Abstract
Simple retroviruses present a unique opportunity for examining the host-virus relationship. Following exogenous infection and integration into the germ line, copies of these viruses can become fixed within the genome. The resulting endogenous proviral “fossils” represent a record of past retroviral infections and forms. Previous work in our laboratory has been directed at dissecting the extensive nonecotropic murine leukemia virus content of the mouse genome. One such provirus, hortulanus endogenous murine leukemia virus (HEMV), found in a single copy in the genome of Mus spicilegus, was remarkable for characteristics that suggested that it was ancient and related to the hypothetical common ancestor of murine leukemia viruses (MLVs) and other gammaretroviral species. In the present study, we have analyzed its functional properties. Transfection of a molecular clone of the HEMV provirus into mouse-derived cell lines revealed that it is replication competent. Furthermore, host range and interference studies revealed a strictly ecotropic host range and the use of a receptor distinct from those used by other classical MLVs. The identity of nucleotide sequence of the long terminal repeats (LTRs) further suggested that HEMV is a relatively recent insertion into the M. spicilegus genome at the distal end of chromosome 7. Although unique to M. spicilegus, its presence in a homozygous state in three individuals obtained from different regions implies that it has been present long enough to become fixed in this species. Exhaustive phylogenetic analysis of all regions of the HEMV genome supported the previously assigned ancestral position of HEMV relative to other MLV-related viruses. Thus, HEMV is a relatively recent introduction into the Mus germ line but is representative of a relatively ancestral MLV group.
Simple retroviruses present a unique opportunity to study the evolution of a parasitic species within the context of its host. These viruses are characterized by a small RNA genome that becomes covalently linked, in the form of proviral DNA, with the genome of the host during an obligatory integration step in their life cycle. Rarely, proviruses can become fixed within the genome of the host population, with each endogenous proviral “fossil” representing a snapshot of a past retroviral infection. The information present in endogenous proviruses not only can provide evidence for their age and lineage but can also further our understanding of the host-virus relationship.
Gammaretroviruses are simple retroviruses that are widespread, both as exogenous infectious agents and as endogenous proviruses in mice, cats, baboons, and other mammalian, avian, and reptilian species (30). A major group of these viruses, the murine leukemia viruses (MLVs) of mice, makes an ideal subject for study of the recent evolution of the host-virus relationship. There are currently five recognized major subgroups of MLV distributed into two broader classes based on their host ranges and sequence relationships: ecotropic and nonecotropic. The nonecotropic viruses include the endogenous polytropic, modified polytropic, and xenotropic viruses as well as exogenous amphotropic and 10A1 viruses. Despite their distinctive receptor usage (43), these viruses all have highly related env genes, with regions critical for receptor interaction, known as variable region A (VRA) and variable region B (VRB), exhibiting receptor-specific variation within an otherwise largely conserved framework. The overall diversity of sequence in VRA and VRB among the subgroups indicates that MLV has had a very active evolutionary history and continues to evolve to adapt to use different receptors at a rapid rate. This evolution is most likely driven by the appearance of host animals that have become resistant due to polymorphisms in receptor genes or to the presence of endogenous proviruses whose env genes can block receptor access. Analysis of the endogenous proviruses in different species of mice, particularly their receptor utilization, should reveal important aspects of the coevolution of virus and host. Fortunately, evolution of the host, mice of the genus Mus, has similarly been the focus of extensive research for many years (2, 5, 6, 35). Until recently, however, attempts to resolve the interplay of virus and host have been hampered by the number and complexity of proviruses present in the genomes of wild mice.
In prior work, we developed the use of specific oligonucleotide probes and PCR primers aimed at polymorphisms within the env gene and U3 region to comprehensively identify endogenous MLVs and isolate individual proviruses themselves (14, 40, 45, 46). A catalogue of the nonecotropic proviral MLV content of wild mice based upon U3 region polymorphisms led to the description of nine sub-subgroups: four xenotropic related and five polytropic related. These data further allowed the parsimonious construction of MLV phylogenies for both env and U3 sequences. A novel endogenous provirus found in the distantly related Mus spicilegus (formerly M. hortulanus) appeared to occupy a relatively central position in both the U3 and env trees, near the inferred common ancestor of MLV-related viruses, including those in nonmouse species like cats and baboons (46).
The sequence of a partial clone of the hortulanus endogenous murine leukemia virus (HEMV) provirus, including the env gene and the complete U3 region, revealed some unusual features. Based on hybridization with specific oligonucleotide probes, HEMV was originally identified as a member of the X-IV class of endogenous MLVs, the most extensively represented group in the many mouse species tested. This relationship suggested that the X-IV viruses, like HEMV, were active within a deeply ancestral mouse population. The phylogenetic analysis also placed HEMV on a branch separate from the X-IV viruses and other MLVs, although this split occurred very early in the MLV history. Confirming HEMV's distinctiveness, an oligonucleotide probe designed to identify only HEMV-like U3 regions failed to hybridize to other X-IV loci. HEMV is specifically resident in the genome of M. spicilegus only, possibly indicating that it independently evolved into its current form within this Mus branch.
Despite its central phylogenetic position among MLV-like proviruses, the HEMV env gene differs in significant ways from that of related retroviruses. Although it lacks nonsense or frameshift mutations in its open reading frame (ORF), HEMV has a significantly truncated VRA region and a very short VRB region. For this reason, we speculated that the loss of information in these regions might render its Env protein nonfunctional. However, during the course of this study, the sequence and partial tropism data of the virus M813 (an exogenous virus of M. cervicolor originally identified by Benveniste et al. [4]) were published (36). While not identical, M813 is similar to HEMV in VRA and VRB, raising the possibility that the HEMV env gene might encode a functional product.
To test the functional consequences of the unique aspects of the HEMV provirus, we have cloned the proviral DNA and found that, contrary to our speculation, it is intact, is fully capable of replication as a virus, and, despite its apparent age, is a relatively recent insertion into the germ line of M. spicilegus. Consistent with its replication competence, the HEMV Env protein is fully functional and furthermore represents a novel subgroup of MLV, with functional receptors found only on species of Mus, including M. spicilegus. The implications of this more detailed characterization of this otherwise ancestral-appearing MLV on our understanding of the mechanism of retroviral evolution are discussed.
MATERIALS AND METHODS
Cell lines.
NIH 3T3 cells (including the packaging line MPAC and cells chronically infected with MLV) were grown in Dulbecco's modified Eagle's medium (Gibco) with 10% calf serum. 293T, 293 mCAT1, SC1, Mus dunni TF, E36, FEF, D17, Cos1, MPAC (24), and M. spicilegus TF cells were maintained in the same medium except with 10% fetal bovine serum (FBS). REF, Rat1, QT6, and C300 cells were grown in modified Richter's modified Eagle's medium, Tufts formulation (Irvine Scientific), with 10% FBS. CHO-K1 cells were cultured in RPMI 1640 medium with l-glutamine (catalog no. 11875-093; Gibco) plus 10%FBS.
Mouse DNA.
Mus cervicolor popaeus (J53), Mus caroli (J136), Mus cookii (J135), and Mus spicilegus (Halbturn) (J131) genomic DNA preparations were donated by Christine Kozak. Mus famulus (FAM), Mus platythrix (PTX), Mus spicilegus (ZRU), Mus macedonicus (XBS), Mus cervicolor cervicolor (CRV), and Mus musculus bactrianus (BIR) genomic DNA preparations were provided by Francois Bonhomme. Mus dunni DNA was prepared from M. dunni tail fibroblasts using the DNeasy tissue kit (QIAGEN). All other mouse DNAs were purchased from the Mouse DNA Resource at Jackson Laboratories, Bar Harbor, ME.
Plasmids and constructs.
The vectors used in this study include pHEMV18 (HEMV env gene) (46); pSV-Ψ minus-E-MLV (26); pMLVgagpol (structural and enzymatic protein expression), pMCF 247 5′ (polytropic env gene), and pSVA-MLV (amphotropic env gene) (28, 29); pFBXsalf (xenotropic env gene) (44); pMOV-GaLV SEATO (gibbon ape leukemia virus [GALV] env gene) (47); pLacPuro (34); pB6 (replication-competent 10A1 provirus) (33); and pNCS (replication-competent Moloney MLV [MoMLV]) (10). Digestion of pHEMV18 with BsaAI and NheI released the HEMV env gene coding sequence. Digestion of pB6 completely with NheI and partially with BsaAI released the 10A1 env gene. These env genes were cloned into both pSV-Ψ minus-E-MLV (to create pSV-Ψ minus-HEMV-MLV and pSV-Ψ minus-10A1-MLV) and pNCS (to create replication-competent pseudotype HEMV-MoMLV). pSV-Ψ minus-E-MLV and pNCS had been completely digested with PmlI and partially digested with NheI prior to the introduction of the env genes. The entire HEMV provirus was cloned into the EcoRI sites of pCR2.1-TOPO to make pHEMV-TOPO.
PCR and primers.
PCRs were performed with either Taq (Sigma) (original HEMV cloning) or Taq Platinum High Fidelity (Invitrogen) in 50-μl volumes. In general, reactions were set up with the following mixture: 5 pmol each primer, 1 unit polymerase, 2 mM deoxynucleoside triphosphates, and distilled water to a final volume of 50 μl plus buffers (regular Taq polymerase with 5 μl of 10× “genome amp” reaction buffer [100 mM Tris-HCl, pH 8.3, 20 mM Tris-HCl, pH 8.0, 250 mM KCl, 35 mM MgCl2, and 250 nM EDTA] and Taq Platinum with 5 μl of 10× “HiFi buffer” and 2 μl 50 mM MgSO4). Cycling parameters generally followed this protocol: 1 min at 95°, to dissociate the enzyme from its inhibitory antibody, followed by 30 cycles of a 30-second 95° melting step, 30-second (primer Tm, −5°) annealing step, and an extension step (1 min/kb) at 68°. An exception to this cycling protocol was the 47° annealing temperature found to be optimal for the inosine-containing primer Tris2F.
Many primers were chosen using the Primer3 program available at the Whitehead Institute website (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). For primers not chosen due to optimal Tm or other considerations, sequences were screened using the “oligonucleotide properties calculator” at the Northwestern University website (http://www.basic.nwu.edu/biotools/oligocalc.html). Primers used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Length (bp)a | Description | Sequence |
|---|---|---|---|
| KT59 | 26F | X-IV-specific U3 | GGA′AGTTCA′GTT′AGA′GAT′CAA′GGC′TG |
| KT61 | 26F | HEMV-specific U3 | GCC′ATA′AGC′AAG′CTA′GCA′ATA′GTA′AC |
| KT61R | 26R | R&C of KT61 | GTT′ACT′ATT′GCT′AGC′TTG′CTT′ATG′GC |
| UniEnv3 | 20F | Universal nonecotropic env | GGA′TAC′ACG′CCG′CTC′ACG′TA |
| HE5 | 20R | HEMV-specific env | GTT′TAG′CGA′GCA′GGC′AGT′TC |
| Tris2F | 20R | Universal MLV RT | AGI′AGG′TCR′TCI′ACR′TAS′TG |
| HRTIF | 20F | HEMV-specific RT | TTT′GAT′CCT′GCT′GCA′CTA′CG |
| 3LTRR1 | 33R | HEMV-specific U5 EcoRI | ACC′ACC′GAT′CCT′GGG′GGT′CTT′TCA′TTG′AAT′CCC |
| 5LTRR1 | 33F | HEMV-specific U3 EcoRI | GGA′ATT′CAA′TGA′AAG′ACC′CCG′CCA′TAA′GCA′AGC |
| 3LTRF | 24F | HEMV-specific env | CAT′CCG′GCC′TCT′AGA′AGA′ATC′AGT |
| HGAG5R | 20R | HEMV-specific gag | AAG′ATC′GGA′GGT′CGT′AGT′CG |
| HGAG7R | 20R | HEMV-specific gag | TGC′GTT′GAA′CAT′CTC′TCC′AG |
| HE9 | 27F | HEMV-specific env | GTA′GCA′GCA′GAA′GTT′CCC′CAG′GCT′AGG |
| INT5F | 20F | Mouse chromosome 7 specific | GGG′CCA′GAA′AAC′ATC′TTC′AA |
| INT6R | 20R | Mouse chromosome 7 specific | AAC′TCC′TCC′TGG′CCT′TGT′TT |
| LTR5R | 24R | HEMV-specific gag | CAC′AGA′ACG′ATC′GCT′GGA′CAG′CTT |
F, forward; R, reverse sense relative to the RNA genome.
Virus construction.
Pseudotyped single-round infection viruses were obtained from doubly or triply transfected 293T cells. Double transfections combined pSV-Ψ minus MoMLV, HEMV, or 10A1 plasmids with pLacPuro. Triple transfections were done with mixtures of pMLVgagpol and pLacPuro with the env gene constructs pMCF 247 5′, pSVA-MLV, pMOV-GaLV SEATO, and pFBXsalf. Replication-competent 10A1, MoMLV, and HEMV were grown in NIH 3T3 cells. Virus-containing medium was filtered through a 0.45-μm membrane and then either used directly or stored at −70°.
Transfection of 293T cells was modified from a method described previously (32). 293T cells were grown to confluence in 100-mm plates and then split 1:4 the day before to yield 90% confluent cultures. One hour before transfection, the medium was replaced with 9 ml of fresh medium. For each 100-mm plate, 30 μg of total DNA was mixed with sterile distilled water to a final volume of 450 μl. To this mixture, 60 μl of 2 M CaCl2 was added dropwise. This solution was immediately added dropwise to 500 μl 2× HBS (42 mM HEPES, 274 mM NaCl, 10 mM KCl, and 1.8 mM Na2HPO4, pH 7.10) and was allowed to sit for 1 min. This final mixture was added dropwise to the 9 ml of medium on the cells. One hundred microliters of 10 mM chloroquine was then added to the plate and swirled to mix. After 3 to 5 h of incubation at 37° and 5% CO2, this medium was replaced with fresh medium. Lipofectamine Plus transfection was carried out according to the manufacturer's protocol. Assay or virus collection was performed 36 to 48 h later.
Titering of single-round infections/determination of infectious units.
Target cells were plated onto 6-well plates the day before to give 85% to 90% confluency on the day of the infection. Cells were infected with 1 ml of three 10-fold dilutions (100, 10−1, and 10−2) of LacZ+ PuroR replication-defective virus plus 8 μg/ml polybrene. Three to 5 h after the addition of virus, 3 ml of fresh medium was added to each well. After incubation for 36 to 48 h at 37°C and 5% CO2, cells were washed once with phosphate-buffered saline (PBS) (Ca2+ and Mg2+ free) and then fixed with 0.05% glutaraldehyde in PBS for 15 to 20 min. Cells were then washed once with PBS and then stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution [0.1 M NaPO4, pH 7.3, 1.3 mM MgCl2, 3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6, 1 mg/ml X-Gal]. Blue cells were counted by eye using a light microscope.
Interference assay.
NIH 3T3 cells chronically infected with ecotropic (MoMLV), amphotropic, polytropic, 10A1, HEMV, and M813 viruses were plated in 6-well plates at 1 × 105 cells/well the day before the assay. LacZ+ replication-defective viruses were then added, and infectious units were assayed as described above.
Virus growth and RT assay.
NIH 3T3 cells in two wells each (of a 24-well plate) were transfected with 0.3 μg of retroviral construct mixed with 0.1 μg pLacPuro using Lipofectamine Plus. Forty-eight hours after transfection, one well was stained with X-Gal to determine transfection efficiency. Cells were then split 1:5 every 3 days (to achieve 90 to 95% confluence) for the duration of the experiment. Medium was collected at each passage and immediately frozen at −70°C to assay for reverse transcriptase (RT) activity. Other cell lines were treated similarly. At passage 4 for HEMV and passage 5 for 10A1 and MoMLV, 100 μl of 0.45-μm-filtered medium was added to a subconfluent well of a 24-well plate of SC-1 and MMK cells. These cells were split 3 days later, and media representing passage 1 were collected.
Medium collected from virus-producing cells was loaded in triplicate into 96-well round-bottomed plates at 10 μl/well. To each well, 50 μl of assay buffer (50 mM Tris-HCl, pH 8.3, 10 mM β-mercaptoethanol, 10 mM MgCl2, 5.15 mM NaCl, 1 μM ATP, 1 U poly(rA)/oligo(dT), 5 μCi [α-32P]TTP) was added, and then the plates were incubated at 37°C for 1 h. The reaction mixture was filtered through 96-well DEAE filters (Millipore). The filters were washed seven times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and dried, and radioactivity was determined as counts per minute (cpm) in a scintillation counter.
Ligation-mediated PCR, flanking sequence cloning, and integration site occupation.
Ligation-mediated PCR was performed using the “Universal GenomeWalker” kit (BD Biosciences). Internal nested HEMV primers were coupled with the provided adaptor primers. Primers were as follows: 5′ junction HGAG5R and HGAG7R and 3′ KT61 and KT59. The amplification reaction mixture used 2.5 μg of PANCEVO/Ei DNA digested with the blunt cutters DraI, EcoRV, PvuII, and StuI before ligation of the adaptor termini as the template. The amplicons were sequenced starting from the M13 forward and M13 reverse sites flanking the inserts. The location of the HEMV integration was initially discovered by BLASTn (http://www.ncbi.nlm.nih.gov:80/BLAST/) and then confirmed using the Ensemble mouse genome viewer (http://www.ensembl.org/Mus_musculus/).
Based upon the flanking sequence data, a set of primers, Int5F and Int6R, were designed. These primers were then used to investigate site occupation in other species, both across the integration site and with an HEMV env gene-specific primer, 3LTRF, to amplify a possible HEMV 3′ junction.
Phylogenetic analysis.
Sequences used to construct the phylogenies were either determined from samples sent to the Tufts Core Facility or acquired from Entrez. Alignments were initially performed by the ClustalW (version 1.4) algorithm included with the MacVector (version 7.0) program (Oxford Molecular Group) and were then corrected by hand. Trees were constructed by exhaustive parsimony analysis using PAUP (version 4.0b10) (42). Branches were swapped using the TBR algorithm using the steepest descent. Gaps were treated as “missing data.”
Nucleotide sequence accession number.
The nucleotide sequence of the HEMV provirus has been submitted to GenBank under accession number AY818896.
RESULTS
An HEMV env pseudotype can mediate a single round of infection.
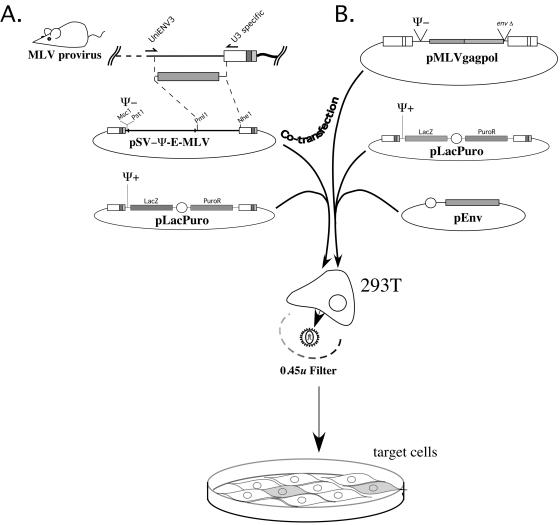
The HEMV env gene differs from that of other MLVs by the presence of a significantly shorter VRA, a region known to be involved in receptor binding. Its VRB is also very short, although its length matches that of the ecotropic MLVs. To determine whether the HEMV-encoded Env protein was functional, we made an HEMV-MLV pseudotype by swapping its env gene into the replication-defective Moloney MLV vector SV-Ψ minus-E-MLV(Fig. 1A). 293T cells were then cotransfected with this construct and the Ψ-plus pLacPuro marker vector. Two days posttransfection, virus was harvested, filtered, and assayed for Lac expression following infection of NIH 3T3 cells. In this initial experiment, we found that the HEMV env gene could lead to production of infectious virus capable of inducing LacZ expression in NIH 3T3 cells at titers comparable to those achieved with the positive-control MoMLV (up to about 107 infectious units/ml).
FIG. 1.
Pseudotype construction and LacZ+ PuroR virus production. (A) The HEMV pseudotype was constructed by PCR amplification and cloning of a region including the env gene from M. spicilegus DNA. The amplified fragment was swapped into the ecotropic packaging-deficient vector pSV-Ψ minus-E-MLV. This construct, pSV-Ψ minus-HEMV-MLV, and the packagable marker vector pLacPuro were transfected into 293T cells. Viral particles produced by these cells were then used to infect tissue culture cells. Infection efficiency was assessed by staining for β-galactosidase. (B) A similar method was used to make other LacZ+ PuroR pseudotypes. A triple transfection method using pMLVgagpol, pLacPuro, and various env gene constructs produced these viruses.
HEMV has a unique ecotropic species tropism.
The results of the initial experiment implied that the HEMV env gene encoded a functional protein capable of recognizing a receptor found on mouse cells. To determine the relationship between this receptor and those of other MLV subgroups and related gammaretroviruses, LacZ+ PuroR viruses pseudotyped with Env proteins of other MLV subgroups were constructed either by swapping their envelopes into pSV-Ψ minus-E-MLV as had been done for HEMV or by triple transfection of 293T cells with an envelope expression construct, pLacPuro, and the viral structural protein encoding plasmid pMLVgagpol (Fig. 1B).
Cell lines used to test for tropism were selected mostly for their species representation but in the case of 293mCAT1, E36, and Mus molossinus kidney (MMK) cells for other properties. 293mCAT1 cells stably express the ecotropic MLV receptor mCAT1 and are therefore infectible by this subgroup of MLV (1). E36 cells are Chinese hamster lung cells that express a variant of the Pit2 protein that allows them to be infected by both amphotropic MLV and GALV as well as the other nonecotropic MLVs (8, 11). MMK cells are also reported to be susceptible to GALV infection (48).
HEMV exhibited an ecotropic MLV-like species tropism, meaning that it could infect only cells derived from the Mus subgenera including M. spicilegus itself (Table 2). However, these results further indicated a tropism distinct from the established ecotropic viruses, which can also infect rat cells. Furthermore, HEMV could not infect the human 293mCAT1 cells that are susceptible to ecotropic subgroup infection. We conclude that although it has an ecotropic host range, HEMV must use a receptor distinct from that used by classical ecotropic MLV.
TABLE 2.
Species tropism of HEMV
| Cell line | Titer (U/ml)a
|
||||||
|---|---|---|---|---|---|---|---|
| HEMV | Ecotropic (MoMLV) | Polytropic (MCF247) | Xenotropic MLV | Amphotropic (4070A MLV) | 10A1 MLV | GALV | |
| Mouse | |||||||
| NIH 3T3 | 5.9 × 105 | 1.8 × 105 | 2.4 × 103 | 8.9 × 102 | 8.2 × 104 | 1.4 × 105 | <1 |
| SC-1 | 3.5 × 105 | 2.0 × 105 | 3.6 × 103 | 1.3 × 103 | 1.3 × 105 | 1.3 × 104 | <1 |
| M. dunni | 3.1 × 106 | 6.2 × 102 | 1.1 × 106 | 7.6 × 105 | 6.5 × 105 | 7.7 × 105 | <1 |
| MMK | 1.7 × 106 | 7.5 × 105 | 1 | 2 | <1 | 2.8 × 104 | <1 |
| M. spicilegus TF | 7.3 × 102 | 1.0 × 103 | 2.1 × 102 | 3.5 × 102 | 1.1 × 103 | ND | 1.2 × 102 |
| Rat | |||||||
| Rat1 | <1 | 2.4 × 105 | 3.0 × 106 | 3.2 × 106 | 1.3 × 105 | 2.8 × 104 | 2.3 × 105 |
| REF | <1 | 1.9 × 102 | 1.6 × 103 | 1.5 × 103 | 3.1 × 103 | 3.9 × 102 | 1.5 × 103 |
| Hamster | |||||||
| E36 | <1 | <1 | 1.6 × 102 | 8 | 1.4 × 103 | 4.4 × 104 | 1.7 × 105 |
| CHO-K1 | <1 | <1 | 3.3 × 102 | <1 | <1 | <1 | <1 |
| Primate | |||||||
| 293T | <1 | <1 | 4.2 × 106 | 3.3 × 106 | 1.6 × 105 | 2.9 × 104 | 2.3 × 105 |
| 293mCAT1 | <1 | 3.5 × 105 | 1.1 × 106 | 1.5 × 106 | 7.3 × 104 | 2.1 × 104 | 1.4 × 105 |
| COS1 | <1 | <1 | 8.3 × 104 | 3.1 × 104 | 1.7 × 104 | 9.0 × 103 | 2.6 × 104 |
| Avian | |||||||
| C300 | <1 | <1 | 10 | 13 | 1.2 × 103 | 2.7 × 102 | 7.7 × 103 |
| QT6 | <1 | <1 | 3.0 × 106 | 1.6 × 106 | <1 | 8.4 × 102 | 6.7 × 104 |
| Cat | |||||||
| FEF AH927 | <1 | <1 | 1.6 × 106 | 3.6 × 106 | 5.2 × 105 | 7.6 × 104 | 1.1 × 106 |
| Dog | |||||||
| D17 | <1 | <1 | 1.1 × 106 | 1.0 × 106 | 6.3 × 104 | 1.2 × 105 | 2.7 × 105 |
Infectious units per ml of MoMLV pseudotypes of MLV Lac plus PuroR with the indicated env genes (Fig. 1). ND, not determined.
Some discrepancies from previous reports occurred in our analysis. First, our GALV pseudotype was unable to infect MMK cells (48). Our xenotropic Env pseudotype was able to weakly infect NIH 3T3 but not CHO-K1 cells, which have been shown to have some susceptibility to the subgroup in the past (27). Conversely, there was weak infection of CHO-K1 cells with MCF 247. The reasons for these minor tropism aberrations were not pursued.
The HEMV provirus appears to be replication competent and of recent origin.
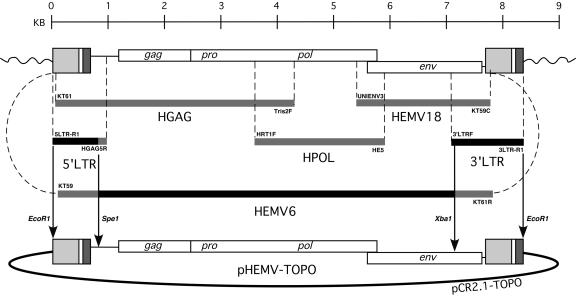
The species tropism results suggested that the HEMV provirus might represent a heretofore-uncharacterized MLV subgroup using a receptor whose susceptible orthologue was limited to species of Mus. To analyze the properties of this unusual provirus further, it was cloned by assembling a collection of partial PCR amplification products (Fig. 2).
FIG. 2.
Construction of the complete HEMV proviral vector pHEMV-TOPO. HEMV was originally cloned and sequenced piecemeal following the successful amplification of the fragments shown from the M. spicilegus genome. The final construct was derived primarily from HEMV6 coupled with the 5′ LTR and 3′ LTR regions. The final construct was ligated into the vector pCR2.1-TOPO. Amplicons acquired and used for initial analysis and sequencing are colored light grey. Amplicons acquired and utilized, or partially utilized, in the final construction of the vector are colored black. The final vector construct was made by ligating together the indicated sequences cut with the indicated restriction endonucleases. Primers used to acquire each amplicon are shown associated with each sequence.
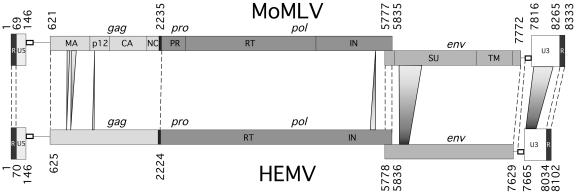
The sequence of the cloned HEMV DNA was consistent with an intact, potentially replication-competent MLV-like provirus. The HEMV provirus from integration site to integration site is 8,542 bases in length, predicting a packaged genome of 8,102 nucleotides (Fig. 3). It is free of nonsense codons and other obvious mutations that might render it defective. Furthermore, the long terminal repeats (LTRs) are identical, which, when combined with its apparent coding competence, would argue that HEMV is a recent addition to the M. spicilegus genome. The structure of the HEMV provirus is very similar to those of other MLV-related viruses (9). It utilizes tRNAPRO as a primer for reverse transcription. Like other MLVs, the gag and pro-pol genes are in the same open reading frame, separated by an amber codon followed by a credible pseudoknot sequence that could promote readthrough suppression. The gag ORF starts at base 625, with a cryptic CTG site at base 361 for translation of the glycosylated form of Gag, and terminates at base 2224. Following gag, the pro-pol ORF starts at base 2212 and terminates at base 5836. The likely splice donor/acceptor pairs for env are located at bases 207/208 and 5489/5490, respectively. The env gene region starts with the leader peptide ATG at base 5778 and ends at base 7629. A polypurine tract follows soon after. A polyadenylation signal, AATAAA, is readily apparent in R.
FIG. 3.
Comparison of the HEMV and MLV genomes. The HEMV genome is shown aligned with that of MoMLV (39). The two genomes differ by 232 bases, most of which are in two regions: the foreshortened VRA region in SU and the enhancer region of U3 (46). Two other differences in the otherwise-well-aligned genomes are worth noting: the HEMV MA protein is 6 amino acids shorter than Moloney MLV MA, and there are insertions of 4 amino acids in the C terminus of the HEMV IN and 1 amino acid in HEMV p12. The IN insertion and 4 of the 6 residues lacking in MA are, at this time, unique to HEMV.
A full proviral HEMV clone is replication competent.
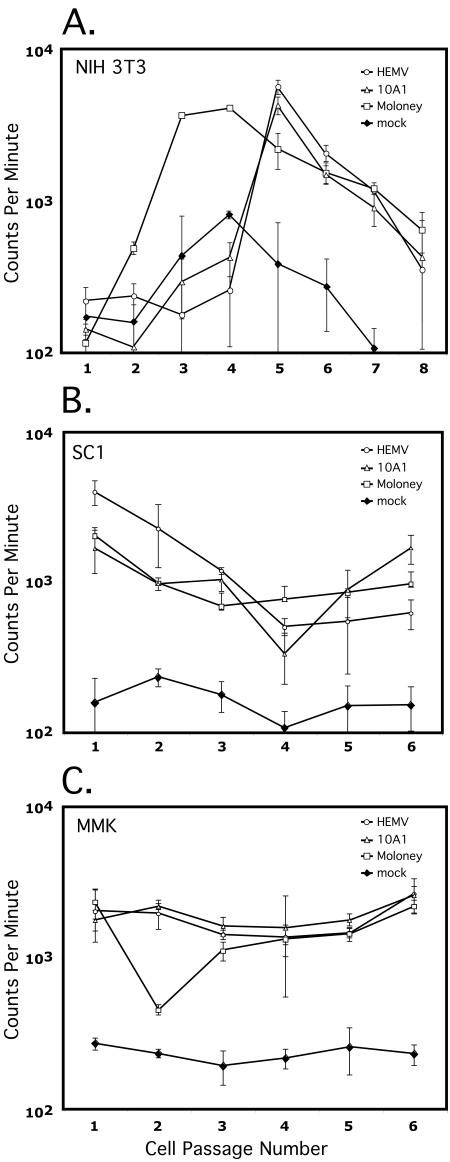
To determine if the cloned HEMV provirus was replication competent, we transfected NIH 3T3 cells with the construct HEMV-TOPO. Spread of the virus through the culture was monitored by assaying for reverse transcriptase activity in the medium at every cell passage and then compared with levels obtained for cells transfected with other replication-competent MLVs. Cells were split 1:5 every 3 days for 4 weeks.
The results showed that HEMV is replication competent, matching the rise to peak RT activity of 10A1 but lagging 1 week behind the MoMLV culture (Fig. 4A). This difference in kinetics may reflect either stochastic differences in the initial transfection or more efficient replication of the laboratory-adapted strain. A notable drop in RT activity towards the end of the experiment perhaps reflected the increased viral burden and was mirrored by a drop in cellular growth rate. Despite the apparent successful spread of HEMV throughout the NIH 3T3 cells in the previous experiment, there remained a remote possibility that the rise in RT activity observed in these HEMV-transfected cells was the result of stable production of defective virus. To determine if exogenous HEMV could initiate and support an active infection, virus collected from the peaks of RT activity (passage 5 for HEMV and 10A1 and passage 4 for MoMLV) was filtered to remove cellular debris and used to initiate infection of the other mouse cells lines, SC-1 and MMK (Fig. 4B and C). HEMV readily and rapidly infected these cells as well, as indicated by the high levels of RT activity.
FIG. 4.
The complete HEMV clone can yield virus infectious for NIH 3T3 cells. (A) NIH 3T3 cells were transfected with complete proviral constructs of HEMV, Moloney MLV, and 10A1. Virus production and spread were followed by reverse transcriptase activity in the culture medium, as determined by the amount of incorporated [32P]TTP into poly(dT) using a poly(A)-oligo(dT) primer-template. Samples were collected every 3 days, prior to passaging the cells, and were counted in triplicate. The samples that contained the most RT activity (passage 4 for Moloney MLV and passage 5 for HEMV and 10A1) were filtered and used to infect the other mouse cell lines, SC1 (B) and MMK (C). Mock transfections were treated in exactly the same manner except that no viral DNA was included in the transfection procedure.
HEMV represents a novel MLV subgroup.
Species tropism alone cannot reveal whether different viruses belong to the same subgroup, i.e., use the same receptor for infection. Retroviruses suffer a restriction to entry of cells previously infected with a virus of the same subgroup. The ability of infection with one virus to interfere with infection by a second virus can therefore be used to determine if two viruses utilize the same receptor for entry. Normally, this resistance is very strong, reducing the apparent infectious units of a viral preparation by several orders of magnitude or more. Any deviation within an order of magnitude from the infectious units observed in a positive control is considered to indicate no interference. To test the hypothesis that HEMV is unique in its receptor usage, we performed an interference analysis. NIH 3T3 cells chronically infected with ecotropic, polytropic, and amphotropic MLVs and HEMV rescued from the molecular clone were challenged with LacZ-containing defective viruses of the same subgroups. The results of this cross-interference analysis showed that HEMV is representative of a novel subgroup of murine leukemia virus. It could infect, with little loss of titer, cells chronically infected with the other subgroups, including 10A1 (not shown), and interfered with itself completely (Table 3). All viruses of other subgroups behaved as expected, reducing the infectivity of intrasubgroup challenge viruses to below the limit of detection.
TABLE 3.
Interference analysis
| Preinfected subgroup | Challenge subgroupa
|
||||
|---|---|---|---|---|---|
| Ecotropic (MoMLV) | Amphotropic (1040A MLV) | Polytropic (MCF247 MLV) | 10A1 MLV | HEMV | |
| None | 1 | 1 | 1 | 1 | 1 |
| HEMV | 0.72 | 0.85 | 0.94 | 0.91 | <2.81 × 10−7 |
| Ecotropic | <1.22 × 10−6 | 0.52 | 0.55 | 2.17 | 0.98 |
| Amphtropic | 1.58 | <1.82 × 10−6 | 0.40 | 0.99 | 1.29 |
| Polytropic | 1.07 | 0.51 | <7.14 × 10−4 | 1.84 | 1.18 |
Numbers reflect the ratio of infectious units of LacZpuro MLV pseudotyped with Env proteins from the indicated viruses on NIH 3T3 cells infected with the indicated viruses to infectious units on uninfected cells.
Although HEMV represents a novel subgroup relative to “classical” MLVs, preliminary cross-interference experiments suggest a relationship in receptor usage to M813 (36). A more complete analysis of this relationship will be published separately (C. H. Tipper and J. M. Coffin, unpublished data).
The HEMV provirus is likely integrated at the distal end of M. spicilegus chromosome 7.
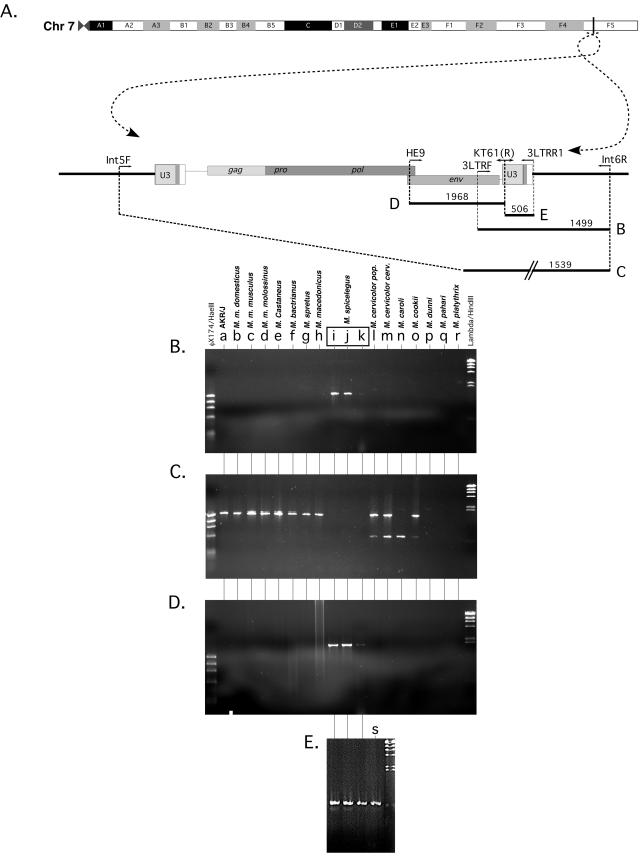
Despite the complete cloning and sequencing of HEMV, we did not know the location of the endogenous copy within the M. spicilegus genome. This information was important pragmatically for verifying the identity of our clone. Furthermore, the location would reveal the 4- to 6-base flanking repeats of the host genome target sequence that are characteristic of an actual integration event, indicating that HEMV arrived via an exogenous infection (or at least by the activity of a retroviral IN). Finally, probes for the chromosomal flanking sequence would allow us to assay for this site's occupation in other species. It was also of interest to determine the proximity of the provirus to any genes or other features of the mouse genome.
The HEMV integration site was cloned using ligation-mediated PCR as described in Materials and Methods (Fig. 5). Comparison of the sequence of the HEMV flanking region with the mouse genome sequence implied that HEMV is integrated in a region corresponding to the distal end of chromosome 7 in M. musculus C57BL/6, specifically at base 125,863,352 (of 137,389,636) according to the Ensemble mouse genome viewer at the Sanger Institute website (http://www.ensembl.org/Mus_musculus/) (Fig. 6A). It is not located in or near any genes, the closest being Ebf3, a transcription factor possibly involved in neuronal differentiation (15), at a distance of about a megabase.
FIG. 5.
Alignment of the HEMV integration junctions with mouse chromosome 7. The HEMV junction fragments were cloned by ligation-mediated PCR and sequenced. The integration site was located in the mouse genome sequence database (http://www.ensembl.org/Mus_musculus/). Data from both the 5′ and 3′ flanking sequences are in agreement that HEMV is integrated into what should be the distal end of M. spicilegus chromosome 7. The genomes of M. spicilegus, M. macedonicus, M cookii, and M castaneus are all slightly polymorphic within 50 bp of the integration site with respect to the reference C57BL/6 sequence. M. spicilegus sequence was confirmed by sequencing at least two independent clones for each junction in both directions. Highlighted are the four host residues that flank the integration site within M. spicilegus aligned with the unoccupied sites within the other genomes as well as the TG and CA dinucleotides that mark the termini of the provirus.
FIG. 6.
Occupation of the HEMV integration site. (A) Schematic of mouse chromosome 7, complete with HEMV provirus integrated at a position corresponding to C57BL/6 residue 125,863,352 (of 137,389,636) and including the locations of primers used in the PCR amplification experiment in panels B, C, D, and E. (B) Amplification with the primer pair 3LTRF and Int6R should detect a 1,499-bp junction fragment only if the HEMV provirus is present at the same site. (C) Amplification with the primer pair Int5F and Int6R was used to detect the 1,539 bp predicted for the unoccupied integration site. The results for M. caroli, M. dunni, M. (coelomus) pahari, and M. (pyromus) platythrix are discussed in the text. The presence of an HEMV-like provirus was detected by PCR amplification with HEMV VRA (D)- and LTR (E)-specific primers only in the three M. spicilegus individuals and the tail fibroblast DNA from M. spicilegus (Pancevo) used in the tropism experiments. φX174/HaeIII markers were run in lane 1, and lambda/HindIII markers were run in lane 20. Lanes: a, AKR/J; b, M. m. domesticus (Zalende); c, M. m. musculus (CzechII), d, M. m. molossinus (MolC); e, M. castaneus (Cast/Ei); f, M. bactrianus (BIR); g, M. spretus (Spret/Ei); h, M. macedonicus (XBS); i, M. spicilegus (Pancevo); j, M. spicilegus (Halbturn); k, M. spicilegus (ZRU); l, M. cervicolor (popaeus); m, M. cervicolor (cervicolor); n, M. caroli; o, M. cookii; p, M. dunni; q, M. (coelomus) pahari; r, M. (pyromus) platythrix; s, M. spicilegus (Pancevo) (tail fibroblast preparation).
The region surrounding the integration site is about 95% identical between M. spicilegus and C57BL/6J (41 individual bases over the 804-bp total aligned sequences, in addition to a 16-bp gap in M. spicilegus 253 bp 3′ of the proviral element). Surprisingly, considering the best alignment with the sequenced C57BL/6 genome, the expected 4-base direct repeat of genomic sequence on either side of the HEMV locus was imperfect, with AGAA on the 5′ side and AAAC on the 3′ side. The corresponding sequence in C57BL/6J is GAAC, and in the more closely related M. macedonicus, it is AAAC. The junctions were independently cloned and sequenced twice with complete concordance of sequence. At this time, we believe that the best explanation for this discordance in flanking repeats is that integration was adjacent to the sequence AAAC and that repair of the 5′ copy was highly error prone (see Discussion).
The HEMV integration site is unoccupied in other Mus species, and an HEMV-like U3 sequence is present only in M. spicilegus.
The determination of the integration site allowed us to test for the presence of HEMV at that genomic location in other species. We did this search by looking for the chromosome/provirus junctions as well as unoccupied integration sites using PCR with primers complementary to flanking as well as proviral sequences. We compared DNA from the laboratory strain AKR/J, from 3 unrelated M. spicilegus individuals, and from 15 other species and subspecies of Mus. As shown schematically in Fig. 6A, PCRs were performed using primers to detect the presence of the provirus at the site identified (Fig. 6B), the unoccupied integration site (Fig. 6C), and related proviruses independent of integration site (Fig. 6D and E). The presence of amplified fragments of the predicted size for integrated provirus only in the four M. spicilegus individuals and of the size expected for the unoccupied site in all mice except M. spicilegus implies that the HEMV provirus on distal chromosome 7 is unique to this species. Furthermore, it must be homozygous in at least three of the four individuals which had been collected from the limits of the species' range, suggesting residence in the species long enough to become fixed in the genome, or nearly so. In addition, no proviruses containing related LTRs were found in any species outside of M. spicilegus. More distantly related mouse species, such as M. caroli, M. dunni, M. (coelomus) pahari, and M. (pyromus) platythrix, failed to yield amplification products of the correct size with any primer pair, most likely due to local genomic polymorphisms that defeated the PCR strategy. All products were sequenced to confirm their identity. The approximately 500-bp fragment in the two M. cervicolor lanes and the one M. caroli lane were determined to be artifacts.
Phylogenetic analysis of the HEMV provirus.
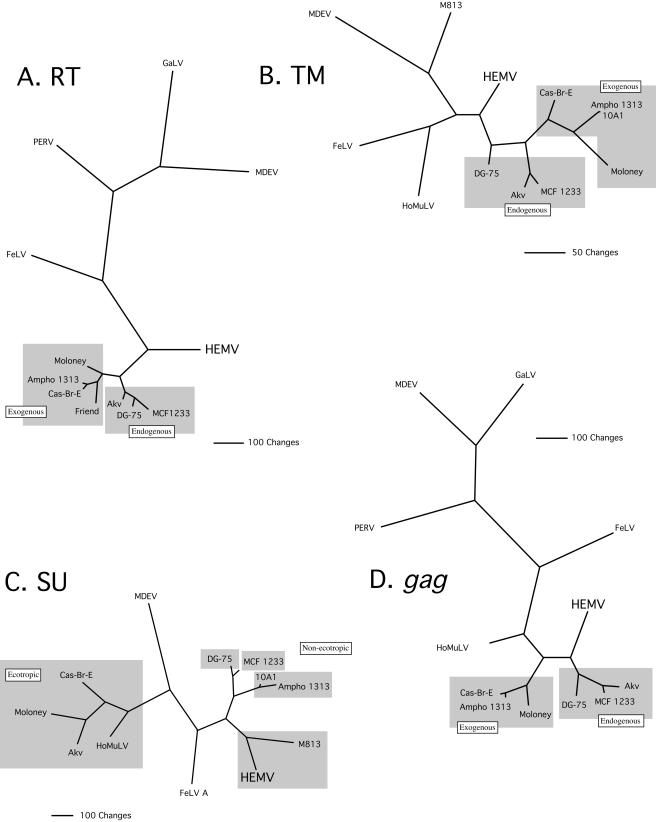
Previous phylogenetic analyses of the U3 regions and env genes of MLV-like viruses suggested that HEMV occupies a relatively central position. The availability of the complete nucleotide sequence of HEMV allowed us to determine this relationship for all other regions.
Trees relating HEMV sequences to those of other MLV-like viruses were constructed for gag, RT, SU, and TM DNA using maximum parsimony (42) (Fig. 7). The RT and TM trees were consistent with those derived for U3 and the env gene (46), establishing HEMV as the sole resident of an evolutionary branch derived from the node that separates the MLVs from other MLV-like viruses. The RT tree indicates that the MLVs are otherwise tightly associated but that there is a clear split of endogenous and exogenous viruses (Fig. 7A). When TM sequences were used for analysis, HEMV again straddled the node between the MLVs and other gammaretroviruses. Interestingly, there was a less-clear division between endogenous and exogenous MLVs in the TM tree (Fig. 7B).
FIG. 7.
Phylogenetic analysis of regions of MLV-related genomes. DNA sequences of the viruses shown were analyzed using maximum parsimony with the exhaustive search option (42). (A) Reverse transcriptase. (B) The TM region of the Env protein. (C) The SU region. (D) The gag region. Boxes in A, B, and D indicate endogenous and exogenous viruses; boxes in C indicate different subgroups. The sequence of the exogenous xenotropic virus DG-75 was used in this analysis, as it is the only complete virus of this subgroup available (37). It should be noted that it is highly possible that despite its distinctly xenotropic env region, this virus is a recombinant between an endogenous xenotropic locus and some other MLV. Regardless, initial phylogenetic analyses that included other established partial xenotropic sequences did not distinguish DG-75, so we consider it to be an example of this endogenous subgroup.
The SU tree shows clear clustering of the MLVs into the recognized subgroups, reflecting the independent evolution of this portion of the genome to the utilization of distinct receptors. Despite their origin from different locales, different species, and endogenous versus exogenous nature, the classical ecotropic SU regions cluster together, as do the polytropic and xenotropic endogenous MLVs and the 10A1 and amphotropic exogenous viruses. There is no obvious separation into mouse and nonmouse clades. Finally, HEMV clusters with M813, an exogenous virus that has a rather different host range (36) (Fig. 7C).
The gag gene tree again preserves the division between endogenous and exogenous viruses and viruses of mouse origin as opposed to those of nonmouse origin, but HEMV is located on a branch that is positioned slightly differently from its location in the RT and TM trees (Fig. 7D).
Overall, the phylogenetic analysis, taken together with our previous report (46), is consistent with HEMV occupying a position distinct from all other gammaretroviruses, with a relatively close relationship to the common ancestor of MLV-like viruses.
DISCUSSION
Unique among infectious agents, simple retroviruses have left behind a fossil record in the form of endogenous proviruses that can be used to infer the evolutionary history of the host-virus relationship (21, 23, 46). To date, endogenous retroviruses, including gammaretroviruses, have been found in all vertebrates studied (30). Although all species contain large numbers of relatively ancient proviruses, only a few, including most members of the genus Mus, have proviruses that were inserted in relatively recent times. Such recently inserted proviruses are polymorphic in location within a species, have accumulated little or no sequence difference between the LTRs, and are often capable of giving rise to infectious virus. In prior work, we have defined distinct features of endogenous MLVs and catalogued their representatives in inbred and wild-derived mouse strains (12-14, 40, 45).
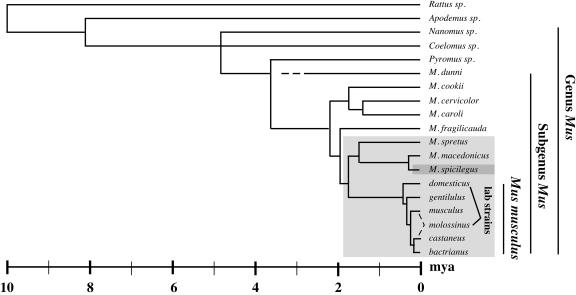
The HEMV provirus was identified in the course of continuing work to identify distinct features of endogenous MLVs and to catalogue their representatives in laboratory and wild-derived mouse strains. Analysis of the U3 regions of multiple endogenous MLV loci isolated from wild-derived mice indicated that they are highly polymorphic and that the previously defined xenotropic, polytropic, and modified polytropic groups could be further subdivided into even more detailed classes, which were labeled X-I through X-IV (xenotropic subgroup-related I-IV) and P-I through P-V (polytropic subgroup related). The class X-IV viruses were the most broadly represented within the Mus subgenus and could be found in many species, including the single provirus in M. spicilegus. This distribution suggests that this class of virus was active in a Mus population that preexisted a period of radiation, termed the West Palearctic split, approximately 2 million years ago and therefore existed prior to all other classes (Fig. 8). The single X-IV provirus within the genome of the distantly related M. spicilegus was christened HEMV for hortulanus endogenous MLV (46) (Mus hortulanus is the prior name for Mus spicilegus).
FIG. 8.
Ten million years of mouse evolution. The cladogram shows recognized mouse taxa and their relationship as inferred by several molecular techniques. M. m. molossinus, endemic to Japan, is a hybrid of M. m. musculus and M. m. castaneus. The myriad strains of laboratory mouse are derived primarily from the fancy mouse breeding fashionable in Europe (M. m. domesticus) and Japan (M. m. molossinus) in the 19th century. The lighter grey box indicates species derived from a common ancestor before the creation of the West Palearctic species M. spretus, M. macedonicus, and M. spicilegus. Endogenous MLVs have been confirmed to reside only in these species. The darker grey box highlights the HEMV host M. spicilegus (this figure is a composite work from references 2, 5, and 35).
HEMV was hypothesized to be an endogenous MLV of some antiquity for four reasons (46). First, it is the sole MLV provirus present within the genome of a mouse species known to have split relatively distantly from the more typical MLV host M. musculus (Fig. 8). Second, hybridization analysis revealed that its closest relatives, class X-IV viruses, are broadly distributed throughout the branches of murine evolution after the split that created M. spicilegus, with many shared proviruses. Third, the HEMV U3 region lacks much of the complexity found in other endogenous MLVs. Variation among U3 regions of these viruses is largely a matter of duplication and acquisition of short sequence motifs, and a scheme based on parsimonious consideration of these events placed HEMV near a putative ancestral state. Finally, phylogenetic trees based on both the U3 region and the env gene placed HEMV on relatively short branches separate from the other MLVs and MLV-like viruses arising near the common ancestor of viruses of mice and other species. Even without consideration of the first three points, this analysis implies that HEMV resembles the theoretical common ancestor for all MLV-like gammaretroviruses more than any other member.
Consistent with the assumption of antiquity, PCR amplification of a virus-host DNA junction fragment showed that HEMV is present and homozygous in three individuals (or their descendants) from different parts of the range of M. spicilegus. This result implies that HEMV was integrated sufficiently long ago to be fixed in the host species. However, PCR analysis and hybridization with an HEMV-specific oligonucleotide probe failed to detect similar elements in M. m. musculus, M. spretus, M. cookii, M. caroli, and M. cervicolor (popaeus). Thus, HEMV appears to be unique to its host species.
Relative to other endogenous MLVs, the env gene of HEMV is also significantly shortened, with notable deletions in the receptor-interacting VRA and possibly VRB regions (46). Like the LTR and pol genes, it also occupies a relatively central location on the MLV tree. Although it lacked other obvious defects, these sizable deletions implied that HEMV might be defective. Somewhat surprisingly, experiments using an MLV pseudotyped with the HEMV Env protein showed that it was fully functional and that HEMV represented a novel subgroup of MLV. Initial results indicated that the host range dictated by the HEMV Env protein resembled that of the ecotropic MLV subgroup in that it could infect only cells derived from the genus Mus. Unlike ecotropic MLV, it could not infect the two strains of rat cell tested. In support of this distinction, HEMV could not infect human cells stably transfected with the ecotropic receptor mCAT1. The fact that HEMV represents a novel subgroup of MLV was confirmed by performing an interference assay using a replication competent-Moloney MLV expressing the HEMV env gene. HEMV did not cross-interfere with the other traditional MLV subgroups. We thus conclude that HEMV represents a novel subgroup of MLV, in that it uses a receptor distinct from those of all other known viruses tested here. The results of a similar study with M813 support this conclusion (see below).
Although the competence of the HEMV Env protein to mediate entry was unexpected due to its unusual primary sequence, it remained possible that the HEMV locus was ancient. There are numerous examples of defective endogenous loci whose env genes have been preserved either as protective against future infections or coopted for host function (19, 20, 22, 25, 31). Inconsistent with a role in protection is the observation that primary tail fibroblast cells derived from a provirus-positive M. spicilegus individual were readily infectible by the HEMV pseudotype, implying that expression of the provirus, or at least the env gene, is not high enough to protect these cells from exogenous HEMV infection. Although levels of expression are yet to be tested directly, it is likely that, analogous to other replication-competent MLVs and ALVs, the HEMV provirus is strongly suppressed by CpG methylation (7, 17).
To assess the replication competence of HEMV, the entire provirus was cloned and sequenced. We found it to be intact, lacking in obvious defects in the form of deletions, nonsense mutations, and frameshift mutations. Furthermore, its LTRs were identical in sequence, consistent with a relatively recent insertion into the M. spicilegus genome. The lack of a mismatch between the LTRs implies that it is younger than the probable appearance of a single nucleotide change. Using a rate for silent substitutions in rodents of 7.9 × 10−9 substitutions/site/year, a single mismatch in a 1,000-base-pair region (i.e., both LTRs) should occur about once every 125,000 years (18, 21, 38). Furthermore, the lack of a provirus at this site in M. macedonicus, a species which split from M. spicilegus within the last 200,000 years, further caps the age of HEMV. The likeliest date of insertion of this element probably lies towards the extreme end of these estimates because it is present and homozygous in the three tested M. spicilegus isolates, all of which were collected from the edges of its range. M. spicilegus is an aboriginal species commonly known as the mound-building mouse, and it has adapted to its environment partially by forming tight-knit kin groups with little migration. It is unlikely, given this social behavior, that HEMV's ubiquity stems from the recent migration of provirus-carrying individuals between the regions. HEMV must therefore have been active as an exogenous virus just after the split from M. macedonicus.
The 5′ and 3′ integration junctions between virus and cell DNA were also cloned. Their sequence revealed that HEMV is located in a region corresponding to the distal end of C57BL/6 chromosome 7. According to the current information provided by the assembled mouse genome and the mouse/endogenous retrovirus linkage map, this region is otherwise devoid of retroviral integration and is not densely packed with host coding regions or even expressed sequence tags that suggest active transcripts (14) (http://www.ensembl.org/Mus_musculus/). Although the ends of the provirus were exactly as predicted, a surprising finding was that the lesions in the host chromosome created by the HEMV IN were not perfectly repaired to create the canonical 4-base direct repeats. However, alignment with the C57BL/6 mouse genome does suggest that there was a 4-base offset to the enzymatic attack. This conclusion is based upon the assumption that the M. spicilegus genome and the C57BL/6 genome are concordant at this site, which is not unreasonable, considering the similarity of genomic regions flanking the HERV integration to C57 DNA. If the HEMV provirus is completely fixed, it may not be possible to determine the sequence of the unoccupied site. In support of this misrepair hypothesis, the sequence of the unoccupied site in the closely related species M. macedonicus is AAAC, the same as the 3′ flanking repeat, suggesting that the mismatch resulted from error-prone repair of the 5′ repeat.
Although apparently intact, the HEMV provirus contained numerous single base changes that made it different from other MLV subgroups. To test its competence for replication, a complete clone of the HEMV provirus was created in a vector that contained no integral eukaryotic promoter. We found that this proviral clone could seed an active infection of NIH 3T3 cells following its initial transfection, as determined by assaying for the appearance of reverse transcriptase activity in the culture supernatant. Furthermore, viruses harvested from this transfection at the peak appearance of RT activity rapidly spread through freshly infected mouse cell cultures and were able to induce superinfection resistance to HEMV-pseudotyped virus. Thus, this provirus has maintained full biological activity since integration into an ancestor of Mus spicilegus, perhaps 100,000 or more years ago.
For the most part, phylogenetic analysis of HEMV supported the relationships suggested by previous consideration of its entire env gene and U3 region (46). Exhaustive maximum-parsimony searches that included only HEMV and its closer MLV-like relatives were performed. Sequences for RT, the entire gag gene, and the entire env gene were included in separate analyses. A key difference in the trees presented in this study is that, unlike the previous work, the env gene was split into SU and TM regions. We analyzed each Env protein subunit separately due to large differences in conservation of the two sequences, because there are multiple examples of env genes that are recombinants of these two regions (41, 45) and because TM and RT tree topologies tend to agree (3). This latter point is useful because complete sequence information for many gammaretroviruses is not available. The SU tree is the only one not to place HEMV at the junction of the MLVs and the other gammaretroviruses, although it did indicate that this region of HEMV shares (along with M813) ancestry with a very early progenitor of the nonecotropic MLVs and one not far removed from a hypothetical ancestor of feline leukemia virus. Unlike trees based on other regions of the genome, which tend to group exogenous and endogenous MLVs separately, the SU tree, not surprisingly, groups viruses by subgroup.
The closest relative of the HEMV SU protein is that of MLV M813, thought to be an exogenous virus from M. cervicolor, a species that last shared a common ancestor with M. spicilegus approximately 2 million years ago (2, 5, 6). The M813 receptor was recently discovered to be the sodium-dependent myo-inositol transporter SMIT1 (16). Recently, we have found that HEMV also uses SMIT1 as a receptor, although the two viruses have quite different host ranges. The conclusions of this research have broader implications and will be detailed in an upcoming paper (Tipper and Coffin, unpublished).
There are two possibilities for the origin of HEMV. One is that it represents a solitary endogenous member of a class of retrovirus that has evolved independently in the M. spicilegus lineage. The presence of HEMV-like X-IV viruses within the M. spretus genome indicates that a putative HEMV progenitor existed within the lineage that established the two species. Alternately, it is possible that the endogenous HEMV locus resulted from infection of M. spicilegus with a virus from a sympatric species. The hypothetical age of the endogenous element puts its integration soon after the split from M. macedonicus, a time when an early M. spicilegus population may not have yet adopted its peculiar social habits. The lack of shared (prespeciation) fixed HEMV loci would better fit this hypothesis. Further data are required to distinguish between these alternatives.
In conclusion, HEMV closely resembles the hypothetical MLV common ancestor by most of the genotypic and phenotypic data reported above. Unlike most other infectious endogenous proviruses of chickens and mice, it has a widespread distribution in the M. spicilegus genome but is fully capable of supporting an exogenous infection.
Acknowledgments
We thank Naomi Rosenberg, David Lazinski, and Cathy Squires for useful advice and the Tufts GRASP center for technical support. We also acknowledge the generosity of many who contributed material to this project, notably Francois Bonhomme and Christine Kozak.
J.M.C. was an American Cancer Society Research Professor of Molecular Biology and Microbiology with support from the F. M. Kirby Foundation. This work was supported by grant R01 CA 89441-01 from the National Cancer Institute.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Auffray, J., A. Orth, J. Catalan, J. Gonzalez, E. Desmarais, and F. Bonhomme. 2003. Phylogenetic position and description of a new species of subgenus Mus (Rodentia, Mammalia) from Thailand. Zoologica Scripta 32:119-127. [Google Scholar]
- 3.Benit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benveniste, R. E., R. Callahan, C. J. Sherr, V. Chapman, and G. J. Todaro. 1977. Two distinct endogenous type C viruses isolated from the Asian rodent Mus cervicolor: conservation of virogene sequences in related rodent species. J. Virol. 21:849-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonhomme, F., and J. Guenet. 1995. The Laboratory mouse and its wild relatives, 3rd ed. Oxford University Press, New York, N.Y.
- 6.Boursot, P., W. Din, R. Anand, D. Darviche, B. Dod, F. Von Deimling, G. P. Talwar, and F. Bonhomme. 1996. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J. Evol. Biol. 9:391-415. [Google Scholar]
- 7.Challita, P. M., and D. B. Kohn. 1994. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc. Natl. Acad. Sci. USA 91:2567-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudry, G. J., K. B. Farrell, Y. T. Ting, C. Schmitz, S. Y. Lie, C. J. Petropoulos, and M. V. Eiden. 1999. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 10.Colicelli, J., and S. P. Goff. 1988. Isolation of an integrated provirus of Moloney murine leukemia virus with long terminal repeats in inverted orientation: integration utilizing two U3 sequences. J. Virol. 62:633-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eglitis, M. A., M. V. Eiden, and C. A. Wilson. 1993. Gibbon ape leukemia virus and the amphotropic murine leukemia virus 4070A exhibit an unusual interference pattern on E36 Chinese hamster cells. J. Virol. 67:5472-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel, W. N., and J. M. Coffin. 1994. Endogenous nonecotropic proviruses mapped with oligonucleotide probes from the long terminal repeat region. Mamm. Genome 5:275-281. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, W. N., J. P. Stoye, B. A. Taylor, and J. M. Coffin. 1989. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J. Virol. 63:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankel, W. N., J. P. Stoye, B. A. Taylor, and J. M. Coffin. 1990. A linkage map of endogenous murine leukemia proviruses. Genetics 124:221-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garel, S., F. Marin, M. G. Mattei, C. Vesque, A. Vincent, and P. Charnay. 1997. Family of Ebf/Olf-1-related genes potentially involved in neuronal differentiation and regional specification in the central nervous system. Dev. Dyn. 210:191-205. [DOI] [PubMed] [Google Scholar]
- 16.Hein, S., V. Prassolov, Y. Zhang, D. Ivanov, J. Löhler, S. R. Ross, and C. Stocking. 2003. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J. Virol. 77:5926-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hejnar, J., J. Plachy, J. Geryk, O. Machon, K. Trejbalova, R. V. Guntaka, and J. Svoboda. 1999. Inhibition of the Rous sarcoma virus long terminal repeat-driven transcription by in vitro methylation: different sensitivity in permissive chicken cells versus mammalian cells. Virology 255:171-181. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 101:1668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda, H., K. Kato, H. Kitani, T. Suzuki, T. Yoshida, Y. Inaguma, N. Yamamoto, J.-G. Suh, B.-H. Hyun, T. Yamagata, T. Namikawa, and T. Tomita. 2001. Virological properties and nucleotide sequences of Cas-E-type endogenous ecotropic murine leukemia viruses in South Asian wild mice, Mus musculus castaneus. J. Virol. 75:5049-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda, H., and H. Sugimura. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J. Virol. 63:5405-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, W. E., and J. M. Coffin. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. USA 96:10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, Y. T., M. S. Lyu, A. Buckler-White, and C. A. Kozak. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J. Virol. 76:8218-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak, C. A., and R. R. O'Neill. 1987. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J. Virol. 61:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loiler, S. A., N. L. DiFronzo, and C. A. Holland. 1997. Gene transfer to human cells using retrovirus vectors produced by a new polytropic packaging cell line. J. Virol. 71:4825-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyu, M. S., A. Nihrane, and C. A. Kozak. 1999. Receptor-mediated interference mechanism responsible for resistance to polytropic leukemia viruses in Mus castaneus. J. Virol. 73:3733-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann, R., R. C. Mulligan, and D. Baltimore. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33:153-159. [DOI] [PubMed] [Google Scholar]
- 27.Marin, M., C. S. Tailor, A. Nouri, S. L. Kozak, and D. Kabat. 1999. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J. Virol. 73:9362-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markowitz, D., S. Goff, and A. Bank. 1988. Construction and use of a safe and efficient amphotropic packaging cell line. Virology 167:400-406. [PubMed] [Google Scholar]
- 29.Markowitz, D., S. Goff, and A. Bank. 1988. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J. Virol. 62:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 32.Muller, A. J., J. C. Young, A. M. Pendergast, M. Pondel, N. R. Landau, D. R. Littman, and O. N. Witte. 1991. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol. Cell. Biol. 11:1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott, D., R. Friedrich, and A. Rein. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J. Virol. 64:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeiffer, J. K., R. S. Topping, N. H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prager, E. M., C. Orrego, and R. D. Sage. 1998. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics 150:835-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prassolov, V., S. Hein, M. Ziegler, D. Ivanov, C. Munk, J. Lohler, and C. Stocking. 2001. Mus cervicolor murine leukemia virus isolate M813 belongs to a unique receptor interference group. J. Virol. 75:4490-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raisch, K. P., M. Pizzato, H. Y. Sun, Y. Takeuchi, L. W. Cashdollar, and S. E. Grossberg. 2003. Molecular cloning, complete sequence, and biological characterization of a xenotropic murine leukemia virus constitutively released from the human B-lymphoblastoid cell line DG-75. Virology 308:83-91. [DOI] [PubMed] [Google Scholar]
- 38.Ridley, M. 1996. Evolution, 2nd ed. Blackwell Science, Cambridge, Mass.
- 39.Shinnick, T. M., R. A. Lerner, and J. G. Sutcliffe. 1981. Nucleotide sequence of Moloney murine leukaemia virus. Nature 293:543-548. [DOI] [PubMed] [Google Scholar]
- 40.Stoye, J. P., and J. M. Coffin. 1988. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J. Virol. 62:168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swofford, D. L. 2002. PAUP: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 43.Tailor, C. S., D. Lavillette, M. Marin, and D. Kabat. 2003. Cell surface receptors for gammaretroviruses. Curr. Top. Microbiol. Immunol. 281:29-106. [DOI] [PubMed] [Google Scholar]
- 44.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozak, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomonaga, K., and J. M. Coffin. 1998. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 72:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomonaga, K., and J. M. Coffin. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J. Virol. 73:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, C., M. S. Reitz, H. Okayama, and M. V. Eiden. 1989. Formation of infectious hybrid virions with gibbon ape leukemia virus and human T-cell leukemia virus retroviral envelope glycoproteins and the gag and pol proteins of Moloney murine leukemia virus. J. Virol. 63:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Comparison of cDNAs encoding the gibbon ape leukaemia virus receptor from susceptible and nonsusceptible murine cells. J. Gen. Virol. 75:1901-1908. [DOI] [PubMed] [Google Scholar]