Abstract
Toroviruses (order Nidovirales) are enveloped positive-strand RNA viruses of mammals. The prototype torovirus, equine torovirus strain Berne (Berne virus [BEV]), uses two different transcription strategies to produce a 3′-coterminal nested set of subgenomic (sg) mRNAs. Its mRNA 2 carries a leader sequence derived from the 5′ end of the genome and is produced via discontinuous transcription. The remaining three sg mRNAs, 3 to 5, are colinear with the 3′ end of the genome and are made via non-discontinuous RNA synthesis. Their synthesis is supposedly regulated by short conserved sequence motifs, 5′-ACN3-4CUUUAGA-3′, within the noncoding intergenic regions that precede the M, HE, and N genes (A. L. van Vliet, S. L. Smits, P. J. Rottier, and R. J. de Groot, EMBO J. 21:6571-6580, 2002). We have now studied the—for nidoviruses unusual—non-discontinuous transcription mechanism in further detail by probing the role of the postulated transcription-regulating sequences (TRSs). To this end, we constructed a synthetic defective interfering (DI) RNA, carrying a 24-nucleotide segment of the intergenic region between the HE and N genes. We demonstrate that this DI RNA, when introduced into BEV-infected cells, directs the synthesis of a sg DI RNA species; in fact, a 16-nucleotide cassette containing the TRS already proved sufficient. Synthesis of this sg DI RNA, like that of mRNAs 3 to 5 of the standard virus, initiated at the 5′-most adenylate of the TRS. An extensive mutational analysis of the TRS is presented. Our results provide first and formal experimental evidence that the conserved motifs within the BEV intergenic sequences indeed drive sg RNA synthesis.
Toro-, corona-, arteri-, and roniviruses are evolutionary related positive-strand RNA viruses, which have been united in the order Nidovirales (5). Despite considerable differences in genome size, virion architecture and host range, the common ancestry of these viruses is apparent from sequence identity in their replicase proteins and from similarities in genome organization, gene order, and replication strategy (5, 6, 9, 32). The 5′-most two-thirds of nidovirus genomes are occupied by two large overlapping open reading frames, ORF1a and -1b. These encode polyproteins from which the various subunits of the viral replicase/transcriptase are derived. Further downstream, a variable number of smaller genes is found, mostly encoding structural proteins, that are typically expressed from a 3′-coterminal nested set of subgenomic (sg) mRNAs.
Over the years, coronaviruses and arteriviruses have received considerable attention because of their remarkable transcription strategy, which entails discontinuous RNA synthesis. Their sg mRNAs are chimeric in that they all possess a 5′ common leader sequence, which is derived from the 5′ terminus of the genomic RNA and fused to the “body” of the transcript (i.e., the 3′-terminal part, which carries the coding information). The leader and mRNA body sequences are joined within a short conserved sequence motif, the transcription-regulating sequence (TRS), which precedes each transcription unit (for reviews, see references 4, 9, 12, 34, and 36). A copy of the TRS is also found immediately downstream of the genomic leader sequence. The sg mRNAs are not transcribed from a full-length minus-strand genomic RNA template but rather from a mirror set of minus-strand sg RNAs (23, 28, 29). According to the prevailing view, the discontinuous transcription step occurs during minus-strand synthesis via a process resembling homology-assisted copy-choice RNA recombination. In this model, originally put forward and championed by Sawicki et al. (22-25) and later refined by Snijder and coworkers (19-21, 37) and Enjuanes and coworkers (42), the TRSs act as signals for transcription attenuation, and the subsequent template switch of the RNA polymerase is guided by the sequence complementarity between the anti-TRS sequence at the 3′ end of the nascent minus-strand RNA and the leader TRS at the 5′ end of the genome. The resulting chimeric sg minus-strand RNA, carrying a 3′-terminal anti-leader sequence, would in turn function as a template for the production of subgenomic positive-strand RNAs (2, 18-29, 37, 42).
Different from what one might expect, discontinuous transcription is not a common trait of nidoviruses. Thus, roniviruses produce two sg mRNA species, neither of which has a leader (7). Toroviruses are even more exceptional in that they combine two different strategies, discontinuous and non-discontinuous RNA synthesis, to generate their set of mRNAs (38).
The prototype torovirus, equine torovirus strain Berne (BEV), has a genome of ∼28 kb (“RNA 1”; S. L. Smits and R. J. de Groot, unpublished data) and produces four sg RNAs of 7, 2, 1.2, and 0.7 kb, designated RNAs 2 to 5, respectively. These serve as mRNAs for the spike (S), membrane (M), hemagglutinin-esterase (HE), and nucleocapsid (N) proteins, respectively (33). mRNAs 3, 4, and 5 do not possess a leader: they are fully colinear with the 3′ end of the viral genome. mRNA 2, however, is chimeric in structure and consists of a sequence identical to the 5′-most 18 nucleotides (nt) of the genome fused to the 3′-most 6,856 nt (38).
The M, HE, and N genes, but not the S gene, are preceded by short noncoding “intergenic” regions, which contain the highly conserved consensus motif 5′-ACN3-4CUUUAGA-3′, a sequence also found at the very 5′ end of the BEV genome. As determined by 5′ RLM-RACE reverse transcription-PCR (RT-PCR) and primer extension analysis of RNAs 3, 4, and 5, transcription in all cases initiates at the 5′-terminal adenylate of this motif (38). Based upon these observations, we assume that the “intergenic” consensus sequence represents the torovirus TRS equivalent. However, in contrast to the TRSs of arteri- and coronaviruses, the torovirus TRSs do not function as sites for homology-assisted template-switching. Rather, they supposedly exert dual key roles: as terminators of transcription during minus-strand sg RNA synthesis and, in the complementary orientation at the 3′ end of a minus-strand sg RNA template, as promoters of positive-strand sg RNA synthesis (38). Thus far, however, formal proof that the toroviral TRSs can direct RNA synthesis is lacking. In the current manuscript, we followed an approach, based upon the use of synthetic defective interfering (DI) RNAs, to produce experimental evidence to this effect.
MATERIALS AND METHODS
Cells and virus.
BEV was grown in equine dermis (Ederm) cells (American Type Tissue Culture Collection); a virus stock, devoid of DI viruses (38), was used in all experiments. Ederm cells were maintained as monolayer cultures in Dulbecco modified Eagle medium (DMEM), containing 20% fetal calf serum, 100 IU penicillin/ml, and 100 μg of streptomycin/ml (DMEM20).
Construction of synthetic pDI1000 and derivatives.
Conventional PCR mutagenesis and recombinant DNA procedures were used to generate plasmid pDI1000, a pUCBM-20-derived expression plasmid, carrying a full-length cDNA copy of a natural BEV DI RNA, DI1000 (30), downstream of the bacteriophage T7 RNA polymerase promoter. The reconstructed DI1000 sequence consists of the 5′-most 604 nt of the BEV genome fused to the 3′-most 242 residues and is extended at the 5′ end by a GG-dinucleotide and at the 3′ end by a 15-nucleotide poly(A) stretch and a unique PacI site.
pDI1000-Xho was constructed from pDI1000 by deleting all sequences derived from the N gene, excluding the termination codon. Concomitantly, a new unique XhoI cloning site was created, which allowed the introduction of double-stranded oligonucleotides, the sequence of which corresponded to the intergenic region between the BEV HE and N genes and variants thereof (Table 1).
TABLE 1.
Oligonucleotide primers
| Oligonucleotide | Sequence (5′ to 3′) | Polarity | Positionsa | Template |
|---|---|---|---|---|
| 1107 | TAATACGACTCACTATAGNNAACGTATCTTTAGAAGTTTATG | + | 1-24 | DI |
| 1283 | GTTAATTAATTTTTTTTTTTTTTTAGCTGCTTT | − | 27984-27992 | DI |
| 1353 | CACAATTTCTCGAGTAAAACCTTTTCAGCAACCTT | + | 27793-27813 | DI |
| 1354 | AGGTTTTACTCGAGAAATTGTGCAAACAACCAAAC | − | 583-603 | DI |
| 294 | CTTACATGGAGACACTCAACCA | − | 27805-27826 | 3′ NTR |
| 1225 | CTCGTTGATTAAATTGTGCAAA | − | 593-604 | DI |
| 1807 | GAAAAGGTTTTACTCGAG | − | 27793-27804 | DI |
| 1131 | CGCGGATCCACCATGAATTCTATGCTTAATCCAAA | + | 27316-27338 | N |
| 1132 | TTTTCTGCAGTTAATTCATAACCACCTTAATAG | − | 27776-27798 | N |
| 2208 | TTTTCTGCAGCCTACTTAATTACTAATACAAAAAC | − | 27252-27276 | HE |
| 2209 | CGGGATCCGATGTTGATTTTTATCCAACTAG | + | 26801-26823 | HE |
| 1801 | TCGACAGACACTATCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1802 | TCGAGCTCTTTCTCTAAAGATAGTGTCTG | − | 27285-27307 | IGR 4/5 |
| 1803 | TCGACACTATCTTTAGAGC | + | 27286-27301 | IGR 4/5 |
| 1804 | TCGAGCTCTAAAGATAGTG | − | 27286-27301 | IGR 4/5 |
| 1805 | TCGACTATCTTTAGAC | + | 27289-27300 | IGR 4/5 |
| 1806 | TCGAGTCTAAAGATAG | − | 27289-27300 | IGR 4/5 |
| 1903 | TCGACAGADACTATCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1904 | TCGACAGACBCTATCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1905 | TCGACAGACADTATCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1906 | TCGAGCTCTTTCTCTAAAGATAGTHTCTG | − | 27285-27307 | IGR 4/5 |
| 1907 | TCGAGCTCTTTCTCTAAAGATAGVGTCTG | − | 27285-27307 | IGR 4/5 |
| 1908 | TCGAGCTCTTTCTCTAAAGATAHTGTCTG | − | 27285-27307 | IGR 4/5 |
| 1936 | TCGACAGACACTCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1937 | TCGACAGACACTTCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1938 | TCGACAGACACTTATCTTTAGAGAAAGAGC | + | 27285-27307 | IGR 4/5 |
| 1940 | TCGACAGACACTTTTATCTTTAGAGAAAGAG | + | 27285-27307 | IGR 4/5 |
| 1941 | TCGAGCTCTTTCTCTAAAGAGTGTCTG | − | 27285-27307 | IGR 4/5 |
| 1942 | TCGAGCTCTTTCTCTAAAGAAGTGTCTG | − | 27285-27307 | IGR 4/5 |
| 1943 | TCGAGCTCTTTCTCTAAAGATAAGTGTCTG | − | 27285-27307 | IGR 4/5 |
| 1945 | TCGAGCTCTTTCTCTAAAGATAAAAGTGTCTG | − | 27285-27307 | IGR 4/5 |
| A | GCTGATGGCGATGAATGAACACTGCGTTTGCTGGCTTTGATGAAA | − | NAb | Adapter |
| B | GCTGATGGCGATGAATGAACACTG | + | NA | Adapter |
Genomic location of BEV-derived sequences.
NA, not applicable.
Preparation of synthetic DI RNA.
pDI1000 and derivatives were linearized with PacI and purified by using the QiaQuick PCR purification kit (Qiagen). In vitro transcription was performed essentially as described by Bredenbeek et al. (3). Reaction mixtures contained ∼1 μg of linearized template; 40 mM Tris-HCl (pH 7.6); 6 mM MgCl2; 2 mM spermidine; 1 mM each of GTP, CTP, ATP, and UTP; 30 U of RNA guard (Amersham Pharmacia Biotech, Inc.); 1 mM 7mGpppG cap analogue (Amersham Pharmacia Biotech, Inc.); and 50 U of T7 RNA polymerase (Invitrogen) in a total volume of 20 μl. Incubation was for 2 h at 37°C.
Transfection of synthetic DI RNA and serial passage experiments.
Ederm cells (∼7 × 105), grown to subconfluency in 35-mm wells, were infected with BEV at a multiplicity of infection (MOI) of >5 50% tissue culture infective doses/cell. At 1 h postinfection (p.i.), the cells were washed once with DMEM and then incubated at room temperature for 10 min with a transfection mixture consisting of 0.2 ml of DMEM, 10 μl of Lipofectin Reagent (Invitrogen), and 20 μl in vitro transcription mixture. Then, 0.8 ml of DMEM was added, and incubation was continued for 4 h at 37°C, after which the tissue culture supernatant was replaced by 1 ml of DMEM20. At 16 h p.i., the tissue culture supernatants were harvested, and 500 μl was used to infect fresh monolayers of Ederm cells (∼1 × 106) grown in 35-mm wells. During serial passages, additional BEV was added to the inoculum to an MOI of 5 50% tissue culture infective doses/cell. Inoculation was for 1 h at 37°C, after which incubation was continued with 1 ml of DMEM20 for 16 h.
Isolation of intracellular BEV RNA and RNA hybridization analyses.
To detect DI1000 genomic and subgenomic RNAs, total intracellular RNA was isolated from BEV-infected cells at 11 or 16 h p.i., essentially as described by Sawicki and Sawicki (23). RNA was separated in 2% agarose gels containing 2.2 M formaldehyde in MOPS buffer (10 mM morpholinepropanesulfonic acid [pH 7], 5 mM sodium acetate, 1 mM EDTA) as described previously (23). Per lane, an amount of RNA extracted from the equivalent 5 × 105 Ederm cells was loaded. Gels were dried under vacuum at 58°C and hybridized with 5′-end-labeled oligonucleotide probes for 16 h at 5°C below the melting temperature (Tm) according to the method of Meinkoth and Wahl (16). Quantitation was performed with a Storm PhosphorImager/Fluorimager 860 (Molecular Dynamics) and ImageQuant software.
RLM-RACE RT-PCR.
Advanced RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE)-RT-PCR analysis of subgenomic DI RNA was performed as described previously (15, 38) with the FirstChoice RLM-RACE kit (Ambion, Austin, TX) according to the manufacturer's instructions. For this procedure, the amount of passage (P)3 intracellular RNA used per reaction corresponded to that extracted from 106 DI-TRS MAX (+)-infected Ederm cells. Oligonucleotide A was used as adapter and ligated to the 5′ end of decapped RNAs. Subsequent RT-PCR was performed with adapter-specific primer B and BEV-specific primer 294 (Table 1).
RESULTS AND DISCUSSION
Admittedly, nidovirus transcription should best be studied in the context of the complete genome (8, 37, 42). However, methods for genetic modification of toroviruses are not yet available. Before the recent advent of reverse genetics techniques for coronaviruses, their mechanism of sg RNA synthesis was effectively studied by using DI viruses (for example, see references 11, 14, 35, 39, and 41), i.e., mutants, which arise spontaneously when the standard virus is passaged in cell culture at high MOIs. The genomes of these DI viruses, DI RNAs, often have suffered large deletions and, as a consequence, have lost most, if not all, of their protein coding capacity. They retain, however, the sequences essential for replication and packaging, which allow them to be propagated by the replication machinery of the standard virus at the expense of the latter (10).
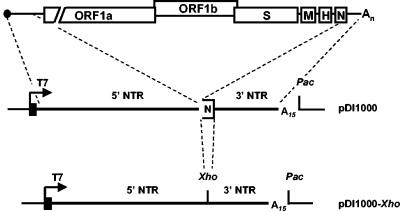
Also during serial passage of BEV in tissue culture cells, DI RNAs rapidly emerge. Snijder et al. (30) identified a small DI RNA, DI1000, which consisted of the 5′-most 604 nt of the standard virus genome, i.e., only a part of the 821-nt 5′-nontranslated region (5′-NTR), joined to the 3′-most 242 nt, comprising the 45-nt terminal sequences of the N gene and the complete 3′-NTR. To develop a DI-based model system to study BEV replication and transcription, we generated a pUCBM20-derived vector, pDI1000, which contains a full-length cDNA of DI1000, downstream of the bacteriophage T7 RNA polymerase promoter (Fig. 1).
FIG. 1.
Schematic representation of the genome organization of BEV and the structure of pDI1000 and its derivate pDI1000-Xho. The upper panel shows a schematic representation of the BEV genome. Indicated are the cap (black dot) and the poly(A) tail (An); open boxes represent genes for the replicase (ORF1a and 1b) and for the spike (S), membrane (M), hemagglutinin-esterase (H), and nucleocapsid (N) proteins. The lower panels depict the structure of plasmids pDI1000 and pDI1000-Xho. cDNA segments derived from the 5′- and 3′-NTRs of the BEV genome are indicated by lines. Sequences derived from the 3′ end of the N-gene are shown as a half-open box. Black boxes with arrows indicate T7 promoters and the direction of transcription, “A15” indicates stretches of fifteen A-residues. Also indicated are restriction sites for PacI and XhoI.
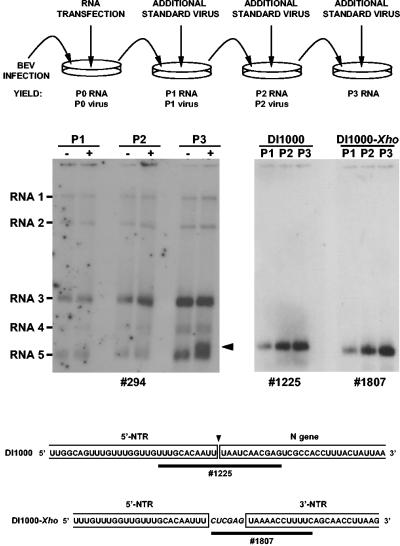
To determine whether synthetic pDI1000-derived transcripts are replication competent, RNA synthesized in vitro was used to transfect BEV-infected Ederm cells by lipofection. At 16 h p.i., tissue culture supernatant was harvested, and the supernatant, containing standard and DI virus, was serially passaged for up to three times in freshly seeded, confluent Ederm cell monolayers. During each inoculation, additional standard virus was added to ensure high MOI. conditions and one-step growth propagation (Fig. 2). Total intracellular RNA was extracted from Ederm cells, infected with passage 0 (P0), P1, and P2 virus (for an explication of the nomenclature used, see Fig. 2), and subjected to hybridization analysis. As a DI-specific probe, we used oligonucleotide 1225, a 22-mer, which spans the junction site of the 5′-NTR and N sequences in DI1000 (Table 1). As shown in Fig. 2, DI1000 RNA was detected in increasing amounts in P1, P2, and P3 RNA preparations. We conclude that synthetic DI1000 RNA is efficiently propagated in BEV-infected cells.
FIG. 2.
Transfection and serial passage of synthetic DI RNAs in BEV-infected Ederm cells. The top panel provides a schematic outline of the experiment and an explanation of the nomenclature used for virus stocks and intracellular RNA preparations. In the middle panel, the analysis of intracellular RNAs is shown. DI1000 and DI1000-Xho RNAs, synthesized in vitro were used to transfect BEV-infected Ederm cells by lipofection. Total intracellular RNA was isolated from BEV-infected cells at 16 h p.i. RNA samples, extracted from the equivalent of ∼5 × 105 cells, were separated in denaturing 2% agarose-formaldehyde gels, which were then dried and subjected to hybridization with 5′-end-labeled oligonucleotide probes. On the left-hand side, the results are shown of a hybridization with oligonucleotide 294 of P1, P2, and P3 RNA, obtained after initial transfection (+) or mock transfection (−) with DI1000 RNA. The arrowhead indicates the position of DI1000. On the right-hand side, the results are shown of a hybridization with DI-specific oligonucleotide probes 1225 and 1807 of P1, P2, and P3 RNAs, obtained after initial transfection with DI1000 or DI1000-Xho RNA, respectively. The bottom panel explains the structure of the DI-specific oligonucleotide probes 1225 and 1807. Shown are (partial) nucleotide sequences of DI1000 and DI1000-Xho, comprising the regions where 5′-NTR sequences are fused to sequences of the N gene or of the 3′-NTR, respectively. Viral sequences are boxed; the fusion site in DI1000 is indicated by an arrowhead pointed downwards. The sequence CUCGAG, nonboxed and shown in italics, corresponds to the XhoI site newly introduced into pDI1000-Xho. The binding sites of oligonucleotide probes 1225 and 1807 are indicated by black bars.
We next designed a DI1000 derivative, DI1000-Xho, to create a platform for transcription studies (Fig. 1). Sequences derived from the BEV N gene, corresponding to the 3′-terminal 42 residues, excluding the termination codon (30), were deleted. Concomitantly, a unique XhoI cloning site was created at the resulting junction of 5′- and 3′-NTR sequences, to allow convenient insertion of BEV TRS derivatives via cassette mutagenesis. DI1000-Xho RNA and its derivatives (see below) were specifically detected with oligonucleotide probe 1807, an 18-mer, comprising a sequence complementary to the 5′-most 12 nt of the 3′-NTR (including the termination codon of the N gene) 3′-terminally extended by the nonviral sequence 5′-CTCGAG-3′ (Table 1). As shown in Fig. 2, DI1000-Xho accumulated to similar amounts as DI1000 during serial passage experiments. These observations show that the N gene-derived sequences in DI1000 are not essential for DI RNA replication. Apparently, in the BEV genome, all sequences required for positive- and minus-strand genomic RNA synthesis and, presumably also for packaging (30), reside within the 5′-terminal 604 residues of the 5′-NTR and within the 3′-NTR.
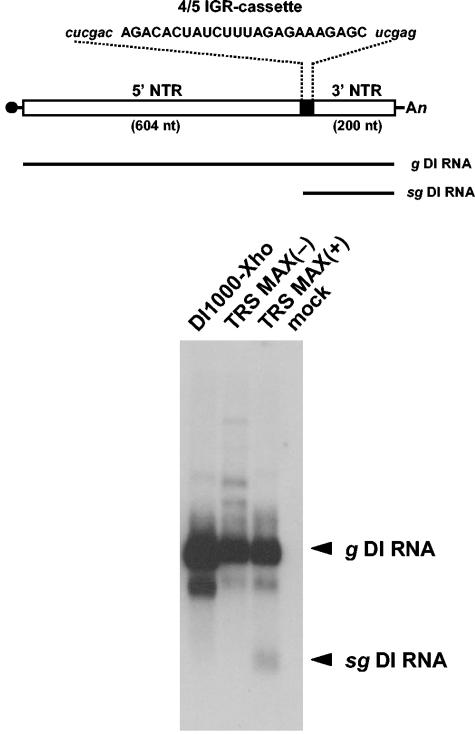
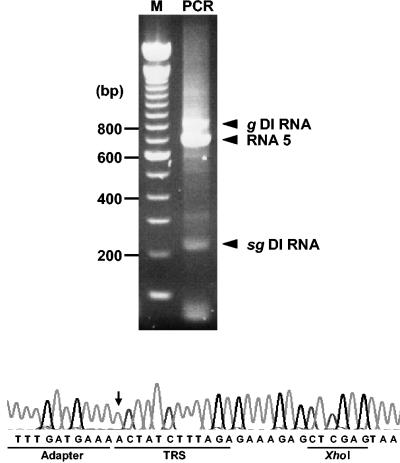
To determine whether the noncoding intergenic region preceding the N gene (IGR 4/5) is sufficient to drive the synthesis of subgenomic RNA, we inserted (part of) this sequence as a 24-mer double-stranded oligonucleotide cassette into pDI1000-Xho. The resulting plasmids, designated pDI-TRS MAX(+) and pDI-TRS MAX(−), carry the cassette in the positive and negative orientation, respectively (Fig. 3). Synthetic RNAs were transfected in BEV-infected cells, followed by serial passage. Hybridization was performed with oligonucleotide probe 1807 (Table 1), which was designed to specifically detect both genomic and subgenomic DI1000-Xho RNAs. IGR 4/5-driven transcription should give rise to a sg DI RNA species of ∼225 nt, excluding the poly(A) tail. Indeed, upon analysis of P3 RNA, an sg DI RNA species of this size was detected in samples from DI-TRS-MAX(+)-infected cells but not in those from cells infected with DI1000-Xho or with DI-TRS MAX(-) (Fig. 3). To study whether this RNA species indeed resulted from IGR 4/5-driven sg RNA synthesis, we subjected P3 RNA extracted from DI-TRS-MAX(+)-infected cells to 5′ RLM-RACE RT-PCR (15, 38) with adaptor-specific oligonucleotide B and the DI-specific oligonucleotide 294 as primers (Table 1). As shown in Fig. 4, three major products of 219, 698, and 823 bp were obtained. The larger two products were identified by sequence analysis to be derived from the genomic DI RNA and from BEV RNA 5 (not shown). The smallest species, however, corresponded to a transcript, which had initiated, as predicted, at the 5′-terminal adenylate of the 5′-ACUAUCUUUAGA-3′ consensus motif within the IGR 4/5 sequence in DI-TRS MAX(+) (Fig. 4).
FIG. 3.
Detection of an sg DI RNA in DI-TRS MAX(+)-infected cells. The upper panel shows a schematic representation of the structure of synthetic DI RNA, DI-TRS MAX(+). 5′- and 3′-NTR sequences are indicated by white boxes, the oligonucleotide cassette by a black box. The black circle represents the cap structure, and “An” indicates the poly(A) tail. IGR 4/5-derived sequences (capital case) and flanking nonviral residues (lowercase, italics) are given. Black lines below depict the structures of the genomic DI RNA and the predicted sg DI RNA. The lower panel shows the hybridization analysis of intracellular P3 RNAs, extracted from BEV-infected Ederm cells coinfected with DI viruses DI1000-Xho, DI-TRS MAX(+), DI-TRS MAX(−), or no DI virus (mock). RNA electrophoresis and hybridization with oligonucleotide probe 1807 were as described in Fig. 2. Genomic and subgenomic DI RNAs are indicated (g DI RNA and sg DI RNA, respectively).
FIG. 4.
Detection of TRS-driven sg DI RNA by 5′ RLM-RACE RT-PCR analysis. Total intracellular P3 RNA extracted from BEV/DI-TRS MAX(+)-infected cells was subjected to 5′ RLM-RACE RT-PCR. The resulting amplicons were separated in 1.5% agarose gels alongside the 100-bp marker (Invitrogen). Molecular weights (in base pairs) are indicated on the left. Amplicons corresponding to the 5′ ends of genomic DI-TRS MAX(+) RNA (g DI RNA), BEV mRNA 5 (RNA 5) and subgenomic DI RNA (sg DI RNA) are indicated. Shown are the electropherogram and the deduced nucleotide sequence of the 219-bp sg DI RNA amplicon. Sequences corresponding to the adapter oligonucleotide A, the TRS, and the diagnostic TRS-MAX(+)-specific XhoI site are underlined. The arrow indicates the 5′-terminal residue of the sg DI RNA.
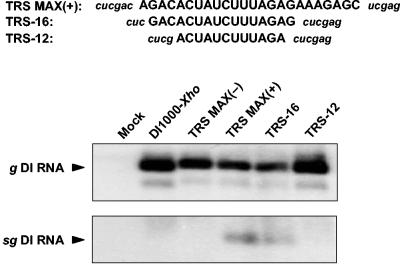
Subsequent deletion mutagenesis experiments indicated that the essential signals for sg DI RNA synthesis indeed reside within the consensus motif and in its immediate flanking sequences. When trimmed back to a 16-nt segment spanning the consensus motif (TRS-16), the IGR 4/5 sequence still directed the synthesis of sg DI RNA, although transcription was reduced to ∼60% of that of seen for TRS-MAX(+) (calculated from the ratios of the genomic to subgenomic DI RNAs as measured by STORM PhosphorImager analysis). Further removal of the 5′-flanking three nucleotides and the 3′-flanking G residue (TRS-12) reduced transcription below the detection limit (Fig. 5).
FIG. 5.
Mutagenesis analysis of the IGR 4/5 promoter sequence. The upper panel shows the sequences of oligonucleotide cassettes present in TRS MAX(+), TRS-16, and TRS-12. IGR 4/5-derived sequences are in uppercase letters; flanking sequences are in lowercase italic letters. The lower panel shows the results from hybridization analysis with oligonucleotide probe 1807 of intracellular P3 RNAs, obtained after initial (mock) transfection with DI1000-Xho, DI-TRS MAX(−), DI-TRS MAX(+), DI-TRS-16, or DI-TRS-12. Genomic and subgenomic DI RNAs are indicated (g DI RNA and sg DI RNA, respectively).
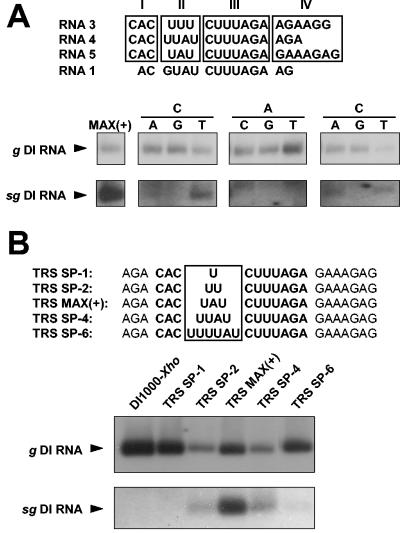
When torovirus intergenic regions are compared, four subelements can be distinguished, namely, a strictly conserved CAC-trinucleotide of which the adenylate represents the transcription initiation site, an AU-rich spacer of variable length, the CUUUAGA core and a purine-rich region (going from 5′ → 3′; Fig. 6A). Of the CAC-trinucleotide, the adenylate seems crucial for efficient sg RNA synthesis: in the context of the IGR 4/5 TRS, substitution of this residue by any other nucleotide diminished transcription below detection limits. As shown in Fig. 6A, even after prolonged exposure, no sg DI RNA was found. Substitution of the downstream C residue by G also led to loss of detectable RNA synthesis, but some production of the sg DI RNA was observed when this residue was changed to A or U (Fig. 6A). The C residue, upstream of the transcription initiation site, also seems to be important for transcription as substitution by either A or G resulted in loss of detectable sg DI RNA synthesis. However, a C→U mutation apparently was allowed. The AU spacer was more tolerant of modifications, a finding consistent with its variable length in natural BEV TRSs. Reducing the triple-nucleotide UAU-spacer in TRS-MAX(+) to a dinucleotide UU sequence did not seem to affect transcription, nor did the insertion of an additional U residue [transcription levels were estimated to be ∼90% of that of TRS-MAX(+) as calculated from the ratios of the genomic to subgenomic DI RNAs]. Even an AU-spacer extended to 6 nt still directed the synthesis of sg DI RNA, although with reduced efficiency (∼20%) compared to the wild-type sequence. However, when the spacer was reduced to a single uridylate, sg RNA synthesis was no longer detectable (Fig. 6B).
FIG. 6.
Mutagenesis analysis of the IGR 4/5 promoter sequence. (A) Effect of mutations in the TIS element and the conserved upstream C residue. The upper panel shows the sequence of the genomic (RNA 1) TRS and of those of sg RNAs 3 through 5. The different TRS subelements are boxed and numbered as follows: (I) the inititating adenylate and flanking conserved cytidylates, (II) variable AU-rich spacer, (III) conserved TRS core sequence, and (IV) purine-rich region. The results are shown below of hybridization analysis with oligonucleotide probe 1807 of P3 RNAs, obtained after initial transfection with DI-TRS MAX(+) or with derivatives thereof. The CAC residues in TRS subelement I were each replaced by any other nucleotide. The genomic and subgenomic DI RNAs are indicated (g DI RNA and sg DI RNA, respectively). (B) Effect of insertions and deletions in the AU-rich spacer element II. The upper panel shows the sequences of oligonucleotide cassettes as present in TRS MAX(+), TRS SP-1, TRS SP-2, TRS SP-4, and TRS SP-6. IGR 4/5-derived sequences are in uppercase letters with the TRS sequences in boldface. Below are shown the results of hybridization analysis with oligonucleotide probe 1807 of P3 RNAs, obtained after initial transfection with DI-TRS MAX(+) or with the various derivatives thereof. Genomic and subgenomic DI RNAs are indicated (g DI RNA and sg DI RNA, respectively).
The combined results, described here, provide first and formal experimental evidence that the conserved sequences within the BEV intergenic regions are essential and sufficient to drive sg RNA synthesis. Dissection of these TRS sequences by mutational analysis demonstrated in particular the importance of the 5′-most adenylate of this motif. This finding is consistent with our observation that all sg mRNAs (38) and also the sg DI RNA (see below) initiate at this residue. For efficient transcription, the adenylate should be flanked preferably by cytidylates, but it may be spatially separated from the CUUUAGA core sequence by a spacer of 3 to up to 6 nt.
In our current working model for torovirus transcription (38), the TRSs are thought to act both as minus-strand termination signals and as transcription initiation signals for positive-strand synthesis, with the subgenomic minus-strand RNAs serving as templates. Such a mechanism would resemble in many aspects the premature termination model for subgenomic mRNA transcription in noda- and tombusviruses (for an elegant, comprehensive review, see reference 40). Thus far, however, we have only shown TRS-driven synthesis of positive-strand sg DI RNA. Repeated attempts to also identify minus-strand sg DI RNAs have not been successful, but the detection of these RNA species may well have been precluded by their low abundance. By using Northern blot analysis and 3′-RLM RACE RT-PCR, we have, however, recently obtained evidence for the existence of a 5′-coterminal nested set of minus-strand sg RNAs that is complementary to the nested set of mRNAs in BEV-infected cells (A. L. W. van Vliet, S. L. Smits, and R. J. de Groot, unpublished observations).
Our findings show that synthetic DI RNAs are useful tools also for studying torovirus transcription. However, this approach has its limitations. It is laborious and will certainly not be suitable to study all aspects of torovirus sg RNA synthesis. Of most concern, the ratio of sg DI to genomic DI RNA differs significantly from that seen for the standard virus and is almost reversed compared to the ratio of mRNA 5 to viral genome. Similar observations were made when DI RNAs were used to explore coronavirus sg mRNA synthesis (14, 35). The phenomenon may be due to the small size of DI RNAs and simply be related to the amount of time required to replicate a particular length of RNA. Thus, TRS-driven synthesis of sg DI RNA (i.e., transcription) might occur as efficiently as that of the mRNAs of the standard virus, but because of a more efficient replication relative to that of the genome of the standard virus, the DI genome might become over-represented. Alternatively, DI RNAs may have been selected for optimal genome multiplication and inherently represent poor templates for sg RNA synthesis. For example, DI RNAs may lack particular signals required for efficient recruitment into transcriptase complexes. However, it is also quite possible that the TRS inserted into pDI1000 merely represents the minimal promoter element required for sg RNA synthesis and that flanking sequences and/or overall genome context affect transcription efficiency. In coronaviruses, there is indeed evidence that TRS-flanking sequences influence the efficacy of sg RNA production (1, 8, 17). The potential complexities of transcription regulation are perhaps most poignantly illustrated by recent findings made for the tombusvirus Tomato bushy stunt virus. Here, efficient synthesis of the two sg mRNAs critically depends on long-range RNA-RNA interactions involving three different sequence elements that are dispersed throughout the genome (13). Future studies of torovirus transcription should therefore ideally be undertaken through reverse genetics of the standard virus, that is, once protocols to this end become available. Interestingly, the BEV HE gene is nonfunctional because of a large deletion (31), but its dedicated mRNA, mRNA 4, is still produced, albeit in reduced quantities compared to RNAs 3 and 5 (33, 38). For torovirus promoter studies in the context of the complete viral genome, the TRS of mRNA 4 would thus be the perfect target.
Acknowledgments
We are grateful to Peter J. M. Rottier and Jolanda D. F. de Groot-Mijnes for critical reading of the manuscript.
REFERENCES
- 1.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bredenbeek, P. J., A. F. Noten, M. C. Horzinek, and W. J. Spaan. 1990. Identification and stability of a 30-kDa nonstructural protein encoded by mRNA 2 of mouse hepatitis virus in infected cells. Virology 175:303-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian, D. A., and W. J. Spaan. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 6.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81:1473-1484. [DOI] [PubMed] [Google Scholar]
- 7.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:927-935. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, K. M., B. Yount, A. C. Sims, and R. S. Baric. 2004. Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus. J. Virol. 78:6061-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland, J. J. 1991. Defective viral genomes, p. 151-165. In B. N. Fields and D. M. Knipe (ed.), Fundamental virology. Raven Press, New York, N.Y.
- 11.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, H. X., and K. A. White. 2004. A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J. 23:3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruyama, K., and S. Sugano. 1994. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138:171-174. [DOI] [PubMed] [Google Scholar]
- 16.Meinkoth, J., and G. Wahl. 1984. Hybridization of nucleic acids immobilized on solid supports. Anal. Biochem. 138:267-284. [DOI] [PubMed] [Google Scholar]
- 17.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasternak, A. O., A. P. Gultyaev, W. J. Spaan, and E. J. Snijder. 2000. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 74:11642-11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2004. Regulation of relative abundance of arterivirus subgenomic mRNAs. J. Virol. 78:8102-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawicki, D., T. Wang, and S. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 23.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 25.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 26.Schaad, M. C., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169-8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sethna, P. B., and D. A. Brian. 1997. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J. Virol. 71:7744-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snijder, E. J., J. A. den Boon, M. C. Horzinek, and W. J. Spaan. 1991. Characterization of defective interfering RNAs of Berne virus. J. Gen. Virol. 72:1635-1643. [DOI] [PubMed] [Google Scholar]
- 31.Snijder, E. J., J. A. den Boon, M. C. Horzinek, and W. J. Spaan. 1991. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology 180:448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snijder, E. J., and M. C. Horzinek. 1993. Toroviruses: replication, evolution and comparison with other members of the coronavirus-like superfamily. J. Gen. Virol. 74:2305-2316. [DOI] [PubMed] [Google Scholar]
- 33.Snijder, E. J., M. C. Horzinek, and W. J. Spaan. 1990. A 3′-coterminal nested set of independently transcribed mRNAs is generated during Berne virus replication. J. Virol. 64:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 35.van der Most, R. G., R. J. de Groot, and W. J. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription and RNA recombination, p. 11-31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, Inc., New York, N.Y.
- 37.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., and X. Zhang. 2000. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J. Virol. 74:10571-10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in positive-strand RNA viruses. Virology 304:147-154. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., C. L. Liao, and M. M. Lai. 1994. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J. Virol. 68:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]