to a novel cellular protein, DIPA (5), and the expression of DIPA was shown to affect HDV RNA replication; however, the nature and significance of this protein has so far not been characterized. In sum, HDAg interacts with several pol II transcription factors and nucleolar proteins, raising the possibility that HDAg may be associated with cellular pol I and pol II transcription machineries.
L-HDAg has been shown to inhibit HDV RNA replication when it is expressed, even in a small amount relative to S-HDAg, at the beginning of HDV replication (9). Theoretically, this property would make it impossible for HDV to initiate replication in natural infection, since the viral RNA is complexed with almost equal amounts of L- and S-HDAg in the virion (55). Fortunately for the virus, L-HDAg inhibits only genomic, not antigenomic, RNA synthesis (45), thus allowing at least the initial round of viral RNA replication at the beginning of infection. Whether L-HDAg inhibits HDV RNA replication late in the replication cycle of HDV and thus regulates the steady-state level of viral RNA is still under debate. It has been shown that the presence or absence of L-HDAg did not affect the steady-state level of HDV RNA late in the viral replication cycle (41). However, excessive RNA editing was shown to result in the inhibition of RNA replication of some HDV genotypes (27, 56). It is not clear whether this inhibition was caused by L-HDAg per se or the editing-induced hypermutations of the HDV genome (42). Regardless of whether L-HDAg does or does not inhibit HDV RNA replication, its role in HDV RNA replication is of considerable interest, as L-HDAg is usually colocalized with S-HDAg (12) and has been found to be in the PML body of the nucleus (2), which is an active transcription site in the nucleus and may be near the site of HDV RNA replication (unpublished observations). Curiously, L-HDAg can activate a variety of transcription promoters (DNA-templated transcription) in a contransfection assay (71), although the functional significance of this finding is currently unknown.
ENZYMOLOGY OF HDV RNA REPLICATION: POL I AND POL II
It has long been assumed that the replication of HDV RNA, similar to that of plant viroids, is mediated by cellular polymerases. However, the evidence for this hypothesis is still very tenuous. For plant viroids, it has not been resolved whether the plant RdRP, pol II, or even pol I is involved in this process. For HDV replication, it was first suggested that pol II carries out HDV RNA replication based on a nuclear run-on experiment which showed that HDV RNA synthesis could be inhibited by a low concentration of α-amanitin (1 to 5 μg/ml) (39). The most dramatic report came from Taylor's laboratory and showed that hepatocyte nuclear extracts were able to replicate the 1.7-kb genomic and antigenomic RNA faithfully in vitro without any extraneous factors (including HDAg) (18). Furthermore, the reaction could be inhibited by an antibody against pol II and by α-amanitin. However, this claim is inconsistent with the rolling-circle mechanism, and this report has never been reproduced. Another report demonstrated that pol II was able to use a partial HDV antigenomic-sense RNA hairpin fragment as a template for an elongation reaction (elongation from the 3′ end of a cleavage fragment of the template) in vitro (17). Significantly, the elongation reaction was stimulated by S-HDAg (76). S-HDAg was also able to promote elongation in a DNA-dependent RNA transcription reaction. However, it is not clear whether such an elongation reaction (creating a genomic-antigenomic RNA chimeric molecule) is a biologically relevant process in HDV replication. Nevertheless, this was the most convincing evidence so far showing the capability of pol II in replicating HDV RNA in some fashion. Both S- and L-HDAg have also been shown to affect the cellular pol II-mediated, DNA-dependent RNA transcription either positively or negatively (36, 71). This effect further suggests the intimate association of HDAg with the cellular transcription machineries.
The evidence that HDV RNA replication in the cells is mediated by pol II came from a study of the effects of long-term α-amanitin treatment in a hepatocyte cell line after HDV RNA transfection (47). This study showed that HDV mRNA transcription and RNA replication could be inhibited by α-amanitin at 1 to 5 μg/ml, consistent with the α-amanitin sensitivity of pol II (39, 47). Most significantly, when HDV RNA replication in a cell line expressing an α-amanitin-resistant pol II was carried out, HDV RNA replication became resistant to α-amanitin (47). These data indicate strongly that pol II mediates HDV RNA replication. Unexpectedly, these studies also showed that the syntheses of the genomic and antigenomic strands have different sensitivities to α-amanitin. The genomic RNA synthesis (from the antigenomic RNA template), similar to mRNA transcription (from the genomic RNA template), is exquisitely sensitive to α-amanitin (1 to 5 μg/ml); in contrast, the antigenomic RNA strand synthesis (from the genomic RNA template) is surprisingly resistant (inhibited at >100 μg/ml) (40, 43, 47). The latter finding suggests the involvement of other polymerases, such as pol I. Alternatively, the antigenomic RNA synthesis may occur in a compartment not permeable to α-amanitin. In either case, there appear to be separate transcription machineries for genomic and antigenomic RNA syntheses. This interpretation is further supported by the other differences in metabolic requirements between the syntheses of these two strands (Table 1). It should be noted that there was another recent report claiming that there was no evidence for the involvement of polymerases other than pol II in HDV RNA replication (48). However, that report actually examined only the genomic RNA synthesis. Thus, the evidence so far supports the novel idea that the genomic and antigenomic HDV RNA syntheses are carried out separately in different transcription machineries. For viroid RNA replication, there was also an earlier report suggesting that pol I and pol II mediate the synthesis of the separate RNA strands of potato spindle tuber viroid (62).
The involvement of two separate transcription machineries in genomic and antigenomic RNA syntheses is consistent with the fates of these two RNA strands. The genomic RNA is exported to the cytoplasm immediately after synthesis, whereas the antigenomic RNA is retained in the nucleus (40). The pol II transcription machinery, which makes HDV genomic RNA, is known to be coupled to the nuclear export machinery (14, 52). In contrast, HDV antigenomic RNA synthesis is postulated to occur in the nucleolus, where rRNA synthesis takes place; correspondingly, the antigenomic RNA, similar to rRNA, is not immediately transported to the cytoplasm. Furthermore, HDAg binds not only pol II transcription factors but also nucleolar proteins (24). HDAg has been found in both the nucleoplasm and the nucleolus (74). Most of the published studies examining HDV RNA replication in the cells are clouded by the fact that the genomic RNA synthesis is at least 10 times more robust than the antigenomic RNA synthesis; therefore, previous studies examining total HDV RNA synthesis in the cells may have overlooked the antigenomic RNA synthesis. More recent studies using BrUTP labeling have localized the newly synthesized genomic RNA in the nucleoplasm (near the PML body) and the antigenomic RNA near the nucleolus (Y.-J. Li and M. M. C. Lai, unpublished observation).
If HDV RNA synthesis is mediated by DNA-dependent RNA polymerases, how do these enzymes recognize RNA as a template? Conceivably, HDAg may convert the structure of HDV RNA (double-stranded form?) into the double-strand DNA conformation. HDAg binds to HDV RNA and serves as an RNA chaperone to alter the RNA conformation (25). Interestingly, the double-stranded cDNA of HDV RNA contains a bidirectional promoter activity for pol II (38, 64); this promoter coincides very closely with the promoter determined for the natural HDV RNA (1), further supporting the interchangeability of the DNA-RNA forms of the HDV genome. This promoter is localized to cDNA sequences corresponding to the end of the rodlike structure of HDV RNA. Another possibility is that HDAg may serve as a critical activator or coactivator in the HDV RNA transcription complex, conferring template specificity to either pol I or pol II for recognition of HDV RNA. This possibility is consistent with the finding that HDAg interacts with pol II (76) and possibly other transcription factors.
Another interesting issue is how HDV RNA is differentially transported to the different subnuclear compartments to carry out different replication and transcription functions. The different modified forms of HDAg may carry out these diverse functions (34, 50, 65). Significantly, the methylation-defective mutant of S-HDAg failed to aggregate in the speckle structure in the nucleus (34); correspondingly, the genomic RNA synthesis, which is postulated to occur in the nucleoplasm, failed to take place. In contrast, the antigenomic RNA synthesis, which is postulated to take place in the nucleolus, can be mediated by this methylation-defective mutant (34). Thus, there is a correlation between the HDAg localization and the replication of the separate HDV RNA strands. In addition, certain HDV RNA sequences may also determine the fate of the RNA, as HDV RNA can be shuttled in the absence of HDAg (66). Finally, another intriguing question is whether HDV RNA replication involves a cytoplasmic phase. The genomic RNA, but not antigenomic RNA, is exported to the cytoplasm right after synthesis (40). This fact is consistent with the eventual packaging of the genomic RNA into virions. However, the RNA export happens well before the virus assembly takes place. Also, HDV RNA can go back into the nucleus (66). Whether certain steps of HDV replication cycle, e.g., RNA ligation, take place in the cytoplasm and whether shuttling of HDV RNA between the cytoplasm and the nucleus is necessary for successful RNA replication are important questions awaiting answers.
PERSPECTIVES
HDV RNA replication represents a new facet of cellular transcription machinery. HDV relies on cellular DNA-dependent RNA polymerases and their transcription machineries, in conjunction with a viral protein, HDAg, to carry out RNA-dependent RNA replication. Many aspects of this replication process are yet to be elucidated; chief among them is the nature of the enzymes involved. Pol II has been suspected for a long time, but pol I has now been implicated in the antigenomic RNA synthesis. There is still a remote possibility that a previously unidentified RNA polymerase is responsible for these syntheses, although the human genome does not contain the RdRP orthologs (63). Regardless, the ability of cells to replicate an exogenous HDV RNA without an exogenous polymerase implies that mammalian cells have an innate capacity to replicate other exogenous RNA or their own cellular RNA. RNA-dependent RNA synthesis in certain mammalian cells has been documented on several occasions (68). Recent small interfering RNA studies further suggest that certain RNA species may be amplified by RNA-dependent RNA replication in the cells. HDV RNA replication thus provides a potential test model for such an unorthodox and unexplored frontier of human cells.
Acknowledgments
I thank Tom Macnaughton for help in preparing figures and Stephanie Chang for editorial assistance.
REFERENCES
- 1.Beard, M. R., T. B. Macnaughton, and E. J. Gowans. 1996. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J. Virol. 70:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, P., R. Brazas, D. Ganem, and G. G. Maul. 2000. Hepatitis delta virus replication generates complexes of large hepatitis delta antigen and antigenomic RNA that affiliate with and alter nuclear domain 10. J. Virol. 74:5329-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bichko, V. V., and J. M. Taylor. 1996. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J. Virol. 70:8064-8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNAs. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 5.Brazas, R., and D. Ganem. 1996. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science 274:90-94. [DOI] [PubMed] [Google Scholar]
- 6.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: Specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, F.-L., P.-J. Chen, S.-J. Tu, C.-J. Wang, and D.-S. Chen. 1991. The large form of hepatitis δ antigen is crucial for assembly of hepatitis δ virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, M.-F., S. C. Baker, L. H. Soe, T. Kamahora, J. G. Keck, S. Makino, S. Govindarajan, and M. M. C. Lai. 1988. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 62:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao, M., S.-Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, P.-J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. L. Gerin, and J. Taylor. 1986. Structure and replication of the genome of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cogoni, C., and G. Macino. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166-169. [DOI] [PubMed] [Google Scholar]
- 12.Cunha, C., J. Monjardino, D. Chang, S. Krause, and M. Carmo-Fonseca. 1998. Localization of hepatitis delta virus RNA in the nucleus of human cells. RNA 4:680-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543-553. [DOI] [PubMed] [Google Scholar]
- 14.Dimaano, C., and K. S. Ullman. 2004. Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 24:3069-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elena, S. F., J. Dopazo, R. Flores, T. O. Diener, and A. Moya. 1991. Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis δ virus RNA. Proc. Natl. Acad. Sci. USA 88:5631-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferre-D'Amare, A. R., K. Zhou, and J. A. Doudna. 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395:567-574. [DOI] [PubMed] [Google Scholar]
- 17.Filipovska, J., and M. M. Konarska. 2000. Specific HDV RNA-templated transcription by pol II in vitro. RNA 6:41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, T.-B., and J. Taylor. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 67:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glenn, J. S., J. A. Watson, C. M. Havel, and J. M. White. 1992. Identification of a prenylation site in delta virus large antigen. Science 256:1331-1333. [DOI] [PubMed] [Google Scholar]
- 20.Govindarajan, S., K. P. Chin, A. G. Redeker, and R. L. Peters. 1984. Fulminant B viral hepatitis: role of delta agent. Gastroenterology 86:1416-1420. [PubMed] [Google Scholar]
- 21.Gudima, S. O., J. Chang, and J. M. Taylor. 2004. Features affecting the ability of hepatitis delta virus RNAs to initiate RNA-directed RNA synthesis. J. Virol. 78:5737-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh, S.-Y., M. Chao, L. Coates, and J. Taylor. 1990. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J. Virol. 64:3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh, S.-Y., and J. Taylor. 1991. Regulation of polyadenylation of hepatitis delta virus antigenomic RNA. J. Virol. 65:6438-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, W. H., B. Y. Yung, W. J. Syu, and Y. H. Lee. 2001. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 276:25166-25175. [DOI] [PubMed] [Google Scholar]
- 25.Huang, Z. S., and H. N. Wu. 1998. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 273:26455-26461. [DOI] [PubMed] [Google Scholar]
- 26.Hwang, S. B., and M. M. C. Lai. 1994. Isoprenylation masks a conformational epitope and enhances trans-dominant function of the large hepatitis delta antigen. J. Virol. 68:2958-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayan, G. C., and J. L. Casey. 2002. Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 76:3819-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeng, K. S., A. Daniel, and M. M. C. Lai. 1996. A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J. Virol. 70:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Merde, and H. Schellekens. 1986. The hepatitis delta (δ) virus possesses a circular RNA. Nature 323:558-560. [DOI] [PubMed] [Google Scholar]
- 30.Kuo, M. Y.-P., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai, M. M. C. 1995. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 64:259-286. [DOI] [PubMed] [Google Scholar]
- 32.Lee, C. H., S. C. Chang, C. J. Chen, and M. F. Chang. 1998. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 273:7650-7656. [DOI] [PubMed] [Google Scholar]
- 33.Lee, C.-Z., P. J. Chen, and D. S. Chen. 1995. Large hepatitis delta antigen in packaging and replication inhibition: role of the carboxyl-terminal 19 amino acids and amino-terminal sequences. J. Virol. 69:5332-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Y.-J., M. R. Stallcup, and M. M. C. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78:13325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo, K., S. B. Hwang, R. Duncan, M. Trousdale, and M. M. C. Lai. 1998. Characterization of mRNA for hepatitis delta antigen: exclusion of the full-length antigenomic RNA as an mRNA. Virology 250:94-105. [DOI] [PubMed] [Google Scholar]
- 36.Lo, K., G.-W. Sheu, and M. M. C. Lai. 1998. Inhibition of cellular RNA polymerase II transcription by delta antigen of hepatitis delta virus. Virology 247:178-188. [DOI] [PubMed] [Google Scholar]
- 37.Luo, G., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macnaughton, T. B., M. R. Beard, M. Chao, E. J. Gowans, and M. M. C. Lai. 1993. Endogenous promoters can direct the transcription of hepatitis delta virus RNA from a recircularized cDNA template. Virology 196:629-636. [DOI] [PubMed] [Google Scholar]
- 39.Macnaughton, T. B., E. J. Gowans, S. P. McNamara, and C. J. Burrell. 1991. Hepatitis δ antigen is necessary for access of hepatitis δ virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology 184:387-390. [DOI] [PubMed] [Google Scholar]
- 40.Macnaughton, T. B., and M. M. C. Lai. 2002. Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macnaughton, T. B., and M. M. C. Lai. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J. Virol. 76:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macnaughton, T. B., Y. I. Li, A. L. Doughty, and M. M. Lai. 2003. Hepatitis delta virus RNA encoding the large delta antigen cannot sustain replication due to rapid accumulation of mutations associated with RNA editing. J. Virol. 77:12048-12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macnaughton, T. B., Y.-J. Wang, and M. M. C. Lai. 1993. Replication of hepatitis delta virus RNA: effect of mutations of the autocatalytic cleavage sites. J. Virol. 67:2228-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modahl, L. E., and M. M. Lai. 2000. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J. Virol. 74:7375-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modahl, L. E., and M. M. C. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. C. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20:6030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mu, J.-J., D. S. Chen, and P.-J. Chen. 2001. The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 75:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mu, J.-J., Y. G. Tsay, L. J. Juan, T. F. Fu, W. H. Huang, D. S. Chen, and P. J. Chen. 2004. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 319:60-70. [DOI] [PubMed] [Google Scholar]
- 51.Mu, J.-J., H.-L. Wu, B.-L. Chiang, R.-P. Chang, D.-S. Chen, and P.-J. Chen. 1999. Characterization of the phosphorylated forms and the phosphorylated residues of hepatitis delta virus delta antigens. J. Virol. 73:10540-10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed, R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15:326-331. [DOI] [PubMed] [Google Scholar]
- 53.Reid, C. E., and D. W. Lazinski. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 97:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. K. Shih, R. H. Purcell, and J. L. Gerin. 1980. δ agent: association of δ antigen with hepatitis B surface antigen and RNA in serum of δ-infected chimpanzees. Proc. Natl. Acad. Sci. USA 77:6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ryu, W.-S., H. J. Netter, M. Bayer, and J. Taylor. 1993. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 67:3281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato, S., C. Cornillez-Ty, and D. W. Lazinski. 2004. By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J. Virol. 78:8120-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiebel, W., B. Haas, S. Marinkovic, A. Klanner, and H. L. Sanger. 1993. RNA-directed RNA polymerase from tomato leaves. I. Purification and physical properties. J. Biol. Chem. 268:11851-11857. [PubMed] [Google Scholar]
- 58.Schiebel, W., T. Pelissier, L. Riedel, S. Thalmeir, R. Schiebel, D. Kempe, F. Lottspeich, H. L. Sanger, and M. Wassenegger. 1998. Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10:2087-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharmeen, L., M. Y.-P. Kuo, and J. Taylor. 1989. Self-ligating RNA sequences on the antigenome of human hepatitis delta virus. J. Virol. 63:1428-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheu, G.-T., and M. M. C. Lai. 2000. Recombinant hepatitis delta antigen from E. coli promotes hepatitis delta virus RNA replication only from the genomic strand but not the antigenomic strand. Virology 278:578-586. [DOI] [PubMed] [Google Scholar]
- 61.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465-476. [DOI] [PubMed] [Google Scholar]
- 62.Spiesmacher, E., H.-P. Muhlbach, M. Tabler, and H. L. Sanger. 1985. Synthesis of (+) and (−) RNA molecules of potato spindle tuber viroid (PSTV) in isolated nuclei and its impairment by transcription inhibitors. Biosci. Rep. 5:251-265. [DOI] [PubMed] [Google Scholar]
- 63.Stein, P., P. Svoboda, M. Anger, and M. Schultz. 2003. RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tai, F.-P., P.-J. Chen, F.-L. Chang, and D.-S. Chen. 1993. Hepatitis delta virus cDNA can be used in transfection experiments to initiate viral RNA replication. Virology 197:137-142. [DOI] [PubMed] [Google Scholar]
- 65.Tan, K. P., K. N. Shih, and S. J. Lo. 2004. Ser-123 of the large antigen of hepatitis delta virus modulates its cellular localization to the nucleolus, SC-35 speckles or the cytoplasm. J. Gen. Virol. 85:1685-1694. [DOI] [PubMed] [Google Scholar]
- 66.Tavanez, J. P., C. Cunha, M. C. A. Silva, E. David, J. Monjardino, and M. Carmo-Fonseca. 2002. Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA 8:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor, J. M. 1999. Human hepatitis delta virus: an agent with similarities to certain satellite RNAs of plants. Curr. Top. Microbiol. Immunol. 239:107-122. [DOI] [PubMed] [Google Scholar]
- 68.Volloch, V., B. Schweitzer, and S. Rits. 1996. Antisense globin RNA in mouse erythoid tissues: structure, origin, and possible function. Proc. Natl. Acad. Sci. USA 93:2476-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, H.-W., H.-L. Wu, D.-S. Chen, and P.-J. Chen. 1997. Identification of the functional regions required for hepatitis D virus replication and transcription by linker-scanning mutagenesis of viral genome. Virology 239:119-131. [DOI] [PubMed] [Google Scholar]
- 70.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta viral genome. Nature (London) 323:508-514. (Erratum, 328: 456.) [DOI] [PubMed] [Google Scholar]
- 71.Wei, Y., and D. Ganem. 1998. Activation of heterologous gene expression by the large isoform of hepatitis delta antigen. J. Virol. 72:2089-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong, S. K., and D. W. Lazinski. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 99:15118-151123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, H.-N., Y.-J. Lin, F.-P. Lin, S. Makino, M.-F. Chang, and M. M. C. Lai. 1989. Human hepatitis δ virus RNA subfragments contain an autocleavage activity. Proc. Natl. Acad. Sci. USA 86:1831-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xia, Y.-P., C.-T. Yeh, J.-H. Ou, and M. M. C. Lai. 1992. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol. 66:914-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamaguchi, Y., S. Delehouzee, and H. Handa. 2002. HIV and hepatitis delta virus: evolution takes different paths to relieve blocks in transcriptional elongation. Microbes Infect. 4:1169-1175. [DOI] [PubMed] [Google Scholar]
- 76.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293:124-127. [DOI] [PubMed] [Google Scholar]