Abstract
The foamy virus (FV) genome contains two promoters, the canonical long terminal repeat (LTR) promoter, containing three consensus AP-1 binding sites, and an internal promoter (IP) within the env gene. We investigated the regulation of the two promoters in lytic and persistent infections and found that in the presence of a constitutive source of the viral transactivator protein Tas, transactivation of the LTR promoter and that of the IP differ. In lytic infections, both the LTR promoter and the IP are efficiently transactivated by Tas, while in persistent infections, the IP is efficiently transactivated by Tas, but the LTR promoter is not. Analysis of proteins expressed from the LTR promoter and the IP during infection indicated that IP transcription is more robust than that of the LTR promoter in persistently infected cells, while the opposite is true for lytically infected cells. Coculture experiments also showed that LTR promoter transcription is greatest in cells which support lytic replication. Replacement of much of the LTR promoter with the IP leads to increased viral replication in persistent but not lytic infections. We also found that the induction of persistently infected cells with phorbol 12-myristate 13-acetate (PMA) greatly enhanced viral replication and transcription from the SFVcpz(hu) (new name for human FV) LTR promoter. However, mutation of three consensus AP-1 binding sites in the FV LTR promoter did not affect viral replication in lytically or persistently infected cells, nor did the same mutations affect LTR promoter transactivation by Tas in PMA-treated cells. Our data indicate that differential regulation of transcription is important in the outcome of FV infection but is unlikely to depend on AP-1.
Foamy viruses (FVs) are unique among retroviruses in their establishment of life-long persistent infections without any accompanying pathologies. Infection is characterized by the presence of viral DNA in a large number of organs (9, 42), without detectable levels of viral RNA or protein expression (6, 9, 42, 44). Indeed, viral transcription has been detected only in the oral mucosa of a single infected animal (9). However, virus can be recovered readily by coculturing of infected tissues, peripheral blood, or throat swab specimens with susceptible cell lines (6, 18, 42, 44, 46, 49). Thus, in most locations in vivo, FV replication is latent; however, when the virus is removed from such a context, replication can proceed. In contrast to the in vivo situation, FV replication in vitro can result in either lytic or persistent infection (13, 41, 53). Infection of many cell types in vitro is often accompanied by cytopathic effects (CPE) and rapid cell killing. Since such infections do not mimic the in vivo situation, we sought to develop a tissue culture system in which there is little viral replication. For these studies we used the prototypic human FV (HFV) clone HFV13 (29). HFV has recently been renamed SFVcpz(hu) to more clearly indicate that the original HFV isolate is a chimpanzee FV isolated from a human-derived cell culture (17). It has been previously shown that several human hematopoietic cell-derived lines can be infected with SFVcpz(hu), but with low levels of viral replication and no CPE (33, 53). We examined the role of viral transcription in regulating virus production in these cell lines.
Several characteristics of FV transcription may allow for different types of viral replication, such as lytic and persistent infections. One factor which could be involved in regulating viral replication is the presence of an internal promoter (IP) (27) in addition to the conventional long terminal repeat (LTR) promoter. The IP has low basal activity and drives the expression of the requisite transcriptional transactivator, tas (25). Tas is a DNA binding protein which transactivates both the IP and the LTR promoter (15, 25–27). Interestingly, the Tas protein binds to distinct sequences in the LTR promoter and the IP which share no homology, indicating that Tas may transactivate the two promoters via different mechanisms (8, 20, 23). In addition, Tas has a higher affinity for the IP than for the LTR promoter (20). These facts, coupled with the lack of basal LTR promoter transcription in the absence of Tas, provide a number of possible ways to regulate FV replication. It is generally thought that after infection and integration into a new host cell, the low basal activity of the IP drives the expression of Tas which, due to its higher affinity for its own promoter, drives the expression of additional Tas via a positive-feedback loop (25, 26, 28). Once sufficient levels of Tas are attained, LTR promoter transcription can proceed and viral replication can commence. This bimodal, temporal pattern of transcription could be regulated at (i) Tas-independent, basal transcription of the IP, (ii) Tas-dependent transactivation of the IP, or (iii) Tas-dependent transactivation of the LTR promoter. A better understanding of how FVs achieve persistence in vitro may provide a better understanding of how they persist in vivo.
We have examined the relationship between the SFVcpz(hu) LTR promoter and the IP in a variety of lytic and persistent infections in vitro. We have shown that the SFVcpz(hu) IP is more efficiently transactivated in persistently infected cells, while the LTR promoter is more efficiently transactivated in lytically infected cells. Activation of persistently infected cells with the phorbol ester phorbol 12-myristate 13-acetate (PMA) resulted in greatly enhanced LTR promoter transcription and viral replication. However, mutation of three consensus AP-1 binding sites in the SFVcpz(hu) LTR promoter had little or no effect on lytic replication or PMA-induced viral replication in persistently infected cells. Our findings suggest that the regulation of the two FV promoters is important in determining the outcome of FV infection.
MATERIALS AND METHODS
Cells and viruses.
Virus titers were determined using the previously described FAB indicator cell line (52). Diploid human embryonic lung (HEL) cells (ATCC CCL-137), baby hamster kidney (BHK-21) cells (ATCC CCL-10), and FAB cells were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics. Human erythroleukemia (H92) cells (ATCC TIB-180), Raji cells (ATCC CRL-2367), U937 cells (ATCC CRL 1593.2), and Jurkat cells (ATCC TIB-152) were grown in RPMI-1640 medium supplemented with 10% FBS. Infection of H92, Raji, Jurkat, and U937 cells by coculturing was performed as previously described (33). Green fluorescent protein (GFP) indicator lines H92–5Lg and Jurkat-5Lg were transduced with retroviral vector LN-5Lg (see below) pseudotyped with vesicular stomatitis virus glycoprotein (VSV-g) as previously described (4). H92 and Jurkat cells were selected in 600 and 1,200 μg of G418 (Life Technologies, Inc.)/ml, respectively.
Plasmids.
The infectious molecular clone pSFVcpz(hu)13 was provided by R. Flugel (29). A cytomegalovirus (CMV)-driven SFVcpz(hu) vector, pC-SFVcpz(hu), was generated such that the 5′ end of the viral RNA begins at the same nucleotide as in wild-type SFVcpz(hu). The U3 region of the SFVcpz(hu) LTR promoter was excised from pSub1 (3) by digestion with EagI and partial digestion with XbaI. A linker containing EagI and XbaI ends was cloned into pSub1, yielding pCMVsub1. The CMV immediate-early (CMV-IE) promoter was amplified from vector pCR3.0 (Invitrogen) using the forward primer CMVEagI (5′GATCGATCGGCCGGCGCGCGTTGADATTGATTATTG3′) and the reverse primer CMVXbaI (5′ATCTAGACTCGAAGGCTTATATAGACCTCCCACCGTACACG3′) (positions of introduced restriction enzyme sites EagI and XbaI are underlined). The PCR product was digested with EagI and XbaI and cloned into pCMVsub1. pC-SFVcpz(hu) was generated by subcloning an EagI/SwaI fragment from pCMVsub1 into pSFVcpz(hu)13.
The vector LN-5Lg was constructed as follows. pEGFP-1 (Clontech) was digested with AflII and blunt ended with Klenow and subsequently digested with PstI. The resulting 1,005-bp fragment containing the enhanced green fluorescent protein (GFP) and SV40 polyadenlyation signal was subcloned into pHSRV5LG (52) digested with PstI and StuI and named pHSRV5LGFP. pHSRV5LGFP was then digested with SmaI and AvrII, and the resulting 1,544-bp fragment was subcloned into LNSX digested with NruI and AvrII. The SV40 polyadenlyation signal was removed by digestion with NotI and AvrII followed by Klenow treatment and vector recircularization.
The promoterless luciferase reporter construct pGL3 (Promega) was used to construct LTR-luc and IP-luc. A 1,274-bp fragment containing the complete SFVcpz(hu) 5′ LTR was generated by digesting pSFVcpz(hu)13 with KpnI and AvrII. This fragment was cloned into pGL3 digested with KpnI and NheI, yielding LTR-luc. IP-luc was constructed as follows. A 467-bp fragment was amplified using oligonucleotides 8971SmaI (GATCCCGGGATATGTTCCTAGCATCGTGAC) and 9438NcoI (5′AATCCATGGTACAATCTTAAATATAAGAATAACC3′), which creates an NcoI restriction site overlapping the start codon for tas (shown in bold type). The product was cloned into SmaI- and NcoI-digested pGL3. Vector pCMV-tas was constructed by amplifying the entire tas gene by PCR using oligonucleotides 5′ATCTCTAGACTCGAGCCAGCCATGGATTCCTACGAAAAAGAAG3′ and 5′CCCTCTAGATTATAAAACTGAATGTTCACC3′. The product was digested with XbaI, underlined, and cloned into XbaI-digested pCR3.0 (Invitrogen). The luciferase expression constructs pLTRΔ1, pLTRΔ23, and pLTRΔ123 were constructed as follows. Plasmid LTR-luc was used as a template for site-directed mutagenesis with a Quickchange site-directed mutagenesis kit (Stratagene). The AP-1 binding sequence, 5′TGACTCAG3′, was mutated at the first position using the forward primer AP1m1F (5′CATTGACAGAGATGACCCAAGATGAAATTAGAAAAAGG3′) and the reverse primer AP1m1R (5′CCTTTTTCTAATTTCATCTTGGGTCATCTCTGTCAATG3′), yielding pLTRΔ1 (locations of mutated AP-1 binding sequences are underlined). The second and third AP-1 sites were mutated using the forward primer AP1m23F (5′GTGACCCCTTCATCGATTCCGGAAGCGATTCCGATGGACCCTTC3′) and the reverse primer AP1m23R (5′GAAGGGTCCATCGGAATCGCTTCCGGAATCGATGAAGGGGTCAC3′), yielding pLTRΔ23. All three AP-1 binding sites were mutated using pLTRΔ1 as the template for a second round of mutagenesis with primers AP1m23F and AP1m23R. The resulting plasmid was termed pLTRΔ123. None of the mutations disrupted the bel2 open reading frame (ORF). All clones were sequenced to confirm the presence of the desired mutations and to confirm the absence of unwanted mutations.
Vector pC-SFVcpz(hu)-ΔAP1 was generated as follows. A minimal BstEII/SacI fragment containing the mutated AP-1 binding sites was excised from pLTRΔ123 and subcloned into pSub5 (3), yielding pSub5-ΔAP123. pC-SFVcpz(hu)-Δ AP1 was generated by cloning a BlpI/SalI fragment from pSub5-ΔAP123 into BlpI- and SalI-digested pC-SFVcpz(hu).
Plasmid pBS-IP was constructed as follows. A 246-bp fragment from positions 9061 to 9307 of the SFVcpz(hu) DNA genome was amplified using oligonucleotides 9061BamHI (5′ACTGGATCCCTTTGAGCCACGACTGCC3′) and 9307EcoRV (5′ ACTGATATCCAATTCCTTGTAGAGCAGAAGC3′). The resulting product was cloned into BamHI- and EcoRV-digested pBS-SKII(+). Plasmid pGAPDHBS, containing the human glyceraldehyde phosphate dehydrogenase gene (GAPDH), was provided by Mark Groudine, Fred Hutchinson Cancer Research Center.
Luciferase reporter assays.
BHK-21 cells were seeded at 2 × 104 cells per well in 48-well plates. The following day, cells were transfected with 1 μg of DNA and 2 μl of Fugene (Roche) per well according to the manufacturer's instructions. Each reaction contained 0.3 μg of reporter construct, 0.3 μg of pUC19, and 0.1 μg of CMV–β-galactosidase vector to monitor transfection efficiency; 0.3 μg of pCMV-tas or an additional 0.3 μg of pUC19 was added to appropriate reactions. After 48 h, lysates were prepared with 250 μl of passive lysis buffer (Promega). Ten microliters of cleared lysate was analyzed with the firefly luciferase system (Promega) and Autolumat LB 953 instrumentation (Berthold). β-Galactosidase expression was measured at 420 nm using o-nitrophenyl-β-d-galactopyranoside (2). Nonadherent cell lines were transfected with DMRIE-C reagent (Life Technologies). For each reaction, 4 μl of DMRIE-C and 250 μl of DMEM were mixed with 2 μl of DNA in 100 μl of DMEM and incubated at 37°C for 30 min. Cells were counted, washed, and resuspended in DMEM at 2 × 106 cells/ml, and 250 μl of cells was added to the DNA–DMRIE-C mixture and incubated for 4 h at 37°C. Then, 500 μl of RPMI-1640 medium supplemented with 22% FBS was added. In some cases, Jurkat cells were treated with 50 nM PMA at 20 h posttransfection. At 48 h posttransfection, cells were pelleted and lysates were prepared with 250 μl of passive lysis buffer. A 25-μl portion of cleared lysate was analyzed for luciferase activity. A second 25-μl portion of lysate was monitored for β-galactosidase activity using a Galacto-Light Plus system according to the manufacturer's instructions (Tropix, Inc.).
Western blotting.
Western blot analysis was performed essentially as previously described (33). Briefly, Jurkat cells persistently infected with SFVcpz(hu) were plated in T-75 flasks (Falcon) at 5 × 105 cells/ml and treated with 50 nM PMA. At various times, 6 ml of cells was harvested and pelleted by low-speed centrifugation. Lysates were prepared with 250 μl of Ab buffer, and genomic DNA was sheared by passage through a 23-gauge needle. Lysates were cleared by high-speed centrifugation before loading on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels and detection using enhanced chemiluminescence (Amersham).
RIPA.
BHK-21 cells (2 × 106) were plated on 10-cm dishes, infected with SFVcpz(hu) at a multiplicity of infection (MOI) of 0.5, and grown until extensive syncytium formation was observed (about 40 h). Growth medium was removed and replaced with 5 ml of DMEM lacking cysteine and methionine but containing 400 μCi of 35-S Express protein label (NEN). Persistently infected H92, Raji, Jurkat, and U937 cells (5 × 106) were harvested and labeled as described above. After 4 h, cells were harvested in 1 ml of Ab buffer containing 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 μg of pepstatin A/ml, 0.57 mM phenylmethylsulfonyl fluoride (Sigma), and 1 μg of Pefablock (Roche)/ml (protease inhibitors). Genomic DNA was sheared and cleared by centrifugation. Lysates were precleared by incubation with 50 μl of protein A-Sepharose for 1 h at 4°C. BHK-21 cell lysate (100 μl) and 600 μl of H92, Raji, Jurkat, or U937 cell lysate were immunoprecipitated with 4 μl of anti-Tas antiserum and 2 μl of anti-Gag antiserum overnight at 4°C in a total volume of 1 ml. The following day, 100 μl of protein A-Sepharose (75 mg/ml in phosphate-buffered saline) was added to each reaction and mixed for 4 h at 4°C. Reactions were washed twice with radioimmunoprecipitation (RIPA) buffer (10 mM Tris, 150 mM NaCl, 1% Nonidet P-40, 1% deoxycholic acid, 0.1% SDS, 0.5% aprotinin [pH 7.4]), once with high-salt buffer (10 mM Tris, 2 M NaCl, 1% Nonidet P-40, 1% deoxycholic acid [pH 7.4]), and again with RIPA buffer. Samples were then separated on SDS–10% polyacrylamide gels and visualized by autoradiography. Quantification was done by phosphorimaging with ImageQuant software.
RPA.
An RNase protection assay (RPA) was performed using a Direct Protect kit (Ambion). Jurkat cells were treated as in the Western blotting procedure, and total nucleic acid was isolated in 400 μl of lysis solution. A 25-μl portion of cleared lysate was used in each hybridization. pBS-IP was linearized with BamHI, and T7 RNA polymerase was used to generate a 305-bp 32P-dUTP-labeled probe and, after RNase protection, 246- and 110-bp products indicating LTR promoter and IP transcripts, respectively. Plasmid pGAPDHBS was linearized with HindIII, and T7 runoff transcripts produced an unprotected 590-bp product and, after RNase protection, a protected 546-bp product.
RESULTS
The viral LTR promoter and IP are differentially regulated in lytic and persistent infections in vitro.
We hypothesized that differential regulation of the LTR promoter and IP may play a role in determining lytic or persistent infection in vitro. Lytic SFVcpz(hu) infection is generally observed in fibroblast-derived cells, such as BHK-21 cells and HEL cells, and is characterized by titers of >105/ml, CPE, extensive cytoplasmic vacuolation, and cell death. In contrast, persistent infection by SFVcpz(hu) generally occurs in but is not limited to cells of human hematopoietic lineages, in which there are few or no adverse effects on cell replication. Titers vary widely from >103/ml in the human erythroleukemia cell line H92 and the Burkitt's lymphoma-derived Raji cell line to <102/ml in the monocytic lymphoma-derived U937 cell line and the T-cell lymphoma-derived Jurkat cell line (Table 1). In a previous study (53), Raji cells were unable to be productively infected, but in the current study, using coculture methods, we were able to efficiently infect these cells.
TABLE 1.
FV titers in lytically and persistently infected cells
| Cells | Source | Infectiona | Titerb |
|---|---|---|---|
| HEL | Normal lung fibroblast | Lytic | 1.7 × 106 (2) |
| H92 | Erythroleukemia | Persistent | 1.7 × 104 (4) |
| Raji | Burkitt's lymphoma | Persistent | 1.2 × 103 (5) |
| U937 | Monocytic lymphoma | Persistent | 1.0 × 102 (8) |
| Jurkat | Acute T-cell leukemia | Persistent | 5.9 × 101 (16) |
Classification of cells infected with SFVcpz(hu). Lytic infection is characterized by syncytium formation, cytoplasmic vacuolization, and cell death. Persistent infection is characterized by lack of any notable CPE or impairment of normal cell growth.
Titers were determined as described in Materials and Methods, and are reported as the mean. Values in parentheses indicate number of independent experiments performed.
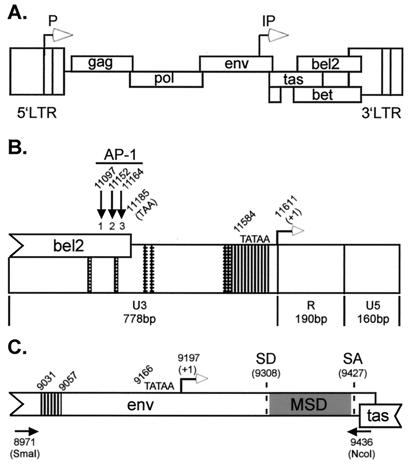
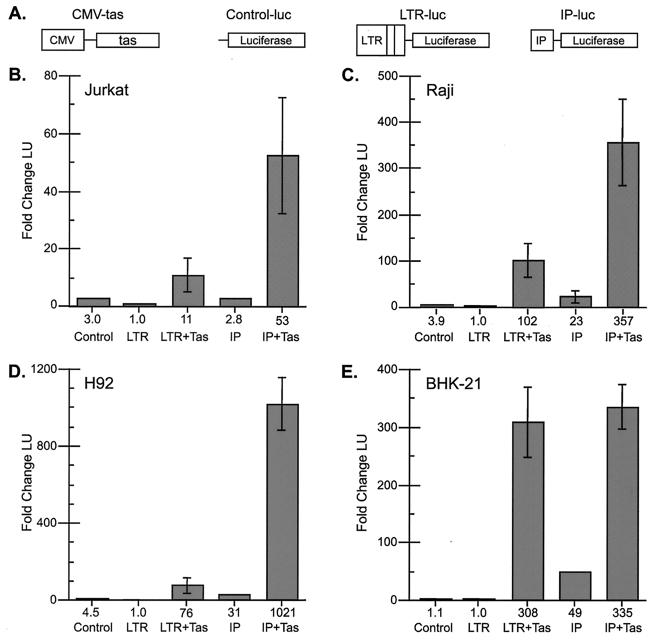
Salient features of the SFVcpz(hu) genome and the viral LTR promoter and IP are shown in Fig. 1A, B, and C, respectively. To analyze the activity of the LTR promoter and the IP in cell types which support either lytic or persistent infection, reporter constructs were generated which express firefly luciferase (luc) from either the LTR promoter or the IP (Fig. 2A). Transient transfection of LTR-luc or IP-luc allowed us to determine the basal transcriptional activity for the LTR promoter and the IP in the various cell types. In agreement with previous studies, the basal activity of the LTR promoter was lower than that of a promoterless luciferase control vector (Fig. 2B to E, compare LTR and Control). Similarly, the basal activity of the IP was low but was significantly higher than that of the promoterless control vector in all cell types except Jurkat cells (Fig. 2B to E, compare IP and Control). Thus, only in Jurkat cells could the difference in the basal activity of the IP account for the difference between lytic and persistent infections.
FIG. 1.
Schematic representation of the SFVcpz(hu) DNA genome, LTR promoter, and IP. Open arrows, LTR promoter and IP; closed arrows, AP-1 consensus site; vertical bands, Tas binding site; cross-hatched area, Tas-responsive element. (A) 11,955-bp SFVcpz(hu) DNA genome and known ORFs. (B) SFVcpz(hu) 3′LTR promoter. U3, R, and U5 regions, TATAA box, region of bel2 which overlaps U3, and locations of the three consensus AP-1 binding sites are indicated. (C) IP. The env and overlapping tas ORFs are shown. IP splice donor (SD) and splice acceptor (SA) sites, membrane-spanning domain (MSD), and locations of PCR primers (black arrows) used to amplify the IP are noted.
FIG. 2.
Transactivation of the LTR promoter and IP by transient transfection. (A) Schematic diagrams of constructs used in transfections. (B to E) Transactivation results. Control, promoterless control luciferase vector; LTR, LTR-luc; LTR+Tas, LTR-luc plus CMV-tas; IP, IP-luc; IP+Tas, IP-luc plus CMV-tas. The fold change in luciferase units (LU) relative to the value for the LTR promoter is shown below each column. (B) Jurkat cells, 1 LU = 131 raw LU (RLU). (C) Raji cells, 1 LU = 914 RLU. (D) H92 cells, 1 LU = 843 RLU. (E) BHK-21 cells, 1 LU = 39,656 RLU. All values are based on at least three independent experiments and are reported as the mean and standard error of the mean.
When CMV-tas was transiently transfected in addition to LTR-luc or IP-luc, differences in promoter transcription became apparent in lytically and persistently infected cell types. While the expression of Tas had no effect on the promoterless control construct (data not shown), in all cells the IP was efficiently transactivated in the presence of Tas (Fig. 2B to E, compare IP and IP+Tas). However, the change in the level of expression of the IP upon the addition of Tas was significantly higher in all three persistently infected cell types than in the lytically infected BHK-21 cell type. Tas-mediated transactivation of the IP was 19-fold in Jurkat cells, 15-fold in Raji cells, 32-fold in H92 cells, but only 7-fold in BHK-21 cells. Differences in Tas-mediated transactivation were also apparent for the LTR promoter. Transactivation of the LTR promoter by Tas was observed in all cell lines (Fig. 2B to E, compare LTR and LTR+Tas). However, the LTR promoter was transactivated to a lesser extent in the persistently infected cell types than in the lytically infected BHK-21 cell type. Tas-mediated transactivation of the LTR promoter was approximately 11-fold in Jurkat cells, approximately 102-fold in Raji cells, approximately 76-fold in H92 cells, but 308-fold in BHK-21 cells. Similar levels of LTR promoter and IP transactivation were observed when the IP, instead of CMV, was used to drive the expression of Tas (data not shown). These data indicate that in the presence of excess Tas, LTR promoter transcription may be limiting in persistently infected cells.
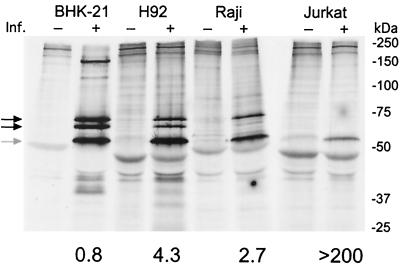
To confirm that the differences seen in promoter activity in transient transfections were reflected in infected cells, RIPA analysis with antisera against Gag and Tas was used to measure the activity of the LTR promoter and the IP, respectively. Because the anti-Tas antiserum reacts with Bet, which is produced in much larger quantities than Tas, immunoprecipitated Bet protein was used as a measure of IP activity. Persistently infected Jurkat, Raji, or H92 cells and lytically infected BHK-21 cells, undergoing extensive syncytium formation, were radiolabeled with 35S-cysteine-methionine. Gag and Bet proteins were then immunoprecipitated using a mixture of anti-Gag and anti-Tas polyclonal antisera (Fig. 3). To ensure efficient immunoprecipitation, excess amounts of each antiserum were used (data not shown). The amount of BHK-21 cell lysate assayed was approximately six times smaller than that used for the persistently infected cells. IP activity, leading to Bet expression, was evident in BHK-21, H92, Raji, and Jurkat cells (Fig. 3, grey arrow). LTR promoter activity, leading to Gag synthesis, was evident in BHK-21, H92, and Raji cells (Fig. 3, black arrows). Similar to the results of the transient transfection assays, IP activity, measured by Bet protein expression, was higher, relative to that measured by Gag protein expression, in persistently infected cells than in lytically infected BHK-21 cells. In contrast, LTR promoter activity, measured by Gag protein expression, was significantly higher in lytically infected BHK-21 cells than in persistently infected cells.
FIG. 3.
Promoter activity in lytic and persistent infections. RIPA was performed with the indicated uninfected and infected cell types using a mixture of anti-Gag and anti-Bet polyclonal antisera. Black arrows indicate 74- and 70-kDa Gag proteins. The grey arrow indicates the 52-kDa Bet protein. Locations of molecular mass markers are shown on the right. Immunoprecipitated Gag and Bet proteins were quantified by phosphorimaging after subtraction of the background from adjacent uninfected-cell lanes. The resulting values were then normalized to total 35S incorporated, and the ratio of Bet to Gag proteins was calculated (shown below the lanes). Values derived for Bet are considered IP activity, while values for Gag are considered LTR promoter activity. Inf., infection.
Phosphorimaging analysis of the Gag and Bet proteins in Fig. 3 was performed and normalized to total 35S incorporation. The relative activities of the IP and the LTR promoter were determined by taking the ratio of the normalized IP (Bet) and LTR promoter (Gag) values. The ratio of IP to LTR promoter activities was significantly higher in persistently infected cells than in lytically infected cells (Fig. 3, number under each set of lanes). For Jurkat cells, this ratio was >200 because the sensitivity of RIPA was not sufficient to detect any Gag protein. Taken together, these data suggest that in persistently infected cells, despite an efficient positive feedback loop at the IP, whereby substantial amounts of IP-based Tas are produced, there is inefficient transactivation of the LTR promoter. In contrast, in lytically infected BHK-21 cells, Tas produced by the IP efficiently transactivates the LTR promoter, resulting in a higher level of Gag expression and higher virus titers.
Fusion of HEL cells with persistently infected cells allows for LTR promoter transcription.
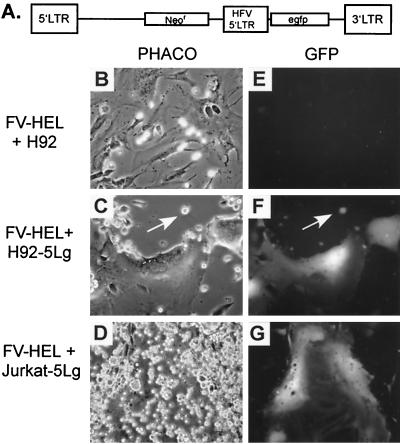
To address whether LTR promoters integrated in persistently infected cells are capable of efficient transcription given the appropriate cellular environment, fusion experiments were performed with either H92 or Jurkat cells and infected HEL cells. Uninfected H92 and Jurkat cells were transduced with the murine leukemia virus-based vector LN-5Lg, which expresses GFP under the control of the SFVcpz(hu) LTR promoter (Fig. 4A). G418-resistant populations were obtained and named H92–5Lg and Jurkat-5Lg, respectively. These cells were then infected by coculturing with lytically infected HEL cells. During coculturing with infected HEL cells, interaction of the SFVcpz(hu) receptor on the H92–5Lg or Jurkat-5Lg cells with the SFVcpz(hu) envelope expressed on the surface of the HEL cells permits fusion between the two cell types. During this process, we observed that the infected HEL cells expressed large amounts of GFP, while very few H92–5Lg and Jurkat-5Lg cells expressed GFP (Fig. 4F and G). This observation indicates that upon fusion with infected HEL cells, the genome of H92–5Lg or Jurkat-5Lg cells enters an environment suitable for LTR promoter transcription, resulting in GFP expression.
FIG. 4.
GFP expression of H92–5Lg or Jurkat-5Lg cells upon coculturing with lytically infected HEL cells (FV-HEL cells). PHACO, phase-contrast light microscopy. GFP, fluorescent imaging of GFP protein expression. All images in panels B to G were captured at a magnification of ×200. (A) Schematic diagram of the LN-5Lg vector used to transduce H92 and Jurkat cells. (B and E) PHACO and GFP images, respectively, of FV-HEL cells cocultured with naive H92 cells. (C and F) PHACO and GFP images, respectively, of FV-HEL cells cocultured with H92–5Lg cells. (D and G) PHACO and GFP images, respectively, of FV-HEL cells cocultured with Jurkat-5Lg cells. egfp, enhanced GFP.
A possible explanation for the poor LTR promoter expression in H92–5Lg and Jurkat-5Lg cells is that only small fractions of these cells were infected with SFVcpz(hu). However, single-cell cloning of infected H92 cells and subsequent proviral detection by PCR indicated that over 70% of the H92 cells were infected using this method (data not shown). Jurkat cells were also efficiently infected using coculturing. When an SFVcpz(hu) vector expressing GFP (33) was used, over 30% of Jurkat cells were infected using this method (data not shown). Thus, despite efficient infection by SFVcpz(hu), LTR promoter transcription is limited in these persistently infected cell types. However, when introduced into a permissive environment, LTR promoter transcription can proceed. From these experiments we cannot conclude whether the lack of LTR promoter transcription in H92 and Jurkat cells is due to the absence of a necessary factor or the presence of an LTR promoter-specific inhibitor.
PMA treatment enhances viral replication in persistently infected cells.
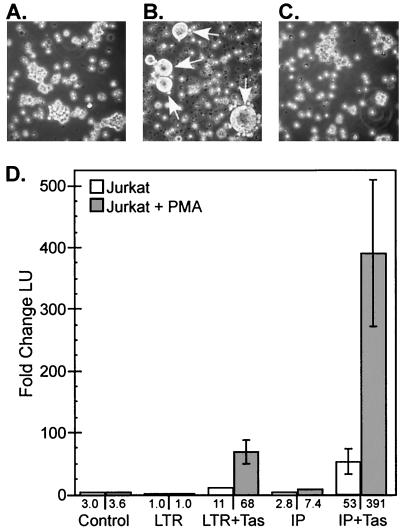
We have shown that in persistently infected cells, despite significant IP activity, there is an unexpected lack of LTR promoter activity. Based on these data, we hypothesized that factors which were necessary for efficient LTR promoter transactivation in persistently infected cells were missing. To examine this hypothesis, we treated persistently infected cells with a variety of cell activators in an effort to supply whatever factors might be necessary for efficient LTR promoter activity. We found that of the cell activators used, PMA had the most profound effect on viral titers (data not shown). Jurkat cells were significantly more responsive to PMA stimulation than the other cell types (Table 2). In Jurkat cells, PMA treatment resulted in syncytium formation and cell death, indicating a switch from persistent to lytic infection (Fig. 5A to C). The molecular clone of SFVcpz(hu) used in all of our experiments has a 646-bp deletion in U3 relative to the original HFV isolate or SFVcpz (17, 45). PMA treatment of Jurkat cells infected with SFVcpz resulted in increases in titers similar to those seen with SFVcpz(hu) (data not shown), indicating that the deleted region of the SFVcpz(hu) LTR promoter has no effect on PMA induction in Jurkat cells.
TABLE 2.
Induction of SFVcpz(hu) in FV-infected cells
| Cells | Fold inductiona of:
|
|
|---|---|---|
| SFVcpz(hu) | SFVcpz(hu)-ΔAP1 | |
| BHK-21 | 1.3 ± 0.2 | ND |
| H92 | 8.5 ± 2.5 | ND |
| Raji | 12.3 ± 4.8 | ND |
| U937 | 32.5 ± 11 | ND |
| Jurkat | 423 ± 77 | 205 ± 3.8 |
Cells persistently infected with SFVcpz(hu) or SFVcpz(hu)-ΔAP1 were treated with 50 μg of PMA per ml for 48 h, and their titers were compared to those of untreated cells. Values shown are the fold induction in titer for PMA-treated versus untreated cells and are reported as the mean and standard deviation. ND, not done.
FIG. 5.
PMA induction of SFVcpz(hu) in Jurkat cells. (A) Light microscopy of untreated SFVcpz(hu)-infected Jurkat cells. (B) SFVcpz(hu)-infected Jurkat cells treated with 50 nM PMA. Arrows indicate multinucleated syncytia. (C) Uninfected Jurkat cells treated with 50 nM PMA. (D) PMA induction of the SFVcpz(hu) LTR promoter and IP in Jurkat cells, as measured by transient transfection. The same constructs as those shown in Fig. 2A were used. Values shown below the white columns represent the fold change in luciferase units (LU) relative to the value for the LTR promoter with no PMA stimulation; 1 LU = 131 raw LU (RLU). Values shown below the grey columns represent the fold change in LU relative to the value for the LTR promoter with PMA stimulation; 1 LU = 102 RLU. All values are based on at least three independent experiments and are reported as the mean and standard error of the mean.
Luciferase reporter assays were performed to directly examine whether PMA stimulation affected the LTR promoter and/or the IP and to determine if Tas was required for PMA-mediated transactivation. Jurkat cells were transfected with the same constructs as those used in the experiment shown in Fig. 2, and a duplicate set of transfections was treated with 50 nM PMA. In the absence of Tas, PMA was unable to transactivate the LTR promoter (Fig. 5D, LTR). However, PMA treatment increased the basal activity of the IP in the absence of Tas (Fig. 5D, IP). PMA treatment of cells transfected with IP-luc or LTR-luc and CMV-tas showed that the IP was stimulated 6.2-fold and the LTR promoter was stimulated 7.4-fold (Fig. 5D, IP+Tas and LTR+Tas, respectively). These results indicate that the IP is slightly stimulated by PMA in the absence of Tas but that both the IP and the LTR promoter are more effectively stimulated by PMA in the presence of Tas.
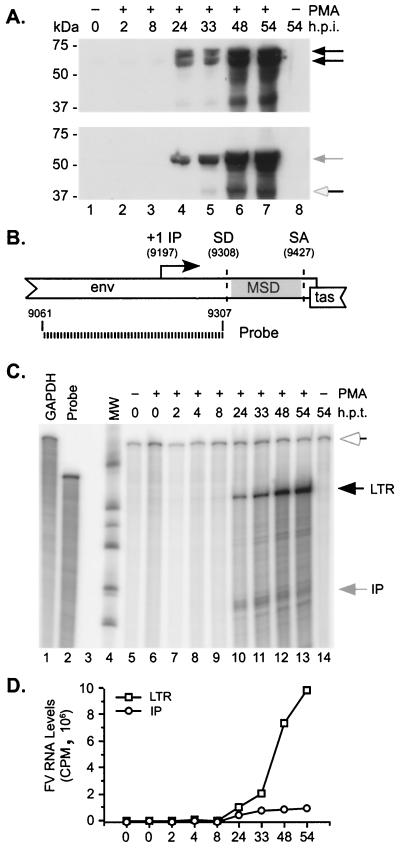
Western blotting was used to determine at what time after PMA treatment induction of the LTR promoter and the IP occurs. Persistently infected Jurkat cells were treated with 50 nM PMA, and cells were harvested at various times (Fig. 6). Western blotting was then performed using either anti-Gag antiserum to measure LTR promoter induction or anti-Tas antiserum to measure IP induction. Both the LTR promoter and the IP were induced between 8 and 24 h posttreatment (Fig. 6A). Recent experiments have shown that by 12 h after PMA treatment, Gag protein is readily detectable (data not shown). RPA analysis was used to determine if the induction of transcription from the LTR promoter and the IP was similar to that observed for protein expression. A single probe was designed to detect both LTR promoter- and IP-based transcripts (Fig. 6B). The 246-bp probe spans the region surrounding the IP transcription start site at position 9197 in the SFVcpz(hu) DNA genome. It ends immediately prior to the splice donor site used in the recently described env-bet transcripts (12, 24). Protected 246-bp transcripts derived from the LTR promoter include gag, pol, env, env-bet, and full-length genomic RNA. Protected 110-bp transcripts derived from the LTR promoter include tas, bet, and the putative bel2 transcript. Infected Jurkat cells were treated with 50 nM PMA, and RNA was harvested at various times (Fig. 6C). Both LTR promoter and IP transcripts were apparent at 24 h (Fig. 6C, black and grey arrows). IP transcripts appeared as three bands migrating near 110 bp, indicating heterogeneous start sites for the IP-driven mRNA, as previously reported (26) (Fig. 6C, grey arrow). Loading was monitored by the expression of GAPDH mRNA (Fig. 6C, white arrow).
FIG. 6.
Characterization of PMA induction in SFVcpz(hu)-infected Jurkat cells. (A and C) Naive or persistently infected Jurkat cells were treated with 50 ng of PMA/ml, and cells were harvested at the indicated times (h.p.i., hours postinfection; h.p.t., hours posttreatment. (A) Western blot analysis of protein expression using anti-Gag (top) or anti-Tas or -Bet (bottom) antisera. Black arrows, Gag; grey arrow, Bet; white arrow, Tas. (B) Schematic diagram showing the IP region and the RNase protection probe used to distinguish between LTR promoter and IP transcripts. (C) Unprotected probes are shown in lanes 1 and 2. Lanes 5 to 14 show RPA analysis at the indicated times after PMA treatment. White arrow, protected 546-bp GAPDH transcripts; black arrow, protected 246-bp LTR promoter transcripts; grey arrow, protected ∼110-bp IP transcripts. (D) Relative RNA levels determined by phosphorimaging quantitation of LTR promoter and IP transcripts in panel C, normalized to GAPDH expression. The x axis shows hours posttreatment.
Phosphorimaging analysis was performed to quantitate expression from the IP and the LTR promoter. Values were normalized to GAPDH expression and plotted. IP activity peaked at about 33 h posttreatment and declined thereafter, while LTR promoter activity peaked at about 48 h and declined by 54 h after PMA treatment (Fig. 6D). By 48 h after PMA treatment, cell viability had decreased to 32% that of untreated controls. These data indicate that PMA treatment results in an environment which supports Tas-mediated transcription from both the LTR promoter and the IP.
Consensus AP-1 binding sites are not required for LTR promoter transactivation or lytic viral replication.
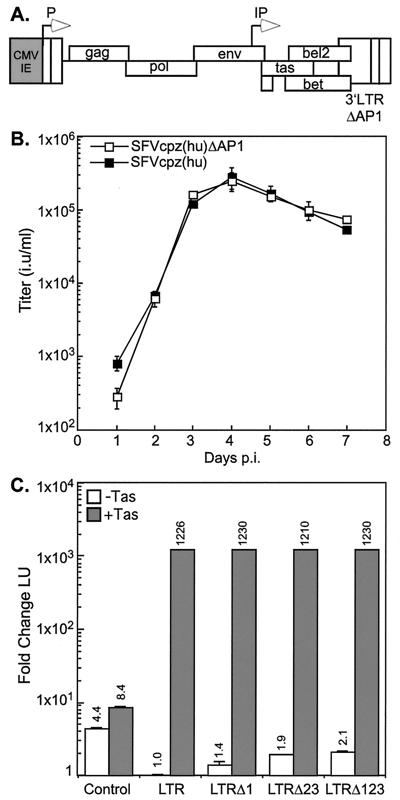
The presence of three consensus AP-1 binding sites in the SFVcpz(hu) LTR (Fig. 1B) suggests a possible role for AP-1 in modulating LTR promoter-based transcription. The three AP-1 binding sites found in the SFVcpz(hu) LTR promoter were previously shown by Maurer et al. to specifically bind recombinant c-Jun–v-Fos complexes (32). These authors also demonstrated that HeLa and BHK-21 cell extracts, both of which contain high levels of AP-1 family members (30, 54), were able to bind an LTR promoter fragment containing AP-1 binding sequences but not an LTR promoter fragment with the AP-1 binding sequences mutated. Given that cells such as BHK-21, which undergo lytic infection, express high levels of AP-1 (54) and that cells such as Jurkat express little or no AP-1 (21, 30), we were interested in the role of the three AP-1 binding sites in regulating lytic and persistent infections. Vector pC-SFVcpz(hu)-ΔAP1 is expressed from the CMV-IE promoter, which was used to replace the U3 region of the 5′ SFVcpz(hu) LTR promoter (Fig. 7A). The 3′ LTR promoter contains specific mutations in all three AP-1 binding sites without disturbing the overlapping bel2 ORF. The CMV-IE promoter directs the initial expression of the viral genome upon transfection, but after a single round of reverse transcription, the 3′ LTR promoter containing the mutated AP-1 sites is copied to the 5′ end of the provirus. Thus, subsequent rounds of viral expression are mediated by the mutated LTR promoter. The use of the CMV-IE promoter also obviates the possibility that recombination will result in viruses with unmutated LTR promoters.
FIG. 7.
Replication kinetics and LTR promoter transactivation of SFVcpz(hu)-ΔAP1 in BHK-21 cells. (A) pC-SFVcpz(hu)-ΔAP1 vector. The U3 region of the 5′ LTR promoter is replaced with the CMV-IE promoter (grey box). Three AP-1 binding sites were mutated in the 3′ LTR promoter. See Materials and Methods for details. (B) BHK-21 cells were infected at day 0 at an MOI of 0.1. Titers of SFVcpz(hu) and SFVcpz(hu)-ΔAP1 were measured at the indicated times postinfection (p.i.). Results are reported as the mean and standard deviation. (C) Transactivation of wild-type SFVcpz(hu) and mutated SFVcpz(hu) LTR promoters by Tas. Luciferase reporter constructs were constructed, and experiments were performed as described in the legend to Fig. 2. LTR, wild-type SFVcpz(hu) LTR promoter; LTRΔ1, SFVcpz(hu) LTR promoter with the first AP-1 binding site mutated; LTRΔ23, SFVcpz(hu) LTR promoter with the second and third AP-1 binding sites mutated; LTRΔ123, SFVcpz(hu) LTR promoter with all three AP-1 binding sites mutated.
We were interested in comparing the replication kinetics of SFVcpz(hu) and SFVcpz(hu)-ΔAP1 during lytic infection. For these experiments, virus stocks were generated by transfecting plasmids pC-SFVcpz(hu) and pC-SFVcpz(hu)-ΔAP1 into BHK-21 cells and harvesting cell-free supernatants when cells showed evidence of significant CPE. The titers of these stocks on FAB cells were then determined, and equivalent amounts of infectious virus were used to infect naive BHK-21 cells at an MOI of 0.1. Every day for 7 days, infected BHK-21 cells and supernatants were collected. Titration of SFVcpz(hu) and SFVcpz(hu)-ΔAP1 virus stocks on FAB cells indicated that there were no differences in the replication kinetics between the two viruses (Fig. 7B). We next examined whether mutating the AP-1 binding sites in the SFVcpz(hu) LTR promoter had any effect on Tas-mediated transactivation of the promoter. The wild-type LTR promoter as well as LTR constructs with AP-1 site 1, AP-1 sites 2 and 3, or all three AP-1 sites mutated were transactivated by Tas in BHK-21 cells equally well (Fig. 7C). These data are in accordance with those of Maurer et al. (32). These results, in addition to our data showing only minimal increases in viral titers when infected BHK-21 cells are treated with PMA, indicate that AP-1 family members are unlikely to mediate virus production in lytically infected cells.
AP-1 binding sites are not required for PMA-mediated induction of viral replication in persistently infected Jurkat cells.
In contrast to the situation in lytically infected cells, virus production in persistently infected Jurkat cells is greatly augmented upon treatment with PMA (Table 2) as well as after cross-linking with anti-CD3 and anti-CD28 monoclonal antibodies (data not shown). We speculated that the presence of the three AP-1 binding sites in the SFVcpz(hu) LTR promoter may be important for viral replication only in specific situations, such as T-cell activation. Many proteins bind AP-1 consensus sequences; these include family members such as c-Jun and c-Fos as well as AP-1-related proteins, such as CREB. In resting T cells, c-Jun is constitutively expressed, but the other AP-1 family members, such as c-Fos, are absent (19). Since phorbol ester treatment and T-cell activation by anti-CD3 and anti-CD28 monoclonal antibodies are known to increase the levels of AP-1 family members dramatically (19, 21), we wanted to determine whether AP-1 binding sites are necessary for the increased viral replication observed in such circumstances.
To address this question, BHK-21 cells were lytically infected with either SFVcpz(hu) or SFVcpz(hu)-ΔAP1. After extensive CPE had developed, equal numbers of uninfected Jurkat cells were cocultured with the lytically infected BHK-21 cells. At the time of coculturing, titers were 1 × 106 and 5.3 × 105 IU per ml for SFVcpz(hu) and SFVcpz(hu)-ΔAP1, respectively. After all of the BHK-21 cells had been lysed, the infected Jurkat cells were subcultured for 3 weeks and the LTR promoter region was sequenced to confirm the presence of the AP-1 mutations (data not shown). SFVcpz(hu)- and SFVcpz(hu)-Δ AP1-infected Jurkat cells were then treated with PMA, and viral titers were determined. Surprisingly, both SFVcpz(hu) and SFVcpz(hu)-ΔAP1 showed similar increases in titers upon PMA stimulation (Table 2). These data indicate that the presence of the three AP-1 binding sites is not necessary for PMA induction of the SFVcpz(hu) LTR promoter.
DISCUSSION
In the current study, we examined promoter activity in a number of persistently infected cell lines because persistent infections more closely approximate FV replication in vivo than do lytic infections of fibroblast-derived cell lines. Viral replication is significantly lower in these cells, particularly Jurkat T cells, than in lytically infected cells. We demonstrate here that LTR promoter transcription is more robust in lytically infected cells, while IP transcription is more robust in persistently infected cells. We show that SFVcpz(hu) replication and transcription in Jurkat cells can be enhanced greatly by PMA treatment. We also demonstrate that mutation of three consensus AP-1 binding sites in the viral LTR promoter has little effect on lytic, persistent, or PMA-enhanced replication. Taken together, our data suggest that the lower replication in persistently infected cells is due to a lack of an LTR promoter-specific transcription factor(s) in these cells.
Natural, experimental, and zoonotic FV infections are life-long, persistent infections without any accompanying pathologies. Although in vitro FV infection can lead to the accumulation of large numbers of integrated proviruses in individual cells (33), no FV-associated cancers have ever been reported. The lack of enhancer elements in the SFVcpz(hu) LTR promoter and the absence of basal LTR promoter transcription in persistently infected cells could account for this observation. One defining characteristic of in vivo FV infection is the lack of detectable viral replication despite the ability to recover virus by coculturing of infected tissues with susceptible cells in vitro (6, 7, 9, 11, 38, 41, 42, 53). The reason for this characteristic is unknown but is likely to be complex. Viral replication may be limited by aspects of the innate immune response, such as an interferon response, which is known to dramatically inhibit FV replication in vitro (10, 39, 40). Gamma interferon produced from phorbol ester-activated peripheral blood mononuclear cells strongly inhibits FV replication (10). Thus, it is interesting that PMA treatment of infected Jurkat cells dramatically induced viral replication (Table 2), despite the fact that such treatment also induced gamma interferon expression (14, 48). Furthermore, inhibition of gamma interferon in PMA-treated cells did not result in increased viral titers (data not shown). FV infection is also known to elicit a robust humoral response (1, 16, 34, 42, 46, 47), but its role in limiting viral replication is unknown. There are no published reports on the role of a cell-mediated immune response in limiting FV replication in vivo.
Apart from host-specific factors which could limit FV replication in vivo, several studies have suggested a number of mechanisms for achieving persistent infection in vitro. In most investigations, the Bet protein was thought to be a key player. One potential role for Bet in the regulation of viral infection was suggested by Bock et al., who found that overexpression of Bet could prevent infection by SFVcpz(hu) (5). Bet arises from a spliced message comprised of the first 88 amino acids of Tas and the entire bel2 ORF (35). Normally, the spliced bet message is derived from the IP (27), but at some frequency, LTR promoter transcripts are apparently spliced using the bet splice donor and acceptor sites, resulting in full-length SFVcpz(hu) genomes which can transcribe only bet and not tas (43). These defective genomes, termed SFVcpz(hu)Δtas, have been suggested to play a role in mediating persistent infection in cells which normally undergo cytolytic infection (41). Cells harboring SFVcpz(hu)Δtas genomes produce large quantities of Bet upon infection with wild-type virus, providing an interesting link between Bet expression and the maintenance of persistent infection. It has been suggested that SFVcpz(hu)Δtas acts as a defective interfering virus (22, 41, 43). The presence of large amounts of this form of FV in infected animals (9) and the accumulation of this viral form during experimental infection of rabbits (42) indicate that SFVcpz(hu)Δtas may play an important role in FV persistence in vivo. Deletion forms of FV are also abundant during persistent infection of Dami megakaryocytic cells (51). Interestingly, in that study, FV replication could be stimulated by treatment of cells with 5-iodo-2′-deoxyuridine, indicating a possible role for promoter methylation in persistent infection. We were unable to induce FV replication in infected Jurkat cells by treatment with either 5-azacytidine or trichostatin A (data not shown). This result indicates that at least in our system, promoter inactivation by methylation or histone acetylation is not a factor in persistent infection. Furthermore, in contrast to work with systems which support lytic replication, it was previously found that in infected H92 cells, there was no evidence for a role of SFVcpz(hu)Δtas in persistent infection (33, 53).
Our work suggests that differential regulation of the two FV promoters may help explain persistent infection in vitro. Transcription factors which act in concert with Tas to mediate LTR promoter and IP transcription have not been identified, but detailed analyses of the SFVcpz(hu) LTR promoter and IP have identified Tas binding sites and a number of Tas-responsive elements within these promoters (Fig. 1B and C) (8, 20, 23). The only putative transcription factor binding sites within the SFVcpz(hu) and SFVcpz LTR promoters are two Ets-1 sites and three perfect consensus AP-1 binding sites (Fig. 1B) (8, 31, 37, 45). DNase footprint analysis clearly showed that these three AP-1 sites were occupied when cell extracts from HeLa and BHK-21 cells were used, but mutation of these three sites showed that they are dispensable for Tas-mediated transactivation in cells which normally undergo lytic infection (23, 32). The authors also noted a small, two- to threefold increase in phorbol ester-mediated LTR promoter transactivation that was obviated by mutation of the AP-1 sites (32). We observed similar levels of phorbol ester stimulation in our lytically infected cells but much greater effects in cells which support persistent infection (Table 2). Interestingly, our data indicate that a virus lacking all three AP-1 sites replicates like the wild-type virus in BHK-21 cells and is induced by PMA in Jurkat cells. Although the AP-1 sites in the SFVcpz(hu) LTR promoter may not be important in PMA-mediated induction, additional transcription factors upregulated by PMA treatment may augment the increases in transcription that we observed in this study.
Experiments with protein synthesis inhibitors indicated that de novo protein synthesis is required for LTR promoter transcription following PMA treatment (data not shown). Further evidence that transcription factors other than AP-1 mediate PMA induction of the SFVcpz(hu) LTR promoter arises from the observation that the kinetics of Fos and Jun induction in PMA-treated Jurkat cells are not consistent with the kinetics of LTR promoter transcription. Fos and Jun are transcribed within 1 h after PMA treatment (21), but RPA analysis showed that LTR promoter transcription is not observed until 8 to 12 h posttreatment. RPA analysis also showed substantial PMA induction of the IP, although there are no consensus AP-1 sites near the IP. This result does not exclude the possibility that the AP-1 sites in the LTR promoter enhance transcription from the IP. Lochelt et al. demonstrated that the activity of the IP is greater when the SFVcpz(hu) LTR promoter is present in cis (25).
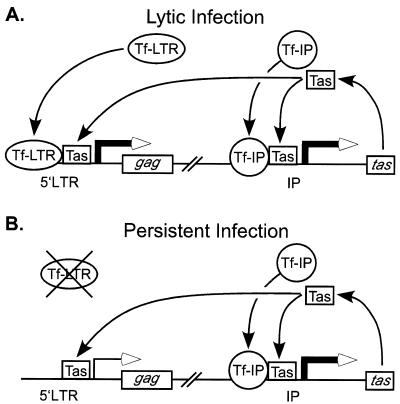
The idea that different transcription factors may mediate LTR promoter and IP transcription is directly supported by experiments with LTR promoter and IP expression constructs from SFVagm (36). In that study, Renne et al. (36) demonstrated through promoter competition experiments that the two promoters in SFVagm are regulated by different mechanisms. In Fig. 8 we propose a similar model to explain the differences in transcription in persistent and lytic infections. In this model, LTR promoter- and IP-specific transcription factors (Tf-LTR and Tf-IP, respectively) are abundant in lytically infected or PMA-treated cells, allowing for transcription from both promoters (Fig. 8A). In contrast, in persistently infected cells, only Tf-IP are present, while Tf-LTR are limiting (Fig. 8B). The missing factors required for LTR promoter transcription in persistently infected cells may be supplied when a cell is activated in vivo. T-cell receptor stimulation of SFVcpz(hu)-infected Jurkat cells with anti-CD3 and anti-CD28 monoclonal antibodies resulted in titer increases comparable to those observed with PMA treatment (data not shown). Because T cells are likely targets for FV infection (7, 50), activation of latently infected T cells provides an intriguing model for the maintenance of persistence in vivo. Activation of latently infected T cells may provide a small burst of viral replication, thereby permitting infection of adjacent resting T cells and further dissemination throughout the host. Such small, transient episodes of viral replication could account for the inability to detect viral replication in infected hosts.
FIG. 8.
Models of persistent and lytic SFVcpz(hu) infection in vitro. (A) Lytically infected or PMA-treated cells are characterized by basal IP transcription and Tas production. Newly synthesized Tas, in combination with unknown Tf-IP, drives the expression of additional Tas. LTR-promoter based transcription is present due to the availability of a distinct set of Tf-LTR. (B) Persistent infection. IP-based transcription mirrors that in panel A, but LTR promoter transcription is absent due to the unavailability of Tf-LTR.
In summary, we propose that distinct sets of transcription factors mediate the relative strengths of the two SFVcpz(hu) promoters in lytic and persistent infections. Our current work clearly shows that the relative level of Bet expression is higher in persistent infections than in lytic infections. Bet was the only protein detectable in infected Jurkat cells. Thus, while in lytic infection SFVcpz(hu)Δtas may be critical in skewing expression toward excess Bet, in persistent infection this can be accomplished by differential promoter regulation. We are currently interested in the identification of the sets of transcription factors which regulate LTR promoter and IP transcription.
ACKNOWLEDGMENTS
This investigation was supported by NIH grant R01 CA18282 to M.L.L. C.D.M. was supported by training grants T32 GM07270 and CA 80416.
We thank Michael Emerman for critical review of the manuscript. We also thank Ottmar Herchenroder for the molecular clone of SFVcpz and Martin Löchelt for the anti-Tas antiserum.
REFERENCES
- 1.Alke A, Schwantes A, Zemba M, Flugel R M, Lochelt M. Characterization of the humoral immune response and virus replication in cats experimentally infected with feline foamy virus. Virology. 2000;275:170–176. doi: 10.1006/viro.2000.0537. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1998. [Google Scholar]
- 3.Baldwin D N, Linial M L. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J Virol. 1999;73:6387–6393. doi: 10.1128/jvi.73.8.6387-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartz S R, Vodicka M A. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 5.Bock M, Heinkelein M, Lindemann D, Rethwilm A. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology. 1998;250:194–204. doi: 10.1006/viro.1998.9362. [DOI] [PubMed] [Google Scholar]
- 6.Brown P, Moreau-Dubois M C, Gajdusek D C. Persistent asymptomatic infection of the laboratory mouse by simian foamy virus type 6: a new model of retrovirus latency. Arch Virol. 1982;71:229–234. doi: 10.1007/BF01314874. [DOI] [PubMed] [Google Scholar]
- 7.Callahan M E, Switzer W M, Matthews A L, Roberts B D, Heneine W, Folks T M, Sandstrom P A. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J Virol. 1999;73:9619–9624. doi: 10.1128/jvi.73.11.9619-9624.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erlwein O, Rethwilm A. BEL-1 transactivator responsive sequences in the long terminal repeat of human foamy virus. Virology. 1993;196:256–268. doi: 10.1006/viro.1993.1474. [DOI] [PubMed] [Google Scholar]
- 9.Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenroder O, Spatz W, Volk B, Bohm N, Toniolo A, Neumann-Haefelin D, Schweizer M. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology. 1999;257:7–14. doi: 10.1006/viro.1999.9634. [DOI] [PubMed] [Google Scholar]
- 10.Falcone V, Schweizer M, Toniolo A, Neumann-Haefelin D, Meyerhans A. Gamma interferon is a major suppressive factor produced by activated human peripheral blood lymphocytes that is able to inhibit foamy virus-induced cytopathic effects. J Virol. 1999;73:1724–1728. doi: 10.1128/jvi.73.2.1724-1728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman M D, Dunnick N R, Barry D W, Parkman P D. Isolation of foamy virus from rhesus, African green and cynomolgus monkey leukocytes. J Med Primatol. 1975;4:287–295. doi: 10.1159/000459871. [DOI] [PubMed] [Google Scholar]
- 12.Giron M L, de The H, Saib A. An evolutionarily conserved splice generates a secreted Env-Bet fusion protein during human foamy virus infection. J Virol. 1998;72:4906–4910. doi: 10.1128/jvi.72.6.4906-4910.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould E A, Hartley J. Comparison of the antigens produced by foamy virus in a cytolytic and a persistent infection of HEp2 cells. J Gen Virol. 1979;44:235–239. doi: 10.1099/0022-1317-44-1-235. [DOI] [PubMed] [Google Scholar]
- 14.Hardy K J, Manger B, Newton M, Stobo J D. Molecular events involved in regulating human interferon-gamma gene expression during T cell activation. J Immunol. 1987;138:2353–2358. [PubMed] [Google Scholar]
- 15.He F, Blair W S, Fukushima J, Cullen B R. The human foamy virus Bel-1 transcription factor is a sequence-specific DNA binding protein. J Virol. 1996;70:3902–3908. doi: 10.1128/jvi.70.6.3902-3908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heneine W, Switzer W M, Sandstrom P, Brown J, Vedapuri S, Schable C A, Khan A S, Lerche N W, Schweizer M, Neumann-Haefelin D, Chapman L E, Folks T M. Identification of a human population infected with simian foamy viruses. Nat Med. 1998;4:403–407. doi: 10.1038/nm0498-403. [DOI] [PubMed] [Google Scholar]
- 17.Herchenroder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 18.Hooks J J, Detrick-Hooks B. Simian foamy virus-induced immunosuppression in rabbits. J Gen Virol. 1979;44:383–390. doi: 10.1099/0022-1317-44-2-383. [DOI] [PubMed] [Google Scholar]
- 19.Hughes C C, Pober J S. Costimulation of peripheral blood T cell activation by human endothelial cells. Enhanced IL-2 transcription correlates with increased c-fos synthesis and increased Fos content of AP-1. J Immunol. 1993;150:3148–3160. [PubMed] [Google Scholar]
- 20.Kang Y, Blair W S, Cullen B R. Identification and functional characterization of a high-affinity Bel-1 DNA binding site located in the human foamy virus internal promoter. J Virol. 1998;72:504–511. doi: 10.1128/jvi.72.1.504-511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvanta A, Kontny E, Jondal M, Okret S, Fredholm B B. Mitogen stimulation of T-cells increases c-Fos and c-Jun protein levels, AP-1 binding and AP-1 transcriptional activity. Cell Signal. 1992;4:275–286. doi: 10.1016/0898-6568(92)90067-i. [DOI] [PubMed] [Google Scholar]
- 22.Lecellier C H, Saib A. Foamy viruses: between retroviruses and pararetroviruses. Virology. 2000;271:1–8. doi: 10.1006/viro.2000.0216. [DOI] [PubMed] [Google Scholar]
- 23.Lee K J, Lee A H, Sung Y C. Multiple positive and negative cis-acting elements that mediate transactivation by bel1 in the long terminal repeat of human foamy virus. J Virol. 1993;67:2317–2326. doi: 10.1128/jvi.67.4.2317-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemann D, Rethwilm A. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J Virol. 1998;72:4088–4094. doi: 10.1128/jvi.72.5.4088-4094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lochelt M, Aboud M, Flugel R M. Increase in the basal transcriptional activity of the human foamy virus internal promoter by the homologous long terminal repeat promoter in cis. Nucleic Acids Res. 1993;21:4226–4230. doi: 10.1093/nar/21.18.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lochelt M, Flugel R M, Aboud M. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lochelt M, Muranyi W, Flugel R M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lochelt M, Yu S F, Linial M L, Flugel R M. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology. 1995;206:601–610. doi: 10.1016/s0042-6822(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 29.Lochelt M, Zentgraf H, Flugel R M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991;184:43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- 30.Maroder M, Farina A R, Vacca A, Felli M P, Meco D, Screpanti I, Frati L, Gulino A. Cell-specific bifunctional role of Jun oncogene family members on glucocorticoid receptor-dependent transcription. Mol Endocrinol. 1993;7:570–584. doi: 10.1210/mend.7.4.8388998. [DOI] [PubMed] [Google Scholar]
- 31.Maurer B, Bannert H, Darai G, Flugel R M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer B, Serfling E, ter Meulen V, Rethwilm A. Transcription factor AP-1 modulates the activity of the human foamy virus long terminal repeat. J Virol. 1991;65:6353–6357. doi: 10.1128/jvi.65.11.6353-6357.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meiering C D, Comstock K E, Linial M L. Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells. J Virol. 2000;74:1718–1726. doi: 10.1128/jvi.74.4.1718-1726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meiering C D, Linial M L. Historical perspective of foamy virus epidemiology and infection. Clin Microbiol Rev. 2001;14:165–176. doi: 10.1128/CMR.14.1.165-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muranyi W, Flugel R M. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renne R, Fleps U, Luciw P A, Neumann-Haefelin D. Transactivation of the two promoters of SFV-3 by different mechanisms. Virology. 1996;221:362–367. doi: 10.1006/viro.1996.0387. [DOI] [PubMed] [Google Scholar]
- 37.Rethwilm A. Regulation of foamy virus gene expression. Curr Top Microbiol Immunol. 1995;193:1–24. doi: 10.1007/978-3-642-78929-8_1. [DOI] [PubMed] [Google Scholar]
- 38.Rethwilm A. Unexpected replication pathways of foamy viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:S248–S253. doi: 10.1097/00042560-199600001-00037. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes-Feuillette A, Lasneret J, Paulien S, Ogunkolade W, Peries J, Canivet M. Effects of human recombinant alpha and gamma and of highly purified natural beta interferons on simian Spumavirinae prototype (simian foamy virus 1) multiplication in human cells. Res Virol. 1990;141:31–43. doi: 10.1016/0923-2516(90)90054-m. [DOI] [PubMed] [Google Scholar]
- 40.Sabile A, Rhodes-Feuillette A, Jaoui F Z, Tobaly-Tapiero J, Giron M L, Lasneret J, Peries J, Canivet M. In vitro studies on interferon-inducing capacity and sensitivity to IFN of human foamy virus. Res Virol. 1996;147:29–37. doi: 10.1016/0923-2516(96)80237-8. [DOI] [PubMed] [Google Scholar]
- 41.Saib A, Koken M H, van der Spek P, Peries J, de The H. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J Virol. 1995;69:5261–5268. doi: 10.1128/jvi.69.9.5261-5268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saib A, Neves M, Giron M L, Guillemin M C, Valla J, Peries J, Canivet M. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology. 1997;228:263–268. doi: 10.1006/viro.1996.8383. [DOI] [PubMed] [Google Scholar]
- 43.Saib A, Peries J, de The H. A defective human foamy provirus generated by pregenome splicing. EMBO J. 1993;12:4439–4444. doi: 10.1002/j.1460-2075.1993.tb06129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santillana-Hayat M, Rozain F, Bittoun P, Chopin-Robert C, Lasneret J, Peries J, Canivet M. Transient immunosuppressive effect induced in rabbits and mice by the human spumaretrovirus prototype HFV (human foamy virus) Res Virol. 1993;144:389–396. doi: 10.1016/s0923-2516(06)80054-3. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt M, Herchenroder O, Heeney J, Rethwilm A. Long terminal repeat U3 length polymorphism of human foamy virus. Virology. 1997;230:167–178. doi: 10.1006/viro.1997.8463. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt M, Niewiesk S, Heeney J, Aguzzi A, Rethwilm A. Mouse model to study the replication of primate foamy viruses. J Gen Virol. 1997;78:1929–1933. doi: 10.1099/0022-1317-78-8-1929. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer M, Falcone V, Gange J, Turek R, Neumann-Haefelin D. Simian foamy virus isolated from an accidentally infected human individual. J Virol. 1997;71:4821–4824. doi: 10.1128/jvi.71.6.4821-4824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw J, Meerovitch K, Elliott J F, Bleackley R C, Paetkau V. Induction, suppression and superinduction of lymphokine mRNA in T lymphocytes. Mol Immunol. 1987;24:409–419. doi: 10.1016/0161-5890(87)90014-9. [DOI] [PubMed] [Google Scholar]
- 49.Swack N S, Hsiung G D. Pathogenesis of simian foamy virus infection in natural and experimental hosts. Infect Immun. 1975;12:470–474. doi: 10.1128/iai.12.3.470-474.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Laer D, Neumann-Haefelin D, Heeney J L, Schweizer M. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology. 1996;221:240–244. doi: 10.1006/viro.1996.0371. [DOI] [PubMed] [Google Scholar]
- 51.Wybier-Franqui J, Tobaly-Tapiero J, Coronel A, Giron M L, Chopin-Robert C, Peries J, Emanoil-Ravier R. Human foamy virus DNA forms and expression in persistently infected Dami megakaryocytic cells. AIDS Res Hum Retrovir. 1995;11:829–836. doi: 10.1089/aid.1995.11.829. [DOI] [PubMed] [Google Scholar]
- 52.Yu S F, Linial M L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6624. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu S F, Stone J, Linial M L. Productive persistent infection of hematopoietic cells by human foamy virus. J Virol. 1996;70:1250–1254. doi: 10.1128/jvi.70.2.1250-1254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yusta B, Somwar R, Wang F, Munroe D, Grinstein S, Klip A, Drucker D J. Identification of glucagon-like peptide-2 (GLP-2)-activated signaling pathways in baby hamster kidney fibroblasts expressing the rat GLP-2 receptor. J Biol Chem. 1999;274:30459–30467. doi: 10.1074/jbc.274.43.30459. [DOI] [PubMed] [Google Scholar]