Abstract
The introduction of West Nile virus (WNV) into North America has been associated with relatively high rates of neurological disease and death in humans, birds, horses, and some other animals. Previous studies identified strains in both genetic lineage 1 and genetic lineage 2, including North American isolates of lineage 1, that were highly virulent in a mouse neuroinvasion model, while other strains were avirulent or significantly attenuated (D. W. C. Beasley, L. Li, M. T. Suderman, and A. D. T. Barrett, Virology 296:17-23, 2002). To begin to elucidate the basis for these differences, we compared a highly virulent New York 1999 (NY99) isolate with a related Old World lineage 1 strain, An4766 (ETH76a), which is attenuated for mouse neuroinvasion. Genomic sequencing of ETH76a revealed a relatively small number of nucleotide (5.1%) and amino acid (0.6%) differences compared with NY99. These differences were located throughout the genome and included five amino acid differences in the envelope protein gene. Substitution of premembrane and envelope genes of ETH76a into a NY99 infectious clone backbone yielded a virus with altered in vitro growth characteristics and a mouse virulence phenotype comparable to ETH76a. Further site-specific mutagenesis studies revealed that the altered phenotype was primarily mediated via loss of envelope protein glycosylation and that this was associated with altered stability of the virion at mildly acidic pH. Therefore, the enhanced virulence of North American WNV strains compared with other Old World lineage 1 strains is at least partly mediated by envelope protein glycosylation.
West Nile virus (WNV) is a member of the Japanese encephalitis virus complex of the family Flaviviridae, genus Flavivirus, which also includes Japanese encephalitis virus, St. Louis encephalitis virus (SLEV), Murray Valley encephalitis virus (MVEV), and others. These viruses are mosquito borne, primarily transmitted by Culex spp., and have wide, overlapping distributions throughout the world (22).
Human infections with WNV had generally been considered to result in a relatively mild febrile illness, although occasional occurrences of encephalitis in humans and horses had been reported. However, beginning in the mid-1990s, large epidemics with significant rates of neurological disease and death were reported in North Africa, Eastern Europe, and Israel. Epidemics of encephalitis among horses were also reported in Italy and France. (For a review of the natural history of WNV in the Old World, see reference 30). In 1999, WNV was identified as the cause of an outbreak of 62 cases of encephalitis in New York City that resulted in seven deaths (31). This emergence was also associated with neurological disease in horses (39) and significant mortality among birds, particularly corvids—a phenomenon that had not been reported during contemporary outbreaks of WNV encephalitis in humans (23). Subsequently, WNV has extended its distribution in the Americas throughout the United States and southern Canada and has recently been isolated in Mexico (12). Serological evidence suggests that the virus is now widely distributed throughout Mexico (6, 12, 21) and also parts of the Caribbean (11, 16, 34). Since 2002, WNV has been responsible for more than 6,300 cases of human meningitis/encephalitis in the United States that resulted in 633 deaths (data from the CDC website, http://www.cdc.gov/westnile, as of 14 December 2004).
The emergence of WNV as a significant agent of arboviral encephalitis has raised several questions regarding possible differences in the virulence properties of WNV strains and the hypothesis that some strains of genetic lineage 1, particularly those circulating in North America and Israel, might have increased virulence for birds and/or mammals (18, 19). Studies by some of us using a mouse neuroinvasion model demonstrated significant differences in the ability of wild-type WNV strains to cause encephalitis-like disease and death following peripheral inoculation. Strains representative of genetic lineages 1 and 2 were found to have virulence phenotypes ranging from highly neuroinvasive, with 50% lethal doses (LD50s) following intraperitoneal (i.p.) inoculation of approximately 1 PFU, to highly attenuated, with LD50s of ≥10,000 PFU (2). Strains isolated in New York City during 1999 were highly neuroinvasive. Other closely related lineage 1 strains that had an attenuated neuroinvasive phenotype were identified; these strains caused scattered mortality in groups of mice across a range of challenge doses. Recently, we and others have reported plaque-purified variants of North American WNV isolates with differing mouse virulence phenotypes that suggest a role for envelope (E) protein glycosylation as a virulence determinant (4, 37).
To elucidate the molecular determinants of virulence differences between these related lineage 1 WNV strains, we sequenced the genome of a mouse attenuated strain, EthAn4766 (designated ETH76a), and compared this sequence with published sequences from North American isolates, particularly the prototype strain 382-99 (designated NY99). By gene substitution and specific mutagenesis of an infectious clone derived from NY99, we have confirmed a critical role for envelope protein glycosylation in the enhanced neuroinvasiveness of NY99 WNV in a mouse model.
MATERIALS AND METHODS
Virus strains.
WNV strains NY99 and ETH76a were obtained from our virus collections held at the Division of Vector-Borne Infectious Diseases, Fort Collins, CO, and the University of Texas Medical Branch, Galveston, TX, respectively. Virus working stocks were grown and plaque titrated in Vero cells.
Construction of WNV NY99 infectious clone and variants.
The WNV NY99 virus-specific infectious cDNA clone was constructed in two plasmids, utilizing a derivative of plasmid pBRUC-139S, as described previously (14). Plasmid pWN-AB contained WNV nucleotides 1 to 2495, which were preceded by restriction sites SstI and MluI and the promoter for T7 polymerase. Plasmid pWN-CG contained WNV nucleotides 2495 to 11029 and an engineered 3′-terminal XbaI site for plasmid linearization just prior to transcription of genomic RNA. Nucleotide sequencing of the NY99 infectious clone (NY99ic) and the parental NY99 stock from which it was derived identified seven differences from the published NY99 sequence (GenBank accession no. AF196835), including two that encoded amino acid substitutions: C1428U, U1855C, C3880U (NS2A-118 His→Tyr), A4922G (NS3-104 Lys→Arg), G7029U, U8811C, and A10851G.
Full-genomic-length cDNA was prepared by cleaving the pWN-AB and pWN-CG plasmids at the natural NgoMIV-2495 site of WNV followed by ligating the two plasmids at this NgoMIV site. No attempt was made to gel purify the in vitro-ligated DNA fragment containing the full-genome-length WNV cDNA. Following in vitro ligation, viral genomic RNA was transcribed by using the AmpliScribe T7 kit (Epicentre Technologies, Madison, WI). Transcription was carried out in the presence of m7-GpppA cap analog for 2 to 3 h at 37°C, and Vero cells were transfected with the transcribed RNA by electroporation as described previously (15).
The premembrane (prM) and envelope (E) genes of WNV strain ETH76a were amplified by reverse transcriptase PCR (RT-PCR) and cloned into pGEM-T for substitution into the pWN-AB plasmid of the NY99 infectious clone. As no suitable restriction sites were available, the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to engineer sites at either end of the cloned ETH76a and NY99 pWN-AB prM-E regions. A BstEII site was engineered at nucleotides 465 to 471 at the 5′ end of prM in both NY99 and ETH76a. At nucleotides 2464 to 2469 at the 3′ end of E, different blunt-end restriction sites were engineered: PmlI in ETH76a and SfoI in NY99. None of these mutations resulted in changes to the encoded amino acid sequence. Complete details of primers used for the mutagenesis are available from the authors on request.
The Quikchange kit was also used to mutate the pWN-AB plasmid to alter amino acid 154 of E from Asn to Ser, thereby removing the potential glycosylation motif of NY99 E protein. Similarly, the pWN-AB plasmid containing prM-E of ETH76a was mutated to restore a functional glycosylation motif in E via a Ser-to-Asn substitution at residue 154. All of the engineered mutations and recombinations of the infectious clone pWN-AB plasmid were confirmed by nucleotide sequencing of the modified plasmids. Transcription and transfection reactions were carried out as described above. Sequences of primers used for mutagenesis of the pWN-AB plasmid are available from the authors on request.
Nucleotide sequencing.
Whole genome and/or prM-E gene sequencing of strain ETH76a and infectious-clone-derived viruses was performed using primers and protocols based on those described by Lanciotti et al. (18) and Beasley et al. (3). Briefly, viral RNA was extracted from culture supernatants of virus-infected Vero cells using the QiaAmp kit (QIAGEN Inc., Valencia, CA), and cDNA products were amplified either using the single-reaction Titan RT-PCR kit according to the manufacturer's recommendations or as a two-step reaction using avian myeloblastosis virus reverse transcriptase (AMV-RT) and Taq polymerase (Titan, AMV-RT and Taq from Roche, Indianapolis, IN). PCR products were directly sequenced in both directions to generate consensus sequences. All sequencing reactions were performed at the University of Texas Medical Branch's Protein Chemistry Core Laboratory and run on an ABI Prism model 3100 DNA sequencer (Applied Biosystems, Foster City, CA). In some cases where RT-PCR amplification was inefficient, products were cloned into the pGEM-T(Easy) vector (Promega, Madison, WI), and three clones for each were sequenced in both directions. Sequences were assembled and analyzed using the Vector NTI suite of programs (Informax Inc., Bethesda, MD).
Mouse virulence testing.
Neuroinvasion and neurovirulence phenotypes of wild-type and infectious-clone-derived viruses were determined by i.p. and intracranial (i.c.) inoculation of 3- to 4-week-old female NIH Swiss mice (Harlan, Indianapolis, IN), respectively. Determination of LD50s was as described previously (2) except that mice were inoculated with doses of virus in the range of 0.1 to 1,000 PFU. To determine the degree of virus replication in mice that survived virus inoculation, surviving animals were challenged at 21 days postinfection (dpi) with 100 PFU (equivalent to ∼100 LD50s) of WNV NY99 delivered i.p. Based upon the number of animals that succumbed to this secondary challenge, we calculated 50% protective doses (PD50s) for each virus strain. Naive control animals of the same age were also inoculated to confirm the infectivity and lethality of the virus challenge dose.
Determination of viremia and neuroinvasion kinetics.
Groups of 26 3- to 4-week-old female NIH Swiss mice were inoculated i.p. with 1,000-PFU doses of either the NY99ic (glycosylated E protein) or NY99/E154 (nonglycosylated) infectious-clone-derived variants. At daily intervals postinfection up to day 7, three mice from each group were sacrificed and blood (for serum) and brains were harvested. Brains were homogenized in minimal essential medium to yield a final volume of 1 ml, and the virus titers in serum and brain homogenates were determined by plaque assay in Vero cells. Five mice from each group were left to progress through the full course of infection, and at 21 days postinfection the surviving animals were challenged i.p. with 100 PFU of WNV NY99.
Other phenotypic characterization of wild-type and clone-derived viruses.
Virus growth kinetics were determined in Vero cells. Confluent 25-cm2 flasks of cells were inoculated with virus at a multiplicity of infection of ∼0.02 for 30 min at room temperature. After incubation, the cell monolayers were rinsed to remove unbound virus, and 8 ml of minimal essential medium supplemented with 2% fetal bovine serum was added. Aliquots of culture supernatants were collected twice daily for 3 days, at which point all monolayers showed significant (>70%) cytopathic effect. Virus titers for each time point sample were determined in duplicate by plaque assay in Vero cells.
In addition, virus titers at some time points were also determined using a quantitative real-time RT-PCR protocol. Briefly, viral RNA was extracted from 70 μl of cell culture supernatant using the QiaAmp kit (QIAGEN), and a ∼100-bp fragment of the 3′ noncoding region (NCR) was amplified using a Taqman One-step RT-PCR (Applied Biosystems) as described elsewhere (40). Virus titers were then determined by comparison with a standard curve. Strains NY99, NY99ic, NY99/ETH, and NY99/E154 had identical sequences at the primer binding sites and through the region being amplified, making direct comparison of results for these strains possible. However, ETH76a differed at one nucleotide in the sense primer binding site and at four other residues within the amplicon, suggesting that results obtained for this strain might not be directly comparable with the others.
The sensitivity of wild-type and clone-derived viruses to acidic pH was assessed using methods similar to those described by McMinn et al. (25). Approximately 105 PFU of virus was treated at pHs ranging between 6.0 and 7.0 for 15 min at room temperature. Samples were then diluted 1:10 in sterile phosphate buffer (pH 8.0) to return the pH to ≥7.4 and plaque titrated in Vero cells to quantitate residual infectivity.
Nucleotide sequence accession number.
The genome sequence of strain ETH76a was submitted to GenBank under accession no. AY603654.
RESULTS
Nucleotide and amino acid sequence differences between ETH76a and NY99.
Comparison of the whole genome sequence of strain ETH76a (GenBank accession no. AY603654) with the previously published sequence for strain NY99 (17) (GenBank accession no. AF196835) revealed a total of 567 nucleotide differences (5.1%) that encoded 22 amino acid differences (0.6%) scattered throughout the polyprotein coding sequence (Table 1). The largest numbers of coding substitutions were contained within the E (five amino acid changes) and NS5 (four changes) protein genes. Each of the other nonstructural (NS) protein genes encoded two amino acid differences, and prM/M encoded one change. The majority of the amino acid differences (16 of 22) were conservative in nature, and many (12 of 22) have been described for other lineage 1 WNV strains (9, 19).
TABLE 1.
Summary of nucleotide and amino acid differences between WNV strains NY99 and ETH76a
| Gene/region | No. of nucleotide differencesa | No. of amino acid differencesa | Specific amino acid differences (NY99 → ETH76a) | Amino acid changes shared withc: |
|---|---|---|---|---|
| 5′ NCR | 0/96 (0%) | n/a | n/a | |
| C | 3/369 (0.8%) | 0/123 (0.0%) | n/a | n/a |
| prM/M | 23/501 (4.6%) | 1/167 (0.6%) | prM-19 Val → Ile | |
| E | 91/1,503 (6.1%) | 5/501 (1.0%) | E-93 Arg → Lys | Eg101 |
| E-126 Ile → Thrb | Fra00; Ita98; VLG4; Rom96; Eg101 | |||
| E-154 Asn → Serd | Manyd | |||
| E-159 Val → Ile | Tun97; Fra00; Ita98; Rom96; Ken98; Eg101 | |||
| E-474 Ile → Val | ||||
| NS1 | 62/1,056 (5.9%) | 2/352 (0.6%) | NS1-109 Glu → Asp | |
| NS1-167 Met → Ile | ||||
| NS2A | 31/693 (4.5%) | 2/231 (0.9%) | NS2A-19 Ala → Val | |
| NS2A-119 His → Tyrb | VLG4 | |||
| NS2B | 27/393 (11.3%) | 2/131 (1.5%) | NS2B-103 Val → Ala | Tun97; Fra00; Ita98; Ken98; Eg101 |
| NS2B-125 Thr → Ser | ||||
| NS3 | 110/1,857 (5.9%) | 2/619 (0.3%) | NS3-249 Pro → Thrb | Tun97; Fra00; Ita98; Ken98 |
| NS3-356 Thr → Ileb | Tun97; Fra00; Ita98; VLG4; Rom96; Ken98; Eg101 | |||
| NS4A | 20/447 (4.5%) | 2/149 (1.3%) | NS4A-85 Ala → Val | Fra00; Ita98; VLG4; Rom96; Ken98; Eg101 |
| NS4A-141 Met → Leu | Eg101 | |||
| NS4B | 44/765 (10.2%) | 2/255 (0.8%) | NS4B-11 Ser → Asn | Eg101 |
| NS4B-23 Val → Ala | Eg101 | |||
| NS5 | 127/2,715 (4.7%) | 4/905 (0.4%) | NS5-177 Arg → Lys | Eg101 |
| NS5-258 Val → Ala | ||||
| NS5-450 His → Tyrb | ||||
| NS5-731 Val → Ala | ||||
| 3′ NCR | 29/634 (4.6%) | n/a | No insertions/deletions |
The number of changes/total residues (% difference) is shown.
Nonconservative amino acid substitution.
Based on whole genome sequence data from Lanciotti et al. (19) and Charrel et al. (9). Eg101, Egypt101 (AF260968); Fra00, France 2000 PaAn001 (AY268132); Ita98, Italy 1998 (AF404757); VLG4, Volgograd 1999 (AF317203); Rom97, Romania 1996 RO9750 (AF260969); Tun97, Tunisia 1997 PaH001 (AY268133); Ken98, Kenya 1998 KN3829 (AY262283); n/a, not applicable.
The indicated mutation results in loss of the E glycosylation motif. Similar mutations at E154 have been reported for fragmentary sequences of several lineage 1 WNV strains (5a, 18). Some other lineage 1 strains, including Eg101, have mutations at E156 that also result in loss of the glycosylation motif.
Ten of the amino acid differences in the ETH76a strain were shared with the Egypt101 strain (Eg101), which has also been reported to have an attenuated neuroinvasion phenotype in mice (2, 5). Of these 10 differences, 6 have also been reported for several European lineage 1 isolates (Table 1), most of which have been associated with epidemics of neurological disease in humans or horses (e.g., Fra00, Ita98, Rom96, VLG4, and Tun97) (9, 19). Four of the five amino acid differences in the E protein were shared with Eg101 and/or other WNV strains. Of particular interest was the Asn→Ser mutation at E154 that abolished the glycosylation motif at residues E154 to 156. Similar mutations at E154 or at E156 (as occurs in Eg101) that prevent E glycosylation have been reported for many Old World lineage 1 WNV strains (19). Two nonconservative differences in NS3, at amino acids 249 and 356, were also shared by many other Old World lineage 1 strains. In addition, ETH76a shared two mutations in NS4B and one in NS5 with Eg101. Other changes that were unique to ETH76a were generally conservative in nature (Table 1).
No nucleotide differences were identified between the 5′ NCR sequences of NY99 and ETH76a, while 29 residues differed in the 3′ NCRs. These included a mutation of the polyprotein stop codon (UAG in NY99 and other North American strains and UAA in ETH76a and many other Old World WNV isolates). No 3′ NCR insertions or deletions were identified, which was consistent with previous partial 3′ NCR sequence data for several lineage 1 strains (1). Of the 29 nucleotide differences identified in the 3′ NCR, 22 were shared with at least one other lineage 1 WNV strain.
Recovery and preliminary characterization of NY99ic and mutant viruses.
To investigate the molecular basis of attenuation of ETH76a, the prM and E protein genes of this virus were substituted into the NY99ic genome to generate a chimeric virus (NY99/ETH). In addition, we had previously reported that most mouse attenuated lineage 1 WNV strains we have studied had a nonglycosylated E protein (5). Therefore, we used site-directed mutagenesis to generate a NY99 virus with a nonglycosylated E protein (NY99/E154). Subsequently, a variant of NY99/ETH encoding a Ser-to-Asn mutation at E154 that permitted glycosylation of E (NY99/ETHgly) was also constructed.
Stocks of the infectious-clone-derived strains (NY99ic, NY99/ETH, NY99/E154, and NY99/ETHgly) were recovered from transfected cells 4 to 5 dpi, all with plaque titers of >106 PFU/ml. These primary stocks were used for all subsequent mouse virulence experiments and other in vitro characterizations. Nucleotide sequencing of the engineered mutants confirmed the presence of introduced genes/mutations in the NY99/ETH and NY99/E154 variants. No other mutations from the NY99ic or ETH76a parent sequences were identified in these variants.
Western blotting of virus-infected Vero cell lysates demonstrated differences in the apparent molecular weights of the E proteins of NY99 and NY99ic compared to ETH76a, NY99/ETH, and NY99/E154 strains consistent with differences in glycosylation (Fig. 1), as has been described elsewhere (14). A subtle difference in the plaque morphology of glycosylated and nonglycosylated strains was noted. At 72 h postinfection, the majority of plaques for glycosylated strains were ≥2 mm in diameter, while the majority of plaques for the nonglycosylated strains were ≤2 mm. However, these differences were not statistically significant (data not shown).
FIG. 1.
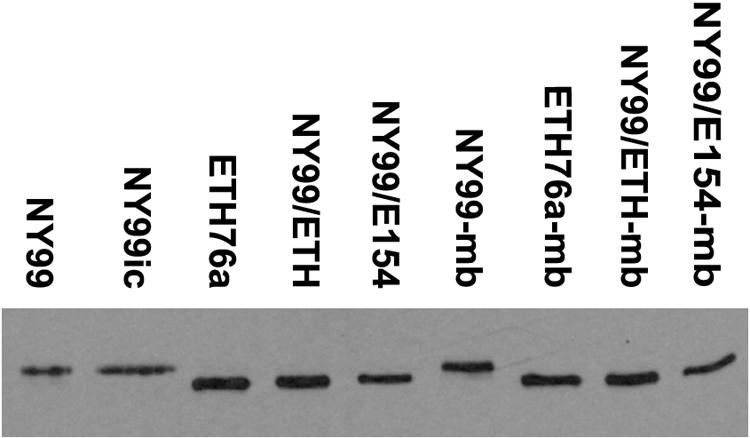
Western blot showing the differing mobilities of E proteins of WNV strains consistent with the presence of Asn-Tyr-Ser glycosylation motif or mutant Ser-Tyr-Ser at residues 154 to 156. Proteins were separated on a 5%/10% discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, electrophoretically transferred to 0.2-μm nitrocellulose, and detected using WNV-specific monoclonal antibody 7H2. The suffix “mb” indicates strains isolated from the brain of a mouse that died following peripheral inoculation with the respective parent strain.
Mouse virulence phenotypes of the wild type and the NY99 infectious-clone-derived WNV strains.
All of the wild-type (NY99 and ETH76a) and infectious-clone-derived (NY99ic, NY99/ETH, and NY99/E154) strains that we tested were highly neurovirulent in 3- to 4-week-old NIH Swiss mice, yielding LD50s between 0.3 and 1.3 PFU after i.c. inoculation (Table 2). Average survival times (ASTs) for mice inoculated with the nonglycosylated strains appeared to be shorter than for NY99 or NY99ic, although only the AST for NY99/ETH was significantly different (Student's t test, P < 0.001).
TABLE 2.
Neuroinvasion and neurovirulence properties of WNV variants in 3- to 4-week-old female NIH Swiss mice
| Virus | i.p. LD50 (PFU) | i.p. ASTa (days ± SD) | i.c. LD50 (PFU) | i.c. ASTa (days ± SD) | PD50b (PFU) |
|---|---|---|---|---|---|
| NY99 | 1.3 | 8.3 ± 1.2 | 0.5 | 6.8 ± 1.1 | 0.8 |
| NY99ic | 1.3 | 8.1 ± 1.0 | 0.3 | 6.7 ± 0.7 | 0.8 |
| ETH76a | 126 | 9.6 ± 1.0 | 1.3 | 5.8 ± 1.9 | 0.1 |
| NY99/ETH | 200 | 11.0 ± 1.9 | 1.1 | 5.3 ± 0.4 | 3.2 |
| NY99/E154 | 126 | 10.2 ± 2.9 | 1.1 | 5.8 ± 0.8 | 2.0 |
| NY99/ETHgly | 2.0 | 8.8 ± 1.7 | 0.8 | 6.7 ± 1.1 | n.d. |
| ETH76a-mbc | 50 | 9.5 ± 1.4 | n.d. | n.d. | 2.0 |
| NY99/ETH-mbc | 32 | 11.4 ± 2.6 | n.d. | n.d. | 3.2 |
| NY99/E154-mbc | 20 | 9.3 ± 1.3 | n.d. | n.d. | 0.8 |
| NY99-mbc | 0.8 | 8.0 ± 1.3 | n.d. | n.d. | 0.8 |
ASTs are for all animals that died.
PD50s were calculated based on a secondary challenge (100 PFU of NY99) of mice that survived the initial i.p. challenge with the respective strain. n.d., not determined.
An “mb”designation identifies strains that were recovered from the brains of dead mice following i.p. challenge with the respective parental strain (i.e. ETH76a-mb is an isolate of ETH76a recovered from the brain of a dead mouse).
As expected, strain NY99 was highly neuroinvasive in our mouse model (i.p. LD50 = 1.3 PFU) (Table 2), and the NY99ic infectious-clone-derived virus retained the virulent phenotype of the parent strain (i.p. LD50 = 1.3 PFU). Consistent with our earlier studies, strain ETH76a had an attenuated neuroinvasion phenotype and caused scattered mortality across the range of virus doses following peripheral inoculation. However, the calculated LD50 of 126 PFU was lower than what we had reported previously for this strain (2). Both the NY99/ETH chimera and the NY99/E154 glycosylation mutant had neuroinvasive properties similar to the attenuated ETH76a strain, with i.p. LD50s of 200 and 126 PFU, respectively. In contrast to the results following i.c. inoculation, ASTs for animals that died following i.p. inoculation with ETH76, NY99/ETH, or NY99/E154 were significantly prolonged compared to NY99 or NY99ic (Student's t test, P < 0.005) (Table 2).
In order to confirm that changes in E glycosylation status were primarily responsible for the observed virulence differences, the NY99/ETHgly mutant (which encoded a glycosylated ETH76a E protein) was constructed and inoculated i.p. and i.c. into mice. This variant was highly virulent, with an i.p. LD50 of 2.0 PFU and an i.c. LD50 of 0.8 PFU (Table 2).
For animals inoculated i.p. with strain NY99, NY99ic, or NY99/ETHgly, the onset and progression of disease were very similar to those described for the hamster (42) and BALB/c mouse (17) models of WNV encephalitis. Mice appeared healthy for about 5 dpi, but between 6 and 8 dpi they began to appear sick, with signs of lethargy, hunched posture, and ruffled fur. In most animals that died, the disease progression was rapid, with death occurring 8 to 24 h after the first signs of illness. Significant signs of neurological involvement—including loss of balance, tremors, and some occurrences of tetany, as well as hind limb paralysis—were observed in many animals. In a small number of animals, bleeding from the mouth and/or nose was observed, as were ocular secretions. A similar disease course was observed for those animals that died following peripheral inoculation with ETH76a, NY99/ETH, or NY99/E154.
PD50s for attenuated strains are comparable to NY99/NY99ic i.p. LD50s.
To determine whether mice that survived inoculation with strain ETH76a, NY99/ETH76, or NY99/E154 were productively infected with those viruses, all survivors were challenged with 100 PFU of NY99 virus, and PD50s were calculated based on the numbers of mice that were protected against this challenge. Naive animals of the same age (6 to 7 weeks) uniformly succumbed to the 100-PFU challenge. PD50s for all of the attenuated strains were around 1 PFU, which were comparable to the LD50 of the neuroinvasive NY99 and NY99ic strains (Table 2). PD50s for NY99 and NY99ic were essentially the same as their LD50s. Eight mice each survived initial challenge with NY99 or NY99ic viruses, primarily at the lowest challenge doses (1 or 0.1 PFU). In each case, one of the eight mice survived secondary challenge with 100 PFU of NY99 virus, thereby showing the potential for mouse survival and immunization following productive infection with these strains. However, the results indicated that productive NY99 or NY99ic infection in these mice generally led to fatal disease.
Virus strains recovered from brains of dead mice retain the virulence phenotype of their parental strain.
To assess whether the death of mice following peripheral inoculation with the attenuated ETH76a, NY99/ETH, or NY99/E154 strain was associated with changes in E glycosylation status of the viruses, we reisolated each of these strains from the brain of a dead mouse (designated “strain name”-mb) and sequenced the prM and E protein genes (2,004 nucleotides). Western blotting of viral proteins yielded results identical to the respective parent strains (Fig. 1), and no reversions to a potentially glycosylated sequence were identified by nucleotide sequencing. Only the NY99/ETH-mb isolate differed in sequence from its parent strain. This strain had a single A→U mutation at nucleotide 2273 that encoded a conservative Thr→Ser change at E436, a residue that lies within a predicted helix of the stem-anchor region. Each of the mouse brain isolates was then inoculated i.p. into 3- to 4-week-old mice to determine LD50s, which showed that the mouse brain isolates each retained the attenuated neuroinvasion phenotypes of their parent strains (Table 2). However, the calculated LD50 for each was approximately two- to sevenfold lower than the respective parent strain, suggesting some further virulence adaptation or selection may have occurred. An NY99-mb isolate retained the highly neuroinvasive phenotype.
Comparison of viremia and neuroinvasion kinetics for glycosylated and nonglycosylated NY99 infectious clone variants.
Comparisons of the replication of NY99 and ETH76a parental strains in 3- to 4-week-old mice had shown that, while NY99 established a strong viremia and was detected in the brains of mice starting around day 4 postinfection, ETH76a caused only a low transient viremia in some animals and was not readily detected in brains of infected mice (5). To confirm that the loss of E protein glycosylation was primarily responsible for this difference, a similar comparison was made using the NY99ic and NY99/E154 infectious-clone-derived variants. As expected, i.p. inoculation with NY99ic produced a strong viremia, peaking around day 2 to 3 postinfection, after which virus was detected in mouse brains starting on day 3 (Table 3). By day 7, all remaining mice infected with NY99ic were sick, and five mice that were not sacrificed for virus titrations died by day 8 postinfection. In contrast, no virus was detected in serum of mice inoculated i.p. with NY99/E154, and only one animal that appeared sick on day 7 was found to have detectable virus in the brain. One of the five mice that were not sacrificed was also sick and died on day 9 postinfection. The four surviving animals were challenged on day 21 postinfection with 100 PFU of NY99, and all survived this challenge.
TABLE 3.
Titers of NY99ic or NY99/E154 infectious-clone-derived WNV variants in the serum and brains of NIH Swiss mice determined at daily intervals following intraperitoneal inoculation
| Day | Animal no. | NY99ic titer in:
|
NY99/E154 titer in:
|
||
|---|---|---|---|---|---|
| Serum (PFU/ml) | Brain (PFU/brain) | Serum (PFU/ml) | Brain (PFU/brain) | ||
| 1 | 1 | 2.0 × 102 | —a | — | — |
| 2 | 7.5 × 102 | — | — | — | |
| 3 | 4.0 × 103 | — | — | — | |
| 2 | 1 | 1.0 × 104 | — | — | — |
| 2 | 2.0 × 103 | — | — | — | |
| 3 | 6.5 × 103 | — | — | — | |
| 3 | 1 | 1.3 × 103 | — | — | — |
| 2 | 9.0 × 102 | — | — | — | |
| 3 | 6.5 × 103 | 5.0 × 101 | — | — | |
| 4 | 1 | — | 5.0 × 102 | — | — |
| 2 | — | 5.5 × 104 | — | — | |
| 3 | — | — | — | — | |
| 5 | 1 | — | 3.5 × 106 | — | — |
| 2 | — | 2.5 × 102 | — | — | |
| 3 | — | 3.3 × 104 | — | — | |
| 6 | 1 | — | 8.8 × 105 | — | — |
| 2 | — | — | — | — | |
| 3 | — | 1.8 × 107 | — | — | |
| 7 | 1 | — | 2.8 × 107 | — | — |
| 2 | — | 6.3 × 103 | — | — | |
| 3 | — | >1.3 × 108 | — | 2.8 × 107 | |
—, no virus was detected. The limit of detection was 25 PFU/ml of serum or 25 PFU/brain.
Attenuated growth of nonglycosylated strains in Vero cells.
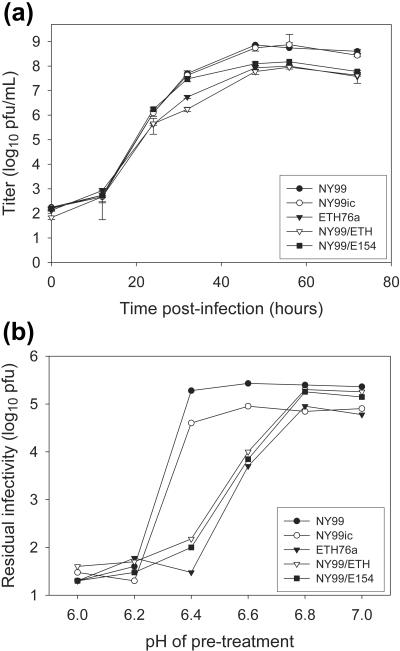
In order to assess the effects of E protein glycosylation on virus growth, in vitro comparisons were made of the growth kinetics of wild-type NY99 and ETH76a, as well as NY99ic, NY99/ETH, and NY99/E154, in Vero cells. Although all strains reached maximal infectivity titers at similar times (around 48 h postinfection), the titers for nonglycosylated ETH76a, NY99/ETH76, and NY99/E154 were 4- to 12-fold lower than for the glycosylated strains (most differences were >5-fold) (Fig. 2a).
FIG. 2.
Comparisons of (a) growth kinetics in Vero cells at an MOI of approximately 0.02 and (b) sensitivity to inactivation at mildly acidic pH for WNV strains (approximately 105 input PFU for each) with glycosylated or nonglycosylated E protein sequences. Error bars in panel a are 1 standard deviation.
Nonglycosylated strains are more sensitive to acidic pH.
Studies with other mosquito-borne flaviviruses have demonstrated that loss of E protein glycosylation was associated with changes in specific infectivity and/or sensitivity to mildly acidic pH (13, 36, 41). Therefore, virus inactivation assays were performed to assess the effects of low-pH treatment on our WNV strains/variants. Aliquots of each virus (approximately 105 PFU) were treated at pHs between 6.0 and 7.0, and residual infectivity was determined by plaque titration in Vero cells (Fig. 2b). Strains NY99 and NY99ic were more stable at mildly acidic pH, with no reduction in virus titer occurring at pH 6.6 or higher and maximal or near-maximal reduction at pH 6.2. In contrast, reduction in infectivity of the nonglycosylated strains was observed at pH 6.6, and maximal inactivation occurred by pH 6.4. Approximately 20 to 80 PFU of residual infectivity was retained for each strain. Consistent with these differences in sensitivity to inactivation at acidic pH, the optimal pH for hemagglutination by these viruses differed, with NY99 and NY99ic having peak hemagglutination activity at pH 6.2 and the nonglycosylated strains having a peak at pH 6.4.
Differences in virus titer determined by quantitative real-time PCR.
To assess whether the observed differences in plaque titer were associated with reductions in overall virus output from cells infected with the nonglycosylated strains/variants or with increased inactivation of secreted virus due to altered stability of the virion, we employed a quantitative real-time RT-PCR protocol (40). Samples for NY99, NY99ic, and the infectious-clone-derived variants collected at the 48- and 56-h time points of the growth curves shown in Fig. 2a were tested. At both time points, titers for strains NY99/ETH and NY99/E154 determined by this method were less than 2.5-fold lower than the RT-PCR titers of NY99 or NY99ic, despite the 4- to 12-fold differences in plaque titers for those samples (Table 4), suggesting that overall virus outputs from infected cells were similar although the proportion of noninfectious virus was higher for the nonglycosylated strains. The presence of five nucleotide mutations in ETH76a within the region targeted for amplification (one in the sense primer binding site and four within the amplicon) suggested that results obtained for that strain might not be directly comparable with the other strains. As expected, titers for ETH76a samples determined by real-time RT-PCR were 100- to 200-fold lower than the plaque titers and more than 500-fold lower than RT-PCR titers for all of the other strains, suggesting that the mutation in the sense primer binding site may have significantly affected the sensitivity of the assay (data not shown).
TABLE 4.
Comparison of titers for glycosylated and nonglycosylated NY99 and infectious-clone-derived WNV variants determined by plaque assay or quantitative Taqman RT-PCR
| Variant | Virus titer at postinfection interval:
|
|||
|---|---|---|---|---|
| 48 ha
|
56 ha
|
|||
| Plaque | Taqman | Plaque | Taqman | |
| NY99 | (7.3 ± 0.4) × 108 | (5.6 ± 0.2) × 108 | (5.5 ± 1.4) × 108 | (6.3 ± 0.3) × 108 |
| NY99ic | (5.5 ± 1.4) × 108 | (5.6 ± 0.2) × 108 | (7.5 ± 1.4) × 108 | (5.4 ± 0.5) × 108 |
| NY99/ETH | (6.0 ± 1.4) × 107 | (2.6 ± 0.1) × 108 | (6.0 ± 0.7) × 107 | (8.4 ± 1.2) × 108 |
| NY99/E154 | (1.3 ± 0.2) × 108 | (4.9 ± 0.2) × 108 | (1.5 ± 0.0) × 108 | (5.0 ± 0.2) × 108 |
The samples for 48- and 56-h time points are the same as those presented in Fig. 2a.
DISCUSSION
The increased incidence of neurological disease associated with recent outbreaks of WNV infection in Europe and North America has led to the hypothesis that some strains of genetic lineage 1 are inherently more virulent than others (18, 19). By comparing the virulent NY99 strain with a mouse attenuated lineage 1 strain, ETH76a, we aimed to identify molecular determinants of enhanced neuroinvasiveness of North American WNV strains. Comparison of genomic nucleotide sequences of NY99 and ETH76a revealed a relatively small number of encoded amino acid differences (Table 1). Of specific interest were amino acid changes within the structural proteins prM and E, particularly a difference at residue 154 of E (Asn in NY99 and Ser in ETH76a) that resulted in the loss of the glycosylation motif.
Many flaviviruses, including members of the Japanese encephalitis serocomplex, dengue viruses, and tick-borne encephalitis virus, have a potential glycosylation motif in structural domain I of the E protein at residues equivalent to E154 to 156 of WNV (35). Analyses of X-ray crystallographic structures of soluble E protein dimers derived from tick-borne encephalitis virus and dengue virus suggested that the carbohydrate moiety at this motif lies over the dimer interface in close proximity to the fusion peptide at the distal end of domain II and may play some role in stabilizing the dimer in its fusion-active form (26, 35). Although no structural data are currently available for the WNV E protein, a cryoelectron microscopy structure of the WNV virion showed a density presumably corresponding to carbohydrate at a similar location in proximity to E154 (29).
The results described here confirm that the highly virulent mouse neuroinvasion phenotype of strain NY99 compared to attenuated strain ETH76a is primarily mediated via the presence of the E protein glycosylation motif in NY99. Substitution of the prM-E genes of ETH76a into the NY99 infectious clone, or specific mutation of residue E154 to abolish glycosylation, resulted in attenuation of that virus to a level comparable to wild-type ETH76a. In contrast, substitution of the ETH76a 3′ NCR into the NY99 infectious clone had no attenuating effect on neuroinvasiveness (data not shown). Furthermore, a mutation that permitted glycosylation of the ETH76a E protein in the NY99 backbone yielded a virus with virulence equivalent to wild-type NY99, confirming that E protein glycosylation mediated the observed differences in virulence. Loss of E protein glycosylation was associated with increased sensitivity to acidic pH (Fig. 2b) and ∼10-fold reductions in peak infectious virus titer (Fig. 2a) in cell culture. However, titers of the nonglycosylated strains determined by quantitative Taqman RT-PCR (i.e., based on RNA genome copy numbers) were comparable to those of the NY99 and NY99ic strains (<2-fold differences) (Table 4), suggesting that the difference in peak titer was probably associated with increased inactivation of nonglycosylated virus during or after release from infected cells.
An earlier study using site-directed mutagenesis of the equivalent E glycosylation motif of a Kunjin virus infectious clone (Kunjin is a subtype of WNV lineage 1) reported similar findings (36). For Kunjin virus, glycosylation of the viral E protein was associated with 10- to 100-fold-greater infectious virus titer in either mammalian or mosquito cell lines compared to the nonglycosylated variant. Although neither variant was strongly neuroinvasive in mice, the presence of the carbohydrate moiety was reported to enhance the peripheral replication of Kunjin virus in mice. Recently, comparisons of plaque-purified variants of North American WNV isolates lacking E protein glycosylation have also suggested a role for this motif in the virulence of WNV (4, 37).
Our results are also similar to comparisons of neuroinvasive and attenuated strains of SLEV and MVEV. Comparison of two wild-type strains of SLEV revealed an association between E protein glycosylation and increased stability at mildly acidic pH (41). The glycosylated SLEV strain was also highly neuroinvasive in mice, while the nonglycosylated strain had an attenuated phenotype similar to what we have observed for WNV strain ETH76a (27). For MVEV, differences in sensitivity to acidic pH were also associated with differences in the mouse neuroinvasive phenotype (24, 25). However, in this case, the altered phenotype was mediated by a single amino acid change at residue E277, which lies in a flexible region between domains I and II that is believed to act as the hinge for the pH-dependent conformational change (26, 35). For the attenuated SLEV and MVEV strains, increased sensitivity to acidic pH resulted in a reduction of infectious virus titers, and this also appeared to be associated with increases in the relative numbers of inactive virus particles produced from infected cells rather than a reduction in the overall rate of viral replication.
Similar effects of envelope protein glycosylation on virion stability and virulence have also been reported for alphaviruses. The alphavirus E1 protein shares little primary sequence homology with flavivirus E but has a strikingly similar tertiary fold (20). Although the alphavirus E1 and E2 proteins are arranged very differently from the flavivirus prM/M and E in the assembled virion, alphavirus fusion with target cell membranes is also a low-pH-driven event. Membrane fusion is mediated via the E1 protein and is associated with significant structural rearrangements (43). Mutation of glycosylation motifs in E1 and/or E2 of Sindbis virus to prevent glycosylation has been associated with decreased plaque size, reduced virus titers in cell culture, and an increase in the virus particle-to-PFU ratio (33, 38). It was proposed that glycosylation acts to significantly stabilize the Sindbis virus virion in its fusion-active form (38).
Earlier comparisons of the viremia kinetics in mice of a New York 1999 isolate (385-99) and ETH76a revealed that ETH76a did not establish a strong viremia following peripheral (i.p.) inoculation and did not invade the brains of most mice (5). Comparisons of NY99 (glycosylated E) and NY99/E154 (nonglycosylated E) showed a similar pattern for viremia and neuroinvasion (Table 3). Our results suggest that this may, at least in part, be due to two factors: (i) decreased stability of ETH76a virions, resulting in lower infectious virus titers in the periphery; and (ii) each dose of “infectious” ETH76a includes approximately 5 to 10 times the dose of noninfectious virions that essentially act as an inactivated antigen and probably provide a significant stimulus to innate and adaptive immune pathways. We hypothesize that in some animals, the reduced specific infectivity of ETH76a most likely allows more rapid control and clearance of virus from the periphery, thereby limiting progression to neuroinvasive disease. Such a mechanism would account for the reduced frequency of neuroinvasion observed in mice infected with ETH76a. However, it is possible that E glycosylation may also affect other aspects of the WNV replication cycle that would impact on the relative virulence of WNV strains, such as target cell tropism, virion assembly and release, or efficiency of E protein fusion with target cell membranes.
Although E protein glycosylation status appears to account for the virulence difference between strains NY99 and ETH76a in our mouse model and is therefore most probably a major determinant of the enhanced virulence of other E glycosylated lineage 1 WNV strains associated with recent epidemics of encephalitis and neurological disease in humans and other animals, it is likely that there are several different determinants of virulence encoded within the genomes of WNV strains. For example, our results are contrary to previously reported data for an attenuated variant of an Israeli lineage 1 WNV strain, WNI (14). In that study, a nonglycosylated variant had increased mouse virulence compared to its parent glycosylated strain. However, additional glycosylated variants derived from these viruses displayed a virulent phenotype, suggesting that another determinant(s) (probably in the nonstructural protein genes) was primarily responsible for the observed virulence differences in that case (8). Other evidence that points to multiple virulence determinants includes the identification of nonglycosylated lineage 1 strains closely related to ETH76a that are almost completely attenuated for neuroinvasion in mice (LD50s ≥ 5,000 PFU) (5), the identification by some of us of mouse attenuated WNV strains isolated in Texas during 2003 that retain the E protein glycosylation motif present in NY99 (10), and the identification of nonglycosylated E variants of a WNV strain isolated in Mexico that appear to be attenuated for neuroinvasiveness and neurovirulence (4). The apparent virulence of any single WNV strain is probably the net result of a combination of determinants that act on the ability of the virus to replicate in the periphery to levels that allow subsequent neuroinvasion.
Two recombinant live, attenuated WNV vaccines currently in development are based on substitution of prM and E genes of NY99 WNV strains into either a yellow fever virus 17D infectious clone (28) or a dengue virus type 4 infectious clone incorporating attenuating mutations (32). Given that the WNV strains with nonglycosylated E proteins studied here were highly immunogenic, it could be expected that chimeric WNV vaccines incorporating a nonglycosylated E protein would have even greater safety without appreciable loss of immunogenicity.
The capacity of WNV strains to cause neuroinvasive disease is most likely based on the interplay of several viral—as well as potential host—determinants. ETH76a shares mutations in the NS2B, NS3, and NS4A genes with other Old World WNV strains (9, 19) (Table 1), including strain KEN98, a glycosylated lineage 1 strain that is highly neuroinvasive in mice (5) but has been reported to have significantly attenuated virulence in American crows compared to NY99 (7). We are continuing to investigate the possible contribution of nonstructural protein mutations to the virulence of WNV.
Acknowledgments
This study was supported by funding from the Centers for Disease Control and Prevention (U50/CCU620538 and U50/CCU620539). D.W.C.B. is supported by the James W. McLaughlin Fellowship fund. B.S.S. is supported by CDC fellowship T01/CCT622892, Training Program in Vector-Borne Infectious Diseases.
REFERENCES
- 1.Beasley, D. W. C., L. Li, M. T. Suderman, and A. D. T. Barrett. 2001. West Nile virus strains differ in mouse neurovirulence and binding to mouse or human brain membrane receptor preparations. Ann. N. Y. Acad. Sci. 951:332-335. [DOI] [PubMed] [Google Scholar]
- 2.Beasley, D. W. C., L. Li, M. T. Suderman, and A. D. T. Barrett. 2002. Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology 296:17-23. [DOI] [PubMed] [Google Scholar]
- 3.Beasley, D. W. C., C. T. Davis, H. Guzman, D. L. Vanlandingham, A. P. A. Travassos da Rosa, R. E. Parsons, S. Higgs, R. B. Tesh, and A. D. T. Barrett. 2003. Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology 309:190-195. [DOI] [PubMed] [Google Scholar]
- 4.Beasley, D. W. C., C. T. Davis, J. Estrada-Franco, R. Navarro-Lopez, A. Campomanes-Cortes, R. B. Tesh, S. C. Weaver, and A. D. T. Barrett. 2004. Genome sequence and attenuating mutation in West Nile virus isolate from Mexico. Emerg. Infect. Dis. 10:2221-2224. [DOI] [PMC free article] [PubMed]
- 5.Beasley, D. W. C., C. T. Davis, M. Whiteman, B. Granwehr, R. M. Kinney, and A. D. T. Barrett. 2004. Molecular determinants of virulence of West Nile virus in North America. In C. H. Calisher and D. E. Griffin (ed.), Emergence and control of zoonotic viral encephalitides. Arch. Virol. Suppl. 18:35-41. [DOI] [PubMed] [Google Scholar]
- 5a.Berthet, F. X., H. G. Zeller, M. T. Drouet, J. Rauzier, J. P. Digoutte, and V. Deubel. 1997. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 78:2293-2297. [DOI] [PubMed] [Google Scholar]
- 6.Blitvich, B. J., I. Fernandez-Salas, J. F. Contreras-Cordero, N. L. Marlenee, J. I. Gonzalez-Rojas, N. Komar, D. J. Gubler, C. H. Calisher, and B. J. Beaty. 2003. Serologic evidence of West Nile virus infection in horses, Coahuila State, Mexico. Emerg. Infect. Dis. 9:853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brault, A. C., S. A. Langevin, R. A. Bowen, N. A. Panella, B. J. Biggerstaff, B. R. Miller, and N. Komar. 2004. Differential virulence of West Nile strains for American crows. Emerg. Infect. Dis. 10:2161-2164. [DOI] [PMC free article] [PubMed]
- 8.Chambers, T. J., M. Halevy, A. Nestorowicz, C. M. Rice, and S. Lustig. 1998. West Nile virus envelope proteins: nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J. Gen. Virol. 79:2375-2380. [DOI] [PubMed] [Google Scholar]
- 9.Charrel, R. N., A. C. Brault, P. Gallian, J. J. Lemasson, B. Murgue, S. Murri, B. Pastorino, H. Zeller, R. de Chesse, P. de Micco, and X. de Lamballerie. 2003. Evolutionary relationship between Old World West Nile virus strains. Evidence for viral gene flow between Africa, the Middle East, and Europe. Virology 315:381-388. [DOI] [PubMed] [Google Scholar]
- 10.Davis, C. T., D. W. C. Beasley, H. Guzman, M. Siirin, R. E. Parsons, R. B. Tesh, and A. D. T. Barrett. 2004. Emergence of attenuated West Nile virus variants in Texas, 2003. Virology 330:342-350. [DOI] [PubMed] [Google Scholar]
- 11.Dupuis, A. P., II, P. P. Marra, and L. D. Kramer. 2003. Serologic evidence of West Nile virus transmission, Jamaica, West Indies. Emerg. Infect. Dis. 9:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrada-Franco, J. G., R. Navarro-Lopez, D. W. C. Beasley, L. Coffey, A. S. Carrara, A. Travassos da Rosa, T. Clements, E. Wang, G. V. Ludwig, A. C. Cortes, P. P. Ramirez, R. B. Tesh, A. D. T. Barrett, and S. C. Weaver. 2003. West Nile virus in Mexico: evidence of widespread circulation since July 2002. Emerg. Infect. Dis. 9:1604-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guirakhoo, F., A. R. Hunt, J. G. Lewis, and J. T. Roehrig. 1993. Selection and partial characterization of dengue 2 virus mutants that induce fusion at elevated pH. Virology 194:219-223. [DOI] [PubMed] [Google Scholar]
- 14.Halevy, M., Y. Akov, D. Ben-Nathan, D. Kobiler, B. Lachmi, and S. Lustig. 1994. Loss of active neuroinvasiveness in attenuated strains of West Nile virus: pathogenicity in immunocompetent and SCID mice. Arch. Virol. 137:355-370. [DOI] [PubMed] [Google Scholar]
- 15.Kinney, R. M., S. Butrapet, G. J. Chang, K. R. Tsuchiya, J. T. Roehrig, N. Bhamarapravati, and D. J. Gubler. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230:300-308. [DOI] [PubMed] [Google Scholar]
- 16.Komar, O., M. B. Robbins, K. Klenk, B. J. Blitvich, N. L. Marlenee, K. L. Burkhalter, D. J. Gubler, G. Gonzalvez, C. J. Pena, A. T. Peterson, and N. Komar. West Nile virus transmission in resident birds, Dominican Republic. Emerg. Infect. Dis. 9:1299-1302. [DOI] [PMC free article] [PubMed]
- 17.Kramer, L. D., and K. A. Bernard. 2001. West Nile virus infection in birds and mammals. Ann. N. Y. Acad. Sci. 951:84-93. [DOI] [PubMed] [Google Scholar]
- 18.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 19.Lanciotti, R. S., G. D. Ebel, V. Deubel, A. J. Kerst, S. Murri, R. Meyer, M. Bowen, N. McKinney, W. E. Morrill, M. B. Crabtree, L. D. Kramer, and J. T. Roehrig. 2002. Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298:96-105. [DOI] [PubMed] [Google Scholar]
- 20.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 21.Lorono-Pino, M. A., B. J. Blitvich, J. A. Farfan-Ale, F. I. Puerto, J. M. Blanco, N. L. Marlenee, E. P. Rosado-Paredes, J. E. Garcia-Rejon, D. J. Gubler, C. H. Calisher, and B. J. Beaty. 2003. Serologic evidence of West Nile virus infection in horses, Yucatan State, Mexico. Emerg. Infect. Dis. 9:857-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie, J. S., A. D. T. Barrett, and V. Deubel. 2002. The Japanese encephalitis serological group of flaviviruses: a brief introduction to the group. Curr. Top. Microbiol. Immunol. 267:1-10. [DOI] [PubMed] [Google Scholar]
- 23.McLean, R. G., S. R. Ubico, D. E. Docherty, W. R. Hansen, L. Sileo, and T. S. McNamara. 2001. West Nile virus transmission and ecology in birds. Ann. N. Y. Acad. Sci. 951:54-57. [DOI] [PubMed] [Google Scholar]
- 24.McMinn, P. C., E. Lee, S. Hartley, J. T. Roehrig, L. Dalgarno, and R. C. Weir. 1995. Murray Valley encephalitis virus envelope protein antigenic variants with altered hemagglutination properties and reduced neuroinvasiveness in mice. Virology 211:10-20. [DOI] [PubMed] [Google Scholar]
- 25.McMinn, P. C., R. C. Weir, and L. Dalgarno. 1996. A mouse-attenuated envelope protein variant of Murray Valley encephalitis virus with altered fusion activity. J. Gen. Virol. 77:2085-2088. [DOI] [PubMed] [Google Scholar]
- 26.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monath, T. P., C. B. Cropp, G. S. Bowen, G. E. Kemp, C. J. Mitchell, and J. J. Gardner. 1980. Variation in virulence for mice and rhesus monkeys among St. Louis encephalitis virus strains of different origin. Am. J. Trop. Med. Hyg. 29:948-962. [DOI] [PubMed] [Google Scholar]
- 28.Monath, T. P., J. Arroyo, C. Miller, and F. Guirakhoo. 2001. West Nile virus vaccine. Curr. Drug Targets Infect. Disord. 1:37-50. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 30.Murgue, B., H. Zeller, and V. Deubel. 2002. The ecology and epidemiology of West Nile virus in Africa, Europe and Asia. Curr. Top. Microbiol. Immunol. 267:195-221. [DOI] [PubMed] [Google Scholar]
- 31.Nash, D., F. Mostashari, A. Fine, J. Miller, D. O'Leary, K. Murray, A. Huang, A. Rosenberg, A. Greenberg, M. Sherman, S. Wong, M. Layton, and the 1999 West Nile Outbreak Response Working Group. 2001. The outbreak of West Nile virus infection in the New York City area in 1999. N. Engl. J. Med. 344:1807-1814. [DOI] [PubMed] [Google Scholar]
- 32.Pletnev, A. G., R. Putnak, J. Speicher, E. J. Wagar, and D. W. Vaughn. 2002. West Nile virus/dengue type 4 virus chimeras that are reduced in neurovirulence and peripheral virulence without loss of immunogenicity or protective efficacy. Proc. Natl. Acad. Sci. USA 99:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quirin, R., M. Salas, S. Zientara, H. Zeller, J. Labie, S. Murri, T. Lefrançois, M. Petitclerc, and D. Martinez. 2004. West Nile Virus, Guadeloupe. Emerg. Infect. Dis. 10:706-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 36.Scherret, J. H., J. S. Mackenzie, A. A. Khromykh, and R. A. Hall. 2001. Biological significance of glycosylation of the envelope protein of Kunjin virus. Ann. N. Y. Acad. Sci. 951:361-363. [DOI] [PubMed] [Google Scholar]
- 37.Shirato, K., H. Miyoshi, A. Goto, Y. Ako, T. Ueki, H. Kariwa, and I. Takashima. 2004. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J. Gen. Virol. 85:3637-3645. [DOI] [PubMed] [Google Scholar]
- 38.Smit, J. S. 2002. Mutational analysis of receptor interaction and membrane fusion activity of Sindbis virus. Ph.D. thesis. University of Groningen, Groningen, The Netherlands. [Online.] http://www.ub.rug.nl/eldoc/dis/medicine/j.m.smit/. (Accessed 28 April 2004.)
- 39.Trock, S. C., B. J. Meade, A. L. Glaser, E. N. Ostlund, R. S. Lanciotti, B. C. Cropp, V. Kulasekera, L. D. Kramer, and N. Komar. 2001. West Nile virus outbreak among horses in New York State, 1999 and 2000. Emerg. Infect. Dis. 7:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanlandingham, D. L., B. S. Schneider, K. Klingler, J. Fair, D. W. C. Beasley, J. Huang, P. Hamilton, and S. Higgs. 2004. Real-time reverse transcriptase-polymerase chain reaction quantification of West Nile virus transmitted by Culex pipiens quinquefasciatus. Am. J. Trop. Med. Hyg. 71:120-123. [PubMed] [Google Scholar]
- 41.Vorndam, V., J. H. Mathews, A. D. T. Barrett, J. T. Roehrig, and D. W. Trent. 1993. Molecular and biological characterization of a non-glycosylated isolate of St Louis encephalitis virus. J. Gen. Virol. 74:2653-2660. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, S. Y., H. Guzman, H. Zhang, A. P. A. Travassos da Rosa, and R. B. Tesh. 2001. West Nile virus infection in the golden hamster (Mesocricetus auratus): a model for West Nile encephalitis. Emerg. Infect. Dis. 7:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]