Abstract
Background
Soil bacteria often form close associations with their host plants, particularly within the root compartment, playing a significant role in plant growth and stress resilience. Vachellia tortilis subsp. raddiana, (V. tortilis subsp. raddiana)a leguminous tree, naturally thrives in the harsh, arid climate of the Guelmim region in southern Morocco. This study aims to explore the diversity and potential plant growth-promoting (PGP) activities of bacteria associated with this tree.
Results
A total of 152 bacterial isolates were obtained from the rhizosphere of V. tortilis subsp. raddiana. Rep-PCR fingerprinting revealed 25 distinct genomic groups, leading to the selection of 84 representative strains for further molecular identification via 16 S rRNA gene sequencing. Seventeen genera were identified, with Bacillus and Pseudomonas being predominant. Bacillus strains demonstrated significant tolerance to water stress (up to 30% PEG), while Pseudomonas strains showed high salinity tolerance (up to 14% NaCl). In vitro studies indicated variability in PGP activities among the strains, including mineral solubilization, biological nitrogen fixation, ACC deaminase activity, and production of auxin, siderophores, ammonia, lytic enzymes, and HCN. Three elite strains were selected for greenhouse inoculation trials with V. tortilis subsp. raddiana. Strain LMR725 notably enhanced various plant growth parameters compared to uninoculated control plants.
Conclusions
The findings underscore the potential of Bacillus and Pseudomonas strains as biofertilizers, with strain LMR725 showing particular promise in enhancing the growth of V. tortilis subsp. raddiana. This strain emerges as a strong candidate for biofertilizer formulation aimed at improving plant growth and resilience in arid environments.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00611-3.
Keywords: Vachellia tortilis subsp. raddiana, Rhizosphere, Rhizobacteria, PGP traits, Stress tolerance, Plant inoculation
Background
Among abiotic stresses, water scarcity and low intensity soil nutrients are the main characteristics of semiarid and arid environments, which are generally caused by irregular rains. Additionally, a significant component of the hydrological cycle is evaporation, which is constrained by lower groundwater infiltration and is affected by low rainfall [1]. Arid regions are highly prone to degradation, resulting in a decrease in critical soil physical properties, including soil structure, plant nutrient availability, organic matter content, and microbial activity. This degradation culminates in the destabilization of natural plant communities [2]. In response to these circumstances, leguminous plants have emerged as viable candidates for inclusion in ecological and sustainable programs designed to mitigate the deleterious effects of soil degradation and climate change.
In this respect, Vachellia species (formerly Acacia species) are considered pioneer plants in projects aimed at the rehabilitation, restoration, and revegetation of degraded areas [3]. This is due to their extended global distribution, their ability to fix nitrogen, and their ability to improve soil fertility. Additionally, their capacity to reduce evapotranspiration has an effect on the establishment, growth, and survival of other plants [4–6]. Vachellia trees also exert a notable influence on the biodiversity and soil ecology of arid ecosystems, modifying solar radiation and soil moisture dynamics [7]. Moreover, they are recognized for their ability to increase the availability of nutrients to both plants and animals in the soil [8, 9].
The new genus name Vachellia was proposed by Kyalangalilwa et al. [10] following deep molecular and phylogenetic studies. The genus encompasses 73 species in Africa, 60 species in the Americas, 36 species in South Asia, and 9 species in the tropical north of Australia, establishing its extensive presence, particularly in the sub-Saharan region [11]. It is considered a particular entity and a capital element in the pantropical distribution.
In Morocco, three Vachellia species grow naturally: V. gummifera, V. tortilis subsp. raddiana, and V. ehrenbergiana [11]. Vachellia tortilis subsp. raddiana is the most common tree in the Saharan areas of Morocco. It is crucial for maintaining the balance of arid and desert ecosystems and combating desertification in all desert regions of North Africa. The different parts of this tree are used in several fields, such as food, feed, and traditional medicine [12].
In addition to the adaptive morphological and physiological mechanisms displayed by this species [3], its symbiotic association with nitrogen-fixing rhizobia [13], arbuscular mycorrhizal fungi [14], and rich rhizospheric microflora found at the root level can improve the adaptation and growth of plants in arid areas. The rhizosphere is defined as the area surrounding plant roots, where a variety of beneficial microorganisms flourish close to the host plant roots [15]. Among all soil compartments, the rhizosphere has the highest concentration of microbial populations that are attracted by the abundant and diverse nutrients secreted by plants at the root level [16]. Some of these microbes, known as rhizobacteria, promote plant development by stimulating plant growth and controlling diseases. These beneficial bacteria, called plant growth-promoting rhizobacteria (PGPRs), increase plant development and soil quality by increasing nitrogen uptake [17, 18] and contribute to soil fertility and the availability of mineral nutrients, in addition to helping plants overcome biotic and abiotic challenges. In fact, beneficial bacteria are present in the bulk soil, rhizosphere (on the root surface), and endo-rhizosphere (within the root surface). All of these factors contribute to sustainable crop production through different mechanisms, such as biological nitrogen fixation, phosphorus solubilization, phytohormone synthesis (mainly auxin, cytokinin, and gibberellins), phytopathogen inhibition, siderophore production, and ACC deaminase activity [19, 20]. The most well-known PGPR strains belong to the genera Acinetobacter, Agrobacterium, Arthobacterium, Azospirillum, Bacillus, Bradyrhizobium, Burkholderia, Pseudomonas, Rhizobium, and Serratia [21].
Given the important role of rhizobacteria in the growth, protection, and adaptation of plants, it is of scientific and practical interest to study this type of bacteria in depth, especially those associated with native plants growing in infertile, degraded soils in arid and/or desert areas. These bacteria may constitute valuable bioresources for the formulation of bioinoculants for biofertilization, biostimulation, bioprotection, and/or adaptation of plants in areas impacted by the stresses linked to the current climatic change phenomenon. In this context, we isolated bacteria from the rhizosphere of the arid-tolerant tree V. tortilis subsp. raddiana, which thrives in the soils of the Guelmim region in southern Morocco. Following rep-PCR fingerprinting and identification through 16 S rRNA gene sequencing, representative strains were examined for in vitro tolerance to salt and drought stresses. Additionally, their plant growth-promoting (PGP) activities, including auxin and siderophore production, as well as phosphate, zinc, and potassium solubilization, were evaluated along with their lytic enzymatic activities. Elite strains exhibiting multiple PGP traits were further assessed in planta through inoculation into V. tortilis subsp. raddiana seedlings under controlled conditions. Plant growth parameter measurements have made it possible to identify promising strains that could be used in the formulation of biofertilizers for this important desert tree species.
Methods
Isolation of bacteria from rhizosphere soil
Soil samples were taken from the soil surrounding the roots of six distant V. tortilis subsp. raddiana trees growing near the locality of Taidalt (a village located near Guelmim in the extreme southwestern part of the Anti-Atlas Mountains in Morocco). Details about the climatic characteristics and predominant vegetation of the sampling area were reported previously [22]. The soil samples were returned to the laboratory and distributed in previously disinfected plastic pots. To isolate rhizospheric bacteria, V.tortilis raddiana seeds were subjected to scarification with 98% sulfuric acid for 2 h, as described by Sakrouhi et al. [23] and subsequently, the seeds were thoroughly washed with sterile water, transferred to Petri dishes containing 0.7% (w/v) agar, and allowed to germinate in darkness at 28 °C for three to five days. Four uniform seedlings were aseptically placed directly into each pot containing the soil samples. After six months of growth under natural conditions, the plants were meticulously uprooted. The roots were vigorously shaken to remove excess soil, and the soil adhering to the roots was identified as rhizosphere soil following the methodology outlined by Fu et al. [24].
Rhizobacterial isolates were obtained via the serial dilution plate method via NB medium (nutrient broth). One gram of rhizosphere soil was suspended in 10 mL of sterilized saline solution and serially diluted to 10−−4, after which 100 µL of the suspension was inoculated into NB plates. After incubation at 28 ± 2 °C for 3 to 4 days, bacterial colonies exhibiting variations in size, shape, and color were isolated and subjected to a purification process through repeated streaking to ensure the purity of the isolates. Following purification, the isolates were preserved in a 50% glycerol solution at -86 °C.
Diversity and molecular identification of rhizospheric isolates
DNA isolation
The bacterial strains were incubated for 4 days at 28 °C in tryptone yeast extract (TY) medium. Genomic DNA extraction followed the protocol outlined by Chen and Kuo [25]. The concentration and purity of the DNA samples were subsequently assessed via a Nanodrop™ spectrophotometer. The DNA samples were then stored at -20 °C until further use.
Rep-PCR fingerprinting
Genetic diversity analysis of the strains was performed via repetitive extragenic palindromic polymerase chain reaction (Rep-PCR) fingerprinting with the REP1 R-1 and REP2-I primers, in accordance with the methodology described by de Bruijn [26]. The obtained data were analyzed through the unweighted pair group with arithmetic mean (UPGMA) clustering method via GelCompar II software (version 2.5, Applied Maths, Belgium).
Amplification and sequencing of the 16S rRNA gene
Total DNA extraction was performed on one representative strain from each REP cluster via a previously described method. The 16 S rRNA gene was subsequently amplified via the fD1/rD1 universal primers [27] and the following PCR program: an initial denaturation step of 5 min at 95 °C, followed by 35 cycles of 1 min at 94 °C, 1 min and 30 s at 62 °C, and 10 min at 72 °C, with a final extension step of 10 min at 72 °C. The PCR products were purified and subjected to sequencing via GenoScreen (Lille, France).
Phylogenetic analyses
After quality checking via Chromas Lite (version 2.1), the sequences were aligned via Ugene (version 2.7) [28] and then subjected to MEGA X for phylogenetic analyses [29]. A phylogenetic tree with maximum likelihood phylogenetic analysis was inferred from distance calculations according to Kimura’s two-parameter model [30].
The accession numbers for the nucleotide sequences utilized in this study are depicted in the phylogenetic tree, and the bootstrap support for each node was assessed with 1000 replicates.
Physiological characterization
Plant growth-promoting traits
Auxin production
For the qualitative assessment of indole-3-acetic acid (IAA) production, YEM-agar plates [31] supplemented with tryptophan (0.5 g L− 1) were inoculated with 5 µL of bacterial culture and incubated at 28 °C for 3 days. Subsequently, Whatman filter paper soaked with Salkowski reagent was placed on the bacterial culture, and the plates were kept in the dark for 30 min. Bacteria producing IAA were identified by the emergence of pink halos around the colonies.
The quantitative determination of auxin production by each strain followed Salkowski’s method [32]. Bacteria were cultivated in YEM liquid medium containing 500 µg mL-1 tryptophan for 48 h at 28 °C in a shaking incubator at 180 rev min-1. After centrifugation for 15 min at 8000 rpm, the cultures were filtered through a 0.2 μm Millipore filter. The supernatants were transferred to new test tubes, mixed with Salkowski’s reagent, and stored at room temperature in the dark for 25 min. The optical density (OD) at 540 nm was measured, and the amount of auxin was calculated via an IAA standard curve [33].
Inorganic phosphate solubilization
The Pikovskaya method [34] was employed to assess the ability of the strains to solubilize inorganic phosphate. For each strain, 10 µL of a fresh bacterial culture was spotted on solid Pikovskaya medium (PVK) comprising glucose (C₆H₁₂O₆) 10 g; Ca₃(PO₄)₂ (tricalcium phosphate) 5 g; yeast extract 0.5 g; ammonium sulfate ((NH₄)₂SO₄) 0.5 g; potassium chloride (KCl) 0.2 g; sodium chloride (NaCl) 0.2 g; magnesium sulfate (MgSO₄) 0.1 g; trace amounts of ferrous sulfate (FeSO₄) and manganese sulfate (MnSO₄); agar 15 g; and distilled water 1 L. The pH value was adjusted to 7.0 ± 0.2 before sterilization. The inoculated plates were maintained at 28 °C for four days. The presence of halo zones around bacterial colonies was considered indicative of phosphate solubilization by strains utilizing tricalcium phosphate (TCP) as the sole phosphate source. The halo diameter to colony diameter ratio was calculated for each strain.
The amount of phosphate solubilized by the strains was determined following the vanadate‒molybdate colorimetric method [35]. Strains were first grown in 250 mL Erlenmeyer flasks containing modified liquid PVK media, where the P source was 0.05 g of rock phosphate instead of tricalcium phosphate. The initial OD600 of the bacterial cultures was set to 0.5. The cultures were incubated for 2 days at 28 °C in a rotary shaker at 180 rpm. The pH and OD600 of the medium were regularly determined, and a sample of each bacterial culture was centrifuged for 15 min at 8000 rpm. The OD400 of the supernatant was subsequently measured after the addition of the vanadate-molybdate mixture. The amount of solubilized phosphate was determined via a KH2PO4 standard curve.
Biological nitrogen fixation and nifH gene amplification
Bacteria were streaked on Burk’s nitrogen (N)-free agar [36]. The growth of bacteria on LB agar plates was used as a positive control. The plates were incubated at 30 °C for 7–10 days. As suggested by Revillas et al. [37], the colonies that appeared were subcultured twice in a row on new Burk’s N-free agar plates. Only bacteria that formed colonies on the third series of plates were considered free nitrogen fixers.
The ability of positive strains to fix nitrogen was confirmed by searching for the presence of the nifH gene (781 bp). The gene was amplified via PCR via two pairs of primers, namely, H1 and nifH2 [38], and nifHf/nifHi (TACGGNAARGGSGGNATC GGCAA/AGC ATG TCYYCSAGYTCNTCCA) [39]. The PCR products were subjected to 1% agarose gel electrophoresis in TBE buffer (0.089 M Tris, 0.089 M boric acid, and 0.002 M EDTA, pH 8).
Siderophore production
Siderophore production was evaluated through the chrome azurol sulfonate (CAS) assay, following the methodology outlined by Schwyn and Neilands [40]. This assay was performed according to the method described by Arora and Verma [41] and involved spot-inoculating bacterial strains onto CAS agar plates, followed by an incubation period at 30 °C for 5‒7 days. The negative controls included noninoculated plates. Bacteria exhibiting siderophore production were identified by the formation of a colored halo around their colonies, as indicated by Louden et al. [42].
Siderophore production was quantified for positive strains via a modified version of the CAS method. The protocol was modified to use 96-well microplate cultures. Modi medium was used to grow the bacteria in triplicate, and a control test consisting of noninoculated Modi medium was performed. After incubation at 28 °C for 7 days, the bacterial cultures were centrifuged at 10,000 rpm for 10 min, and 100 µL of the supernatant was mixed with an equal volume of CAS reagent. The OD630 was then measured via a Biotek elx 808 spectrophotometer for 30 min. The percent siderophore unit (psu) was calculated via the following formula [43]:
Siderophore production = (Ar − As) × 100/Ar.
where Ar = the absorbance of the reference (CAS solution and uninoculated Modi medium), and.
As = absorbance of the sample (CAS solution and cell-free supernatant of a bacterial culture).
Ammonium production
The method of Cappucino and Sherman was adopted for studying ammonium production by the isolates. Fresh bacterial cultures were inoculated in tubes containing 10 mL of peptone water. The negative control consisted of noninoculated medium tubes. After 48 h of incubation in a shaker (150 rpm) at 28 °C, ammonium production was revealed by adding 0.5 mL of Nessler reagent to each tube. The appearance of a brown to yellow color was considered a positive reaction.
Potassium solubilization
K solubilization ability was analyzed via Aleksandrov medium (pH 7.2 ± 0.2), which is composed of 5.0 g of glucose, 0.5 g of MgSO4, 0.005 g of FeCl3, 0.1 g of CaCO3, 2 g of Ca3(PO4)2, 2 g of K-bearing minerals, 15 g of agar, and 1 L of distilled water [44]. An appropriate amount of phenol red was added to the medium before autoclaving. Control plates containing Aleksandrov medium without dye solution were also included. The medium was poured into Petri plates, which were then spot inoculated with fresh bacterial cultures. After incubation for 72 h, the size of the halo zones and the diameter of the colonies were measured. The halo zone size was calculated by subtracting the colony diameter from the total diameter [45].
Zinc solubilization
The ability of the selected strains to solubilize insoluble Zn compounds was tested. Tris-mineral agar medium was used [46], or 0.1% (w/v) insoluble zinc was added in the form of zinc sulfate (ZnSO4) [47]. The pH was adjusted to 7.00 ± 0.25 before autoclaving [48]. Fresh bacterial cultures were spot inoculated on each medium and incubated at 30 °C for 10 days. The appearance of halo areas around bacterial colonies indicated mineral zinc solubilization.
ACC-deaminase activity
The 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase activity of the isolates was studied via the method described by Palaniyandi et al. [49]. Strains were grown in the minimal medium reported by Dworkin and Foster [50], which was supplemented with 3 mmol L− 1 ACC. The same minimal medium supplemented with ammonium sulfate ((NH4)2SO4) was used as a positive control, whereas for the negative control, no nitrogen source was added. The experiment was conducted in 96-well microplates, with each strain tested in triplicate. Blank wells containing minimal medium without bacteria were also included on the outermost edges of the plate. After incubation at 28 °C for 7 days, the OD630 was measured to compare the growth of the isolates under the three different conditions. The strains that were able to utilize ACC as a nitrogen source (ACC deaminase positive) exhibited growth similar to that on (NH4)2SO4, whereas the strains that could not utilize ACC presented growth similar to that of the free nitrogen medium.
Stress tolerance
Salt and drought tolerance
The tests consisted of cultivating the bacteria in YEM medium containing 0–14% NaCl to simulate salt stress or 10%, 20%, or 30% PEG 6000 to mimic drought stress by creating different osmotic potentials as described by Michel and Kaufmann, and Sandhya et al. [51, 52]. The media were inoculated with 1% bacterial cultures grown overnight in YEM medium. The control tests consisted of assessing bacterial growth in the same media in the absence of stress. Three replicates for each isolate at each concentration were included. After 48 h of incubation at 28 °C in a shaking incubator (120 rpm), the OD630 was measured via a microplate reader to estimate bacterial growth under the different conditions tested.
Exopolysaccharide production
EPS production was evaluated via the method described by Paulo et al. [53]. Briefly, liquid cultures of bacteria with an OD600 of 0.8 were used to inoculate 5 mm diameter sterile filter paper discs deposited in Petri dishes containing mineral salt medium supplemented with 10% sucrose. The plate was incubated at 28 °C for 24 h, after which the presence of mucoid colonies around the discs indicated the presence of EPS-producing bacteria.
Biocontrol activities
Hydrogen cyanide (HCN) production
The bacterial isolates were streaked onto LB media supplemented with glycine at a concentration of 0.4% (w/v). An alkaline picric acid solution, prepared by dissolving 2% sodium carbonate in 0.5% picric acid, was applied to Whatman filter paper, which was placed on the surface of the agar. The Petri dishes were sealed with parafilm and incubated at 28 °C for 4‒5 days. The color change of the filter paper from yellow to red‒brown is an indicator of HCN production [54].
Extracellular enzyme activities
Fresh bacterial cultures were spot inoculated on LB agar plates supplemented with either 1% carboxymethyl cellulose (CMC) for cellulase production [55] or skim milk agar plates for protease production [56]. Both types of plates were incubated at 28 °C for 4 days. For cellulase activity, the corresponding plates were stained with 0.1% Congo red for 15 min and then incubated with 1 M NaCl for 15 min. The presence of yellowish zones surrounding colonies indicated positive results. In the case of protease activity, the appearance of a clear zone surrounding the colonies is an indication of positive protease activity.
Chitinolytic activity was assessed on chitin agar plates comprising 1 g of colloidal chitin, 0.1% K2HPO4, 0.05% MgSO47H2O, 1.5% agar, and 2% phenol red. The bacterial suspensions were aliquoted onto the plates, which were then incubated at 30 °C for 7 days. Positive chitinase activity was identified by the development of clear zones surrounding the spots where the inoculants were placed [57].
Effects of selected rhizosphere isolates on Vachellia growth
Seed germination, seedling inoculation, and growth conditions
V. tortilis subsp. raddiana seeds collected in the Guelmim region were pregerminated aseptically via the protocol described by Sakrouhi et al. [23]. Briefly, the seeds were scarified with 98% sulfuric acid for 2 h, washed several times with sterilized distilled water, and germinated in the dark in Petri dishes containing 0.7% agar (w/v). Germinated seeds were then planted in seedling tanks containing 45 alveoli previously filled with a mixture of autoclaved sand and vermiculite (2:1). The seedlings were inoculated with the selected bacterial inoculants (Strains LMR725 and LMR731 were selected for their ability to solubilize phosphate and produce siderophores. Additionally, they can amplify the nifH gene and produce hydrogen cyanide (HCN). In contrast, strain LMR777 was identified as the most proficient isolate for the production of auxin and ACC deaminase) at a rate of 109 cells per seedling. Bacteria were previously grown for 48 h in nutrient broth media until the exponential growth phase [58, 59]. Noninoculated seedlings were used as controls. For each treatment, 10 replicates were used. The plants were grown in a greenhouse under natural light conditions and watered once or twice a week with the mineral mixture described by Hoagland and Arnon [60].
Plant harvest and measured parameters
Following a 6-month growth period, the plants were meticulously uprooted, and their roots were gently washed with tap water to eliminate any residual sand or vermiculite particles. The following parameters were subsequently measured: root dry weight (RDW; g/plant), shoot dry weight (SDW; g/plant), average shoot length (SL; cm), and average root length (RL; cm).
Statistical analysis
The data obtained from the inoculation trials were analyzed using XLSTAT software (version 03.05.2014) to detect significant differences. An analysis of variance (ANOVA) followed by Tukey’s Honest Significant Difference (HSD) test was conducted to determine the variance, with a significance threshold of P ≤ 0.05 for the mean values.
Results
A total of 152 strains were isolated from the rhizosphere of the roots of V. tortilis subsp. raddiana; these strains were analyzed via rep-PCR fingerprinting to evaluate their genetic diversity and to group those that presented a high percentage of similarity. In the present study, a threshold of 80% was established, resulting in the identification of 84 distinct Rep-PCR patterns (marked in bold) among the isolates, which were divided into 25 genomic groups at 65% similarity. (Figure S1).
Phylogenetic analysis of the 16S rRNA gene
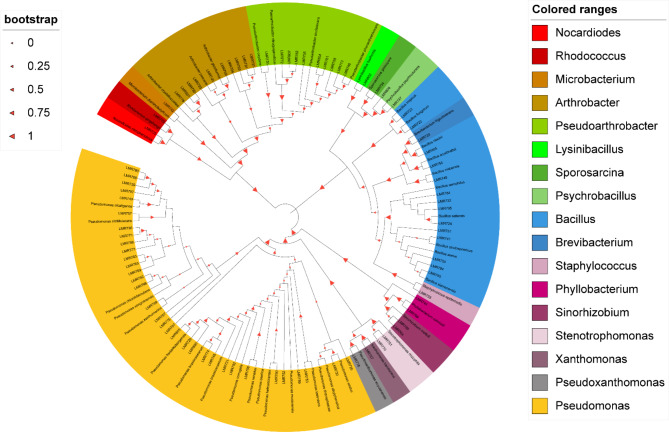
The 84 representative strains were identified via 16 S rRNA gene sequencing. The sequences are available in the GenBank database under the accession numbers shown in Table S1. Most of the sequences presented similarities greater than 98% with 16 S rRNA gene sequences found in the NCBI database, which allows good strain identification, especially at the genus level. The most abundant genus was Pseudomonas, with 33 strains, followed by Bacillus (14 strains), Arthrobacter and Pseudoarthrobacter (10 strains each). The genera Phyllobacterium, Sinorhizobium, Stenotrophomonas, and Psychrobacillus were represented by 2 strains each, whereas only one strain was identified for Rhodococcus, Microbacterium, Nocardiodes, Lysinibacillus, Sporosarcina, Brevibacterium, Staphylococcus, Xanthomonas and Pseudoxanthomonas.
A phylogenetic tree was constructed via the maximum likelihood function of Mega. This analysis involved nucleotide sequences of 84 strains, including reference strains and strains closely related to those sequenced in the present study (Fig. 1). The cladogram shows good individualization of the clades containing strains related to the Pseudomonas and Bacillus genera. The strains of the genus Arthrobacter were well grouped with A. crystallopoites, A. pascens and A. globiformis, which share the same gene pool and are closely related to each other. Similarly, 10 strains were closely related to each other and to Pseudoarthrobacter sp.
Fig. 1.
Maximum likelihood phylogeny of 16 S rRNA gene sequences of endophytic strains isolated from V. tortilis. subsp. raddiana rhizosphere. The analysis was based on 1461 nucleotides Isolates are denoted in bold. Bootstrap values are indicated as percentages derived from 1000 replications
Phytobeneficial traits and stress tolerance of rhizosphere strains
PGP traits
The 84 isolates representing the bacterial diversity in the rhizosphere of V. tortilis subsp. raddiana were tested for their in vitro plant growth-promoting activities (Table 1). Globally, half of the Pseudomonas strains produced auxin (3.24–58.97 µg mL− 1) and siderophores (21.39–77.63%), whereas inorganic phosphate with P values between 12.13 and 43.18 µg mL− 1 was detected in only 7 isolates. Only four isolates were effective potassium solubilizers, and just seven strains were fixed with atmospheric nitrogen.
Table 1.
Plant growth-promoting characteristics of representative rhizosphere isolates
| Strain code | NCBI species | AIA production (µg mL− 1) |
Phosphate production (µg mL− 1) |
Potassium solubilization (%) |
Zinc solubilization (ZnO) | Siderophores index (%) |
ACC deaminase | Ammonium production | NFB | NifH gene |
|---|---|---|---|---|---|---|---|---|---|---|
| LMR724 | Bacillus sp. | - | - | - | - | - | + | + | +++ | |
| LMR725 | Nocardioides sp. | - | 45.82 | - | - | 17.96 | - | - | - | +++ |
| LMR726 | Pseudomonas sp. | - | 17.21 | - | - | 21.39 | - | - | - | - |
| LMR727 | Xanthomonas sp. | - | - | - | - | - | - | - | + | |
| LMR729 | Pseudomonas sp. | - | 36.65 | - | + | 16.27 | - | + | - | - |
| LMR730 | Pseudomonas sp. | - | 43.18 | - | - | 57.27 | - | + | - | - |
| LMR731 | Pseudomonas sp. | - | 38.13 | - | + | 75.14 | - | + | - | +++ |
| LMR732 | Bacillus sp. | - | - | - | - | 32.12 | - | - | - | - |
| LMR734 | Sporosarcina sp. | - | - | 75 | - | 58.99 | - | - | + | - |
| LMR736 | Pseudomonas sp. | 5.27 | - | - | - | 53.72 | - | + | + | - |
| LMR739 | Pseudomonas sp. | 58.97 | - | - | - | - | +++ | - | - | - |
| LMR740 | Arthrobacter sp. | - | - | 50 | - | - | - | - | - | +++ |
| LMR742 | Microbacterium sp. | - | - | - | - | - | - | + | - | - |
| LMR743 | Bacillus sp. | - | - | - | - | - | - | - | - | - |
| LMR744 | Beijerinckia sp. | 58.77 | - | - | ++ | - | +++ | - | + | - |
| LMR745 | Pseudomonas sp. | 24.26 | - | - | + | 77.63 | +++ | ++ | - | +++ |
| LMR748 | Bacillus sp. | 3.69 | 9.75 | - | - | - | - | + | + | - |
| LMR750 | Bacillus sp. | - | - | - | - | - | - | + | + | - |
| LMR752 | Bacillus sp. | - | - | 66.66 | - | - | - | - | - | +++ |
| LMR754 | Pseudomonas sp. | 4.74 | 14.71 | - | - | - | - | ++ | - | - |
| LMR758 | Pseudomonas sp. | 9.58 | - | - | - | - | - | ++ | + | - |
| LMR762 | Pseudarthrobacter sp. | 15.11 | - | 66.66 | - | - | +++ | - | - | - |
| LMR764 | Pseudomonas sp. | 12.75 | - | 75 | - | - | - | ++ | + | - |
| LMR765 | Sinorhizobium sp. | 26.57 | - | 75 | - | - | - | - | - | - |
| LMR767 | Pseudomonas sp. | 40.27 | - | - | + | - | +++ | - | + | - |
| LMR768 | Pseudomonas sp. | 36.9 | - | - | - | - | +++ | - | + | - |
| LMR771 | Stenotrophomonas sp. | - | - | - | - | - | - | + | + | - |
| LMR775 | Pseudomonas sp. | - | 19.92 | 55.55 | - | 76.6 | - | - | - | - |
| LMR777 | Arthrobacter sp. | 101.04 | - | - | - | - | +++ | - | - | - |
| LMR778 | Pseudomonas sp. | 13.89 | - | - | - | 73.57 | - | + | + | - |
| LMR780 | Rhodococcus sp. | - | - | - | - | - | - | ++ | - | - |
| LMR782 | Stenotrophomonas sp. | - | 17.17 | 60 | - | - | - | - | - | - |
| LMR783 | Pseudomonas sp. | 9.22 | - | 69.23 | - | 76 | - | - | + | - |
| LMR787 | Sinorhizobium sp. | - | - | - | - | - | - | - | - | - |
| LMR789 | Pseudomonas sp. | - | 12.13 | - | - | - | - | - | - | - |
| LMR792 | Pseudarthrobacter sp. | 12.35 | - | - | - | 73.16 | - | - | - | - |
| LMR793 | Pseudomonas sp. | - | - | - | - | 50.04 | - | - | - | - |
| LMR796 | Bacillus sp. | - | - | - | - | 78.96 | - | ++ | + | - |
| LMR798 | Pseudomonas sp. | 5.31 | - | 75 | - | 51.11 | - | + | - | - |
| LMR806 | Bacillus sp. | - | - | 63.63 | - | - | - | - | - | - |
+++ Luxuriant growth/strong activity, ++ good growth/moderate activity, + poor growth/weak activity, − no growth/no activity
For potassium solubilization:
Solubilization efficiency (SE) % = (Z-C/C)*100
where
Z = diameter of solubilization zone
C = colony diameter
For the siderophore index:
The percent siderophore unit (psu) = (Ar − As) × 100/Ar
where
Ar = absorbance of reference (CAS solution and uninoculated Modi medium),
and As = absorbance of the sample (CAS solution and cell-free supernatant of a sample)
Importantly, the Bacillus strains presented low PGP activities, with only one strain exhibiting low auxin production and phosphate solubilization (Bacillus strain LMR748) and one strain solubilizing potassium and fixing molecular nitrogen (LMR752). For the latter activity, another strain (LMR724) also showed good growth in NFB media. In addition, two other strains exhibited good siderophore production. Among the other genera, only one Arhtrobacter strain (LMR777) produced auxin at a relatively high rate (101.04 µg mL− 1), while the other possessed both potassium solubilization and nitrogen fixation activities (LMR740). In addition, two strains belonging to the genus Pseudarthrobacter presented different PGP traits: LMR792 produced auxin and siderophores and fixed nitrogen, whereas LMR762 exhibited auxin production and potassium solubilization activities.
For the remaining genera, isolates possessed little or no activity at all. The most important strains displaying more than one activity were LMR725, LMR765, LMR782, and LMR734 (Nocardioides, Sinorhizobium, Stenotrophomonas, and Sporosarcina, respectively).
Concerning zinc solubilization, none of the strains tested could solubilize zinc sulfate or zinc carbonate, and only 5 strains in the collection solubilized zinc oxide (Pseudomonas LMR731 and LMR745, LMR729, LMR 767 and Beijerinckia LMR744). Moreover, the nifH gene was detected in only 6 isolates (Pseudomonas LMR731 and LMR745, Bacillus LMR724, Nocardioides LMR725, Arhtrobacter LMR740, and Bacillus LMR752). Among the whole tested collection, only 17 isolates presented the capacity to produce ammonium when cultivated in the corresponding medium.
Stress tolerance
Among the Pseudomonas strains, only three were unable to grow in the presence of 10% PEG (LMR788, LMR778, and LMR768), while 14 out of 33 strains were able to tolerate up to 30% PEG (Table 2). For bacteria with a maximum tolerated concentration (MTC) of 10% PEG, a minimum growth reduction of 30.25% was observed for strain LMR788, whereas the maximum growth reduction was 43.93% for strain LMR768. At an MTC of 20% PEG, the minimum growth reduction was 65.10% for strain LMR757, and the maximum growth reduction was 87.77% for strain LMR766. At an MTC of 30%, the minimum growth reduction was 54.81% for strain LMR739, and the maximum growth reduction was 87.47% for strain LMR801.
Table 2.
Stress tolerance of selected rhizosphere isolates
| Strains code | NCBI species | PEG MTC (%) |
% of reduction of growth at PEG MTC | NaCl MIC (%) | EPS production |
|---|---|---|---|---|---|
| LMR722 | Bacillus sp. | 10 | 62.71 | 5 | - |
| LMR723 | Bacillus sp. | 30 | 71.06 | 9 | - |
| LMR724 | Bacillus sp. | 30 | 58.23 | 14 | - |
| LMR725 | Nocardioides sp. | 10 | 47.42 | 9 | - |
| LMR726 | Pseudomonas sp. | 30 | 80.26 | 2 | - |
| LMR727 | Xanthomonas sp. | 30 | 86.45 | 14 | - |
| LMR728 | Brevibacterium sp. | 30 | 77.03 | 10 | - |
| LMR729 | Pseudomonas sp. | 30 | 74.39 | 10 | - |
| LMR730 | Pseudomonas sp. | 30 | 65.74 | 4 | - |
| LMR731 | Pseudomonas sp. | 30 | 58.93 | 10 | - |
| LMR732 | Bacillus sp. | 10 | 47.72 | 14 | - |
| LMR733 | Staphlycoccus sp. | 20 | 43.04 | 10 | - |
| LMR734 | Sporosarcina sp. | 30 | 72.44 | 10 | - |
| LMR735 | Pseudarthrobacter sp. | 30 | 75.03 | 10 | - |
| LMR736 | Pseudomonas sp. | 20 | 85.49 | 14 | - |
| LMR737 | Pseudarthrobacter sp. | 30 | 44.50 | 5 | - |
| LMR738 | Pseudarthrobacter sp. | 20 | 78.80 | 4 | - |
| LMR739 | Pseudomonas sp. | 30 | 54.81 | 10 | - |
| LMR740 | Arthrobacter sp. | 30 | 64.23 | 2 | - |
| LMR741 | Bacillus sp. | 20 | 61.31 | 14 | - |
| LMR742 | Microbacterium sp. | 30 | 15.31 | 5 | - |
| LMR743 | Bacillus sp. | 30 | 60.70 | 10 | - |
| LMR744 | Beijerinckia sp. | 20 | 85.17 | 5 | - |
| LMR745 | Pseudomonas sp. | 30 | 77.93 | 5 | - |
| LMR746 | Pseudarthrobacter sp. | 30 | 68.61 | 10 | - |
| LMR747 | Psychrobacillus sp. | 20 | 78.69 | 1 | - |
| LMR748 | Bacillus sp. | 10 | 51.15 | 10 | +++ |
| LMR749 | Pseudomonas sp. | 20 | 83.03 | 10 | +++ |
| LMR750 | Bacillus sp. | 30 | 84.01 | 14 | - |
| LMR751 | Bacillus sp. | 30 | 81.89 | 10 | - |
| LMR752 | Bacillus sp. | 20 | 75.15 | 14 | - |
| LMR753 | Pseudomonas sp. | 20 | 82.25 | 10 | +++ |
| LMR754 | Pseudomonas sp. | 30 | 72.84 | 4% | - |
| LMR755 | Bacillus sp. | 30 | 32.55 | 10 | - |
| LMR756 | Pseudarthrobacter sp. | 20 | 44.26 | 2 | - |
| LMR757 | Pseudomonas sp. | 20 | 65.10 | 10 | - |
| LMR758 | Pseudomonas sp. | 20 | 75.36 | 10 | - |
| LMR759 | Pseudarthrobacter sp. | 20 | 85.97 | 10 | - |
| LMR760 | Pseudomonas sp. | 30 | 77.36 | 4 | - |
| LMR761 | Pseudomonas sp. | 20 | 78.25 | 10 | - |
| LMR762 | Pseudarthrobacter sp. | 30 | 69.77 | 10 | - |
| LMR763 | Paenarthrobacter sp. | 30 | 65.54 | 10 | - |
| LMR764 | Pseudomonas sp. | 20 | 78.89 | 10 | - |
| LMR765 | Sinorhizobium sp. | 20 | 79.21 | 10 | - |
| LMR766 | Pseudomonas sp. | 20 | 87.77 | 10 | - |
| LMR767 | Pseudomonas sp. | 20 | 82.82 | 10 | - |
| LMR768 | Pseudomonas sp. | 10 | 43.93 | 10 | - |
| LMR769 | Phyllobacterium sp. | 30 | 73.76 | 10 | +++ |
| LMR770 | Arthrobacter sp. | 20 | 78.10 | 4 | - |
| LMR771 | Stenotrophomonas sp. | 30 | 74.30 | 2 | - |
| LMR772 | Pseudomonas sp. | 30 | 83.62 | 10 | - |
| LMR773 | Paenarthrobacter sp. | 30 | 93.28 | 10 | - |
| LMR774 | Pseudarthrobacter sp. | 30 | 83.58 | 5 | - |
| LMR775 | Pseudomonas sp. | 20 | 67.77 | 5 | - |
| LMR776 | Pseudomonas sp. | 30 | 77.93 | 10 | - |
| LMR777 | Arthrobacter sp. | 30 | 77.34 | 5 | - |
| LMR778 | Pseudomonas sp. | 10 | 34.78 | 1 | - |
| LMR779 | Pseudoxanthomonas sp. | 30 | 42.83 | 2 | +++ |
| LMR780 | Rhodococcus sp. | 20 | 45.38 | 2 | +++ |
| LMR781 | Pseudomonas sp. | 20 | 69.86 | 1 | - |
| LMR782 | Stenotrophomonas sp. | 20 | 78.79 | 2 | - |
| LMR783 | Pseudomonas sp. | 30 | 79.75 | 10 | - |
| LMR784 | Arthrobacter sp. | 30 | 59.84 | 5 | - |
| LMR785 | Bacillus sp. | 30 | 50.93 | 2 | - |
| LMR786 | Bacillus sp. | 20 | 59.97 | 10 | - |
| LMR787 | Sinorhizobium sp. | 20 | 29.87 | 2 | - |
| LMR788 | Pseudomonas sp. | 10 | 30.25 | 2 | +++ |
| LMR789 | Pseudomonas sp. | 10 | 73.25 | 5 | +++ |
| LMR790 | Pseudomonas sp. | 30 | 76.45 | 5 | - |
| LMR791 | Arthrobacter sp. | 20 | 77.13 | 5 | +++ |
| LMR792 | Pseudarthrobacter sp. | 20 | 44.26 | 2 | - |
| LMR793 | Pseudomonas sp. | 30 | 70.99 | 5 | - |
| LMR794 | Arthrobacter sp. | 20 | 75.38 | 2 | - |
| LMR795 | Bacillus sp. | 20 | 41.67 | 14 | - |
| LMR796 | Bacillus sp. | 30 | 50.69 | 12 | - |
| LMR797 | Pseudarthrobacter sp. | 20 | 57.86 | 1 | - |
| LMR798 | Pseudomonas sp. | 20 | 70.33 | 10 | +++ |
| LMR799 | Pseudomonas sp. | 20 | 83.53 | 2 | - |
| LMR800 | Pseudomonas sp. | 20 | 80.67 | 10 | - |
| LMR801 | Pseudomonas sp. | 30 | 87.47 | 10 | - |
| LMR802 | Arthrobacter sp. | 30 | 85.71 | 10 | - |
| LMR803 | Lysinibacillus sp. | 30 | 66.29 | 5 | - |
| LMR804 | Pseudomonas sp. | 30 | 82.67 | 10 | - |
| LMR805 | Pseudarthrobacter sp. | 20 | 83.58 | 10 | - |
| LMR806 | Bacillus sp. | 10 | 91.83 | 5 | - |
| LMR807 | Psychrobacillus sp. | 30 | 83.27 | 1 | - |
| LMR808 | Paenarthrobacter sp. | 10 | 29.30 | 5 | - |
MTC Maximum tolerated concentration
MIC Minimal Inhibiting Concentration
+++ Luxuriant growth/strong activity, ++ good growth/moderate activity, + poor growth/weak activity, − no growth/no activity
(Growth reduction = 1 - (ODstrain/ODControl) ×100)
ODstrain= OD600 at the MTCPEG
ODControl= OD600 at 0% PEG
When exposed to saline stress, the growth of Pseudomonas strains is generally reduced by concentrations of NaCl ranging from 2 to 14%. This stress condition also affected exopolysaccharide (EPS) production, as only five of the tested strains were capable of producing EPS.
Sixteen strains of Bacillus were evaluated for their ability to grow in the presence of 10%, 20%, or 30% PEG. Only 7 strains were able to grow in media containing 30% PEG, while the growth reduction of the other strains ranged from 32.55 to 91.83% depending on the PEG concentration. The Bacillus strains were found to be tolerant to high NaCl concentrations, with 6 isolates able to grow at a concentration of 14%. Only one Bacillus isolate produced exopolysaccharides (LMR748).
All the Pseudarthrobacter and Arthrobacter strains were able to grow in media containing 10% PEG, while growth reduction percentages ranging from 44.26 to 85.97% were observed at concentrations of 20% and 30% PEG. The results for other genera varied greatly, with strains very sensitive to osmotic stress, such as Beijerinckia LMR744, which presented 65.06% growth reduction at 10% PEG, and very tolerant strains, such as Microbacterium LMR742, which presented only 15.31% growth reduction at 30% PEG.
Regarding salinity tolerance, some strains of Paenarthrobacter and Sinorhizobium exhibited higher tolerance compared to others, although these strains did not produce exopolysaccharides. Conversely, Staphylococcus sp. strain LMR733 and Sporosarcina strain LMR734 were highly sensitive to elevated salt concentrations and also lacked the capacity to produce exopolysaccharides (Table 2).
Biocontrol traits
The results of the production of extracellular enzymes and hydrogen cyanide (HCN) by selected rhizosphere isolates are shown in Table 3. Two strains showed three activities: cellulase, chitinase, and HCN production by the Bacillus strain LMR724 and cellulase, protease, and HCN production by the Bacillus strain LMR796. Ten strains presented two different activities, whereas the remaining strains presented only one activity.
Table 3.
Extracellular enzymes and HCN production of selected rhizosphere isolates
| Strains code | NCBI species | Cellulase | Chitinase | Protease | Hydrogen cyanide |
|---|---|---|---|---|---|
| LMR724 | Bacillus sp. | +++ | + | - | ++ |
| LMR725 | Nocardioides sp. | - | - | - | +++ |
| LMR726 | Pseudomonas sp. | - | - | + | ++ |
| LMR727 | Xanthomonas sp. | +++ | - | - | ++ |
| LMR729 | Pseudomonas sp. | - | - | - | + |
| LMR730 | Pseudomonas sp. | - | - | - | - |
| LMR731 | Pseudomonas sp. | - | - | + | + |
| LMR732 | Bacillus sp. | +++ | - | - | - |
| LMR734 | Sporosarcina sp. | - | ++ | - | + |
| LMR736 | Pseudomonas sp. | - | - | - | + |
| LMR739 | Pseudomonas sp. | +++ | - | - | - |
| LMR740 | Arthrobacter sp. | - | - | + | - |
| LMR742 | Microbacterium sp. | - | - | - | - |
| LMR743 | Bacillus sp. | - | - | + | - |
| LMR744 | Beijerinckia sp. | - | - | - | - |
| LMR745 | Pseudomonas sp. | - | - | - | ₊ |
| LMR748 | Bacillus sp. | - | - | - | |
| LMR750 | Bacillus sp. | +++ | - | - | ++ |
| LMR752 | Bacillus sp. | - | - | - | + |
| LMR754 | Pseudomonas sp. | - | - | - | + |
| LMR758 | Pseudomonas sp. | - | - | - | +++ |
| LMR762 | Pseudarthrobacter sp. | - | - | - | + |
| LMR764 | Pseudomonas sp. | - | - | - | + |
| LMR765 | Sinorhizobium sp. | ++ | - | + | - |
| LMR767 | Pseudomonas sp. | - | - | - | + |
| LMR768 | Pseudomonas sp. | - | - | - | + |
| LMR771 | Stenotrophomonas sp. | - | - | + | ++ |
| LMR775 | Pseudomonas sp. | - | - | + | - |
| LMR777 | Arthrobacter sp. | - | - | - | - |
| LMR778 | Pseudomonas sp. | - | + | - | ++ |
| LMR780 | Rhodococcus sp. | - | - | - | + |
| LMR782 | Stenotrophomonas sp. | - | - | - | + |
| LMR783 | Pseudomonas sp. | + | - | - | + |
| LMR787 | Sinorhizobium sp. | - | + | - | - |
| LMR789 | Pseudomonas sp. | - | - | - | - |
| LMR792 | Pseudarthrobacter sp. | - | - | - | - |
| LMR793 | Pseudomonas sp. | - | - | - | - |
| LMR796 | Bacillus sp. | +++ | - | ++ | + |
| LMR798 | Pseudomonas sp. | - | - | ++ | + |
+++ Luxuriant growth/strong activity, ++ good growth/moderate activity, + poor growth/weak activity, − no growth/no activity
Global analysis of bacterial beneficial activities
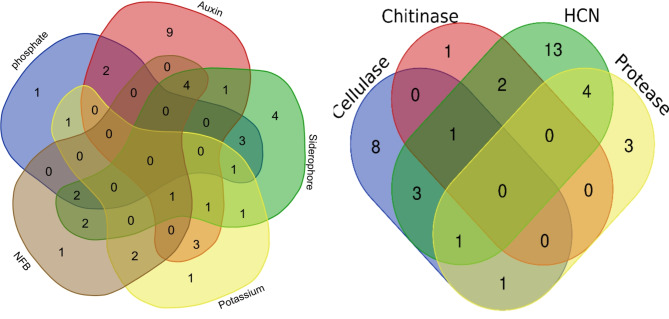
The analysis of the Venn diagram of PGP activities (Fig. 2a) revealed that many strains presented more than one activity. Notably, only the strain LMR798 exhibited 4 activities (auxin and siderophore production, nitrogen fixation, and potassium solubilization), whereas 8 strains presented 3 activities: LMR725 and LMR731 (phosphate solubilization, siderophore production, and nitrogen fixation); LMR745, LMR778, LMR792, and LMR772 (nitrogen fixation, AIA and siderophore production); LMR 783 (potassium solubilization, auxin and siderophore production); and LMR775 (phosphate and potassium solubilization, and siderophore production).
Fig. 2.
Venn diagrams showing the PGP activities (a), extracellular enzyme production and HCN (b) of a collection of 84 rhizospheric strains isolates obtained from V. tortilis. subsp. raddiana rhizosphere (http://bioinformatics.psb.ugent.be/webtools/Venn/)
The second Venn diagram, which grouped extracellular enzyme production and HCN activity (Fig. 2b), revealed that no strain possessed all the activities analyzed.
Effect of isolate inoculation on V. tortilis subsp. raddiana growth under greenhouse conditions
When the effects of the three bacterial strains were compared, LMR725 was found to be particularly effective in promoting the growth of V. tortilis subsp. raddiana. Inoculation with this strain resulted in aerial parts that were equivalent in length to those of the control plants but significantly greater than those of the two other strains tested. In terms of root length and dry shoot weight, Nocardioides strain LMR725 was also the best performing strain, with values significantly greater than those of Arthrobacter LMR731 but equivalent to those of Pseudomonas LMR777 and the control uninoculated plants. However, for root dry weight, no significant differences were detected among the inoculation treatments and the control (Table 4). Overall, inoculation with the LMR725 strain clearly improved the growth parameters of V. tortilis subsp. Raddiana.
Table 4.
Effects of inoculation with selected strains on the shoot and root dry weights of V. Tortilis subsp. raddiana plants grown under greenhouse conditions
| Treatment | Closest species | SL (cm) | RL (cm) | SDW (g) | RDW (g) | ||||
|---|---|---|---|---|---|---|---|---|---|
| LMR725 | Nocardioides sp. | 23.950a ± 4.15 | 15.875ab ± 1.58 | 1.088ab ± 0.23 | 0.489a ± 0.12 | ||||
| LMR731 | Pseudomonas sp. | 17.857bc ± 2.97 | 12.000c ± 2.51 | 0.818bc ± 0.27 | 0.429a ± 0.20 | ||||
| LMR777 | Arthrobacter sp. | 16.911c ± 2.24 | 16.144a ± 2.21 | 0.587c ± 0.23 | 0.356a ± 0.18 | ||||
| Control | - | 20.875abc ± 1.88 | 13.525abc ± 1.63 | 0.577c ± 0.17 | 0.347a ± 0.09 | ||||
The values are the means of six independent replicates (n = 6). Means followed by the same letter(s) within the same column are not significantly (p ≤ 0.05) different according to Tukey’s HSD
Control: Noninoculated plants
SDW: shoot dry weight (g/plant); RDW: root dry weight (g/plant).
SL: Shoot length (cm); RL: Root length (cm)
Discussion
Owing to their nature, rhizobacteria typically populate the rhizosphere or rhizoplane and, occasionally, the roots of growing plants. They are known as the most numerous and versatile of all microorganisms connected to higher plants [61]. It is well documented that a greater density of microbes are found in the rhizosphere than in the bulk soil. Moreover, the structure of soil microbial communities, their metabolic activities, and biogeochemical process rates have been reported to be substantially impacted by plant roots [62].
To date, few studies have investigated the microbiota of the rhizosphere of Vachellia (formerly the Acacia genus) species. In a previous study, Yuan et al. reported results concerning the roots and soil rhizosphere of five Acacia species (Acacia crassicarpa, Acacia cincinnata, Acacia melanoxylon, Acacia mangium, and Acacia mearnsil) [63]. The phylum Acidobacteria was the most abundant phylum in the rhizosphere, followed by Proteobacteria and Actinobacteria. On the other hand, Kamutando et al. reported that Alphaproteobacteria and Gammaproteobacteria (bacterial classes) and Pezizomycetes and Agaricomycetes (fungal classes) were enriched in the rhizosphere of Acacia dealbata compared with bulk soils [64]. In general, most of the research on the microbiota associated with V. tortilis subsp. raddiana has focused on nodule isolates [13, 23, 65–71]. In a recent study, we reported a comprehensive inventory of nonsymbiotic endophytic bacteria isolated from nodules of this tree species. The identified bacterial strains included six strains closely related to 6 different Bacillus species, nine strains associated with 4 Pseudomonas species, two strains related to 2 Microbacterium species, three strains linked to Klebsiella sp., and one strain related to each genus, namely, Stenotrophomonas sp. and Streptomyces sp [72].
In the present study, we isolated and studied the bacteria associated with the roots of V. tortilis subsp. raddiana growing in desert soils collected in the Guelmin region (southern Morocco). Genetic diversity was assessed via Rep-PCR, and representative isolates were identified via 16 S rRNA sequencing. The rhizobacterial community analyzed was diverse, and several genera previously reported in the rhizosphere of other Acacia species, such as Pseudomonas, Bacillus, and Stenotrophomonas, were also identified here [63, 64]. The 16 S rRNA gene is usually sequenced for strain identification; however, in many cases, resolution for identification at the species level is lacking, depending on the genus analyzed [73]. Sequencing of housekeeping genes such as gyrB, rpoB, atpD, and recA is a better and pertinent approach for species identification [74, 75], and more recently, whole-genome sequencing techniques have also been increasingly used for this purpose [76, 77]. For these reasons, the 32 putative species of rhizobacteria identified in our study are given as indicators and should be confirmed via the abovementioned techniques. These genera are affiliated with 17 genera dominated by Pseudomonas and Bacillus. Many studies have reported that Pseudomonas species are the most abundant Gram-negative bacteria present in rhizosphere soils [78, 79]. This genus is also frequently found in the rhizosphere and nodules of legumes [80]. Several Bacillus species have also been found to adapt well to the rhizoplane and rhizosphere of different plants. In particular, Bacillus subtilis and B. mycoides have been reported to be closely associated with the roots of established tea plants [81], while Bacillus cereus was the second most abundant species in the rhizosphere of Acacia plants [82]. This species is known to produce spores and is highly tolerant to environmental stresses [83], which is the case in the soils of the Guelmim region, from which strains were isolated. In addition to Pseudomonas and Bacillus, other predominant genera in the rhizosphere, such as Arthrobacter, Pseudoarthrobacter, Mycobacterium, Burkholderia, Stenotrophomonas and different members of Proteobacteria, Actinobacteria, and Bacteroidetes, were identified in other studies [84, 85]. The diversity of the bacterial community in the rhizosphere is known to be influenced by various factors, such as plant species, soil type, and environmental conditions. These bacteria are believed to play important roles in plant growth, disease suppression, and nutrient acquisition [86]; thus, manipulating this bacterial community is a potential way to improve plant growth and health [87]. Indeed, more studies on the role of these bacteria in the rhizosphere of V. tortilis subsp. raddiana and their potential for biotechnological applications are needed. Future studies should also consider the influence of abiotic factors such as soil type, pH, and moisture on the structure of the bacterial community in the rhizosphere of V. tortilis subsp. Raddiana.
PGP traits of V. tortilis subsp. raddiana rhizospheric bacteria
All the bacteria recovered from the rhizosphere of V. tortilis subsp. raddiana were tested for their main PGP activities, auxin and siderophore production, phosphate solubilization, ACC deaminase activity, ammonium production, nitrogen fixation and nifH genes, and stress tolerance (salt and drought). The most interesting isolates were subjected to another round of screening concerning zinc and potassium solubilization and the production of HCN, chitinase, and protease.
Auxin, siderophore, and ACC deaminase production
Among the rhizobacterial collection, the best producers of IAA were the strains Arthrobacter LMR777 and Beijerinckia LMR744. However, a greater number of strains producing IAA were recorded among those affiliated with the Pseudomonas genus, which is not surprising given the number of previous studies that reported this genus as an efficient IAA producer [78, 88]. In contrast, only one Bacillus strain exhibited positive IAA activity. The ability of microorganisms to produce and release auxins has long been known and is a common feature of different bacteria isolated from the rhizosphere [89]. This phytohormone is a key regulator and determinant of plant development and is involved in many different processes, such as cell differentiation, communication, and division [90]. IAA may also serve as a signal in bacterial‒bacterial communication (quorum sensing) and bacterial‒plant communication [91].
The production of siderophores is one of the major PGP activities among rhizobacteria. In our study, the strains Bacillus LMR796 and Pseudomonas LMR745 were the largest siderophore producers. Different bacterial genera, such as Pseudomonas, Aeromonas, Azotobacter, Bacillus, Rhizobium, Azadirachta, Streptomyces, Burkholderia, and Serratia, have been reported to produce siderophores [92–94]. Siderophores play a vital role in growth promotion, such as binding Fe to leghemoglobin in legume nodules and helping increase enzymatic activities and plant water and mineral uptake [95]. In addition, bacteria that produce siderophores can protect plants from pathogens through competition for iron in the rhizosphere [96].
The rhizobacterial isolates were also screened for ACC deaminase activity, which is known to contribute to the stress adaptation of plants. ACC deaminase cleaves 1-aminocyclopropane-1-carboxylate (ACC), a compound involved in the synthesis of the plant hormone ethylene, leading to a decrease in ethylene production and an increase in plant growth under stress conditions [97]. Some of the strains belonging to the genera Pseudomonas, Arthrobacter, Beijerinckia and Pseudarthrobacter were positive for ACC deaminase activity, which is in accordance with previous studies [16, 98, 99].
Minerals bioavailability
Phosphorus is the second most significant limiting nutrient for plants after nitrogen. This is due to its high rate of fixation and complexation with other soil elements, which makes P not readily available to plants [100]. By participating in the soil phosphorus cycle, PGPR are crucial actors in making soil-immobilized phosphorus accessible to plants [101]. Different genera are known as P solubilizers, such as Azotobacter, Bacillus, Beijerinckia, Burkholderia, Enterobacter, Erwinia, Flavobacterium, Microbacterium, Pseudomonas, Rhizobium and Serratia [102]. In our study, we identified several P-solubilizing strains belonging mainly to the genus Pseudomonas but also to Bacillus and Stenotrophomonas. However, the best P solubilizer in our collection was strain LMR 725, which has 98.5% similarity with Nocardioides nitrophenolicus (Table S1), a p-nitrophenol-degrading actinobacteria initially isolated from industrial wastewater in Korea [103]. To our knowledge, this is the first report on the isolation of strains belonging to this species from desert soils and their ability to solubilize rock phosphate.
Potassium is the third essential macronutrient for plants [104]. It is also present abundantly in insoluble rocks and minerals (approximately 90%), while the amount of soluble potassium in the soil is always low, and only a small fraction (1–2%) is available to plants [105]. Therefore, a system of continuous potassium replenishment in a soil solution is needed to ensure its adequate availability to crop plants [106]. Species of the genera Pseudomonas and Bacillus present potassium solubilization capacity [107–109]. In the present study, only 11 strains among the entire collection demonstrated significant potassium solubilization activity, and interestingly, over half of them belonged to the Bacillus and Pseudomonas genera.
Among soil minerals, zinc is an essential micronutrient involved in various metabolic activities, the stimulation of many enzymes and processes such as photosynthesis and respiration. Thus, it is an essential mineral for the development of plants, humans, and animals. In the rhizosphere, zinc-solubilizing bacteria can convert inorganic Zn to available forms for plant nutrition; thus, these bacteria are potential alternatives for Zn supplementation [110]. Moreno-Jiménez et al. [111] concluded that aridity negatively affects the availability of essential micronutrients, particularly iron and zinc, in dryland soils, posing threats to ecosystems and food production. This trait seems not to be widespread among the desert rhizobacteria isolated in our study, with only five strains related to Pseudomonas and Beijerinckia showing in vitro solubilization of zinc oxide, while no strain was able to solubilize 2 other Zn sources tested. Karnwal et al. also reported that Pseudomonas strains are zinc-solubilizing bacteria [112].
Biological nitrogen fixation is among the most important PGP traits of rhizobacterial strains. Here, we used two approaches to detect these strains: bacterial culture on free nitrogen-selective media and selective amplification of the nifH gene, which is involved in the biosynthesis of nitrogenase, the nitrogen-fixing enzyme. The first method identified 14 strains growing in the absence of a nitrogen source, whereas six isolates presented nifH gene amplification. The ability of rhizospheric bacteria to fix atmospheric nitrogen has been reported for different genera: Pseudomonas [113, 114], Bacillus [115, 116], and Arthrobacter [117, 118]. Notably, some strains that grew in NFB media did not exhibit amplification of the nifH gene. This apparent contradiction may be explained by the diversity of the nifH gene; the use of two pairs of primers is no longer sufficient to achieve satisfying and sharp results. Effectively, as shown by Gaby and Buckley [119], many supposedly universal primer sets miss significant portions of known nifH diversity. Many recent studies have demonstrated the potential of various strains of the genus Pseudomonas sensu stricto to fix nitrogen, including P. stutzeri A1501, P. stutzeri DSM4166, P. azotifigens 6HT33bT, and Pseudomonas sp. K1 [120–122]. Additionally, qRT‒PCR analysis demonstrated the expression of the Pseudomonas strains CY4 and CN11 nifH genes in sugarcane. As a result, the presence of the nifH gene indicates the presence of diazotrophs, and the expression of the gene suggests the presence of BNF [123].
More globally, our results demonstrate that some rhizobacterial strains exhibit many PGP activities at the same time, which is in accordance with previous studies that reported similar results for isolates from the genera Sinorhizobium, Stenotrophomonas, Nocardioides and Sporosarcina [124–127]. Moreover, Pseudomonas strains reportedly possess different PGP traits, such as phosphate solubilization, siderophore, IAA, HCN, antibiotic and ammonium production, nitrogen fixation, ACC deaminase activity, and biological control. These strains are used as biofertilizers that can increase crop yield through direct and indirect mechanisms [128]. In this context, a Pseudomonas putida strain has been reported as a PGPR that improves not only soil enzyme activities but also the growth of kasumbha [129]. Moreover, strains belonging to the Pseudomonas genus are considered efficient PGPR for many crops [130].
Salt and osmotic stress tolerance and exopolysaccharide production
Joshi et al. (2021) reported that some PGPR, including Pseudomonas, Enterobacter, Bacillus, Variovorax, Klebsiella, Burkholderia, Azospirillum, Serratia, and Azotobacter, support plant growth and development under normal and stressful circumstances [131]. Thus, certain PGPR strains can be used with agricultural plants to mitigate the stress caused by climate change, as they are effective at increasing stress tolerance, adaptation, and response mechanisms in plants. Salinity, drought, and low and high temperatures are the most common abiotic stressors in arid and desert areas, and according to Müller et al., drought affects 64% of the worldwide land area, whereas salinity affects only 6% [132].
In the present study, we tested representative rhizobacterial strains for their tolerance to salt by increasing the NaCl concentration in the medium and their tolerance to water stress induced by increased concentrations of PEG6000. The use of PEG to select bacteria that are tolerant to osmotic stress and the effects of these bacteria on the tolerance of plants to drought stress are well documented [133–135]. Among the dominant genera identified, all the Pseudomonas strains were able to grow in the presence of 10% PEG, and half of them were highly tolerant to osmotic stress (up to 30% PEG). The Pseudomonas strains were also highly tolerant to salinity (between 2% and 14% NaCl). Similar results have been reported for isolates belonging to the genus Pseudomonas isolated from maize [136] and rice [137]. It has been reported that significantly higher ROS (Reactive Oxygen Species) production in Pseudomonas putida exposed to water potentials less than 0.5 MPa is among the mechanisms implicated in such a high level of tolerance [138]. The second most abundant genus in the present study, Bacillus, was also highly tolerant to water stress, and 6 strains, up to 14% NaCl, tolerated high salinity stress. Exopolysaccharide production has also been reported as an important characteristic that can be implicated in stress tolerance. This high-molecular-weight biopolymer composed of monosaccharide residues is synthesized by a wide range of PGPR and plays a vital role in enhancing soil fertility and crop yield. In the present study, strains producing EPS belong to different genera, especially Pseudomonas, Bacillus, Arthrobacter, Phyllobacterium, Rhodococcus, and Pseudoxanthomonas. Previous studies reported that EPS production is widespread among PGPR such as Azotobacter vinelandii, Agrobacterium sp., Bacillus drentensis, Rhizobium leguminosarum, Enterobacter cloacae, and Xanthomonas sp [139]. However, in our study, not all the osmotolerant strains produced EPS, which indicates that for these strains, other mechanisms may be involved in osmotolerance.
The importance of osmotolerant plant growth-promoting rhizobacteria (PGPR) strains lies in their potential application for inoculation under water and/or saline stress conditions. In this regard [140], demonstrated notable improvements in chickpea-rhizobia symbiotic performance and plant growth under potted conditions with saline soil. This positive outcome was observed when plants were inoculated with various nonrhizobial endophytic bacteria isolated from chickpea nodules, including B. cereus NUU1, Achromobacter xylosoxidans NUU2, B. thuringiensis, NUU3 and B. subtilis NUU4.
Biocontrol activities of V. tortilis subsp. raddiana rhizospheric bacteria
The production of lytic enzymes, such as chitinases, lipases, proteases, cellulases, and β-1,3 glucanases, by beneficial microbes has been shown to inhibit the growth of pathogenic fungi, including Botrytis, Rhizoctonia, Sclerotium, Phytophthora, Pythium, and Fusarium [141]. Among the strains tested in our study, two belonging to the Bacillus genus (LMR796 and LMR724) presented three of the following four activities: cellulase, protease, chitinase, and HCN production. The strain LMR724 is very closely related to Bacillus safensis RGM2450, a strain that reportedly possesses genomic characteristics that can lead to the biosynthesis of secondary metabolites responsible for its antagonistic activity against phytopathogens, particularly competition for macronutrients, and permeabilization and suppression of membrane biosynthesis [142]. The LMR796 strain is closely related to the Bacillus xiamenensis strain PM14, a rhizosphere strain with in vitro biocontrol properties and the ability to activate red rot disease resistance in sugarcane [143].
Importantly, the production of extracellular enzymes and hydrogen cyanide (HCN) by bacteria in the rhizosphere can significantly affect the growth and development of plants. These substances can affect various plant processes, such as nutrient acquisition and defense against pathogens [144]. For example, cellulase and chitinase production can increase the decomposition of plant biomass and increase nutrient availability [145], whereas protease production can improve the solubilization of organic matter and stimulate plant growth [146]. The production of hydrogen cyanide (HCN) by rhizosphere bacteria can exert both positive and negative effects on plants. Low levels of HCN may stimulate plant growth and enhance defense mechanisms, while elevated levels can be toxic and inhibit growth. In our study, it was observed that the majority of HCN-producing strains are classified within the genera Pseudomonas and Bacillus (see Table 3). This distribution suggests that these genera might play a significant role in modulating plant responses through their HCN production [144].
To ensure the safe application of these bacterial strains in agricultural settings, it is essential to consider their potential for antibiotic resistance. Evaluating the antibiotic susceptibility of these strains is crucial for preventing the spread of antibiotic resistance genes. To this end, we plan to perform whole genome sequencing of the three bacterial strains selected for inoculation. This approach will enable a comprehensive analysis of their genomic content, with a particular focus on identifying and assessing antibiotic resistance genes. By taking these measures, we aim to minimize any associated risks and ensure the responsible use of these strains in biofertilizer development.
Effects of PGPR inoculation on V. tortilis subsp. raddiana growth under greenhouse conditions
Among the collection of rhizobacteria identified and characterized in our study, three strains were selected for in planta experimentation (Nocardioides LMR725, Arthrobacter LMR731, and Pseudomonas LMR777). Compared with noninoculated control plants, V. tortilis subsp. raddiana plants inoculated with the strain LMR725 under controlled conditions presented significantly increased plant growth parameters (88.56% shoot dry weight and 40.92% root dry weight). Strain LMR731 induced nonsignificant increases in both parameters (41.76% and 23.63%, respectively), whereas inoculation with LMR777 had no effect on the growth of the plants.
The higher performing strain LMR725 was closely related to Nocardioides nitrophenolicus. It belongs to a bacterial genus containing species commonly found in soil, water sediment, and sludge [103]. Ikunaga et al. isolated Nocardioides strains from the soil and leaves of wheat plants [147]. Other isolated strains of this genera are plant endophytes and have been shown to suppress crop pathogens [148]. However, our results are the first to show an important plant growth-promoting effect of a strain related to Nocardioides nitrophenolicus. This beneficial impact of phytobene could be related to the multiple PGP traits of this strain, as revealed by in vitro tests (phosphate solubilization, siderophore production, biological nitrogen fixation, salt and drought stress tolerance, and HCN production).
The present study is the first to thoroughly investigate the diversity, osmotolerance and beneficial characteristics of bacteria isolated from the rhizosphere of the desert leguminous tree V. tortilis subsp. raddiana. Our results suggest that this tree rhizosphere is a diverse and rich source of bacteria with the potential to promote plant growth. Among three elite selected strains that were inoculated into V. tortilis subsp. raddiana grown under controlled conditions, strain LMR725 demonstrated significant plant growth-promoting effects. These findings suggest that this strain has the potential to be used in the formulation of a biofertilizer for V. tortilis subsp. raddiana. The strain LMR725 was identified by 16 S rRNA sequencing as closely related to Nocardioides nitrophenolicus, a species known for its potential for degrading complex and toxic molecules. Therefore, further identification of this strain via MLSA approaches and/or whole-genome sequencing and further investigations of the mechanisms underlying the plant growth-promoting effects of this strain are needed. Furthermore, it will be necessary to evaluate the survival and saprophytic abilities of this strain under open field conditions and its impact on V. tortilis subsp. raddiana growth under the harsh natural conditions prevailing in desert regions, where abiotic stressors such as drought and high temperatures may limit plant growth. Further research is needed to substantiate their potential to enhance plant resilience to drought or salinity stress. This line of inquiry is a promising direction for future research. We plan to explore these aspects in subsequent studies, which will include RNA sequencing (RNA-seq) and metabolomic analyses to investigate gene expression and metabolic changes under stress conditions. These future studies will provide a more comprehensive understanding of how these bacterial strains may contribute to plant stress resilience.
The use of biofertilizers, especially microbial inoculants, is a promising sustainable approach for enhancing plant growth, improving soil health, and reducing reliance on synthetic fertilizers. These benefits are particularly relevant in the context of climate change, which dramatically affects many arid and desert ecosystems. In this context, V. tortilis subsp. raddiana and its associated microorganisms offer good opportunities for scientific investigation and development, as well as potential socioeconomic benefits for residents of these regions. However, given the complex characteristics of these microorganisms, it is crucial to conduct a comprehensive evaluation of their ecological and conservation implications. In this context, interdisciplinary collaboration and knowledge dissemination are critical for gaining a deep understanding of the potential applications of these microorganisms [149].
Conclusions
Rhizospheric bacteria associated with V. tortilis subsp. raddiana, a leguminous tree species thriving in desert and Saharan regions of Africa, were isolated and identified through 16 S rRNA sequence analysis. These isolates were characterized in vitro for salt and drought stress tolerance, plant growth-promoting (PGP) activities, and certain biocontrol activities. A subset of rhizospheric isolates exhibiting multiple PGP traits was subsequently assessed in planta through inoculation into V. tortilis subsp. raddiana seedlings. Following six months of growth under greenhouse conditions, LMR725, a strain closely related to Nocardioides nitrophenolicus, had positive effects on plant growth. Therefore, this particular strain has emerged as a promising candidate for the formulation of biofertilizers tailored for V. tortilis subsp. raddiana.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the Faculty of Sciences, Mohammed V University in Rabat, for their support and the Moroccan Ministry of Higher Education, Research and Innovation, which funded the PhD thesis of Mohamed Hnini.
Biographies
Mohamed Hnini
holds a PhD in Microbial Biotechnology, Genomics, and Bioinformatics from Mohammed V University of Rabat, Morocco. His work is centered on the genetic diversity and biological activities of microorganisms, including rhizobia, plant growth-promoting rhizobacteria (PGPR), and endophytic microbes. His research also includes salt and drought tolerance tests, biocontrol and antagonistic activity against phytopathogenic fungi, and plant inoculation experiments. Dr. M.H.‘s expertise spans microbiology, soil science, molecular biology, and bioinformatics, with skills in bacterial isolation, molecular identification, DNA extraction, PCR, microbial diversity.
Jamal Aurag
holds a Doctorat d’Etat and is a Full Professor at Mohammed V University in Rabat, Morocco. His research focuses on the diversity and functioning of rhizobia, plant growth-promoting rhizobacteria (PGPR), endophytes, and arbuscular mycorrhizal fungi (AMF), particularly their interactions with plants under stress conditions such as drought, salinity, and heavy metal exposure. Dr. J.A.‘s expertise spans various disciplines including microbiology, plant physiology, and environmental stress physiology. His skills encompass salt tolerance, soil microbiology, biofertilizers, and more., Dr. J.A. has significantly advanced the understanding of plant-microbe interactions in challenging environmental contexts.
Author contributions
Hnini Mohamed :• Data curation, handling and organizing research data.• Active involvement in the investigation process.• Methodological contributions to the study.• Validation of research findings.• Writing the original draft of the manuscript.• Participating in the review and editing of the manuscript. Aurag Jamal :• Conceptualization of the research.• Funding acquisition for the project.• Investigation as part of the research process.• Methodological contributions to the study.• Project administration responsibilities.• Access to necessary resources for the research.• Overall supervision of the project.
Funding
This research was supported by the Moroccan Ministry of Higher Education, Research and Innovation, which funded the PhD thesis of Mohamed Hnini.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable. This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Camacho Suarez VV, Saraiva Okello AML, Wenninger JW, Uhlenbrook S. Understanding runoff processes in a semi-arid environment through isotope and hydrochemical hydrograph separations. Hydrol Earth Syst Sci. 2015;19:4183–99. [Google Scholar]
- 2.Jeffries P, Barea JM. 4 Arbuscular Mycorrhiza: a key component of sustainable plant–soil ecosystems. In: Hock B, editor. Fungal associations. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. pp. 51–75. [Google Scholar]
- 3.Grouzis M, Floc’h E. Un arbre au désert: Acacia raddiana. 2003.
- 4.Abdallah F, Noumi Z, Touzard B, Belgacem AO, Neffati M, Chaieb M. The influence of Acacia tortilis (Forssk.) Subsp. Raddiana (Savi) and livestock grazing on grass species composition, yield and soil nutrients in arid environments of South Tunisia. Flora - Morphology Distribution Funct Ecol Plants. 2008;203:116–25. [Google Scholar]
- 5.Labidi S, Nasr H, Zouaghi M, Wallander H. Effects of compost addition on extra-radical growth of arbuscular mycorrhizal fungi in Acacia tortilis ssp. raddiana savanna in a pre-saharan area. Appl Soil Ecol. 2007;35:184–92. [Google Scholar]
- 6.Noumi Z, Chaieb M. Dynamics of Acacia tortilis (Forssk.) Hayne subsp. raddiana (Savi) Brenan in arid zones of Tunisia. Acta Bot Gallica. 2012;159:121–6. [Google Scholar]
- 7.Abdallah F, Noumi Z, Ouled-Belgacem A, Michalet R, Touzard B, Chaieb M. The influence of Acacia tortilis (Forssk.) ssp. raddiana (Savi) Brenan presence, grazing, and water availability along the growing season, on the understory herbaceous vegetation in southern Tunisia. J Arid Environ. 2012;76:105–14. [Google Scholar]
- 8.Gedda AE. Rangeland evaluation in relation to pastoralists perceptions in the middle Awash valley of Ethiopia. Thesis. University of the Free State; 2003.
- 9.Munzbergova Z, Ward D. Acacia trees as keystone species in Negev desert ecosystems. J Veg Sci. 2002;13:227–36. [Google Scholar]
- 10.Kyalangalilwa B, Boatwright JS, Daru BH, Maurin O, van der Bank M. Phylogenetic position and revised classification of Acacia s.l. (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia: Reclassification of African Acacia Species. Bot J Linn Soc. 2013;172:500–23. [Google Scholar]
- 11.Mhirit O, Et-Tobi M. Les écosystèmes forestiers face au changement climatique: Situation et perspectives d’adaptation au Maroc. 1999;:37.
- 12.Jaouadi W, Mechergui K, Ammari Y, Hamrouni L, Hanana M, Khouja ML. Étude Ethnobotanique et ethnopharmacologique d’Acacia tortilis (Forssk) Hayne subsp. raddiana (Savi) de la steppe arborée du Nord de l’Afrique. Phytothérapie. 2016;14:285–92. [Google Scholar]
- 13.Khbaya null. Neyra null, Normand null, Zerhari null, Filali-Maltouf null. Genetic diversity and phylogeny of rhizobia that nodulate acacia spp. in Morocco assessed by analysis of rRNA genes. Appl Environ Microbiol. 1998;64:4912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdedaiem R, Rejili M, Mahdhi M, de Lajudie P, Mars M. Phylogeny and distribution of arbuscular mycorrhizal fungi associated with Vachellia tortilis ssp. raddiana in relation to soil properties under arid ecosystems of Tunisia. Mycol Progress. 2020;19:495–504. [Google Scholar]
- 15.Bringhurst R, Cardon Z, Gage D, Bringhurst RM, Cardon ZG, Gage DJ. Galactosides in the rhizosphere: utilization by Sinorhizobium meliloti and development of a biosensor. Proc Natl Acad Sci USA 98: 4540–4545. Proceedings of the National Academy of Sciences. 2001;98. [DOI] [PMC free article] [PubMed]
- 16.Glick BR. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res. 2014;169:30–9. [DOI] [PubMed] [Google Scholar]
- 17.Glick BR. Plant growth-promoting Bacteria: mechanisms and applications. Scientifica. 2012;2012:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song M-H, Zheng L-L, Suding KN, Yin T-F, Yu F-H. Plasticity in nitrogen form uptake and preference in response to long-term nitrogen fertilization. Plant Soil. 2015;394:215–24. [Google Scholar]
- 19.Ahmad F, Ahmad I, Khan MS. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res. 2008;163:173–81. [DOI] [PubMed] [Google Scholar]
- 20.Cummings SP. The application of plant growth promoting rhizobacteria (PGPR) in low input and organic cultivation of graminaceous crops; potential and problems*. Environ Biotechnol. 2009;:43–50.
- 21.Goswami D, Thakker JN, Dhandhukia PC. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2016;2:1127500. [Google Scholar]
- 22.Msanda F, El Aboudi A, Peltier JP. Originalité De La Flore et de la végétation de l’anti-Atlas sud‐occidental (Maroc). Feddes Repert. 2002;113:603–15. [Google Scholar]
- 23.Sakrouhi I, Belfquih M, Sbabou L, Moulin P, Bena G, Filali-Maltouf A, et al. Recovery of symbiotic nitrogen fixing acacia rhizobia from Merzouga Desert sand dunes in South East Morocco – Identification of a probable new species of Ensifer adapted to stressed environments. Syst Appl Microbiol. 2016;39:122–31. [DOI] [PubMed] [Google Scholar]
- 24.Fu L, Penton CR, Ruan Y, Shen Z, Xue C, Li R, et al. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol Biochem. 2017;104:39–48. [Google Scholar]
- 25.Chen WP, Kuo TT. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 1993;21:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Bruijn FJ. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–7. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing platforms. Mol Biol Evol. 2018;35:1547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. [DOI] [PubMed] [Google Scholar]
- 31.Vincent JM. A manual for the practical study of the root-nodule bacteria. A manual for the practical study of the root-nodule bacteria. 1970.
- 32.Ehmann AK. The Van urk-salkowski reagent–a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J Chromatogr. 1977;132 2:267–76. [DOI] [PubMed] [Google Scholar]
- 33.Gordon SA, Weber RP. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951;26:192–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pikovskaya RI, Pikovskaya RI. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. 1948.
- 35.Tandon HLS, Cescas MP, Tyner EH. An acid-free vanadate-molybdate reagent for the determination of total phosphorus in soils. Soil Sci Soc Am J. 1968;32:48–51. [Google Scholar]
- 36.Park M, Kim C, Yang J, Lee H, Shin W, Kim S, et al. Isolation and characterization of diazotrophic growth promoting bacteria from rhizosphere of agricultural crops of Korea. Microbiol Res. 2005;160:127–33. [DOI] [PubMed] [Google Scholar]
- 37.Revillas JJ, Rodelas B, Pozo C, Martínez-Toledo MV, González-López J. Production of B-group vitamins by two Azotobacter strains with phenolic compounds as sole carbon source under diazotrophic and adiazotrophic conditions: b-group vitamin production by azotobacter strains. J Appl Microbiol. 2000;89:486–93. [DOI] [PubMed] [Google Scholar]
- 38.Pandey P, Sahgal M, Maheshwari D, Johri BN. Genetic diversity of rhizobia isolated from medicinal legumes growing in the sub-himalayan region of Uttaranchal. Curr Sci. 2004;86:202–7. [Google Scholar]
- 39.Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts the GenBank accession numbers for the sequences reported in this paper are AF217261 through AF217272 for nodC and AF218126, AF275670 and AF275671 for nifH. Microbiology. 2001;147:981–93. [DOI] [PubMed] [Google Scholar]
- 40.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. [DOI] [PubMed] [Google Scholar]
- 41.Arora N, Verma M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech. 2017;7. [DOI] [PMC free article] [PubMed]
- 42.Louden BC, Haarmann D, Lynne AM. Use of Blue Agar CAS Assay for Siderophore Detection. J Microbiol Biol Educ. 2011;12:51–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Payne SM. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993;1:66–9. [DOI] [PubMed] [Google Scholar]
- 44.Hu X, Chen J, Guo J. Two phosphate- and potassium-solubilizing Bacteria isolated from Tianmu Mountain, Zhejiang, China. World J Microbiol Biotechnol. 2006;22:983–90. [Google Scholar]
- 45.Rajawat MVS, Singh S, Tyagi SP, Saxena AK. A modified plate assay for Rapid Screening of potassium-solubilizing Bacteria. Pedosphere. 2016;26:768–73. [Google Scholar]
- 46.Gandhi A, Muralidharan G. Assessment of zinc solubilizing potentiality of Acinetobacter sp. isolated from rice rhizosphere. Eur J Soil Biol. 2016;76:1–8. [Google Scholar]
- 47.Sharma P, Kumawat KC, Kaur KCK, Kaur S. Assessment of Zinc solubilization by Endophytic Bacteria in Legume Rhizosphere. IJAR. 2012;4:439–41. [Google Scholar]
- 48.Khanghahi MY, Ricciuti P, Allegretta I, Terzano R, Crecchio C. Solubilization of insoluble zinc compounds by zinc solubilizing bacteria (ZSB) and optimization of their growth conditions. Environ Sci Pollut Res. 2018;25:25862–8. [DOI] [PubMed] [Google Scholar]
- 49.Palaniyandi SA, Damodharan K, Yang SH, Suh JW. Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of ‘Micro Tom’ tomato plants. J Appl Microbiol. 2014;117:766–73. [DOI] [PubMed] [Google Scholar]
- 50.Dworkin M, Foster JW. Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol. 1958;75:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michel BE, Kaufmann MR. The osmotic potential of polyethylene glycol 6000 1. Plant Physiol. 1973;51:914–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandhya V, Sk Z, Grover A, Reddy M, Venkateswarlu G. Alleviation of drought stress effects in sunflower seedlings by the exopolysaccharides producing Pseudomonas putida strain GAP-P45. Biol Fertil Soils. 2009;46:17–26. [Google Scholar]
- 53.Paulo EM, Boffo EF, Branco A, Valente AMMP, Melo IS, Ferreira AG, et al. Production, extraction and characterization of exopolysaccharides produced by the native Leuconostoc pseudomesenteroides R2 strain. Acad Bras Cienc. 2012;84:495–508. [DOI] [PubMed] [Google Scholar]
- 54.Miller RL, Higgins VJ. Association of cyanide with infection of birdsfoot trefoil by Stemphylium loti. Phytopathology. 1970;60:104–10. [Google Scholar]
- 55.Lin L, Kan X, Yan H, Wang D. Characterization of extracellular cellulose-degrading enzymes from Bacillus thuringiensis strains. Electron J Biotechnol. 2012;15.
- 56.Kumar K, Gupta SC, Baidoo SK, Chander Y, Rosen CJ. Antibiotic uptake by plants from soil fertilized with animal manure. J Environ Qual. 2005;34:2082–5. [DOI] [PubMed] [Google Scholar]
- 57.Goswami D, Dhandhukia P, Patel P, Thakker JN. Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol Res. 2014;169:66–75. [DOI] [PubMed] [Google Scholar]
- 58.Hnini M. Unlocking the diversity and plant-growth promoting potential of the bulk soil bacteria associated with the Saharan tree Vachellia tortilis subsp. raddiana. Soil Science Society of America Journal. 2024;n/a-n/a.
- 59.Korir H, Mungai NW, Wasike VW. Influence of native rhizobacteria co-inoculation and formulation of bacterial inoculants on the growth and yield of common bean (Phaseolus vulgaris L). J Sustainable Agric Environ. 2024;3:e12095. [Google Scholar]
- 60.Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Circular California Agricultural Experiment Station. 1950;347 2nd edit.
- 61.Gray EJ, Smith DL. Intracellular and extracellular PGPR: commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol Biochem. 2005;37:395–412. [Google Scholar]
- 62.Haichar F, el Marol Z, Berge C, Rangel-Castro O, Prosser JI, Balesdent JI. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2:1221–30. [DOI] [PubMed] [Google Scholar]
- 63.Yuan Z, Liu F, He S, Zhou L, Pan H. Community structure and diversity characteristics of rhizosphere and root endophytic bacterial community in different Acacia species. PLoS ONE. 2022;17:e0262909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamutando CN, Vikram S, Kamgan-Nkuekam G, Makhalanyane TP, Greve M, Le Roux JJ, et al. The functional potential of the Rhizospheric Microbiome of an invasive tree species, Acacia dealbata. Microb Ecol. 2019;77:191–200. [DOI] [PubMed] [Google Scholar]
- 65.Ben Romdhane S, Nasr H, Samba-Mbaye R, Neyra M, Ghorbal MH. Diversity of Acacia tortilis rhizobia revealed by PCR/RFLP on crushed root nodules in Tunisia. Ann Microbiol. 2005;55:249–54. [Google Scholar]
- 66.Degefu T, Wolde-meskel E, Frostegård Å. Phylogenetic multilocus sequence analysis identifies seven novel Ensifer genospecies isolated from a less-well-explored biogeographical region in East Africa. Int J Syst Evol MicroBiol. 2012;62 Pt9:2286–95. [DOI] [PubMed] [Google Scholar]
- 67.Fterich A, Mahdhi M, Mars M. Impact of grazing on soil microbial communities along a chronosequence of Acacia tortilis subsp. raddiana in arid soils in Tunisia. Eur J Soil Biol. 2012;50:56–63. [Google Scholar]
- 68.Noureddine N-E, Amrani S, Aïd F. Statut Symbiotique et souches de rhizobia associées à l’ Acacia tortilis subsp. raddiana [ Acacia Raddiana s. s.], mimosoïdée des régions désertiques de l’Algérie. Botany. 2010;88:39–53. [Google Scholar]
- 69.Odee DW, Haukka K, McInroy SG, Sprent JI, Sutherland JM, Young JPW. Genetic and symbiotic characterization of rhizobia isolated from tree and herbaceous legumes grown in soils from ecologically diverse sites in Kenya. Soil Biol Biochem. 2002;34:801–11. [Google Scholar]
- 70.Romdhane SB, Nasr H, Samba-Mbaye R, Neyra M, Ghorbal MH, Lajudie P. Genetic diversity of Acacia tortilis ssp. raddiana rhizobia in Tunisia assessed by 16S and 16S-23S rDNA genes analysis. J Appl Microbiol. 2006;100:436–45. [DOI] [PubMed] [Google Scholar]
- 71.Zakhia F, Jeder H, Domergue O, Willems A, Cleyet-Marel J-C, Gillis M, et al. Characterisation of Wild Legume nodulating Bacteria (LNB) in the Infra-Arid Zone of Tunisia. Syst Appl Microbiol. 2004;27:380–95. [DOI] [PubMed] [Google Scholar]
- 72.Hnini M, Taha K, Aurag J. Molecular identification and characterization of phytobeneficial osmotolerant endophytic bacteria inhabiting root nodules of the Saharan tree Vachellia tortilis subsp. raddiana. Arch Microbiol. 2023;205:45. [DOI] [PubMed] [Google Scholar]
- 73.Plummer E, Twin J. A comparison of three Bioinformatics pipelines for the analysis of Preterm Gut Microbiota using 16S rRNA gene sequencing data. J Proteom Bioinform. 2015;8.
- 74.Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A. 2008. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). International Journal of Systematic and Evolutionary Microbiology. 2008;58:200–14. [DOI] [PubMed]
- 75.Martens M, Delaere M, Coopman R, De Vos P, Gillis M, Willems A. 2007. Multilocus sequence analysis of Ensifer and related taxa. International Journal of Systematic and Evolutionary Microbiology. 2007;57:489–503. [DOI] [PubMed]
- 76.Tindall BJ, Rosselló-Móra R, Busse H-J, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol MicroBiol. 2010;60:249–66. [DOI] [PubMed] [Google Scholar]
- 77.Yassin AF, Hupfer H, Siering C, Schumann P. Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacterium Collins et al. 1982 emend. Lehnen et al. 2006: proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium. Int J Syst Evol MicroBiol. 2011;61:1265–74. [DOI] [PubMed]
- 78.Kumar M, Tomar RS, Lade H, Paul D. Methylotrophic bacteria in sustainable agriculture. World J Microbiol Biotechnol. 2016;32:120. [DOI] [PubMed] [Google Scholar]
- 79.Saxena AK, Kumar M, Chakdar H, Anuroopa N, Bagyaraj DJ. Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol. 2020;128:1583–94. [DOI] [PubMed] [Google Scholar]
- 80.Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ. Bacterial associations with legumes. CRC Crit Rev Plant Sci. 2015;34:17–42. [Google Scholar]
- 81.Pandey A, Palni LM. Bacillus species: the dominant bacteria of the rhizosphere of established tea bushes. Microbiol Res. 1997;152:359–65. [DOI] [PubMed] [Google Scholar]
- 82.Boparai J, Kaur R, Sharma P. Delineating bacterial community structure of rhizosphere metagenome from Kansal Forest. 2019. 10.1016/j.egg.2019.100044
- 83.Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard J-Y. Bacterial spore structures and their protective role in biocide resistance. J Appl Microbiol. 2012;113:485–98. [DOI] [PubMed] [Google Scholar]
- 84.Hawkes CV, DeAngelis KM, Firestone MK. Root interactions with soil microbial communities and processes. The rhizosphere. Elsevier; 2007. pp. 1–29.
- 85.Herman DJ, Johnson KK, Jaeger CH, Schwartz E, Firestone MK. Root influence on nitrogen mineralization and nitrification in Avena barbata rhizosphere soil. Soil Sci Soc Am J. 2006;70:1504–11. [Google Scholar]
- 86.Saeed Q, Xiukang W, Haider FU, Kučerik J, Mumtaz MZ, Holatko J, et al. Rhizosphere Bacteria in Plant Growth Promotion, Biocontrol, and Bioremediation of Contaminated sites: a Comprehensive Review of effects and mechanisms. Int J Mol Sci. 2021;22:10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar A, Dubey A. Rhizosphere microbiome: Engineering bacterial competitiveness for enhancing crop production. J Adv Res. 2020;24:337–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bharucha U, Patel K, Trivedi UB. Optimization of indole acetic acid production by Pseudomonas putida UB1 and its effect as plant growth-promoting rhizobacteria on mustard (Brassica nigra). Agricultural Res. 2013;2:215–21. [Google Scholar]
- 89.Tariq A, Ahmed A. Auxins-Interkingdom Signaling Molecules. Plant Hormones: Recent Advances, New Perspectives and Applications. 2022:3.
- 90.Kuhn BM, Nodzyński T, Errafi S, Bucher R, Gupta S, Aryal B, et al. Flavonol-induced changes in PIN2 polarity and auxin transport in the Arabidopsis thaliana rol1-2 mutant require phosphatase activity. Sci Rep. 2017;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spaepen S, Versées W, Gocke D, Pohl M, Steyaert J, Vanderleyden J. Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense. J Bacteriol. 2007;189:7626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sabet H, Mortazaeinezhad F. Yield, growth and Fe uptake of cumin (Cuminum cyminum L.) affected by Fe-nano, Fe-chelated and Fe-siderophore fertilization in the calcareous soils. J Trace Elem Med Biol. 2018;50:154–60. [DOI] [PubMed] [Google Scholar]
- 93.Sahu GK, Sindhu SS. Disease control and plant growth promotion of green gram by siderophore producing Pseudomonas Sp. Res J Microbiol. 2011;6:735. [Google Scholar]
- 94.Sultana S, Alam S, Karim MM. Screening of siderophore-producing salt-tolerant rhizobacteria suitable for supporting plant growth in saline soils with iron limitation. J Agric Food Res. 2021;4:100150. [Google Scholar]
- 95.Ferchichi N, Toukabri W, Boularess M, Smaoui A, Mhamdi R, Trabelsi D. Isolation, identification and plant growth promotion ability of endophytic bacteria associated with lupine root nodule grown in Tunisian soil. Arch Microbiol. 2019;201:1333–49. [DOI] [PubMed] [Google Scholar]
- 96.Martínez-Hidalgo P, Hirsch AM. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes J. 2017;1:70–82. [Google Scholar]
- 97.Singh R, Shelke G, Kumar A, Jha P. Biochemistry and genetics of ACC deaminase: a weapon to stress ethylene produced in plants. Front Microbiol. 2015;6. [DOI] [PMC free article] [PubMed]
- 98.Li J, Shah S, Moffatt BA, Glick BR. Isolation and characterization of an unusual 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase gene from Enterobacter cloacae UW4. Antonie Van Leeuwenhoek. 2001;80:255–61. [DOI] [PubMed] [Google Scholar]
- 99.Barnawal D, Bharti N, Maji D, Chanotiya CS, Kalra A. ACC deaminase-containing Arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in Pisum sativum. J Plant Physiol. 2014;171:884–94. [DOI] [PubMed] [Google Scholar]
- 100.Guignard MS, Leitch AR, Acquisti C, Eizaguirre C, Elser JJ, Hessen DO et al. Impacts of Nitrogen and Phosphorus: From Genomes to Natural Ecosystems and Agriculture. Frontiers in Ecology and Evolution. 2017;5.
- 101.Tian J, Ge F, Zhang D, Deng S, Liu X. Roles of phosphate solubilizing microorganisms from managing soil phosphorus deficiency to mediating biogeochemical P cycle. Biology. 2021;10:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kour D, Rana KL, Kaur T, Yadav N, Yadav AN, Kumar M, et al. Biodiversity, current developments and potential biotechnological applications of phosphorus-solubilizing and -mobilizing microbes: a review. Pedosphere. 2021;31:43–75. [Google Scholar]
- 103.Yoon J-H, Cho Y-G, Lee ST, Suzuki K, Nakase T, Park Y-H. Nocardioides nitrophenolicus sp. nov., a p-nitrophenol-degrading bacterium. Int J Syst Evol MicroBiol. 1999;49:675–80. [DOI] [PubMed] [Google Scholar]
- 104.Patel P, Gajjar H, Joshi B, Krishnamurthy R, Amaresan N. Inoculation of Salt-Tolerant Acinetobacter Sp (RSC9) improves the sugarcane (Saccharum sp. Hybrids) growth under salinity stress Condition. Sugar Tech. 2022;24:494–501. [Google Scholar]
- 105.Parmar P, Sindhu SS. Potassium solubilization by rhizosphere bacteria: influence of nutritional and environmental conditions. J Microbiol Res. 2013;3:25–31. [Google Scholar]
- 106.Parmar P, Sindhu SS. The novel and efficient method for isolating potassium solubilizing bacteria from rhizosphere soil. Geomicrobiol J. 2019;36:130–6. [Google Scholar]
- 107.Basak B, Biswas D. Modification of waste mica for alternative source of potassium: evaluation of potassium release in soil from waste mica treated with potassium solubilizing bacteria (KSB). LAP LAMBERT Academic Publishing; 2012.
- 108.Sheng XF, Zhao F, He LY, Qiu G, Chen L. Isolation and characterization of silicate mineral-solubilizing Bacillus globisporus Q12 from the surfaces of weathered feldspar. Can J Microbiol. 2008;54:1064–8. [DOI] [PubMed] [Google Scholar]
- 109.Singh G, Biswas DR, Marwaha TS. Mobilization of Potassium from Waste Mica by Plant Growth promoting Rhizobacteria and its assimilation by Maize (zea Mays) and wheat (triticum Aestivum L.): a Hydroponics Study under Phytotron Growth Chamber. J Plant Nutr. 2010;33:1236–51. [Google Scholar]
- 110.Ramesh A, Sharma SK, Sharma MP, Yadav N, Joshi OP. Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in vertisols of central India. Appl Soil Ecol. 2014;73:87–96. [Google Scholar]
- 111.Moreno-Jiménez E, Plaza C, Saiz H, Manzano R, Flagmeier M, Maestre FT. Aridity and reduced soil micronutrient availability in global drylands. Nat Sustain. 2019;2:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karnwal A. Zinc solubilizing Pseudomonas spp. from vermicompost bestowed with multifaceted plant growth promoting properties and having prospective modulation of zinc biofortification in Abelmoschus esculentus L. J Plant Nutr. 2021;44:1023–38. [Google Scholar]
- 113.Ryan J, Ibrikci H, Sommer R, McNeill A. Chapter 2 Nitrogen in Rainfed and Irrigated Cropping systems in the Mediterranean Region. Advances in Agronomy. Academic; 2009. pp. 53–136.
- 114.Zeng Q, Ding X, Wang J, Han X, Iqbal HMN, Bilal M. Insight into soil nitrogen and phosphorus availability and agricultural sustainability by plant growth-promoting rhizobacteria. Environ Sci Pollut Res. 2022;29:45089–106. [DOI] [PubMed] [Google Scholar]
- 115.Luo L, Zhao C, Wang E, Raza A, Yin C. Bacillus amyloliquefaciens as an excellent agent for biofertilizer and biocontrol in agriculture: an overview for its mechanisms. Microbiol Res. 2022;259:127016. [DOI] [PubMed] [Google Scholar]
- 116.Nahar S, Hasan MF, Sikdar B, Islam MA. Effect of rhizosphere microbiome on different crop growing fields in various rice cultivars and its molecular approaches for sustainable agro-ecosystem. J Crop Sci Biotechnol. 2021;24:521–31. [Google Scholar]
- 117.del Orozco-Mosqueda C, Fadiji AE, Babalola OO, Glick BR, Santoyo G. Rhizobiome engineering: Unveiling complex rhizosphere interactions to enhance plant growth and health. Microbiological Research. 2022;263:127137. [DOI] [PubMed]
- 118.Yadav AN, Verma P, Kumar S, Kumar V, Kumar M, Kumari Sugitha TC, et al. Chapter 2 - Actinobacteria from Rhizosphere: molecular diversity, distributions, and potential biotechnological applications. In: Singh BP, Gupta VK, Passari AK, editors. New and Future developments in Microbial Biotechnology and Bioengineering. Elsevier; 2018. pp. 13–41.
- 119.Gaby JC, Buckley DH. A comprehensive evaluation of PCR primers to amplify the nifH Gene of Nitrogenase. PLoS ONE. 2012;7:e42149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mehnaz S. Plant growth-promoting Bacteria Associated with Sugarcane. In: Maheshwari DK, editor. Bacteria in Agrobiology: Crop ecosystems. Berlin, Heidelberg: Springer; 2011. pp. 165–87. [Google Scholar]
- 121.Setten L, Soto G, Mozzicafreddo M, Fox AR, Lisi C, Cuccioloni M, et al. Engineering Pseudomonas protegens Pf-5 for Nitrogen fixation and its application to improve Plant Growth under Nitrogen-deficient conditions. PLoS ONE. 2013;8:e63666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Solanki MK, Wang Z, Wang F-Y, Li C-N, Lan T-J, Singh RK, et al. Intercropping in Sugarcane Cultivation influenced the Soil properties and enhanced the Diversity of Vital Diazotrophic Bacteria. Sugar Tech. 2017;19:136–47. [Google Scholar]
- 123.Akter Z, Pageni BB, Lupwayi NZ, Balasubramanian PM. Biological nitrogen fixation and nifH gene expression in dry beans (Phaseolus vulgaris L). Can J Plant Sci. 2014;94:203–12. [Google Scholar]
- 124.Bushra R, Uzair B, Manzoor S, Ahmed I. Draft genome of Sporosarcina sp. NCCP-2222T isolated from rhizosphere of mangrove plant Ceriops tagal reveals its potential as a plant biostimulant and constitutes a novel species. Biocatal Agric Biotechnol. 2023;47:102543. [Google Scholar]
- 125.Meena KK, Bitla UM, Sorty AM, Singh DP, Gupta VK, Wakchaure GC et al. Mitigation of salinity stress in wheat seedlings due to the application of Phytohormone-Rich Culture Filtrate Extract of Methylotrophic Actinobacterium Nocardioides sp. NIMMe6. Front Microbiol. 2020;11. [DOI] [PMC free article] [PubMed]
- 126.Rosier A, Beauregard PB, Bais HP. Quorum quenching activity of the PGPR Bacillus subtilis UD1022 alters Nodulation Efficiency of Sinorhizobium meliloti on Medicago truncatula. Front Microbiol. 2021;11. [DOI] [PMC free article] [PubMed]
- 127.Alexander A, Singh VK, Mishra A, Jha B. Plant growth promoting rhizobacterium Stenotrophomonas maltophilia BJ01 augments endurance against N2 starvation by modulating physiology and biochemical activities of Arachis hypogea. PLoS ONE. 2019;14:e0222405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ahemad M, Khan M. Assessment of Plant Growth promoting activities of Rhizobacterium Pseudomonas putida under insecticide-stress. Ann Microbiol. 2012;62:1531–40. [Google Scholar]
- 129.Nosheen A, Yasmin H, Naz R, Bano A, Keyani R, Hussain I. Pseudomonas putida improved soil enzyme activity and growth of kasumbha under low input of mineral fertilizers. Soil Sci Plant Nutr. 2018;64:520–5. [Google Scholar]
- 130.Zhang R, Wang Y-H, Jin J-J, Stull GW, Bruneau A, Cardoso D, et al. Exploration of Plastid Phylogenomic conflict yields new insights into the Deep relationships of Leguminosae. Syst Biol. 2020;69:613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Joshi P, Saini A, Banerjee S, Bose R, Bhandari MS, Pandey A, et al. Agriculturally important microbes: challenges and opportunities. In: Soni R, Suyal DC, Bhargava P, Goel R, editors. Microbiological activity for Soil and Plant Health Management. Singapore: Springer; 2021. pp. 1–34. [Google Scholar]
- 132.Müller C, Cramer W, Hare WL, Lotze-Campen H. Climate change risks for African agriculture. Proceedings of the National Academy of Sciences. 2011;108:4313–5. [DOI] [PMC free article] [PubMed]
- 133.Akram M, Asghar H, Zahir Z, Ahmad H, Hussain MB. Isolation and screening of beneficial bacteria to ameliorate drought stress in wheat. Soil Environ. 2015;34:100. [Google Scholar]
- 134.Forchetti G, Masciarelli O, Alemano S, Alvarez D, Abdala G. Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl Microbiol Biotechnol. 2007;76:1145–52. [DOI] [PubMed] [Google Scholar]
- 135.Vardharajula S, Zulfikar Ali S, Grover M, Reddy G, Bandi V. Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J Plant Interact. 2011;6:1–14. [Google Scholar]
- 136.Montañez A, Abreu C, Gill PR, Hardarson G, Sicardi M. Biological nitrogen fixation in maize (Zea mays L.) by 15 N isotope-dilution and identification of associated culturable diazotrophs. Biol Fertil Soils. 2009;45:253–63. [Google Scholar]
- 137.Pham VTK, Rediers H, Ghequire MGK, Nguyen HH, De Mot R, Vanderleyden J, et al. The plant growth-promoting effect of the nitrogen-fixing endophyte Pseudomonas stutzeri A15. Arch Microbiol. 2017;199:513–7. [DOI] [PubMed] [Google Scholar]
- 138.Chang W-S, van de Mortel M, Nielsen L, Nino de Guzman G, Li X, Halverson LJ. Alginate production by Pseudomonas putida creates a Hydrated Microenvironment and contributes to Biofilm Architecture and stress tolerance under Water-limiting conditions. J Bacteriol. 2007;189:8290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mahmood S, Daur I, Al-Solaimani SG, Ahmad S, Madkour MH, Yasir M et al. Plant Growth promoting Rhizobacteria and Silicon synergistically enhance salinity tolerance of Mung Bean. Front Plant Sci. 2016;7. [DOI] [PMC free article] [PubMed]
- 140.Egamberdieva D, Wirth SJ, Shurigin VV, Hashem A, Abd_Allah EF. Endophytic Bacteria Improve Plant Growth, Symbiotic Performance of Chickpea (Cicer arietinum L.) and induce suppression of Root Rot caused by Fusarium solani under salt stress. Front Microbiol. 2017;8:1887. [DOI] [PMC free article] [PubMed]
- 141.Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol. 2010;60:579–98. [Google Scholar]
- 142.Altimira F, Godoy S, Arias-Aravena M, Araya B, Montes C, Castro JF, et al. Genomic and experimental analysis of the Biostimulant and Antagonistic properties of Phytopathogens of Bacillus safensis and Bacillus siamensis. Microorganisms. 2022;10:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Amna null, Xia Y, Farooq MA, Javed MT, Kamran MA, Mukhtar T, et al. Multi-stress tolerant PGPR Bacillus xiamenensis PM14 activating sugarcane (Saccharum officinarum L.) red rot disease resistance. Plant Physiol Biochem. 2020;151:640–9. [DOI] [PubMed] [Google Scholar]
- 144.Rijavec T, Lapanje A. Hydrogen cyanide in the Rhizosphere: not suppressing plant pathogens, but rather regulating availability of phosphate. Front Microbiol 2016;7. [DOI] [PMC free article] [PubMed]
- 145.Rodrigues AA, Forzani MV, Soares R, de Sibov S, Vieira ST. Isolation and selection of plant growth-promoting bacteria associated with sugarcane. Pesquisa Agropecuária Trop. 2016;46:149–58. [Google Scholar]
- 146.Mahdi I, Fahsi N, Hafidi M, Allaoui A, Biskri L. Plant Growth Enhancement using Rhizospheric Halotolerant phosphate solubilizing bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 isolated from Chenopodium quinoa Willd. Microorganisms. 2020;8:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ikunaga Y, Sato I, Grond S, Numaziri N, Yoshida S, Yamaya H, et al. Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl Microbiol Biotechnol. 2011;89:419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carrer Filho R, Romeiro RS, Garcia FAO. Biocontrole De doenças de parte aérea do tomateiro por Nocardioides Thermolilacinus. Trop Plant Pathol. 2008;33:457–60. [Google Scholar]
- 149.Hnini M, Taha K, Aurag J. Botany, associated microbiota, traditional medicinal uses, and phytochemistry of Vachellia tortilis subsp. raddiana (Savi): a systematic review. J Agric Food Res. 2023;12:100566. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.