Abstract
Reovirus mRNAs are efficiently translated within host cells despite the absence of 3′ polyadenylated tails. The 3′ nontranslated regions (3′NTRs) of reovirus mRNAs contain sequences that exhibit a high degree of gene-segment-specific conservation. To determine whether the 3′NTRs of reovirus mRNAs serve to facilitate efficient translation of viral transcripts, we used T7 RNA polymerase to express constructs engineered with full-length S4 gene cDNA or truncation mutants lacking sequences in the 3′NTR. Full-length and truncated s4 mRNAs were translated using rabbit reticulocyte lysates, and translation product ς3 was quantitated by phosphorimager analysis. In comparison to full-length s4 mRNA, translation of the s4 mRNA lacking the 3′NTR resulted in a 20 to 50% decrease in ς3 produced. Addition to translation reactions of an RNA oligonucleotide corresponding to the S4 3′NTR significantly enhanced translation of full-length s4 mRNA but had no effect on s4 mRNA lacking 3′NTR sequences. Translation of s4 mRNAs with smaller deletions within the 3′NTR identified a discrete region capable of translational enhancement and a second region capable of translational repression. Differences in translational efficiency of full-length and deletion-mutant mRNAs were independent of RNA stability. Protein complexes in reticulocyte lysates that specifically interact with the S4 3′NTR were identified by RNA mobility shift assays. RNA oligonucleotides lacking either enhancer or repressor sequences did not efficiently compete the binding of these complexes to full-length 3′NTR. These results indicate that the reovirus S4 gene 3′NTR contains a translational operator sequence that serves to regulate translational efficiency of the s4 mRNA. Moreover, these findings suggest that cellular proteins interact with reovirus 3′NTR sequences to regulate translation of the nonpolyadenylated reovirus mRNAs.
All viruses must compete for cellular factors to efficiently express and replicate their genomes. Transcription and translation of the mammalian reovirus genome occurs within the cytoplasm of infected cells. The viral transcriptional machinery is contained within the viral core (5, 32); however, translation of viral mRNAs requires host cell factors and occurs in an environment replete with translationally functional cellular transcripts. Reovirus mRNAs lack polyadenylated tails, which serve to promote translation of many cellular mRNAs (10, 24, 30). Therefore, reovirus mRNAs must use alternative strategies to usurp the translational machinery. Viral sequences that enable reovirus mRNAs to achieve translational competence in a milieu of cellular mRNAs are not well understood.
Mammalian reoviruses are nonenveloped, icosahedral viruses formed from two concentric protein shells, termed outer capsid and core (reviewed in reference 26). The reovirus genome consists of 10 discrete segments of double-stranded RNA. After removal of the viral outer capsid during entry of the virus into cells, the core becomes enzymatically active and catalyzes transcription of 10 species of viral mRNA (3, 20, 33). Capped, nonpolyadenylated messages are extruded from the viral core into the cytoplasm, where they are efficiently translated despite an abundance of polyadenylated cellular mRNAs. The s4 mRNA, which encodes the ς3 protein (22, 25), is the most efficiently translated viral transcript in both in vitro translation reactions and reovirus-infected cells (1, 21, 42). Specific nucleotides at the −3 and +4 positions relative to the AUG initiation codon contribute to translational efficiency of some mRNAs (19). However, for the reovirus s4 mRNA, nucleotide polymorphism at these positions does not significantly influence the amount of translation product produced (23), indicating that this region is not the primary determinant of translational efficiency. Sequence elements that influence translational efficiency of the s4 mRNA have not been defined.
Sequences in the 3′ nontranslated regions (3′NTRs) of many viral and cellular transcripts have been shown to regulate translational efficiency. The poly(A) tail augments translation by enhancing mRNA stability and by facilitating ribosome reinitiation through indirect interactions with the 5′NTR via poly(A)-binding protein and translation factor elF-4G (reviewed in reference 29). Specific 3′NTR sequences upstream of the poly(A) tail that influence stability of mRNAs or interact with viral or cellular proteins to either enhance or repress protein synthesis also have been identified. In addition, the 3′NTRs of nonpolyadenylated transcripts also contain sequences that influence translation outcome for this class of mRNAs. For example, deletion of the 3′NTR of the nonpolyadenylated coat protein mRNA of alfalfa mosaic virus decreases the efficiency with which the coat protein transcript is translated when translation factors are limiting (14). This observation suggests that the 3′NTRs of nonpolyadenylated viral mRNAs facilitate efficient translation when competing with polyadenylated mRNAs for the translational machinery.
The 3′NTRs of reovirus gene segments range from 35 to 80 nucleotides in length (9, 15). Analysis of sequence diversity of the reovirus S-class gene segments indicates that sequences at the 5′ and 3′ termini are highly conserved (6, 8, 13, 18). Conserved sequences at the termini of each of these gene segments are not limited to the NTRs and extend into the open reading frames. These observations suggest that there is selection pressure to maintain conservation of these sequences at the nucleotide level. Similar levels of conservation have been observed at the gene-segment termini of other double-stranded RNA viruses, and these conserved sequences have been implicated in regulation of translation and genome replication (27, 28, 38, 40).
To test the hypothesis that conserved sequences in the 3′NTRs of reovirus mRNAs influence translation, we generated full-length and 3′NTR-truncated (3′Δ) s4 mRNAs and introduced these mRNAs into translation reactions using rabbit reticulocyte lysates. Our results indicate that discrete sequences within the S4 3′NTR serve to either enhance or repress translation of the s4 transcript. Additionally, we demonstrate that an RNA oligonucleotide corresponding to the S4 gene 3′NTR forms stable complexes with proteins contained in reticulocyte lysates. These findings indicate that the reovirus S4 3′NTR contains sequences that regulate translational efficiency of the s4 mRNA and suggest that cellular components interact with these regulatory sequences to influence translation.
MATERIALS AND METHODS
Cells and viruses.
Mouse L929 cells were grown in either suspension or monolayer cultures in Joklik's modified Eagle's minimal essential medium (Irvine Scientific, Santa Ana, Calif.) that was supplemented to contain 5% fetal calf serum (Intergen Co., Purchase, N.Y.), 2 mM l-glutamine, 100 U of penicillin G per ml, 100 μg of streptomycin per ml, and 250 ng of amphotericin B per ml (Irvine Scientific). Reovirus strain type 3 Dearing (T3D) is a laboratory stock. A second-passage L-cell lysate stock of twice-plaque-purified T3D was used for the experiments described (36).
Generation of reovirus mRNA expression constructs.
A cDNA corresponding to the reovirus T3D S4 gene was amplified by reverse transcription (RT) and PCR using avian myeloblastosis virus (AMV) reverse transcriptase and Pfu DNA polymerase (Promega Corp., Madison, Wis.) with primers a and b (Table 1). The S4 gene cDNA was ligated into plasmid pCR2.1 (Invitrogen, Carlsbad, Calif.) and then transferred to T7 RNA polymerase expression vector pGEM3z/f± (Promega) following digestion with EcoRI and PstI (New England Biolabs, Beverly, Mass.). Sequences between the T7 RNA polymerase promoter and the 5′ nucleotide of the S4 gene cDNA were deleted by PCR mutagenesis (Exsite; Stratagene, La Jolla, Calif.) with 5′-phosphorylated primers c and d (Table 1) to generate plasmid construct pS4.
TABLE 1.
Primers used to generate s4 mRNAs for studies of reovirus translation
| Primer | Sequence | Nucleotidesa |
|---|---|---|
| a | 5′-GCGAATTCGCTATTTTTGCCTCTTCCCAGA-3′ | 1–22* |
| b | 5′-CATGCCTGCAGATGAATGAAGCCTGTCCCACGTC-3′ | 1173–1196* |
| c | 5′-GCTATTTTTGCCTCTTCCCAGA-3′ | 1–22 |
| d | 5′-TATAGTGAGTCGTATTACAATTC-3′ | 2721–2743* |
| e | 5′-CTGCAGGCATGCAAGCTTGAG-3′ | 48–68* |
| f | 5′-TTAGCCAAGAATCATCGGATCGCC-3′ | 1107–1130 |
| g | 5′-TGTTTAGCCAAGAATCATCGGATC-3′ | 1110–1133 |
| h | 5′-CCGGGCTTGACCAACCTGGTGTGAC-3′ | 1151–1175 |
| i | 5′-ACCAACCTGGTGTGACGTGGGAC-3′ | 1160–1182 |
| j | 5′-TGGGGGTGTTTAGCCAAGAATCATC-3′ | 1115–1139 |
| k | 5′-CAAGCCCGGCGCTGTGAAGATGGG-3′ | 1136–1159 |
| l | 5′-GACAGGCTTCATTCATCTGCAGGC-3′ | 1180–1196* |
| m | 5′-CAGGTTGGTCAAGCCCGGCGCTGTG-3′ | 1144–1168 |
| n | 5′-CATTCATCTGCAGGCATGCAAGCTTG-3′ | 1189–1196* |
| o | 5′-CCACGTCACACCAGGTTGGTCAAGC-3′ | 1155–1179 |
| p | 5′-CTGCAGGCATGCAAGCTTGAGTATTC-3′ | 48–73* |
| q | 5′-CCAGTGATGATTGGCGATCCGATGA-3′ | 1095–1119 |
Sequences refer to nucleotides in the T3D S4 gene cDNA, with the exception of primers designated with an asterisk. These nucleotides correspond to sequences in plasmid pGEM3z/f±. Primers a, b, l, and n each contain nucleotide sequences from the S4 gene and pGEM3z/f±.
Deletions within the S4 gene 3′NTR were generated by PCR mutagenesis using pS4 and the phosphorylated primer sets shown in Table 1: pS4-3′Δ (e and f), pS4-Δ1134-1150 (g and h), pS4-Δ1140-1159 (i and j), pS4-Δ1162-1181 (k and l), pS4-Δ1169-1189 (m and n), and pS4-Δ1181-1195 (o and p). The pS4-3′NTR construct was generated by PCR mutagenesis using pS4 and phosphorylated primers d and q. Mutagenesis was performed according to the manufacturer's instructions as follows. Sixty micrograms of plasmid template per ml, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 100 μg of bovine serum albumin per ml, and 5 U of a Pfu/Taq polymerase blend (Pfu Turbo; Stratagene) were combined in a volume of 25 μl. Reaction temperatures were used as recommended except that annealing temperatures of 52°C were used during the initial cycle and increased to 58°C in the subsequent eight cycles. The 5′ and 3′ termini of the PCR products were ligated, and the resulting plasmids were used to transform XL-1 Blue supercompetent Escherichia coli (Epicurian coli; Stratagene). Plasmids were isolated by Wizard Mini-prep column purification (Promega) and sequenced using T4 DNA polymerase and dideoxy-chain termination (Sequenase; U.S. Biochemical, Cleveland, Ohio).
A cDNA corresponding to the reovirus T3D S1 gene was amplified by RT and PCR using AMV reverse transcriptase and Pfu/Taq DNA polymerase blend with primers aa and bb (Table 2). The S1 gene cDNA was ligated into plasmid pCR2.1 and then transferred to pGEM3z/f± following digestion with EcoRI and PstI. Sequences between the T7 RNA polymerase promoter and the 5′ nucleotide of the S1 gene cDNA were deleted using PCR mutagenesis with 5′-phosphorylated primers cc and dd (Table 2) to generate plasmid construct pS1. The S1 gene 3′NTR was deleted by PCR mutagenesis using pS1 and the phosphorylated primers ee and ff (Table 2). S1 gene sequences were confirmed prior to use.
TABLE 2.
Primers used to generate s1 mRNAs for studies of reovirus translation
| Primer | Sequence | Nucleotidesa |
|---|---|---|
| aa | 5′-GCGAATTCGCTATTGGTCGGATGGATCCTCG-3′ | 1–23* |
| bb | 5′-CATGCCTGCAGATGAAATGCCCCAGTGCCGCGG-3′ | 1394–1416* |
| cc | 5′-GCTATTGGTCGGATGGATCCTCG-3′ | 1–23 |
| dd | 5′-TATAGTGAGTCGTATTACAATTC-3′ | 2721–2743* |
| ee | 5′-CTGCAGGCATGCAAGCTTGAG-3′ | 48–68* |
| ff | 5′-TCACGTGAAACTACGCGGGTACG-3′ | 1358–1380 |
Sequences refer to nucleotides in the T3D S1 gene cDNA, with the exception of primers designated with an asterisk. These nucleotides correspond to sequences in plasmid pGEM3z/f±. Primers aa and bb contain nucleotide sequences from the S1 gene and pGEM3z/f±.
Generation of reovirus mRNA transcripts.
Plasmids were linearized by digestion with 1 U of PstI per μl, and 3′ overhang sequences were removed by incubation with 1 to 2 U of T4 DNA polymerase (Promega) per μl at 11°C for 20 min in the presence of 1 mM Tris-HCl (pH 7.9), 1 mM MgCl2, 5 mM NaCl, 0.1 mM dithiothreitol (DTT), 10 μM deoxynucleoside triphosphate, and 50 μg of bovine serum albumin per in a final volume of 20 μl. Linearized plasmids were gel purified and resuspended in nuclease-free water.
In vitro transcription of capped reovirus mRNAs was performed using linearized plasmid as template and the mMessage mMachine T7 RNA polymerase transcription system (Ambion, Austin, Tex.). Transcription reactions were incubated with or without 1 μl of [α-32P]UTP (3,000 Ci per mmol; Dupont NEN, Wilmington, Del.) at 37°C for 2 h followed by digestion of input plasmid with 250 U of DNase 1 per ml at 37°C for 15 min. Prior to addition of T7 RNA polymerase, linearized plasmid and transcription reagents were incubated with RNase inhibitor RNAsecure (Ambion) at 60°C for 20 min. RNA oligonucleotides were similarly transcribed from plasmid linearized with PstI using the megaShortscript T7 RNA polymerase transcription system (Ambion).
RNA transcripts and oligonucleotides were purified from transcription reactions and rabbit reticulocyte lysates using Tri-reagent (Molecular Research, Cincinnati, Ohio). Transcription reactions were mixed with 3 volumes of Tri-reagent and incubated at room temperature for 5 min. The mixture was vortexed vigorously with 1.5 volumes of chloroform followed by incubation at room temperature for 15 min. Following centrifugation at 12,000 × g, RNA was precipitated in 2.5 volumes of isopropanol and 0.75 volumes of NC buffer (0.8 M sodium citrate, 1.2 M NaCl). RNA concentration was determined using optical density at 260 nm and confirmed by electrophoresis in 5% polyacrylamide-urea gels followed by ethidium bromide staining and densitometry (Eagle Eye software; Stratagene).
Sequencing 5′ and 3′ termini of reovirus RNAs.
Sequences of s4 and s1 mRNA 5′ termini were determined by dideoxy-chain termination using reverse transcriptase and primers 5′-ACATACCATATCTGG-3′ and 5′-CTCAAGAGCGATGATTCG-3′, respectively (6). Sequences of 3′ termini were determined by rapid amplification of cDNA ends (RACE)-PCR. Adenosine bases were appended to the 3′ terminus of transcribed RNA by incubation with dATP and terminal transferase (Roche Molecular Biochemicals, Indianapolis, Ind.). RNA was purified by Tri-reagent and subjected to RT using AMV reverse transcriptase and oligo(dT) primer 5′-GACCACGCGTATCGATGTCGAC(T)16N-3′. The resultant cDNA was amplified in a primary round of PCR using Pfu DNA polymerase with oligo(dT) primer and either S4-specific primer 5′-GCTATTTTTGCCTCTTCC-3′ or S1-specific primer 5′-GCTATTGGTCGGATGG-3′. A secondary round of PCR amplification was performed using Taq DNA polymerase with a primer corresponding to a sequence 5′ to the oligo(dT) primer, 5′-GACCACGCGTATCGATGTCGAC-3′, and either S4-specific primer 5′-CGTCGTTTGCATGCATTG-3′ or S1-specific primer 5′-GTCAGTGATGCTCAACTTGCAATCTCC-3′. PCR product from the secondary amplification was ligated into pCR2.1, and sequences were determined by dideoxy-chain termination.
Translation of reovirus mRNAs in rabbit reticulocyte lysates.
Full-length and mutant reovirus mRNAs were translated by incubation with rabbit reticulocyte lysates (Promega) at 30°C for various intervals. Translation reactions were supplemented with 2 to 3 U of RNasin RNase inhibitor (Promega) per μl, 2.25 mM DTT, 0.05 M KCl, 1 mM magnesium acetate 0.2 mM amino acid mixture minus methionine, and 0.5 mCi of Easy Tag Express -[35S] Protein Labeling Mix (Dupont NEN) per ml at 1,175 Ci per mmol in a final volume of 3.12 μl. Reactions were incubated in the presence or absence of various concentrations of globin mRNA (Gibco BRL, Rockville, Md.).
SDS-PAGE and phosphorimager analysis of translation products.
Equal volumes of translation reactions were added to 2× sample buffer (125 mM Tris, 10% β-mercaptoethanol, 4% sodium dodecyl sulfate [SDS], 0.02% bromophenol blue). Samples were boiled for 5 min, loaded into lanes of 12% polyacrylamide gels, and subjected to polyacrylamide gel electrophoresis (PAGE) at 200-V constant voltage for 1 h (2). Following electrophoresis, gels were dried and exposed either to film (Eastman Kodak Company, Rochester, N.Y.) or an imaging plate (Fuji Medical Systems, Inc., Stamford, Conn.) for phosphorimager analysis. Band intensity was quantitated by determining photostimulus luminescence (PSL) units, using a Fuji2000 phosphorimager (Fuji Medical Systems, Inc.).
Statistical analysis.
Differences in translational efficiency of full-length and s4 3′Δ mRNAs in the presence or absence of S4 3′NTR RNA oligonucleotides were analyzed by calculating the area under the curve and performing an equal-variance t test. The panel of mutants was analyzed statistically using a single-factor analysis of variance followed by a pairwise comparison of individual mutants with the full-length s4 mRNA, using a one-tailed t test assuming unequal variance. Statistical analysis was performed using Microsoft Excel 95 (Microsoft, Redmond, Wash.).
RNA gel shift analysis of S4 3′NTR-binding complexes.
Radiolabeled probes for electrophoretic mobility shift assays (EMSAs) were generated using in vitro transcription with the pS4-3′NTR construct as template and T7 RNA polymerase in the presence of 0.0011 mM [α-32P]UTP (3,000 Ci per mmol) with 7.5 mM ATP, CTP, and GTP and 0.0275 mM UTP. Reactions were incubated at 37°C for 1 h, followed by addition of 2 μl of T7 RNA polymerase enzyme mix (MegaShortscript; Ambion). Reactions were incubated at 37°C for an additional 1 h and then terminated by plasmid digestion with DNase I. The 3′NTR probe was purified using Tri-reagent as described above and analyzed for purity by electrophoresis in a 5% polyacrylamide-urea gel and autoradiography.
RNA EMSAs were performed according to previously described techniques (4), with minor modifications. Rabbit reticulocyte lysate at a concentration of 12.5 mg of protein per ml was incubated in the presence or absence of increasing concentrations of unlabeled competitor RNA in 20 mM HEPES–5% glycerol–1 mM EDTA–0.1 mg of yeast tRNA per ml–60 mM KCl–1 mM DTT in a volume of 20 μl. Either 5S rRNA (Gibco BRL) or poly(A) RNA (400 to 600 bp) (Sigma) was added to the lysates as nonspecific competitor RNA. Following incubation on ice for 20 min, radiolabeled probe (105 cpm, 1 ng of probe) was added to the lysates and incubated at room temperature for 20 min. Competitor probes were added in molar excess to the radiolabeled probe. Reactions were incubated in the presence of 10 mg of heparin per ml at room temperature for 10 min, followed by the addition of gel shift loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, 15% Ficoll 400). Protein-RNA complexes were electrophoresed in 30% TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA)–acrylamide (29:1) nondenaturing gels at 180-V constant voltage for 2 h. Following electrophoresis, gels were dried and exposed to film.
RESULTS
Deletion of the reovirus S4 gene 3′NTR reduces translational efficiency of the s4 mRNA.
To determine whether the 3′NTR is required for efficient translation of reovirus mRNAs, we generated a cDNA corresponding to the reovirus S4 gene, which encodes the ς3 protein (12, 22, 25). The S4 gene cDNA was inserted into a plasmid containing a T7 RNA polymerase promoter such that transcription initiated with a 5′-terminal guanosine and terminated with a 3′-terminal cytosine, which correspond to the terminal nucleotides of the S4 gene (12) (Fig. 1A). Plasmid constructs containing the S4 gene cDNA were transcribed in the presence of m7GTP to generate capped transcripts with 5′- and 3′-terminal sequences identical to native reovirus s4 mRNAs. To confirm authenticity of the in vitro-generated s4 transcripts, nucleotide sequences at the 5′ termini of these transcripts were determined by direct RNA sequence analysis, and sequences at the 3′ termini were determined following RACE-PCR. Analysis of these sequences demonstrated that the 5′ and 3′ termini of in vitro-generated s4 mRNAs are identical to native s4 mRNAs (data not shown).
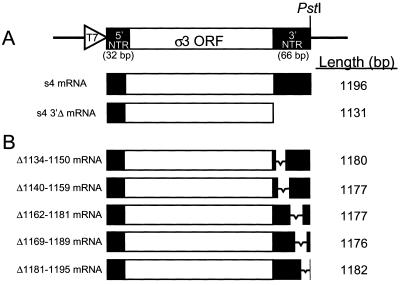
FIG. 1.
Diagram of templates used for transcription of full-length and deletion-mutant s4 mRNAs. (A) The relevant region of plasmid pGEM3z/f± used for cloning a full-length cDNA corresponding to the S4 gene of reovirus strain T3D. Indicated are a T7 RNA polymerase promoter (T7), which was cloned adjacent to the 5′-terminal guanosine of the S4 gene cDNA, the ς3 open reading frame (ORF), and a PstI site, which was engineered adjacent to the 3′ terminus of the S4 gene cDNA. Digestion of plasmid constructs with PstI followed by incubation with T4 DNA polymerase resulted in templates for transcription that initiate with the 5′ guanosine and terminate with the 3′ cytosine of the S4 gene. Capped s4 mRNAs and s4 3′Δ mRNAs were transcribed in vitro and used in translation studies. Nucleotide lengths are indicated on the right. The s4 3′Δ mRNA terminates with UAAC, corresponding to the stop codon of the transcript and the 3′-terminal cytosine of the full-length S4 gene. (B) Schematic of S4 gene 3′NTR deletion constructs generated from cDNA clones by PCR mutagenesis. The viral sequences deleted in each of the mutant S4 gene cDNAs are indicated on the left; the length of each mRNA transcript is indicated on the right.
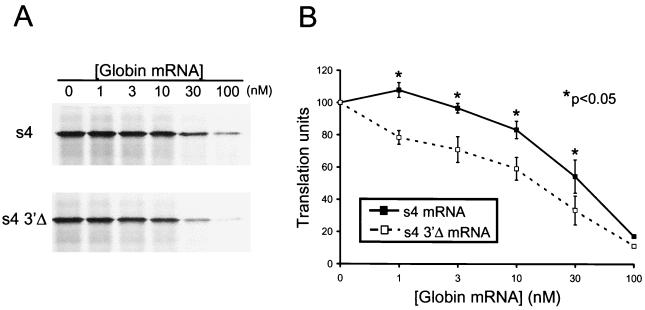
We examined whether sequences in the 3′NTR influence the translation of the reovirus s4 mRNA under conditions in which components of translation were either in excess or limiting. Full-length s4 mRNAs or s4 mRNAs lacking the 3′NTR (s4 3′Δ) at a concentration of 10 nM were incubated in rabbit reticulocyte lysates in the presence of increasing concentrations of polyadenylated globin mRNA from 0 to 100 nM. Translation product ς3 was resolved by SDS-PAGE and quantitated by phosphorimager analysis (Fig. 2). In reticulocyte lysates lacking globin mRNA, full-length s4 and s4 3′Δ mRNAs were translated equivalently. However, as the concentration of globin mRNA added to the translation reactions was increased, ς3 protein produced during translation of the full-length s4 mRNA was greater than that produced during translation of s4-3′Δ mRNA. We found that at concentrations of globin mRNA greater than 1 nM, full-length s4 mRNA was translated 20% more efficiently than s4-3′Δ mRNA (P < 0.05). These findings suggest that the S4 3′NTR confers a translational advantage to the reovirus s4 mRNA under conditions in which components of the translational machinery are limiting.
FIG. 2.
Translational efficiency of full-length s4 and s4 3′Δ mRNAs. Reovirus s4 and s4 3′Δ mRNAs at a concentration of 10 nM were added to rabbit reticulocyte lysates containing increasing concentrations of globin mRNA from 0 to 100 nM. Following incubation at 30°C for 15 min, translation products were resolved by SDS-PAGE (A) and quantitated by phosphorimager analysis (B). Translation units are defined as the PSL units of translation product at the indicated concentration of globin mRNA divided by the PSL units for the s4 mRNA translation product in the absence of globin mRNA. The results are expressed as the mean translation units (×100) determined from three independent experiments performed in triplicate. Error bars indicate standard errors of the means.
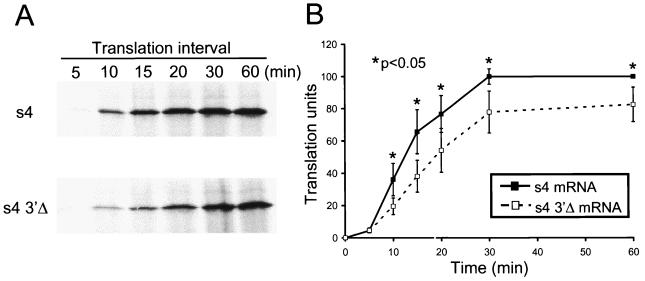
To determine whether the kinetics of translation of full-length s4 mRNA differ from those of s4 3′Δ mRNA, transcripts at a concentration of 40 nM were incubated in reticulocyte lysates in the presence of 10 nM globin mRNA for intervals from 5 to 60 min (Fig. 3). After 5 min of incubation, no protein was detected by autoradiography, and very little was detected by phosphorimager analysis. However, after 10 min of incubation, significantly more ς3 was produced following translation of full-length s4 mRNA than following translation of s4 3′Δ mRNA. The magnitude of this difference in translation efficiency remained constant at each interval tested and was statistically significant (P < 0.05). Translation products of both transcripts approached a plateau by 30 min of incubation, with little change in steady-state levels of ς3 produced after that time. This result indicates that full-length s4 mRNAs are translated at an increased rate relative to s4 mRNAs lacking the 3′NTR, resulting in synthesis of significantly greater protein product.
FIG. 3.
Translation kinetics of full-length s4 and s4 3′Δ mRNAs. Reovirus s4 and s4 3′Δ mRNAs at a concentration of 40 nM were added to rabbit reticulocyte lysates containing 10 nM globin mRNA. Reactions were incubated at 30°C for the times shown followed by SDS-PAGE (A) and phosphorimager analysis (B). Translation units are defined as the PSL units of translation product at the indicated time divided by the PSL units for the s4 mRNA translation product at 60 min. The results are expressed as the mean translation units (×100) determined from two independent experiments performed in triplicate. Error bars indicate standard errors of the means.
The S4 gene 3′NTR alters translational efficiency when provided in trans.
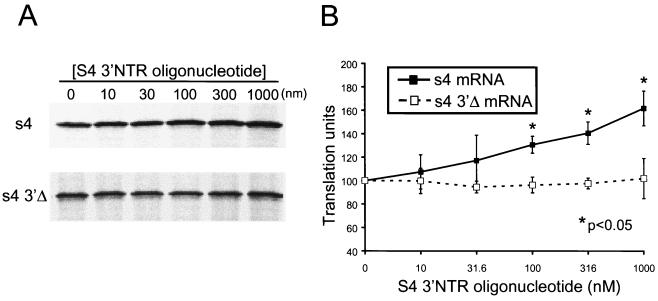
To test whether translational enhancement of the s4 mRNA provided by the 3′NTR could be altered by supplying S4 3′NTR sequences in trans, we incubated full-length s4 mRNA and s4 3′Δ mRNA in reticulocyte lysates containing RNA oligonucleotides corresponding to the 3′-terminal 100 nucleotides of the S4 gene (Fig. 4). In an analysis of S4 gene nucleotide sequences of 16 reovirus strains, sequence variability within this region is restricted to only six nucleotides (18). We observed no significant change in basal levels of translation of s4 3′Δ mRNA in the presence of increasing concentrations of exogenous S4 3′NTR RNA oligonucleotide. In contrast, translation of full-length s4 mRNA was significantly enhanced in the presence of the S4 3′NTR RNA oligonucleotide. At a 100-fold molar excess of 3′NTR oligonucleotide to s4 mRNA, translation of the full-length s4 mRNA was increased by approximately 60% in comparison to mRNA translated in the absence of exogenous S4 3′NTR oligonucleotide. This finding indicates that when provided in trans, the S4 gene 3′NTR blocks inhibition of translation of the full-length s4 mRNA, which suggests that the S4 3′NTR contains sequences that confer translational repression.
FIG. 4.
Translation of full-length s4 and s4 3′Δ mRNAs in the presence of an RNA oligonucleotide corresponding to the S4 3′NTR supplied in trans. Reovirus s4 and s4 3′Δ mRNAs at a concentration of 10 nM were incubated at 30°C for 15 min in rabbit reticulocyte lysates containing 10 nM globin mRNA. An RNA oligonucleotide corresponding to the 3′-terminal 100 nucleotides of the s4 mRNA was added at the concentrations shown prior to incubation. Translation products were resolved by SDS-PAGE (A) and quantitated by phosphorimager analysis (B). Translation units are defined as the PSL units of translation product at the indicated S4 3′NTR oligonucleotide concentration divided by the PSL units of either the s4 or s4 3′Δ mRNA translation product in the absence of exogenous 3′NTR. The results are expressed as the mean translation units (×100) determined from a representative experiment performed in triplicate. Error bars indicate standard deviations of the mean.
To determine the specificity of the S4 3′NTR RNA oligonucleotide on translational efficiency, we generated plasmid constructs to facilitate transcription of full-length s1 and 3′Δ s1 mRNAs. Sequence analysis confirmed that the 5′ and 3′ termini of in vitro-generated s1 mRNAs are identical to those of native s1 mRNAs (data not shown). Reovirus s1 mRNAs were translated in reticulocyte lysates in the presence of 10 nM globin mRNA and increasing concentrations of the S4 3′NTR RNA oligonucleotide. We found no effect on translation of either the s1 or s1-3′Δ mRNA with increasing concentrations of oligonucleotide (data not shown). In addition, we observed no translational enhancement when nonviral, polyadenylated globin mRNA was incubated in reticulocyte lysates containing the S4 3′NTR RNA oligonucleotide (data not shown). These results suggest that the translational enhancement provided by addition of the S4 3′NTR to translation reactions is specific to the s4 mRNA.
Discrete regions of the S4 gene 3′NTR differentially influence translational efficiency.
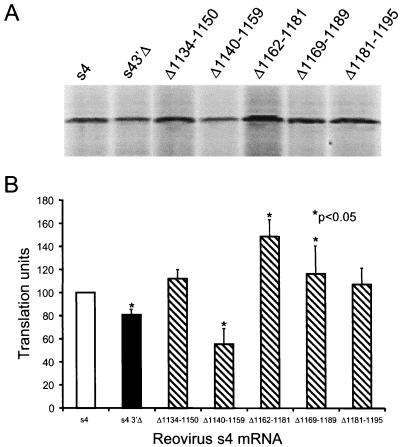
Our data show that deletion of the 3′NTR reduces translational efficiency of the reovirus s4 mRNA, yet addition of these sequences in trans augments translation of the same transcript. These results suggest that the S4 3′NTR has independent sequence elements that differentially influence translation of the s4 mRNA. We examined whether such functions could be genetically dissected by using s4 mRNAs lacking small regions of sequence within the 3′NTR (Fig. 1B). Translation of full-length s4 and s4 3′Δ mRNAs was compared to five additional s4 mRNAs with 3′NTR deletions ranging from 15 to 21 nucleotides in length (Fig. 5). Deletion of nucleotides 1140 to 1159 reduced translational efficiency approximately 40% relative to full-length s4 mRNA (P < 0.05), which suggests that this sequence region contains an enhancer of translation. In contrast, deletion of nucleotides 1162 to 1181 enhanced translational efficiency greater than 50% relative to full-length s4 mRNA (P < 0.05). Deletion of nucleotides 1169 to 1189 also enhanced translational efficiency, but the effect was more modest, approximately 10 to 15% (P < 0.05). These findings demonstrate that sequences encompassing nucleotides 1162 to 1181 contain a repressor of translation.
FIG. 5.
Effect of deletions within the S4 3′NTR on translation of the s4 mRNA. Full-length and deletion-mutant s4 mRNAs at a concentration of 10 nM were incubated in rabbit reticulocyte lysates containing 10 nM globin mRNA at 30°C for 15 min. Translation products were resolved by SDS-PAGE (A) and quantitated by phosphorimager analysis (B). Translation units are defined as the PSL units of translation product of the indicated s4 mRNA divided by the PSL units of translation product of full-length s4 mRNA. The results are expressed as the mean translation units (×100) determined from three independent experiments performed in triplicate. Error bars indicate standard errors of the means.
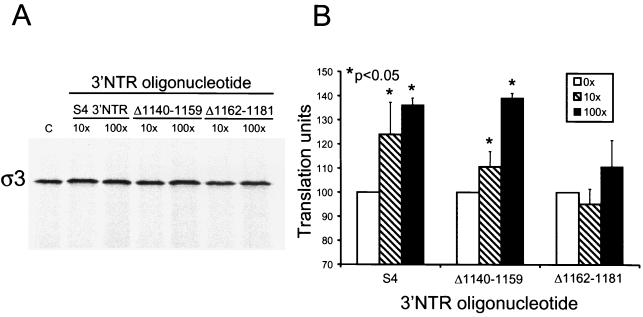
To test whether deletion of the S4 3′NTR sequences that mediate enhancer and repressor effects alters the enhancement activity observed when an oligonucleotide corresponding to the S4 3′NTR is added in trans, we generated RNA oligonucleotides corresponding to the 3′NTRs of s4 mRNA deletion mutants Δ1140-1159 and Δ1162-1181 and determined their effects on translational outcome. Full-length s4 mRNA was incubated in reticulocyte lysates with oligonucleotides corresponding to the full-length S4 3′NTR, the Δ1140-1159 3′NTR, or the Δ1162-1181 3′NTR, and translation product ς3 was resolved by SDS-PAGE and quantitated by phosphorimager analysis (Fig. 6). As shown previously, translation of s4 mRNA in reticulocyte lysates in the presence of a 100-fold molar excess of the S4 3′NTR RNA oligonucleotide resulted in a 50% increase in ς3 protein produced in comparison to translation of s4 mRNA alone (P < 0.05). When s4 transcripts were translated in reticulocyte lysates in the presence of increasing concentrations of the RNA oligonucleotide corresponding to the Δ1140-1159 3′NTR, production of ς3 was similarly enhanced in the presence of both 10- and 100-fold molar excess oligonucleotide, reaching an approximately 50% increase in comparison to ς3 produced from s4 mRNA alone (P < 0.05). However, when s4 mRNA was translated in reticulocyte lysates in the presence of an RNA oligonucleotide corresponding to the Δ1162-1181 3′NTR, no significant enhancement of ς3 synthesis was observed. This finding indicates that an S4 3′NTR oligonucleotide with a deletion of nucleotides 1162 to 1181 does not enhance translation of the s4 mRNA in trans, which suggests that nucleotides 1162 to 1181 of the 3′NTR titrate a translational repressor activity.
FIG. 6.
Translation of full-length s4 mRNA in the presence of full-length and internally truncated S4 3′NTR RNA oligonucleotides supplied in trans. Reovirus s4 mRNA at a concentration of 1 nM was incubated at 30°C for 15 min in rabbit reticulocyte lysates containing 1 nM globin mRNA. Increasing concentrations (×1 nM) of RNA oligonucleotides corresponding to the 3′-terminal 100 nucleotides of the s4 mRNA (S4) or mutant RNA oligonucleotides corresponding to the 3′-terminal 100 nucleotides of mutant s4 mRNAs Δ1140–1159 and Δ1162–1181 were added prior to incubation. Translation products were resolved by SDS-PAGE (A) and quantitated using phosphorimager analysis (B). Translation units are defined as the PSL units of translation product of s4 mRNA in the presence of 3′NTR RNA oligonucleotide divided by the PSL units of translation product in the absence of 3′NTR RNA oligonucleotide. The results are expressed as the mean translation units (×100) determined from a representative experiment performed in triplicate. Error bars indicate standard deviations of the mean. C, control without oligonucleotide.
Deletion of the reovirus S4 gene 3′NTR does not alter s4 mRNA stability.
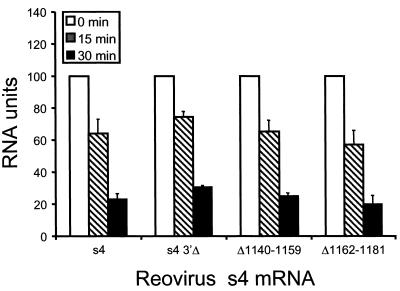
To determine whether deletion of the 3′NTR affects stability of the s4 mRNA, full-length s4 and s4 3′Δ mRNAs were quantitated following incubation in reticulocyte lysates. Radiolabeled transcripts were incubated in reticulocyte lysates from 5 to 60 min, isolated by Tri-reagent extraction, and subjected to electrophoresis in denaturing polyacrylamide gels. The relative amounts of mRNA present were determined by phosphorimager analysis (Fig. 7 and data not shown). Full-length s4 and s4 3′Δ mRNAs were equivalently stable in reticulocyte lysates, with greater than 60% of input RNA lost by 30 min of incubation and greater than 80% of input mRNA lost by 60 min of incubation. The kinetics of degradation of the full-length s4 mRNA and the s4 3′Δ mRNA were identical (data not shown). Therefore, differences in the translational efficiency of these mRNAs are not attributable to differences in relative stability.
FIG. 7.
Effect of deletions within the s4 3′NTR on RNA stability. Radiolabeled full-length s4 mRNA (s4), s4 3′Δ mRNA (s4 3′Δ), and Δ1140–1159 and Δ1162–1181 mRNAs were incubated in rabbit reticulocyte lysates and purified at the times shown. Purified transcripts were analyzed by electrophoresis in denaturing polyacrylamide gels and quantitated by phosphorimager analysis. RNA units are defined as the PSL units of mRNA at the indicated time divided by the PSL units of the homologous s4 mRNA at time zero. The results are expressed as the mean RNA units (×100) determined from two independent experiments performed in triplicate. Error bars indicate standard errors of the means.
Although full-length and 3′Δ s4 mRNAs showed identical degradation kinetics, we thought it possible that independent regions within the 3′NTR might differentially regulate transcript stability. To test this hypothesis, we compared the stability of mutant s4 reovirus mRNAs Δ1162-1181 and Δ1140-1159 with the stability of the full-length and s4-3′Δ mRNAs. We found that smaller deletions within the 3′NTR were not associated with alterations in the kinetics of mRNA decay (Fig. 7). These findings indicate that alterations in translational efficiency observed for s4 mRNAs containing internal deletions in the 3′NTR occur independently of RNA stability, which suggests that the S4 3′NTR mediates translational enhancement by an alternative mechanism.
The reovirus S4 gene 3′NTR forms complexes with proteins in reticulocyte lysates.
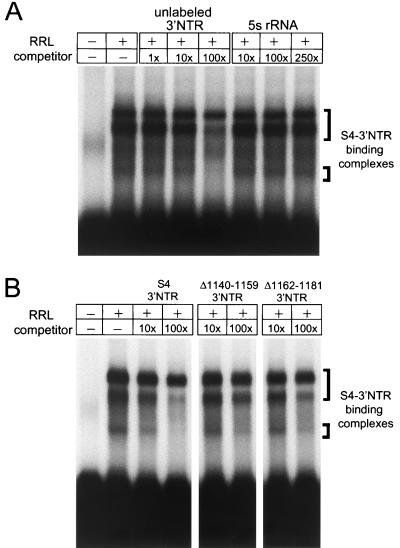
To determine whether proteins contained in rabbit reticulocyte lysates bind specifically to the S4 3′NTR, RNA EMSAs were performed with 32P-labeled S4 3′NTR as probe (Fig. 8A). Following incubation of reticulocyte lysate with the S4 3′NTR probe, three probe-containing protein complexes were observed after electrophoresis in nondenaturing polyacrylamide gels. Specificity of the interactions was determined by competition analysis using unlabeled S4 3′NTR as specific competitor and unlabeled 5S rRNA, a 120-nucleotide RNA oligonucleotide, as nonspecific competitor. Band intensity of each of the three 3′NTR-containing complexes was diminished in the presence of excess unlabeled 3′NTR, with competition of each complex occurring with different efficiencies. In contrast, no change was observed in the intensity of the protein-RNA complexes in the presence of excess unlabeled 5S rRNA. As an additional test of specificity, an unlabeled poly(A) oligonucleotide was assessed for the capacity to compete the binding of the labeled S4 3′NTR to the reticulocyte protein complexes. Similar to the results for 5S rRNA, poly(A) did not compete the binding of the S4 3′NTR to these complexes (data not shown). We next tested whether the mutant S4 3′NTR oligonucleotides Δ1140-1159 and Δ1162-1181 could compete the binding of the S4 3′NTR to protein complexes in reticulocyte lysates (Fig. 8B). Both of the mutant oligonucleotides were substantially less efficient than the full-length S4 3′NTR in competing for reticulocyte components that bind the S4 3′NTR. We found no change in the band intensity of the slowest-migrating complex and only a modest decrease in band intensities of the two faster-migrating complexes, even at the highest concentration of competitor used (250-fold). These findings demonstrate that cellular proteins in rabbit reticulocyte lysates bind specifically to the reovirus S4 3′NTR.
FIG. 8.
The reovirus S4 3′NTR is bound by cellular proteins. (A) EMSA of rabbit reticulocyte lysate (RRL) using reovirus S4 3′NTR as probe. Lysates were incubated with 32P-labeled S4 3′NTR RNA oligonucleotide, and incubation mixtures were resolved by acrylamide gel electrophoresis, dried, and exposed to film. S4 3′NTR-binding complexes are indicated. Specificity is shown by incubation with 1-, 10-, or 100-fold, excess unlabeled 3′NTR as specific competitor and 10-, 100-, or 250-fold excess 5S rRNA as nonspecific competitor. Competitor RNAs were incubated with reticulocyte lysates on ice for 20 min prior to addition of radiolabeled S4 3′NTR probe. (B) Competition analysis of S4 3′NTR-binding complexes using unlabeled mutant S4 3′NTR RNA oligonucleotides. Lysates were incubated with either 10- or 100-fold excess unlabeled 3′NTR, Δ1140–1159 3′NTR, or Δ1162–1181 3′NTR, and EMSAs were performed as for panel A.
DISCUSSION
The 5′ and 3′NTRs of reovirus gene segments are highly conserved (6, 8, 13, 18, 34, 39). This level of sequence conservation suggests that the NTRs contain important cis regulatory elements that determine the fate of nascent viral RNA. We used full-length and truncated mRNA transcripts to determine the role of the 3′NTR in translation of the reovirus s4 mRNA. Our results indicate that the S4 gene 3′NTR contains discrete regions of sequence that differentially influence translational efficiency. A region of sequence including nucleotides 1140 to 1159 enhances translation, while a second region including nucleotides 1162 to 1181 represses translation. Furthermore, the S4 gene 3′NTR interacts with protein components of reticulocyte lysates, which suggests that cellular proteins mediate the regulatory properties of the S4 3′NTR.
Differences in translational efficiency of full-length and 3′Δ s4 mRNAs were determined in vitro using rabbit reticulocyte lysates in the presence and absence of polyadenylated globin mRNA as competitor. We found that the S4 3′NTR facilitates efficient translation of reovirus transcripts in an environment in which reovirus mRNAs must compete with cellular mRNAs for components of the translational machinery. Although the effects observed in the in vitro assays used were modest, the translational enhancement provided by the S4 3′NTR is likely to be substantial during the many cycles of viral replication that occur in an infected host. Importantly, the observed differences in translational efficiency were found to be independent of RNA stability, thus excluding the possibility that the S4 3′NTR regulates translation via preservation of RNA structural integrity. A similar finding has been made in studies of alfalfa mosaic virus. Deletion of the coat protein mRNA 3′NTR results in approximately 50% less translation product in comparison to that produced following translation of a full-length transcript when translated in the presence of an equimolar ratio of globin mRNA (14). However, equivalent yields of coat protein are synthesized from full-length and 3′NTR-truncated mRNAs in the absence of competitor mRNA (14). Thus, our studies of reovirus s4 mRNA in conjunction with studies of alfalfa mosaic virus translational control define a paradigm in which 3′NTRs contain sequences that enhance translation in an environment where cellular transcripts are in excess and components of the translational machinery are limited.
Sequences in the 3′NTRs of many transcripts have been shown to enhance translation via interactions with the translational machinery (29). We hypothesized that if a component of the translational machinery bound the S4 3′NTR to augment translation, addition of exogenous 3′NTR might compete for interactions with such components and reduce translational efficiency of the s4 mRNA. Surprisingly, we found that addition to translation reactions of an RNA oligonucleotide corresponding to the S4 3′NTR augmented translation of the full-length s4 mRNA and had no effect on translation of the s4 3′Δ mRNA. Additionally, the S4 3′NTR does not alter translational efficiency of other RNAs, including the reovirus s1 mRNA, indicating that the observed effects are specific to the s4 transcript. These findings suggest that the S4 3′NTR contains a repressor sequence that when added in trans is capable of relieving translational repression by titrating a repressor activity, resulting in translational enhancement. We postulate that the S4 3′NTR also contains an enhancer sequence, but the effect of the repressor sequence is dominant when the 3′NTR is added to translation reactions in trans. Such a scenario would be possible if either the translational enhancer activity was in molar excess of repressor activity or translational enhancement required sequences elsewhere in the transcript in addition to sequences in the 3′NTR.
We directly tested whether enhancer and repressor sequences are present in the S4 3′NTR by assessing translational efficiency of s4 mRNAs containing small deletions in the 3′NTR. Findings from these experiments confirm the existence of distinct sequences in the 3′NTR that differentially regulate translation. Nucleotides 1142 to 1159 are required for translational enhancement, whereas nucleotides 1162 to 1181 are required for translational repression. We also observed a modest increase in translation of an mRNA lacking nucleotides 1169 to 1189, which suggests that the repressor region may include those sequences as well. Opposing enhancer and repressor regions in the 3′NTRs of eukaryotic mRNAs have been previously described. The 3′NTR of the β-F1-ATPase mRNA contains a translational repressor that is bound by an inhibitory protein found in fetal rat liver extracts. The 3′NTR of this transcript also contains a translational enhancer that is capable of titrating enhancement activity when added to translation reactions in trans (17), a situation similar in some respects to the translational regulatory region in the reovirus s4 mRNA.
As an additional confirmatory test of whether the reovirus S4 gene 3′NTR contains discrete enhancer and repressor elements, we tested the effect of RNA oligonucleotides with deletions of either enhancer or repressor sequences on translational efficiency of the full-length s4 mRNA when added to translation reactions in trans. We hypothesized that if enhancer sequences were deleted from an S4 3′NTR RNA oligonucleotide, only proteins that repress translation would be capable of binding the NTR oligonucleotide, and translational inhibition of the s4 mRNA would be relieved, similar to the effect observed with the intact S4 3′NTR. As anticipated, we found that addition to translation reactions of an RNA oligonucleotide lacking 3′NTR nucleotides 1140 to 1159 (the enhancer region) enhanced translation. However, we found that addition to translation reactions of an RNA oligonucleotide lacking 3′NTR nucleotides 1162 to 1181 (the repressor region) did not diminish translation of the s4 mRNA. It is possible that proteins mediating the enhancer function are in vast excess of the RNA oligonucleotide. Alternatively, sequences within the enhancer region may be involved in intramolecular interactions, with sequences elsewhere in the s4 mRNA required for efficient assembly of the translational machinery. Consistent with this idea, RNA secondary structure algorithms suggest that the full-length s4 mRNA folds such that the 5′ and 3′ termini base pair to form an extended region of duplex RNA (reference (18) and data not shown).
Sequences in the 3′NTRs of many viral and cellular transcripts have been shown to interact with components of the translational machinery to influence translational efficiency (11, 16, 28, 37). We performed RNA EMSAs to test whether cellular proteins are capable of binding the S4 3′NTR. The results show that three complexes contained in reticulocyte lysates bind specifically to a probe corresponding to the S4 3′NTR. The binding of the labeled S4 3′NTR to each of these complexes was inhibited by preincubation with unlabeled S4 3′NTR but not with nonspecific competitors or mutant S4 3′NTRs. The competition by the unlabeled S4 3′NTR does not follow first-order kinetics, which is what would be expected if multiple complexes are bound by the labeled S4 3′NTR. It is not apparent from our studies whether these protein complexes serve to enhance or repress translation, but these results are consistent with a model in which cellular proteins are important for mediating translational control. Studies are now in progress to identify the cellular proteins that interact with the S4 3′NTR and determine whether these interactions occur in cultured cells.
Regulation of gene expression for mammalian reoviruses is not well understood. Since there is little evidence to support control of gene expression at the level of transcription or mRNA stability, the primary opportunity for regulation occurs during translation. The existence of a translational operator sequence in the S4 gene 3′NTR suggests that translation of the s4 mRNA is a tightly controlled process. We hypothesize that this region determines appropriate expression of the ς3 protein by enhancing or repressing translation at optimal times during infection or in appropriate cell types. Careful assessment of this hypothesis awaits development of a means to introduce targeted mutations into the reovirus genome. In addition, our results suggest that cellular proteins serve both regulatory and catalytic functions during translation of reovirus transcripts. It will be interesting to explore the precise biochemical mechanisms and biological roles of the translational program of reovirus. These studies will contribute to an enhanced understanding of protein-RNA interactions and mechanisms of translational control in eukaryotic cells.
ACKNOWLEDGMENTS
We are grateful to Margo Brinton, Susan Low, and Erica White for expert advice. We thank Christopher Coffey for assistance with statistical analysis and Michelle Becker, Jim Chappell, Ron Emeson, Neil Green, Jacek Hawiger, and Jim Patton for careful review of the manuscript.
This work was supported by Public Health Service awards F31 GM17208 from the National Institute of General Medical Sciences (M.M.-G.) and AI32539 from the National Institute of Allergy and Infectious Diseases, the Turner Scholars Program (T.S.D.), and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards CA68485 for the Vanderbilt Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center.
Footnotes
Dedicated to the memory of George C. Lamb, Jr.
REFERENCES
- 1.Atwater J A, Munemitsu S M, Samuel C E. Biosynthesis of reovirus-specified polypeptides. Efficiency of expression of cDNAs of the reovirus S1 and S4 genes in transfected animal cells differs at the level of translation. Virology. 1987;159:350–357. doi: 10.1016/0042-6822(87)90473-9. [DOI] [PubMed] [Google Scholar]
- 2.Baer G S, Dermody T S. Mutations in reovirus outer-capsid protein ς3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A K, Shatkin A J. Transcription in vitro by reovirus-associated ribonucleic acid-dependent polymerase. J Virol. 1970;6:1–11. doi: 10.1128/jvi.6.1.1-11.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsa J, Graham A F. Reovirus RNA polymerase activity in purified virions. Biochem Biophys Res Commun. 1968;33:895–901. doi: 10.1016/0006-291x(68)90396-3. [DOI] [PubMed] [Google Scholar]
- 6.Chappell J D, Goral M I, Rodgers S E, dePamphilis C W, Dermody T S. Sequence diversity within the reovirus S2 gene: reovirus genes reassort in nature, and their termini are predicted to form a panhandle motif. J Virol. 1994;68:750–756. doi: 10.1128/jvi.68.2.750-756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constable A, Quick S, Gray N K, Hentze M W. Modulation of the RNA-binding activity of a regulatory protein by iron in vitro: switching between enzymatic and genetic function? Proc Natl Acad Sci USA. 1992;89:4554–4558. doi: 10.1073/pnas.89.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dermody T S, Nibert M L, Bassel-Duby R, Fields B N. Sequence diversity in S1 genes and S1 translation products of eleven type 3 reovirus strains. J Virol. 1990;64:4842–4850. doi: 10.1128/jvi.64.10.4842-4850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dermody T S, Schiff L A, Nibert M L, Coombs K M, Fields B N. The S2 gene nucleotide sequences of prototype strains of the three reovirus serotypes: characterization of reovirus core protein ς2. J Virol. 1991;65:5721–5731. doi: 10.1128/jvi.65.11.5721-5731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 11.Gallie D R, Lewis N J, Marzluff W F. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giantini M, Seliger L S, Furuichi Y, Shatkin A J. Reovirus type 3 genome segment S4: nucleotide sequence of the gene encoding a major virion surface protein. J Virol. 1984;52:984–987. doi: 10.1128/jvi.52.3.984-987.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goral M I, Mochow-Grundy M, Dermody T S. Sequence diversity within the reovirus S3 gene: reoviruses evolve independently of host species, geographic locale, and date of isolation. Virology. 1996;216:265–271. doi: 10.1006/viro.1996.0059. [DOI] [PubMed] [Google Scholar]
- 14.Hann L E, Webb A C, Cai J M, Gehrke L. Identification of a competitive translation determinant in the 3′ untranslated region of alfalfa mosaic virus coat protein mRNA. Mol Cell Biol. 1997;17:2005–2013. doi: 10.1128/mcb.17.4.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison S J, Farsetta D L, Kim J, Noble S, Broering T J, Nibert M L. Mammalian reovirus L3 gene sequences and evidence for a distinct amino-terminal region of the λ1 protein. Virology. 1999;258:54–64. doi: 10.1006/viro.1999.9707. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Tahara S M, Lai M M. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izquierdo J M, Cuezva J M. Control of the translational efficiency of βF1-ATPase mRNA depends on the regulation of a protein that binds the 3′ untranslated region of the mRNA. Mol Cell Biol. 1997;17:5255–5268. doi: 10.1128/mcb.17.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedl R, Schmechel S, Schiff L. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J Virol. 1995;69:552–559. doi: 10.1128/jvi.69.1.552-559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin D H, Mendelsohn N, Schonberg M, Klett H, Silverstein S, Kapuler A M. Properties of RNA transcriptase in reovirus subviral particles. Proc Natl Acad Sci USA. 1970;66:890–897. doi: 10.1073/pnas.66.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin K H, Samuel C E. Biosynthesis of reovirus-specified polypeptides: purification and characterization of the small-sized class mRNAs of reovirus type 3. Coding assignments and translational efficiencies. Virology. 1980;106:1–13. doi: 10.1016/0042-6822(80)90216-0. [DOI] [PubMed] [Google Scholar]
- 22.McCrae M A, Joklik W K. The nature of the polypeptide encoded by each of the ten double-stranded RNA segments of reovirus type 3. Virology. 1978;89:578–593. doi: 10.1016/0042-6822(78)90199-x. [DOI] [PubMed] [Google Scholar]
- 23.Munemitsu S M, Samuel C E. Biosynthesis of reovirus-specified polypeptides: effect of point mutation of the sequences flanking the 5′-proximal AUG initiator codons of the reovirus S1 and S4 genes on the efficiency of messenger RNA translation. Virology. 1988;163:643–646. doi: 10.1016/0042-6822(88)90309-1. [DOI] [PubMed] [Google Scholar]
- 24.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mustoe T A, Ramig R F, Sharpe A H, Fields B N. Genetics of reovirus: identification of the dsRNA segments encoding the polypeptides of the μ and ς size classes. Virology. 1978;89:594–604. doi: 10.1016/0042-6822(78)90200-3. [DOI] [PubMed] [Google Scholar]
- 26.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1557–1596. [Google Scholar]
- 27.Patton J T, Wentz M, Xiaobo J, Ramig R F. cis-acting signals that promote genome replication in rotavirus mRNA. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 30.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 31.Schlegl J, Gegout V, Schlager B, Hentze M W, Westhof E, Ehresmann C, Ehresmann B, Romby P. Probing the structure of the regulatory region of human transferrin receptor messenger RNA and its interaction with iron regulatory protein-1. RNA. 1997;3:1159–1172. [PMC free article] [PubMed] [Google Scholar]
- 32.Shatkin A J, Sipe J D. RNA polymerase activity in purified reoviruses. Proc Natl Acad Sci USA. 1968;61:1462–1469. doi: 10.1073/pnas.61.4.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skehel J J, Joklik W K. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology. 1969;39:822–831. doi: 10.1016/0042-6822(69)90019-1. [DOI] [PubMed] [Google Scholar]
- 34.Tarlow O, McCorquodale J G, McCrae M A. Molecular cloning and sequencing of the gene (M2) encoding the major virion structural protein μ1-μ1C of serotypes 1 and 3 of mammalian reovirus. Virology. 1988;164:141–146. doi: 10.1016/0042-6822(88)90629-0. [DOI] [PubMed] [Google Scholar]
- 35.Thomson A M, Rogers J T, Leedman P J. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int J Biochem Cell Biol. 1999;31:1139–1152. doi: 10.1016/s1357-2725(99)00080-1. [DOI] [PubMed] [Google Scholar]
- 36.Virgin H W, IV, Bassel-Duby R, Fields B N, Tyler K L. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z F, Ingledue T C, Dominski Z, Sanchez R, Marzluff W F. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol. 1999;19:835–845. doi: 10.1128/mcb.19.1.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wentz M J, Patton J T, Ramig R F. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiener J R, Joklik W K. The sequences of the reovirus serotype 1, 2, and 3 L1 genome segments and analysis of the mode of divergence of the reovirus serotypes. Virology. 1989;169:194–203. doi: 10.1016/0042-6822(89)90055-x. [DOI] [PubMed] [Google Scholar]
- 40.Xu Z K, Anzola J V, Nalin C M, Nuss D L. The 3′-terminal sequence of a wound tumor virus transcript can influence conformational and functional properties associated with the 5′-terminus. Virology. 1989;170:511–522. doi: 10.1016/0042-6822(89)90443-1. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Q, Guo Y, Liu Y. Regulation of the stability of heat-stable antigen mRNA by interplay between two novel cis elements in the 3′ untranslated region. Mol Cell Biol. 1998;18:815–826. doi: 10.1128/mcb.18.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zweerink H J, Joklik W K. Studies on the intracellular synthesis of reovirus-specified proteins. Virology. 1970;41:501–518. doi: 10.1016/0042-6822(70)90171-6. [DOI] [PubMed] [Google Scholar]