Abstract
Hepatitis C virus (HCV) encodes two viral envelope glycoproteins. E1 contains 4 or 5 N-linked glycosylation sites and E2 contains up to 11, with most of the sites being well conserved, suggesting that they play an essential role in some functions of these proteins. For this study, we used retroviral pseudotyped particles harboring mutated HCV envelope glycoproteins to study these glycans. The mutants were named with an N followed by a number related to the relative position of the potential glycosylation site in each glycoprotein (E1N1 to E1N4 for E1 mutants and E2N1 to E2N11 for E2 mutants). The characterization of these mutants allowed us to define three phenotypes. For the first group (E1N3, E2N3, E2N5, E2N6, E2N7, and E2N9), the infectivities of the mutants were close to that of the wild type. The second group (E1N1, E1N2, E1N4, E2N1, and E2N11) contained mutants that were still infectious but whose infectivities were reduced to <50% that of the wild type. The third group (E2N2, E2N4, E2N8, and E2N10) contained mutants that had almost totally lost infectivity. The absence of infectivity of the E2N8 and E2N10 mutants was due to the lack of incorporation of the E1E2 heterodimer into HCVpp, which was due to misfolding of the heterodimer, as shown by immunoprecipitation with conformation-sensitive antibodies and by a CD81 pull-down assay. The absence of infectivity of the E2N2 and E2N4 mutants indicated that these two glycans are involved in controlling HCV entry. Altogether, the data indicate that some glycans of HCV envelope glycoproteins play a major role in protein folding and others play a role in HCV entry.
Hepatitis C virus (HCV) is a major cause of chronic liver diseases, including chronic hepatitis, cirrhosis, and hepatocellular carcinoma (34). HCV is a small enveloped virus that belongs to the Hepacivirus genus in the Flaviviridae family (30). Its genome is supposed to be directly translated after uncoating of the particle and encodes a single polyprotein precursor of ∼3,010 amino acid residues. The HCV polyprotein is synthesized on endoplasmic reticulum (ER)-associated ribosomes and is cleaved co- and posttranslationally by cellular and viral proteases to yield at least 10 mature products (reviewed in reference 30). The two envelope glycoproteins, E1 and E2, are released from the polyprotein by signal peptidase cleavages. HCV glycoproteins are type I transmembrane proteins with N-terminal ectodomains and C-terminal hydrophobic anchors (41). During their synthesis, the ectodomains of HCV glycoproteins are targeted to the ER lumen, where they are modified by N-linked glycosylation. After their synthesis, HCV glycoproteins E1 and E2 assemble as noncovalent heterodimers which are mainly retained in the ER (41). HCV envelope glycoproteins contain 4 or 5 potential glycosylation sites (E1) or up to 11 glycosylation sites (E2) (21). Most of these glycosylation sites are well conserved (21, 58), suggesting that they play an essential role in some functions of HCV envelope glycoproteins.
N-linked glycosylation is one of the most common types of protein modification. N-linked oligosaccharides are added to specific asparagine residues in the context of the consensus sequence Asn-X-Ser/Thr (28). The sugar composition of oligosaccharides and the number and size of branches in the sugar tree vary widely (28). However, the glycans are homogeneous and relatively simple when they are initially added in the ER to growing nascent polypeptides. In the early secretory pathway, glycans play a role in protein folding, quality control, and certain sorting events. Viral envelope proteins usually contain N-linked glycans that can play a major role in their folding, in their entry functions, or in modulating the immune response (22, 39, 40, 53, 54).
A characterization of the HCV glycoprotein E1 has shown that in the absence of E2, different glycoforms of E1 are produced and the glycosylation of E1 is improved by the coexpression of E2 in cis (13). Site-directed mutagenesis has also shown that the presence of some glycans in E1 is necessary for the formation of noncovalent E1E2 heterodimers (36). More recently, the glycosylation process of the HCV envelope glycoprotein E1 was analyzed in the context of a Man-P-Dol-deficientcell line (B3F7) and was shown to occur posttranslationally (17). It has also been reported that the presence of multiple N-linked glycans is required for the proper folding of E2 and for the formation of its antigenic structure (38, 49). However, these studies were done with a truncated form of E2, which does not necessarily have the same properties in the absence of E1 (6).
Due to the lack of a robust cell culture system to amplify HCV, all studies on the glycosylation of HCV envelope glycoproteins have focused on the role of glycans in the biogenesis of these proteins. Recently, infectious pseudotyped particles (HCVpp) that are assembled by displaying unmodified HCV envelope glycoproteins on retroviral core particles have been successfully generated and now enable studies of HCV entry (2, 12, 24). For this study, we used HCVpp harboring mutated envelope glycoproteins to determine the role of N-linked glycans in the folding and entry functions of HCV envelope glycoproteins. Interestingly, some mutations altered the folding of HCV envelope glycoproteins, whereas other mutations affected HCVpp infectivity, indicating that some glycans of HCV envelope glycoproteins play a major role in protein folding or HCV entry.
MATERIALS AND METHODS
Cell culture.
Huh-7 human hepatoma cells (37) and 293T human embryo kidney cells (293tsA1609neo) obtained from the American Type Culture Collection (Manassas, Va.) were grown in Dulbecco's minimum essential medium (Invitrogen) supplemented with 10% fetal bovine serum.
Site-directed mutagenesis.
The DNA sequences used to construct HCV glycosylation mutants were derived from the H strain (genotype 1a) (19). The fragments encoding the glycosylation mutants of E1 were excised from pTM1/ΔCE1[N1 to N4]E2p7 with AscI and EcoNI (36) and ligated into an AscI/EcoNI-digested phCMV-E1E2 plasmid (2). For E2 mutants, the AAT or AAC codon for the asparagine residue (Asn) of the consensus sequence Asn-X-Ser/Thr was replaced with the CAA codon for a glutamine residue (Gln). The mutations were constructed by sequential PCR steps as described previously (1), using the high-fidelity Deep Vent DNA polymerase (New England Biolabs). The mutants were then assembled by a second PCR amplification. For mutants E2N1 to E2N5, the amplicons were digested with BclI and AscI and ligated into a BclI/AscI-digested phCMV-E1E2 plasmid. For mutants E2N6 to E2N11, the amplicons were digested with AscI and BglII and ligated into an AscI/BglII-digested phCMV-E1E2 plasmid. The positions of the glycosylation sites are indicated in Fig. 1A and 2A. The sequences of all constructs were verified.
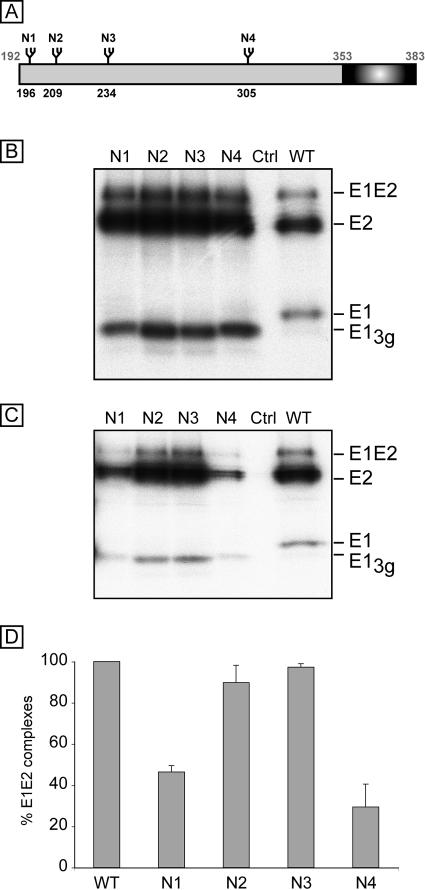
FIG. 1.
Analysis of E1 glycosylation mutants and their effects on the formation of E1E2 complexes. (A) Schematic representation of N-glycosylation sites in HCV glycoprotein E1. The positions of the glycosylation sites are indicated above the box corresponding to the E1 protein. The numbers correspond to the positions in the polyprotein of reference strain H (GenBank accession no. AF009606). The mutants are named with an N followed by a number related to the position of the glycosylation site on the map. The transmembrane domain of E1 is indicated with a dark box. (B and C) Role of E1 glycans in the formation of E1E2 complexes. A plasmids encoding a wild-type (WT) or mutant E1 protein in the context of an E1E2 polyprotein was transfected into 293T cells. A control experiment was performed with 293T cells transfected with a plasmid encoding the feline endogenous retrovirus RD114 glycoprotein (Ctrl) (46). At 24 h posttransfection, the cells were pulse labeled with [35S]methionine-[35S]cysteine for 30 min and then chased for 4 h. Cell lysates were used for immunoprecipitation with the MAbs A4 (anti-E1) (B) and H53 (anti-E2) (C). Immunoprecipitates were separated by SDS-PAGE (10% polyacrylamide) under reducing conditions. E1-3g corresponds to E1 lacking one glycan. The HCV envelope proteins E1 and E2 as well as SDS-resistant E1E2 complexes are indicated on the right. (D) Percentages of noncovalent E1E2 heterodimers. The intensities of the bands corresponding to the E2 proteins precipitated by MAbs H53 and A4 were measured by phosphorimaging for three independent experiments, and the mean percentage of noncovalent E1E2 heterodimers was cal-culated for each mutant as follows: (amount of E2 protein from the mutant precipitated by MAb H53/amount of E2 from the mutant precipitated by MAb A4)/(amount of wild-type E2 precipitated by MAb H53/amount of wild-type E2 precipitated by MAb A4).
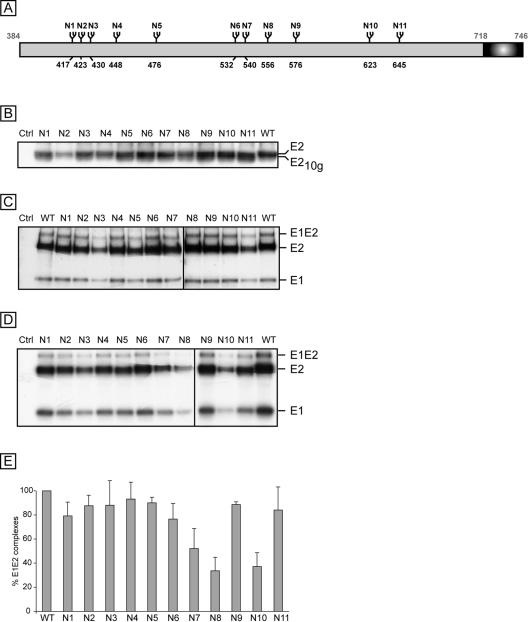
FIG. 2.
Analysis of E2 glycosylation mutants and their effects on the formation of E1E2 complexes. (A) Schematic representation of N-glycosylation sites in HCV glycoprotein E2. The positions of the glycosylation sites are indicated above the box corresponding to the E2 protein. The numbers correspond to the positions in the polyprotein of reference strain H (accession no. AF009606). The mutants are named with an N followed by a number related to the position of the glycosylation site on the map. The transmembrane domain of E2 is indicated with a dark box. (B) Analysis of N-linked glycosylation mutants of E2. Plasmids containing the sequences encoding mutants E2N1 to E2N11 were transiently expressed in 293T cells. At 16 h posttransfection, cell lysates were separated by SDS-PAGE (10% polyacrylamide) and the proteins of interest were revealed by Western blotting with an anti-E2 (3/11) MAb. A cell lysate containing the wild-type E1 and E2 proteins (WT) was run in parallel. E2-10g corresponds to E2 lacking one glycan. (C and D) Role of E2 glycans in the formation of E1E2 complexes. A plasmid encoding a wild-type (WT) or mutant E2 protein in the context of an E1E2 polyprotein was transfected into 293T cells. A control experiment was performed with 293T cells transfected with a plasmid encoding the RD114 retroviral envelope protein (Ctrl) (46). At 24 h posttransfection, the cells were pulse labeled with [35S]methionine-[35S]cysteine for 30 min and then chased for 4 h. Cell lysates were used for immunoprecipitation with the MAbs 3/11 (C) and H53 (D), and the immunoprecipitates were separated by SDS-PAGE (10% polyacrylamide) under reducing conditions. HCV envelope proteins E1 and E2, as well as SDS-resistant E1E2 complexes, are indicated on the right. (E) Percentages of noncovalent E1E2 heterodimers. The intensities of the bands corresponding to the E2 proteins precipitated by MAbs H53 and 3/11 were measured by phosphorimaging for three independent experiments, and the mean percentage of noncovalent E1E2 heterodimers was calculated for each mutant as follows: (amount of mutant E2 protein precipitated by MAb H53/amount of mutant E2 precipitated by MAb 3/11)/(amount of wild-type E2 precipitated by MAb H53/amount of wild-type E2 precipitated by MAb 3/11).
Antibodies.
The monoclonal antibodies (MAbs) A4 (anti-E1) (14), H2 (anti-E2) (11), H53 (anti-E2) (7), 3/11 (anti-E2; kindly provided by J. McKeating) (18), and R187 (anti-murine leukemia virus [MLV] capsid; ATCC CRL1912) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. An anti-calnexin antibody (SPA-860) was supplied by Stress Gen.
Production of HCVpp and infection assays.
The production of HCVpp and infection assays have been described previously (2, 42). For this work, we used a cytomegalovirus (CMV)-Gag-Pol-murine leukemia virus (MLV) packaging construct, which encodes the MLV gag and pol genes, and the MLV-Luc plasmid, which encodes an MLV-based transfer vector containing a CMV-Luc internal transcriptional unit. To analyze the incorporation of HCV envelope glycoproteins into pseudotyped particles, we pelleted HCVpp by centrifugation through a 30% sucrose cushion and analyzed it by Western blotting. Within a given preparation of virions, similar amounts of virion-associated MLV capsid proteins were detected for HCVpp generated with the different mutants. However, as previously observed, important differences in the absolute quantities of virion-associated capsids could be noticed when two independent preparations of HCVpp were compared (29). Thus, to minimize artifacts due to differences in the quality of preparations, we conducted each evaluation experiment by using concurrently generated HCVpp. A normalization of pseudotyped vector stocks was therefore performed by using luciferase activities determined with Huh-7 cells as previously described (29).
Immunoprecipitation.
Monolayers of 293T cells grown in six-well plates were transfected with different glycosylation mutants. At 16 h posttransfection, the cells were washed and incubated in Dulbecco's modified medium without l-methionine and l-cysteine (Invitrogen) for 30 min. Transfected cells were pulse labeled for 30 min with 100 μCi per ml of Promix ([35S]methionine-[35S]cysteine; Amersham) as described previously (14). The cells were washed twice with medium containing a 10-fold excess of methionine and cysteine, followed by a 4-h chase. The cells were lysed with 1 ml of 0.5% Igepal CA-630 in phosphate-buffered saline, and immunoprecipitation was performed as previously described (15). Immunoprecipitates were eluted from protein A-Sepharose beads in 30 μl of 4× Laemmli sample buffer by heating for 10 min at 70°C and were analyzed by 10% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gels were treated with sodium salicylate, dried, and exposed at −70°C to preflashed Hyperfilm-MP (Amersham).
Western blotting.
After separation by SDS-PAGE, protein preparations were transferred to nitrocellulose membranes (Hybond-ECL; Amersham) by use of a Trans-Blot apparatus (Bio-Rad) and were detected with a specific MAb followed by rabbit anti-mouse (Dako) or goat anti-rat (Biosource) immunoglobulin conjugated to peroxidase (diluted 1/1,000). The proteins of interest were analyzed by enhanced chemiluminescence detection (Amersham) as recommended by the manufacturer.
CD81 pull-down assay.
CD81 pull-down experiments were performed as previously described (6). Recombinant fusion proteins containing the large extracellular loop of human or murine CD81 fused to glutathione S-transferase (GST) were preadsorbed onto glutathione-Sepharose 4B beads according to the manufacturer's recommendations (Pharmacia Biotech, Uppsala, Sweden). Precipitates were separated by SDS-PAGE (10% polyacrylamide), followed by Western blotting with an anti-E1 (A4) or anti-E2 (3/11) MAb.
RESULTS
Identification of functional N-linked glycosylation sites in HCV envelope glycoproteins.
To determine which of the potential glycosylation motifs of the HCV envelope glycoproteins are used, we mutated the asparagines to which glycans are potentially added to glutamines, which is known to prevent glycosylation without affecting the physicochemical properties of the sequon. The mutants were named with an N followed by a number related to the relative position of the potential glycosylation site in each glycoprotein (E1N1 to E1N4 for E1 mutants and E2N1 to E2N11 for E2 mutants) (Fig. 1A and 2A). In most HCV isolates, the E1 sequence contains five Asn-X-Ser sequons (21). However, one of them contains a Pro immediately downstream of the sequon, which is known to be unfavorable for N-linked glycosylation. Site-directed mutagenesis of E1 sequons has indeed confirmed that this site is not glycosylated, whereas the other sites can be glycosylated (36) (Fig. 1). In our study, the E1 mutants, named E1N1 to E1N4, were introduced into the phCMV/E1E2 expression vector so that E1 and E2 were coexpressed as a polyprotein. The expression of E1 mutants was analyzed by immunoprecipitation after the transfection of 293T cells with the appropriate plasmids. As shown in Fig. 1, E1 mutants migrated faster than the wild-type glycoprotein. This faster migrating form of E1 was called E1-3g because it contained three glycans instead of four. In addition, after the removal of the glycans by endoglycosidase H, all of the mutants comigrated with the deglycosylated form of wild-type E1 (data not shown), confirming that the migration shift of the nondeglycosylated proteins was due to the lack of glycan in the mutant proteins and not to changes in the protein backbone. E1N1 and E1N4 had a slightly lower migration rate than the other mutants, which was not observed after deglycosylation (data not shown). This suggests that there might be differences in the trimming of the associated glycans of these mutants, which might be due to misfolding of E1N1 and E1N4. Altogether, these data indicate that glycans are present at positions 1 to 4 in wild-type E1, as previously shown (36).
Like the case for the majority of HCV isolates, the E2 sequence used for this work contains 11 potential N-linked glycosylation sites (21) (Fig. 2A). To determine which of the 11 predicted N-linked glycosylation sites were used for the modification of E2, we mutated each site individually in the context of the phCMV/E1E2 expression vector. The mutants were named E2N1 to E2N11 and numbered from the N terminus of E2 (Fig. 2). The expression of the E2 mutants was analyzed by Western blotting after the transfection of 293T cells with the appropriate plasmids. As shown in Fig. 2B, all of the mutants migrated slightly faster than the wild-type protein. This faster migrating form of E2 was called E2-10g because it contained 10 glycans instead of 11. In addition, after removal of the glycans by endoglycosidase H, all of the mutants comigrated with the deglycosylated form of wild-type E2 (data not shown), confirming that the migration shift of the nondeglycosylated proteins was due to the lack of glycan in the mutant proteins and not to changes in the protein backbone. Together, these data indicate that glycans are present at positions 1 to 11 in wild-type E2.
Role of N-linked glycans in folding of HCV envelope glycoproteins.
In the absence of conformation-sensitive MAbs directed against E1, it is not easy to directly evaluate E1 folding. We therefore analyzed the formation of E1E2 complexes with a conformation-sensitive MAb. We used an immunoprecipitation assay with MAb H53, which has been shown to specifically precipitate the noncovalent E1E2 complex (7, 16). Cells expressing mutated HCV proteins were pulse labeled with [35S]methionine-[35S]cysteine for 30 min and chased for 4 h. These conditions have been shown to be appropriate for detecting the peak of heterodimer formation (11). Immunoprecipitation with a conformation-insensitive anti-E1 MAb (A4) was performed in parallel to compare the levels of expression of the E1 and E2 proteins. As shown in Fig. 1C, noncovalent E1E2 heterodimers were immunoprecipitated with H53 for all of the mutants. However, compared to the case for immunoprecipitation with the anti-E1 MAb A4, the relative amount of noncovalent E1E2 complexes was strongly reduced for the E1N1 and E1N4 mutants. Indeed, a quantitative analysis indicated that MAb H53 precipitated only 46% and 29% of complexes for the E1N1 and E1N4 mutants, respectively. In addition, these data also suggest that the E1N2 and E1N3 mutants do not dramatically alter E1E2 assembly. However, we cannot exclude some misfolding of E1 in a domain that is not involved in the E1E2 interaction. These results are in agreement with an earlier study of glycosylation mutants of E1 (36).
A similar approach was developed to analyze the ability of E2 glycosylation mutants to form E1E2 heterodimers. Mutant proteins were immunoprecipitated with the conformation-sensitive MAb H53, and a control immunoprecipitation assay with a conformation-insensitive anti-E2 MAb (3/11) was performed in parallel to compare the levels of expression of the E1 and E2 proteins. As shown in Fig. 2D, noncovalent E1E2 heterodimers were immunoprecipitated with H53 for all of the mutants. However, compared to the case for immunoprecipitation with the anti-E2 MAb 3/11, the relative amount of noncovalent E1E2 complexes was strongly reduced for the E2N7, E2N8, and E2N10 mutants. Indeed, a quantitative analysis indicated that MAb H53 precipitated only 51%, 33%, and 36% of complexes for the E2N7, E2N8, and E2N10 mutants, respectively. In addition, these data also suggest that the E2N1, E2N2, E2N3, E2N4, E2N5, E2N6, E2N9, and E2N11 mutants do not dramatically alter E1E2 assembly. A similar alteration in the recognition of E1E2 heterodimers was observed for the E2N7, E2N8, and E2N10 mutants when another conformation-sensitive MAb was used (H2) (11; data not shown).
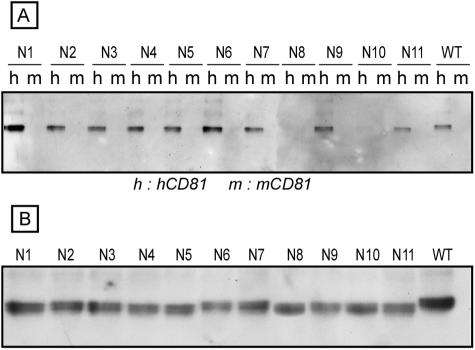
CD81 is a putative receptor or coreceptor for HCV (2, 9, 57), and it has been shown that the CD81-E2 interaction is dependent on the conformation of E2 (18). We therefore used human CD81 as another probe to further characterize our glycosylation mutants. For this purpose, we analyzed E1 and E2 mutants in a CD81LEL-GST pull-down assay as previously described (6). An interaction between CD81LEL-GST and E2 was revealed in a pull-down assay followed by Western blotting with an anti-E2 antibody. The murine CD81LEL protein was used as a negative control because it does not interact with E2 in this pull-down assay (6, 18, 43). As shown in Fig. 3A, E2 was precipitated with human CD81LEL for nine of the E2 glycosylation mutants (E2N1, E2N2, E2N3, E2N4, E2N5, E2N6, E2N6, E2N7, E2N9, and E2N11), and the intensities of the bands corresponding to E2 were similar to that for wild-type E2. Interestingly, no signal was detected above the background for the E2N8 and E2N10 mutants, in spite of their expression levels being similar to those of wild-type E2 (Fig. 3B), suggesting an alteration in the formation of the CD81-binding domain in these mutants. This is in agreement with the results of immunoprecipitation with conformation-sensitive MAbs, indicating an alteration in E2 folding when the N8 or N10 glycosylation site was mutated. It is worth noting that the E2N7 mutant behaved differently than the E2N8 and E2N10 mutants. Indeed, mutation of the E2N7 glycosylation site did not reduce the E2 interaction with human CD81LEL (Fig. 3), whereas its recognition by conformation-sensitive MAbs was reduced (Fig. 2 and data not shown). These data suggest that partial folding occurred in the E2N7 mutant, leading to the formation of the CD81-binding domain.
FIG. 3.
Interaction of HCV glycosylation mutants with human CD81LEL. A plasmid encoding a wild-type (WT) or mutant E2 protein in the context of an E1E2 polyprotein was transfected into 293T cells. At 24 h posttransfection, the cells were lysed and analyzed in a CD81LEL-GST pull-down assay (A) using human (h) or murine (m) CD81LEL fused to GST. The presence of the E2 glycoprotein in the precipitates was revealed by Western blotting with an anti-E2 MAb (3/11). The expression of mutant proteins was verified by direct Western blotting of cell lysates (B).
We also analyzed the E1 mutants in our CD81LEL-GST pull-down assay. However, none of the mutations led to an alteration in the E2 interaction with CD81 (data not shown), indicating that mutation of the glycosylation sites in E1 does not affect the formation of the CD81-binding domain in E2.
Together, these data indicate that the glycans at positions E2N7, E2N8, and E2N10 play a major role in the folding of E2 and, consequently, in the formation of noncovalent E1E2 heterodimers.
Interestingly, calnexin, an ER chaperone that binds to monoglucosylated N-linked oligosaccharides (23), has been shown to interact with HCV envelope glycoproteins (5, 15, 35). However, E2 proteins mutated at positions E2N7, E2N8, and E2N10 were still recognized by calnexin (data not shown), indicating that the alteration in E2 folding was not due to a lack of recognition by this chaperone, as previously shown for E1N1 and E1N4 mutants (36).
Role of E1 glycans on HCVpp infectivity.
HCVpp that are assembled by displaying unmodified HCV envelope glycoproteins on retroviral core particles have recently been reported and now enable studies of HCV entry (2, 24). We therefore analyzed our glycosylation mutants in the HCVpp model to determine their effects on HCVpp infectivity. For a precise determination of the effects of the mutations on the entry function, one possibility would be to normalize the infectivity relative to E1 and E2 protein levels in HCVpp. This was, however, not possible in our case because the E1/E2 ratio varied for some mutants. For this reason, we measured the global infectivities of our mutants and analyzed the levels of incorporation of envelope glycoproteins into HCVpp. This allowed us to discriminate between mutations that affected folding or assembly and those that affected entry.
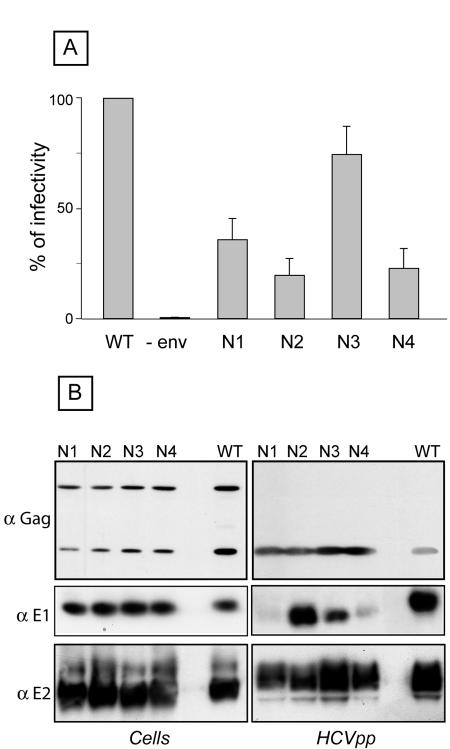
HCVpp were generated for each glycosylation mutant, and their infectivity was compared to that of HCVpp generated in the presence of wild-type HCV envelope glycoproteins. Particles produced in the absence of envelope proteins were used as a negative control to evaluate the background level of luciferase activity. As shown in Fig. 4A, a strong reduction in infectivity was observed for the E1N1, E1N2, and E1N4 mutants. Indeed, compared to wild-type E1E2, these mutants showed 36%, 20%, and 23% infectivity, respectively. In contrast, the infectivity of the E1N3 mutant was only slightly affected (74.5%).
FIG. 4.
Role of E1 glycans in HCVpp infectivity. (A) Plasmids encoding E1 mutants in the context of an E1E2 polyprotein were used to generate HCVpp (E1N1 to E1N4). Control experiments in the absence of HCV envelope proteins were also performed (−env). Infection assays with the luciferase reporter gene were performed by using target Huh-7 human hepatocarcinoma cells. Similar inputs of viral particles were used in each experiment, and this was confirmed by comparing the amounts of capsid protein incorporated into HCVpp (see panel B, anti-Gag). The results are expressed as percentages of infectivity. For each mutant, the percentage of infectivity was calculated as the luciferase activity of HCVpp produced with mutant envelope proteins divided by the luciferase activity of HCVpp produced with wild-type E1 and E2 (WT). The results are reported as means ± standard deviations of six independent experiments. Pseudotyped particles produced in the absence of envelope proteins were used as a control. The mean luciferase activity of such pseudotyped particles produced in the absence of envelope proteins represented <2% of the activity measured for HCVpp. The luciferase activities (in relative light units) in a representative experiment were as follows: WT, 3.6 × 105; N1, 1.5 × 105; N2, 3.4 × 104; N3, 2.7 × 105; and N4, 7.7 × 104. (B) Incorporation of HCV envelope proteins into HCVpp. Particles were pelleted through 30% sucrose cushions and analyzed by Western blotting. HCV envelope glycoproteins and the capsid protein of MLV were revealed with the following specific MAbs: anti-E1 (A4), anti-E2 (3/11), and anti-MLV capsid (R187, anti-Gag). The expression of mutant proteins was verified by direct Western blotting of cell lysates. The proteins analyzed for this figure were separated by 15% SDS-PAGE, and for this reason, the differences in size between the mutant and wild-type proteins were reduced.
To determine whether these changes in HCVpp infectivity were due to indirect effects leading to alterations in particle assembly, we analyzed HCVpp for the presence of viral proteins and compared them to cell lysates expressing these proteins. As shown in Fig. 4B, the levels of expression of E1, E2, and Gag were very similar in cell lysates for all of the E1 mutants. The levels of expression of E2 and Gag were similar in HCVpp; however, strong differences were observed for E1 expression. Indeed, the levels of E1 incorporation into HCVpp were dramatically reduced for the E1N1 and E1N4 mutants. The level of E1N2 incorporation into HCVpp was similar to that of wild-type E1, whereas the incorporation level was reduced for the E1N3 mutant (Fig. 4B). The alteration of E1 incorporation into HCVpp observed for the E1N1 and E1N4 mutants suggests that these mutations alter the folding of E1, as mentioned previously (36). The partial reduction in the incorporation of E1N3 into HCVpp did not seem to affect the infectivity of this mutant. Interestingly, the strong reduction in HCVpp infectivity of the E1N2 mutant, together with the normal incorporation of envelope proteins into HCVpp, suggests that the absence of a glycan at this position had a direct effect on HCVpp entry.
Altogether, these results indicate that two E1 glycans (E1N1 and E1N4) play a role in folding and incorporation into HCVpp, whereas a third one (E1N2) modulates the entry functions of HCV envelope glycoproteins.
Role of E2 glycans in HCVpp infectivity.
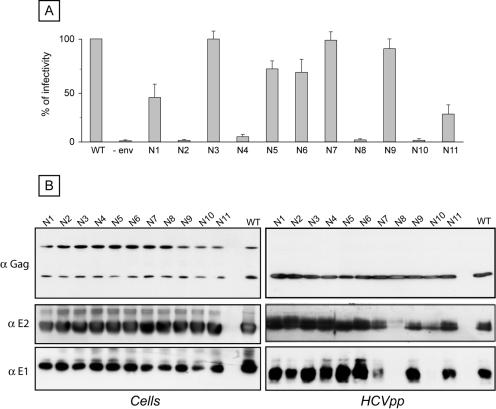
HCVpp were also generated for E2 glycosylation mutants, and their infectivities were also compared to that of HCVpp generated in the presence of wild-type HCV envelope glycoproteins. The characterization of these glycosylation mutants allowed the definition of three phenotypes (Fig. 5A). For the first group, the infectivities of the mutants were close to that of the wild type (E2N3, 100%; E2N5, 71%; E2N6, 68%; E2N7, 100%; E2N9, 91%). The second group contained mutants that were still infectious but whose infectivities were reduced to <50% that of the wild type (E2N1, 41%; E2N11, 27%). The third group contained mutants that had totally or almost totally lost infectivity, i.e., E2N2, E2N4, E2N8, and E2N10.
FIG. 5.
Role of E2 glycans in HCVpp infectivity. (A) Plasmids encoding E2 mutants in the context of an E1E2 polyprotein were used to generate HCVpp (E1N1 to E1N11). Control experiments in the absence of HCV envelope proteins were also performed (−env). Infection assays with the luciferase reporter gene were performed by using target Huh-7 human hepatocarcinoma cells. Similar inputs of viral particles were used in each experiment, and this was confirmed by comparing the amounts of capsid protein incorporated into HCVpp (see panel B, anti-Gag). The results are expressed as percentages of infectivity. For each mutant, the percentage of infectivity was calculated as the luciferase activity of HCVpp produced with mutant envelope proteins divided by the luciferase activity of HCVpp produced with wild-type E1 and E2 (WT). The results are reported as means ± standard deviations of six independent experiments. Pseudotyped particles produced in the absence of envelope proteins were used as a control. The mean luciferase activity of such pseudotyped particles produced in the absence of envelope proteins represented <2% of the activity measured for HCVpp. The luciferase activities (in relative light units) in a representative experiment were as follows: WT, 4.7 × 105; N1, 2.3 × 105; N2, 1.4 × 103; N3, 4.6 × 105; N4, 1.9 × 104; N5, 2.5 × 105; N6, 2.7 × 105; N7, 4.7 × 105; N8, 4.3 × 103; N9, 4.7 × 105; N10, 1.4 × 103; and N11, 1.3 × 105. (B) Incorporation of HCV envelope proteins into HCVpp. Particles were pelleted through 30% sucrose cushions and analyzed by Western blotting. HCV envelope glycoproteins and the capsid protein of MLV were revealed with the following specific MAbs: anti-E1 (A4), anti-E2 (3/11), and anti-MLV capsid (R187, anti-Gag). The expression of mutant proteins was verified by direct Western blotting of cell lysates.
To determine whether these changes in HCVpp infectivity were due to indirect effects leading to alterations in particle assembly, we analyzed HCVpp for the presence of viral proteins and compared them to cell lysates expressing these proteins. As shown in Fig. 5B, the levels of expression of E1, E2, and Gag were very similar in cell lysates for all of the E2 mutants. The levels of expression of Gag were also very similar in HCVpp. However, some differences were observed for the incorporation of E1 and E2 into HCVpp. The levels of E1 and E2 incorporation into HCVpp were very close to that of the wild type for the E2N1, E2N2, E2N3, E2N4, E2N5, E2N6, E2N9, and E2N11 mutants (Fig. 5B). Interestingly, no E1 protein was detected in association with HCVpp generated with the E2N8 and E2N10 mutants, and the levels of E2 incorporation into HCVpp were very low for these mutants. These results are in agreement with an alteration of the folding and assembly of HCV envelope glycoproteins in the absence of a glycan at position 8 or 10 (Fig. 2 and 3). The lack of infectivity of HCVpp generated with the E2N8 and E2N10 mutants was therefore likely due to misfolding of E2 and/or E1E2, leading to the lack of incorporation into HCVpp.
The E2N7 mutant showed an interesting pattern. Indeed, HCVpp generated with this mutant remained as infectious as wild-type particles, but the incorporation of the E1 and E2 proteins was reduced (Fig. 5B). This reduction of incorporation into HCVpp was likely due to an alteration in the folding of E2 and/or E1E2, as suggested by the results of immunoprecipitation with MAb H53 (Fig. 2). The high level of infectivity of the E2N7 mutant with a reduced incorporation of the HCV envelope glycoproteins into HCVpp is not easy to understand. We cannot exclude the possibility that the glycan at position 7 has two opposite effects. We have shown that its absence reduces E2 folding (Fig. 2), but the lack of a glycan at this position might also increase the efficiency of HCVpp infectivity. An enhancement of HCVpp infectivity can indeed be observed for some mutants of HCV envelope glycoproteins (Y. Ciczora, B. Bartosch, F. L. Cosset, and J. Dubuisson, unpublished data).
The lack of infectivity of HCVpp generated with the E2N2 and E2N4 mutants suggests that the glycans at positions E2N2 and E2N4 play a major role in HCVpp entry. Indeed, our results indicate that the HCV envelope glycoproteins were properly folded (Fig. 2 and 3) and efficiently incorporated into HCVpp (Fig. 5B). The reduced infectivity without an alteration in HCVpp assembly, as observed for some other mutants (E2N1, E2N5, E2N6, and E2N11), suggests that the glycans at these positions might also modulate the entry functions of HCV envelope glycoproteins.
Altogether, these data indicate that two E2 glycans (E2N8 and E2N10) play a major role in the folding and incorporation of HCV envelope glycoproteins into HCVpp. In addition, two other E2 glycans (E2N2 and E2N4) are essential for the entry functions of HCV envelope glycoproteins, whereas other glycans (E2N1, E2N5, E2N6, and E2N11) modulate these functions.
DISCUSSION
The envelope proteins of a virus play a pivotal role in its life cycle. They participate in the assembly of the infectious particle. They also play a crucial role in viral entry by binding to a receptor present on the host cell and inducing fusion between the viral envelope and a membrane of the host cell. Viral envelope proteins usually contain N-linked glycans that can play a major role in their folding, in their entry functions, and in modulating the immune response. In this study, we analyzed the role of N-linked glycans in HCV envelope glycoproteins by site-directed mutagenesis. Importantly, some glycans were shown to play a major role in folding, whereas other glycans were shown to be essential for the entry functions of HCV envelope glycoproteins. Together, these data point out the essential role of envelope glycoprotein glycans in the HCV life cycle.
HCV envelope glycoproteins are heavily glycosylated. The HCV glycoprotein E1 has been shown to contain 4 or 5 glycosylation sites (19, 36), and a global sequence analysis of the potential glycosylation sites in E2 indicated that 9 of the 11 sites are strongly conserved (21, 58). A first study of glycosylation mutants of E2 in which individual glycosylation sites were mutated was reported previously (38), but these mutants were not characterized in terms of glycosylation and no clear conclusions can be drawn from that study. More recently, another study of glycosylation mutants was also reported (49). For this more recent study, the N-linked glycosylation sites were first mutated in the context of a truncated form of E2 ending at position 660. The E2 sequence of the HCV isolate used for the study contained 10 instead of 11 potential glycosylation sites. The site at position 476, corresponding to the E2N5 site in the H strain, was missing. This site is indeed the least conserved potential glycosylation site in E2; it is present in approximately 75% of HCV isolates (21). In a study by Slater-Handshy and coworkers, the last two glycosylation sites, corresponding to the E2N10 and E2N11 sites in the H strain, were not occupied in E2660 (49). However, at least one of these sites was occupied in the context of full-length E2 or another truncated form (E2715). Differences in glycosylation efficiencies have also been observed between full-length and truncated forms of the rabies virus glycoprotein (48). These data indicate that it is more relevant to study N-linked glycosylation in the context of full-length HCV envelope glycoproteins. Here we show that when E2 is expressed as an E1E2 polyprotein, all of its glycosylation sites are occupied.
Although the oligosaccharyltransferase recognizes the Asn-X-Ser/Thr consensus sequence, it has been shown that some residues at position X can modulate the level of glycosylation of the site. Residues like Trp, Leu, Asp, and Glu at position X have been shown to be associated with less efficient core glycosylation in a cell-free system (47). Indeed, large hydrophobic amino acids may inhibit core glycosylation by producing an unfavorable local conformation, and the charge of the X residue may influence the ability of oligosaccharyltransferase to bind simultaneously to the sequon and the negatively charged dolichol-PP-oligosaccharide precursor (4, 26). Although an Asp residue is present at the X position in the E2N3 and E2N6 sites and a Trp residue is present in the E2N11 site, our data indicate that these sites are glycosylated. Interestingly, an Asn-X-Thr sequon is present at the E2N6 and E2N11 sites, and it has been shown that glycosylation is more efficient at an Asn-X-Thr site than at an Asn-X-Ser site (27). The presence of Thr might therefore compensate for the negative effect of Asp and Trp at position X of the E2N6 and E2N11 glycosylation sites, respectively.
The presence of glycans on a protein can potentially affect its folding through the calnexin-calreticulin chaperone system found in the ER (23). Calnexin and calreticulin are lectin-like chaperones which show an affinity for monoglucosylated N-linked oligosaccharides (52). Calnexin has been shown to interact with HCV envelope glycoproteins (5, 15, 35), and this chaperone has been suggested to be involved in the folding of HCV envelope glycoproteins (5). Our data indicate that the misfolding observed for some E2 mutants (E2N8 and E2N10) is not due to their lack of interaction with calnexin. This is not surprising, as several other glycosylation sites are potentially available to interact with this chaperone. It is therefore likely that E2N8 and E2N10 mutations have a direct effect on protein folding. The presence of a large polar saccharide is indeed known to affect folding, at least locally, by orienting polypeptide segments toward the surfaces of protein domains (25, 56).
N-linked glycans of viral envelope proteins can play a role in the entry functions of these proteins. Indeed, certain glycans located near the receptor-binding site on the top of the hemagglutinin protein of influenza virus regulate receptor affinities (39). In addition, other glycans located in the stem region maintain the protein in a metastable conformation required for fusion activity (40). The effects of different N-linked glycans on fusion modulation have also been observed with members of the Paramyxoviridae family (54). In this study, we observed that the mutation of some glycosylation sites in HCV envelope glycoproteins could reduce or abolish HCVpp infectivity without apparently affecting folding and the incorporation of the glycoproteins into the particles. Indeed, the glycans at positions E2N2 and E2N4 were shown to be essential for the entry functions of HCV envelope glycoproteins. In addition, the glycans at positions E1N2, E2N1, E2N5, E2N6, and E2N11 were also shown to modulate HCVpp entry. Further studies will therefore be necessary to determine whether these mutations alter binding to other putative receptors or affect the fusion properties of HCV envelope glycoproteins.
In addition to their involvement in protein folding and virus entry, glycans associated with HCV envelope glycoproteins might also be involved in other functions. Interestingly, the C-type lectins DC-SIGN and L-SIGN have been shown to function as adhesion receptors for several viruses, including human immunodeficiency virus (HIV), Ebola virus, cytomegalovirus, and dengue virus (53), and they have now been shown to interact with HCV envelope glycoproteins (20, 32, 33, 45). DC-SIGN is expressed in dendritic cells, some subsets of macrophages, and the placenta (10, 50, 51). L-SIGN is highly expressed in liver sinusoidal endothelial cells (3, 44). Although DC-SIGN and L-SIGN are not expressed on hepatocytes, HCV interactions with these lectins may contribute to the establishment or persistence of infection both by the capture and delivery of virus to the liver and by modulating dendritic cell functions as previously suggested (8, 31). These lectins recognize glycoproteins that contain mannose residues in their N-linked glycans. Further studies on the roles of individual glycans of HCV envelope glycoproteins in interactions with DC-SIGN and L-SIGN would likely provide interesting information.
N-linked glycans can also play a role by modulating the humoral immune response. Indeed, N-linked glycans of the HIV envelope glycoprotein limit its immunogenicity and, in addition, restrict the binding of certain antibodies to their epitopes on the virion surface. A recent study on the mechanism of HIV escape from antibody-mediated neutralization led to the proposal of an evolving “glycan shield” in which the repositioning of glycans on the HIV envelope limits envelope recognition by neutralizing antibodies while maintaining the ability of the envelope protein to interact with the CD4 and coreceptor molecules on the target cell membrane (55). HCV envelope glycoproteins, despite evolving extremely rapidly through point mutations, show limited sequon variation (58). However, two shifting sites have been identified in E2, suggesting that there is some selective pressure on these sites. Further studies will be necessary to determine whether N-linked glycans can also modulate the humoral immune response to HCV.
Acknowledgments
We thank André Pillez and Sophana Ung for their excellent technical assistance. We are grateful to J. McKeating and M. Andrawiss for providing us with reagents.
This work was supported by EU grant QLRT-2000-01120 and by grants from the “Agence Nationale de Recherche sur le Sida et les Hépatites virales” (ANRS), INSERM, ATC-Hépatite C, and the ARC.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, A. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology, vol. 1. John Wiley and Sons, Inc., New York, N.Y.
- 2.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C pseudo-particles containing functional E1E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashirova, A. A., T. B. Geijtenbeek, G. C. van Duijnhoven, S. J. van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193:671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bause, E., and H. Hettkamp. 1979. Primary structural requirements for N-glycosylation of peptides in rat liver. FEBS Lett. 108:341-344. [DOI] [PubMed] [Google Scholar]
- 5.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel, L., C.-C. Kuo, J. Dubuisson, and S. Levy. 2003. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J. Virol. 77:10677-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocquerel, L., J.-C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormier, E. G., R. J. Durso, F. Tsamis, L. Boussemart, C. Manix, W. C. Olson, J. P. Gardner, and T. Dragic. 2004. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:14067-14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 13.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 275:30605-30609. [DOI] [PubMed] [Google Scholar]
- 14.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 17.Duvet, S., A. Op De Beeck, L. Cocquerel, C. Wychowski, R. Cacan, and J. Dubuisson. 2002. Glycosylation of the hepatitis C virus envelope protein E1 occurs posttranslationally in a mannosylphosphoryldolichol-deficient CHO mutant cell line. Glycobiology 12:95-101. [DOI] [PubMed] [Google Scholar]
- 18.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fournillier-Jacob, A., A. Cahour, N. Escriou, M. Girard, and C. Wychowski. 1996. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J. Gen. Virol. 77:1055-1064. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goffard, A., and J. Dubuisson. 2003. Glycosylation of hepatitis C virus envelope proteins. Biochimie 85:295-301. [DOI] [PubMed] [Google Scholar]
- 22.Hebert, D. N., J. X. Zhang, W. Chen, B. Foellmer, and A. Helenius. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J. Cell Biol. 139:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imperiali, B., and S. E. O'Connor. 1999. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 3:643-649. [DOI] [PubMed] [Google Scholar]
- 26.Imperiali, B., and K. L. Shannon. 1991. Differences between Asn-Xaa-Thr-containing peptides: a comparison of soluble conformation and substrate behavior with oligosaccharyltransferase. Biochemistry 30:4374-4380. [DOI] [PubMed] [Google Scholar]
- 27.Kasturi, L., J. R. Eshleman, W. H. Wunner, and S. H. Shakin-Eshleman. 1995. The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J. Biol. Chem. 270:14756-14761. [DOI] [PubMed] [Google Scholar]
- 28.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 29.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of hepatitis C virus of major genotypes and subtypes. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Lozach, P. Y., A. Amara, B. Bartosch, J. L. Virelizier, F. Arenzana-Seisdedos, F. L. Cosset, and R. Altmeyer. 2004. C-type lectins L-SIGN and DC-SIGN capture and transmit infectious hepatitis C virus pseudotype particles. J. Biol. Chem. 279:32035-32045. [DOI] [PubMed] [Google Scholar]
- 32.Lozach, P. Y., H. Lortat-Jacob, A. De Lacroix De Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high-affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig, I. S., A. N. Lekkerkerker, E. Depla, F. Bosman, R. J. Musters, S. Depraetere, Y. van Kooyk, and T. B. Geijtenbeek. 2004. Hepatitis C virus targets DC-SIGN and L-SIGN to escape lysosomal degradation. J. Virol. 78:8322-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1162. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 35.Merola, M., M. Brazzoli, F. Cocchiarella, J. M. Heile, A. Helenius, A. J. Weiner, M. Houghton, and S. Abrignani. 2001. Folding of hepatitis C virus E1 glycoprotein in a cell-free system. J. Virol. 75:11205-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meunier, J.-C., A. Fournillier, A. Choukhi, A. Cahour, L. Cocquerel, J. Dubuisson, and C. Wychowski. 1999. Analysis of the glycosylation sites of hepatitis C virus (HCV) glycoprotein E1 and the influence of E1 glycans on the formation of the HCV glycoprotein complex. J. Gen. Virol. 80:887-896. [DOI] [PubMed] [Google Scholar]
- 37.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 38.Nakano, I., Y. Fukuda, Y. Katano, and T. Hayakawa. 1999. Conformational epitopes detected by cross-reactive antibodies to envelope 2 glycoprotein of the hepatitis C virus. J. Infect. Dis. 180:1328-1333. [DOI] [PubMed] [Google Scholar]
- 39.Ohuchi, M., R. Ohuchi, A. Feldmann, and H. D. Klenk. 1997. Regulation of receptor binding affinity of influenza virus hemagglutinin by its carbohydrate moiety. J. Virol. 71:8377-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohuchi, R., M. Ohuchi, W. Garten, and H. D. Klenk. 1997. Oligosaccharides in the stem region maintain the influenza virus hemagglutinin in the metastable form required for fusion activity. J. Virol. 71:3719-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 42.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 44.Pohlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 98:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandrin, V., B. Boson, P. Salmon, W. Gay, D. Nègre, R. Le Grand, D. Trono, and F.-L. Cosset. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and non-human primates. Blood 100:823-832. [DOI] [PubMed] [Google Scholar]
- 47.Shakin-Eshleman, S. H., S. L. Spitalnik, and L. Kasturi. 1996. The amino acid at the X position of Asn-X-Ser sequon is an important determinant of N-linked core glycosylation efficiency. J. Biol. Chem. 271:6363-6366. [DOI] [PubMed] [Google Scholar]
- 48.Shakin-Eshleman, S. H., W. H. Wunner, and S. L. Spitalnik. 1993. Efficiency of N-linked core glycosylation at asparagine-319 of rabies virus glycoprotein is altered by deletions C-terminal to the glycosylation sequon. Biochemistry 32:9465-9472. [DOI] [PubMed] [Google Scholar]
- 49.Slater-Handshy, T., D. A. Droll, X. Fan, A. M. Di Bisceglie, and T. J. Chambers. 2004. HCV E2 glycoprotein: mutagenesis of N-linked glycosylation sites and its effects on E2 expression and processing. Virology 319:36-48. [DOI] [PubMed] [Google Scholar]
- 50.Soilleux, E. J., L. S. Morris, B. Lee, S. Pohlmann, J. Trowsdale, R. W. Doms, and N. Coleman. 2001. Placental expression of DC-SIGN may mediate intrauterine vertical transmission of HIV. J. Pathol. 195:586-592. [DOI] [PubMed] [Google Scholar]
- 51.Soilleux, E. J., L. S. Morris, G. Leslie, J. Chehimi, Q. Luo, E. Levroney, J. Trowsdale, L. J. Montaner, R. W. Doms, D. Weissman, N. Coleman, and B. Lee. 2002. Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J. Leukoc. Biol. 71:445-457. [PubMed] [Google Scholar]
- 52.Trombetta, E. S., and A. Helenius. 1998. Lectins as chaperones in glycoprotein folding. Curr. Opin. Struct. Biol. 8:587-592. [DOI] [PubMed] [Google Scholar]
- 53.van Kooyk, Y., and T. B. Geijtenbeek. 2003. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 54.von Messling, V., and R. Cattaneo. 2003. N-linked glycans with similar location in the fusion protein head modulate paramyxovirus fusion. J. Virol. 77:10202-10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 56.Wormald, M. R., and R. A. Dwek. 1999. Glycoproteins: glycan presentation and protein-fold stability. Structure Fold Des. 7:R155-R160. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]