Abstract
We examined the ability of small interfering RNAs (siRNAs) to disrupt infection by coxsackievirus B3 (CVB3). The incorporation of siRNAs dramatically decreased cell death in permissive HeLa cells in parallel with a reduction in viral replication. Three of four siRNAs had potent anti-CVB3 activity. The present study thus demonstrates that the antiviral effect is due to the downregulation of viral replication. In addition, an effective CVB3-specific siRNA had similar antiviral effects in other related enteroviruses possessing sequence homology in the targeted region. Because the CVB3-specific siRNA is effective against other enteroviruses, siRNAs have potential for a universal antienterovirus strategy.
Coxsackievirus B3 (CVB3) is a major causative agent of many human diseases, such as meningioencephalitis and myocarditis (16-18, 20, 21). CVB3 is a member of the Picornaviridae family and consists of a positive single-stranded RNA genome coated with capsid proteins, including VP1-4 (Fig. 1A) (23). Previous studies by our group and others have implied that CVB3 infection causes productive virus replication, which results in cell death in both permissive and target cells (1, 2, 9), a process closely associated with CVB3-related human illness (6, 27, 28). Thus, directly blocking viral replication may be an effective strategy for treating the clinical symptoms of CVB3 infection.
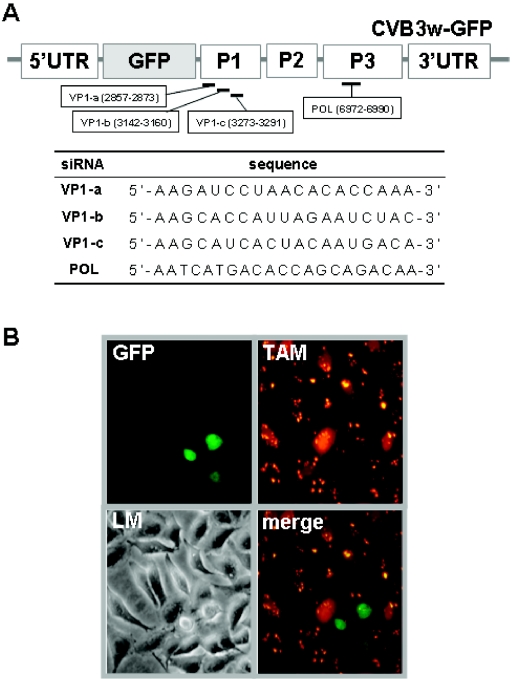
FIG. 1.
Introduction of siRNAs into infected HeLa cells. The CVB3 genome is composed of a 5′UTR; P1, P2, and P3 coding regions; and a 3′ UTR. (A) The sequence and location of synthetic siRNAs perfectly complementary to VP-1 or POL, along with an unmatched control siRNA (5′-AUUCUAUCACUAGCGUGACUU-3′), were designated and synthesized by Dharmacon (Lafayette, CO). (B) Cells with 100 nM POL were infected with CVB3w-GFP for 12 h at a multiplicity of infection of 5. Cells were examined by light (LM) or fluorescence microscopy. Images were captured at an original magnification of ×200. Cells in orange (TAM) represent those expressing siRNA, and cells in green (GFP) are those in which CVB3-GFP has replicated.
RNA interference (RNAi) has emerged as a selective gene-silencing technique for inducing sequence-specific degradation of homologous RNA (8, 10). Posttranscriptional downregulation of gene expression is achieved by a small interfering RNA (siRNA), a 21- to 26-nucleotide RNA duplex (3, 7). Recent studies have reported that RNAi is effective against diverse viruses, such as human immunodeficiency virus and hepatitis C virus (4, 24, 30). To investigate the antiviral potential of RNAi for CVB3 infection, we analyzed the effects of siRNAs specific to various regions of CVB3w (CVB3 woodruff strain) on permissive HeLa cells (Fig. 1A).
Using oligofectamine reagent (Invitrogen, Carlsbad, CA), we transfected the cells with synthetic siRNAs tagged at their 3′ end with tetramethylrhodamine (TAM). We began to detect a fluorescent signal in the cytoplasm within a few hours after addition of siRNA polymerase (POL). Then, the signal reached a plateau 8 to 12 h after transfection (Fig. 1B, TAM). More than 90% of the cells expressed the fluorescence, indicating effective siRNA transport. Infection with CVB3w-green fluorescent protein (GFP) led to a bright green color upon viral replication but in many fewer cells (Fig. 1B, GFP) than with infected cells without siRNA (Fig. 2, virus only). In addition to the CVB3 woodruff genome, CVB3w-GFP encoded GFP directly after the 5′ untranslated region (UTR), followed by a viral protease recognition sequence (Fig. 1A). These results indicate that the siRNA significantly decreased the GFP signal (Fig. 1B, merge). Later we incubated the cells with 100 nM siRNA for 12 h prior to virus infection for all following experiments.
FIG. 2.
Effects of various siRNAs on viral cytotoxicity. Cells were pretreated with or without various siRNAs, including an unmatched control siRNA, prior to infection with CVB3w-GFP under the conditions described in the legend to Fig. 1. Morphological or nuclear changes were examined by using light microscopy (LM) or fluorescence microscopy after staining with Hoechst 33342 for 30 min. The results show that except for VP1-b, CVB3-specific siRNAs reduced cellular (LM column) and nuclear (Hoechst column) morphological changes. Typical changes are indicated by arrowheads. MT, mock treated.
We first investigated the effect of siRNA on viral cytotoxicity. CVB3w-GFP infection was cytopathic within 12 h postinfection, as indicated by rounding up of the cells and by the presence of heavily condensed Hoechst 33342-stained nuclei (Fig. 2, virus only). Preincubation of the cells with siRNAs, except VP1-b and unmatched control siRNA, dramatically reduced the cytopathic effects (Fig. 2).
The pattern of GFP expression in Fig. 2 (middle column) suggests that a decrease in viral replication occurred in the presence of the siRNAs. We quantified the influence of different siRNAs on viral replication by analyzing the production of VP1 protein, progeny virus, and viral genome. For real-time reverse transcription-PCR of the viral genome, viral cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen). Then PCR was performed with the TaqMan PCR master mix (Applied Biosystems, Foster City, CA). Figure 3 shows that the siRNAs lowered the chances of productive infection occurring. VP1 was detected only in the absence of siRNA or in the presence of control or VP1-b siRNAs. A significant reduction of progeny virus production and genome amplification was consistently observed following pretreatment with VP1-a, VP1-c, or POL. A combination of siRNAs did not further downregulate virus or genome production. Together, these findings suggest that CVB3-specific siRNA efficiently protects the HeLa cells from viral challenge by inhibiting viral replication.
FIG. 3.
Effects of siRNA on viral replication. Twelve hours postinfection, cells were analyzed for VP1 expression by immunoblotting (VP1 antibody; Novo, UK) (A), progeny virus production by one-step virus growth and sequential plaque assay (B) and genome amplification by real-time reverse transcription-PCR (C). The results show that VP1-a, VP1-c, and POL siRNA or their use in combination prevented viral RNA synthesis, blocked VP1 production, and reduced the generation of viral progeny. The similar intensities of the actin band (top panel) in all lanes indicate that equal amounts of protein were loaded. MOI, multiplicity of infection.
CVB1-6 belongs to the enterovirus genus, which includes coxsackievirus A and echovirus (Echo) (29). To examine the possibility of using CVB3-specific siRNA for other related enteroviruses, we challenged HeLa cells with various viruses, including the CVB3 Nancy strain (CVB3n), after POL siRNA treatment (Fig. 4). We found that CVB3w-targeting siRNA had anticytopathic effects against CVB1, CVB5, CVB6, coxsackievirus A9, and Echo6, similar to that of CVB3w-GFP. In contrast, protective effects were not observed in CVB2, CVB4, Echo7, or even CVB3n. A comparison of the siRNA target sequence reveals that there were mismatches at several positions that had differential effects on the antiviral potency. A switch from C to T at position 17 seemed to be crucial in this regard. This was confirmed by the finding that a POL siRNA with a C-to-T switch (POL-T) exhibited anticytotoxicity opposite to that of the POL siRNA.
FIG. 4.
Anticytopathic effect of pretreatment with POL siRNA on other enteroviruses. Following treatment with POL siRNA, HeLa cells were challenged with a variety of enteroviruses and examined for cytotoxicity through methyl thiazolyl tetrazolium assay. The POL siRNA sequence was compared with the corresponding sequences of other viruses. The results show that the cytotoxicity of some of the enteroviruses was prevented by preincubation with POL siRNA. In addition, it appeared that C or T at position 17 of the target is closely related to the antiviral effect by POL or POL-T siRNA. CVB3n, CVB3 Nancy strain; +, greater than 80% increase in cell viability compared to that without siRNA; −, less than 20% increase in viability.
Collectively, our results demonstrate that (i) preincubation of HeLa cells with CVB3-specific siRNA prior to infection blocks viral replication and results in a potent antiviral effect; (ii) the interfering effect can vary among siRNAs even if they are perfectly homologous to the target sequence; (iii) combinations of siRNAs do not enhance antiviral potency; and (iv) CVB3-specific siRNA has antiviral effects in other related enteroviruses, depending on the homology of the target sequence. We found that the VP1-b siRNA had nearly no antiviral activity even though the sequence was 100% homologous with the target in CVB3w, whereas the POL siRNA was anticytopathic in other enteroviruses despite the presence of some mismatches. The siRNAs were designed to satisfy properties critical for siRNA functionality (http://www.ambion.com and http://www.oligoengine.com). This means that the ineffectiveness of VP1-b siRNA is not due to improper design (14, 19) or inaccessibility to the target sequence by a stem-loop structure (http://www.bioinfo.rpi.edu/applications/mfold) (31). Thus, VP1-b might have no interfering ability due to other reasons; for example, the inaccessibility of the target site by the binding of RNA-binding protein (7, 25). This also could be due to the unavailability of siRNA to RNA-inducing silencing complex, based on thermodynamic differences (26).
In agreement with reports for other related picornaviruses (5, 11, 13), our study demonstrates that the ability of the CVB3-specific siRNA to inhibit viral replication directly correlated with its antiviral potency. Additionally, a combination of siRNAs had no additive antiviral effect, similar to the results with other picornaviruses (11, 13). Lack of an additive effect might originate from their genome feature, a multicistronic single-stranded RNA genome. Nevertheless, combined treatment might prolong the antiviral effect because it could reduce neutralization by the emergence of siRNA escape mutants. There has been several reports demonstrating that productive CVB3 infection causes diverse human illnesses following substantial losses in functional primary target cells (6, 12, 22). In conclusion, the present study strongly supports the idea that siRNA has an excellent potential for a novel antiviral therapeutic strategy.
The results suggest that siRNAs designed for CVB3 could also have similar antiviral effects in closely related enteroviruses as long as they encode proper target sequences. Thus, a universal and efficient antiviral siRNA against closely related enteroviruses could be generated by careful design and subsequent screening for antiviral characteristics. Maintenance of functional siRNAs in a proper cellular location is a necessary prerequisite. However, synthetic siRNAs are transiently retained, mainly due to constant degradation by lysosomes and cell division (7, 15). We are currently investigating the possibility of overcoming this limitation by an optimal expression of short hairpin siRNA using various gene transfer vehicles.
Acknowledgments
This work was supported by grants 03-PJ1-PG10-20200-0004 from the Ministry of Health & Welfare and R01-2005-000-10668-0 from Korea Science and Engineering Foundation to H. Lee and 01-PJ11-PG9-01BT00B-0019 (the International Mobile Telecommunications 2000 R&D Project) from the Ministry of Information & Communication to Y. K. Kim, Republic of Korea.
REFERENCES
- 1.Ahn, J., J. Choi, C. H. Joo, I. Seo, D. H. Kim, S. Y. Yoon, Y. K. Kim, and H. Lee. 2004. Susceptibility of coxsackievirus B in mouse primary cortical neural cell cultures. J. Gen. Virol. 85:1555-1564. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J., C. H. Joo, I. Seo, D. Kim, H. N. Hong, Y. K. Kim, and H. Lee. 2003. Characteristics of apoptotic cell death induced by coxsackievirus B in permissive Vero cells. Intervirology 46:245-251. [DOI] [PubMed] [Google Scholar]
- 3.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmichael, G. G. 2002. Medicine: silencing viruses with RNA. Nature 418:379-380. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., W. Yan, Q. Du, L. Fei, M. Liu, Z. Ni, Z. Sheng, and Z. Zheng. 2004. RNA interference targeting VP1 inhibits foot-and-mouth disease virus replication in BHK-21 cells and suckling mice. J. Virol. 78:6900-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry, J. D. 1981. Non-polio enteroviruses: coxsackieviruses, echoviruses and enteroviruses, p. 1316-1365. In R. D. Feigin and J. Cherry (ed.), Textbook of pediatric infectious diseases. W. B. Saunders, Philadelphia, Pa.
- 7.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 9.Feuer, R., I. Mena, R. R. Pagarigan, S. Harkins, D. E. Hassett, and J. L. Whitton. 2003. Coxsackievirus B3 and the neonatal CNS: the roles of stem cells, developing neurons, and apoptosis in infection, viral dissemination, and disease. Am. J. Pathol. 163:1379-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 11.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 12.Huber, S. A., R. C. Budd, K. Rossner, and M. K. Newell. 1999. Apoptosis in coxsackievirus B3-induced myocarditis and dilated cardiomyopathy. Ann. N. Y. Acad. Sci. 887:181-190. [DOI] [PubMed] [Google Scholar]
- 13.Kanda, T., Y. Kusov, O. Yokosuka, and V. Gauss-Muller. 2004. Interference of hepatitis A virus replication by small interfering RNAs. Biochem. Biophys. Res. Commun. 318:341-345. [DOI] [PubMed] [Google Scholar]
- 14.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209-216. [DOI] [PubMed] [Google Scholar]
- 15.Lingor, P., U. Michel, U. Scholl, M. Bahr, and S. Kugler. 2004. Transfection of “naked” siRNA results in endosomal uptake and metabolic impairment in cultured neurons. Biochem. Biophys. Res. Commun. 315:1126-1133. [DOI] [PubMed] [Google Scholar]
- 16.Muir, P. 1993. Enteroviruses and heart disease. Br. J. Biomed. Sci. 50:258-271. [PubMed] [Google Scholar]
- 17.Nigrovic, L. E. 2001. What's new with enteroviral infections? Curr. Opin. Pediatr. 13:89-94. [DOI] [PubMed] [Google Scholar]
- 18.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: poliovirus, coxsackieviruses, echoviruses, and newer enteroviruses, p. 723-776. In P. M. Howley and D. M. Knipe (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, New York, N.Y. [Google Scholar]
- 19.Reynolds, A., D. Leake, Q. Boese, S. Scaringe, W. S. Marshall, and A. Khvorova. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22:326-330. [DOI] [PubMed] [Google Scholar]
- 20.Roivainen, M. 1999. Enteroviruses and myocardial infarction. Am. Heart J. 138:S479-S483. [DOI] [PubMed] [Google Scholar]
- 21.Rotbart, H. A. 1995. Enteroviral infections of the central nervous system. Clin. Infect. Dis. 20:971-981. [DOI] [PubMed] [Google Scholar]
- 22.Rotbart, H. A., P. J. Brennan, K. H. Fife, J. R. Romero, J. A. Griffin, M. A. McKinlay, and F. G. Hayden. 1998. Enterovirus meningitis in adults. Clin. Infect. Dis. 27:896-898. [DOI] [PubMed] [Google Scholar]
- 23.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Raven, New York, N.Y.
- 24.Saksela, K. 2003. Human viruses under attack by small inhibitory RNA. Trends Microbiol. 11:345-347. [DOI] [PubMed] [Google Scholar]
- 25.Sharp, P. A. 2001. RNA interference—-2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 26.Tomari, Y., C. Matranga, B. Haley, N. Martinez, and P. D. Zamore. 2004. A protein sensor for siRNA asymmetry. Science 306:1377-1380. [DOI] [PubMed] [Google Scholar]
- 27.Tracy, S., N. M. Chapman, J. Romero, and A. I. Ramsingh. 1996. Genetics of coxsackievirus B cardiovirulence and inflammatory heart muscle disease. Trends Microbiol. 4:175-179. [DOI] [PubMed] [Google Scholar]
- 28.Tracy, S., N. M. Chapman, and Z. Tu. 1992. Coxsackievirus B3 from an infectious cDNA copy of the genome is cardiovirulent in mice. Arch Virol. 122:399-409. [DOI] [PubMed] [Google Scholar]
- 29.van Regenmortel, M. H. V., C. M. Fanquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lomon, J. Maniloft, M. A. Mayo, C. R. Pringe, and R. B. Wickner (ed.). 2000. Virus taxonomy. Academic Press, San Diego, Calif.
- 30.Wang, Q. C., Q. H. Nie, and Z. H. Feng. 2003. RNA interference: antiviral weapon and beyond. World J. Gastroenterol. 9:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshinari, K., M. Miyagishi, and K. Taira. 2004. Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Res. 32:691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]