Abstract
Background
Treatment resistant schizophrenia (TRS) is broadly defined as inadequate response to adequate treatment and is associated with a substantial increase in disease burden. Clozapine is the only approved treatment for TRS, showing superior clinical effect on overall symptomatology compared to other drugs, and is the prototype of atypical antipsychotics. Risperidone, another atypical antipsychotic with a more distinctive dopamine 2 antagonism, is commonly used in treatment of schizophrenia. Here, we conducted a genome-wide association study on patients treated with clozapine (TRS) vs. risperidone (non-TRS) and investigated whether single variants and/or polygenic risk score for schizophrenia are associated with TRS status. We hypothesized that patients who are treated with clozapine and risperidone might exhibit distinct neurobiological phenotypes that match pharmacological profiles of these drugs and can be explained by genetic differences. The study population (n = 1286) was recruited from a routine therapeutic drug monitoring (TDM) service between 2005 and 2022. History of a detectable serum concentration of clozapine and risperidone (without TDM history of clozapine) defined the TRS (n = 478) and non-TRS (n = 808) group, respectively.
Results
We identified a suggestive association between TRS and a common variant within the LINC00523 gene with a significance just below the genome-wide threshold (rs79229764 C > T, OR = 4.89; p = 1.8 × 10−7). Polygenic risk score for schizophrenia was significantly associated with TRS (OR = 1.4, p = 2.1 × 10−6). In a large post-mortem brain sample from schizophrenia donors (n = 214; CommonMind Consortium), gene expression analysis indicated that the rs79229764 variant allele might be involved in the regulation of GPR88 and PUDP, which plays a role in striatal neurotransmission and intellectual disability, respectively.
Conclusions
We report a suggestive genetic association at the rs79229764 locus with TRS and show that genetic liability for schizophrenia is positively associated with TRS. These results suggest a candidate locus for future follow-up studies to elucidate the molecular underpinnings of TRS. Our findings further demonstrate the value of both single variant and polygenic association analyses for TRS prediction.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40246-024-00673-x.
Introduction
Antipsychotics is the cornerstone of schizophrenia treatment, but up to 30% of patients do not respond adequately to standard treatment [1]. These treatment-resistant schizophrenia (TRS) patients suffer from greater symptom burden [1], and clozapine is the only evidence-based treatment for TRS with superior clinical effect on overall positive and negative symptoms [2].
All atypical antipsychotics broadly share antagonism at dopamine 2 (D2) and serotonin 2A (5-HT2A) receptors, but they also have binding capabilities at other receptors [3]. Clozapine has transient and low affinity binding at D2 receptors while favouring antagonism on 5-HT2A receptors [3]. It exhibits the broadest coverage of molecular targets among all antipsychotics, including modulation of glutaminergic, cholinergic, and noradrenergic transmission [3]. Risperidone is a commonly used atypical antipsychotic with a more distinct receptor profile and particularly stronger D2 antagonism than clozapine [3]. Patients who respond adequately to antipsychotics like risperidone are considered as ‘responders’ to D2 receptor antagonism. Since clozapine can only be prescribed after failed response to two other antipsychotics, those who use clozapine may be considered as ‘non-responders’, meaning that symptom improvement is not sufficient with antagonism of these receptors but may require involvement of other neurotransmitter systems.
Currently, knowledge on the underlying neurobiology of TRS is limited. While emerging evidence suggests that TRS has a polygenic architecture [4], previous studies have failed to identify specific gene variants that robustly correlate with TRS [5]. In a genome-wide investigation, Ruderfer et al. identified shared genetic underpinnings between schizophrenia pathophysiology and the mechanisms of action of antipsychotic drugs [6]. Here, we conducted a genome-wide association study (GWAS) on clozapine use, defined as proxy of resistant schizophrenia, vs. risperidone use, defined as proxy for responsive schizophrenia, in patients with confirmed therapeutic concentrations of these two drugs. We both analysed for associations with single variants and polygenic risk score for schizophrenia and hypothesized that genetic differences between patients treated with clozapine and risperidone might reflect aspects underlying the genetic architecture of antipsychotic response.
Methods
Study population
In clinical practice, therapeutic drug monitoring (TDM) may be applied to guide antipsychotic dosing to ensure sufficient treatment intensity, and thus exclude potential ‘pseudo resistance’ and erroneous diagnosis of TRS. The study utilized retrospective data/samples of patients from the routine TDM service at Center for Psychopharmacology, Diakonhjemmet Hospital, Oslo, Norway, who in the period between January 2005 and August 2022 had performed (i) TDM of clozapine and/or risperidone and (ii) genotyping of cytochrome P450 (CYP) enzymes. In Norway, TDM is reimbursed and commonly used as a clinical tool in public psychiatric health service to assess adherence to medication and optimize dosing of drugs for maximizing clinical effect and/or minimizing side effects. Use of CYP genotyping is also reimbursed and increasingly used to guide dose optimization of psychiatric drugs.
The study population consisted of patients of Norwegian inhabitants and is assumed to be of patients with Caucasian ancestry. The main inclusion criterion was the availability of biobanked (residual) blood samples from CYP genotyping and clinical concentration levels of clozapine or risperidone verified by TDM. Patients were phenotyped based on their history of using clozapine or risperidone at clinically relevant concentrations, as determined by their TDM history. The TRS group was defined by clozapine use, while the non-TRS group was defined by risperidone use without any history of clozapine TDM. Further requirements for being included in the risperidone cohort were treatment with ≥ 2 mg/day risperidone at least once (dose range for psychotic disorders) and no previous use/TDM history of clozapine.
In the non-TRS group, 41% of the patients had a TDM history of antipsychotic polypharmacy with risperidone (two or more drugs). Furthermore, in the non-TRS group, TDM history of median number of other, non-clozapine antipsychotics before taking risperidone was 1 (IQR: 0,1), whereas in the clozapine (TRS) group, TDM history of median number of non-clozapine antipsychotics was 2 (IQR; 0,3). All patients were included in the study independent of the clinical setting (out-/inpatient). In Norway, TDM of clozapine is usually performed for dose titration and by routine during ongoing treatment, whereas TDM of risperidone is more often ‘reactive’ due to insufficient clinical effect or suspected side effects. Center for Psychopharmacology is the major laboratory performing the analyses.
The Regional Committee for Medical and Health Research Ethics and the Investigational Review Board at Diakonhjemmet Hospital approved the study. The study used anonymized data and residual blood samples from already performed routine analyses. In these cases, where study inclusion does not pose any burden to the patients, the Health Research Act of Norway allows for using samples and information collecting in clinical routine for research purpose without written informed patient consent. However, it is mandatory to send information to the patients, in advance of starting a project, about their legal rights to reserve against being included. Accordingly, letters were sent to all patients eligible for inclusion, where those requesting to opt-out were excluded from the study.
Serum concentration determination of antipsychotics
Liquid chromatography with mass spectrometry (LC–MS) method with FDA certified and validated analytical assays were applied for determination of serum concentrations of clozapine and risperidone and their metabolites. Lower limit of quantification for clozapine and N-desmethylclozapine was 50 nmol/L, whereas this limit was 1 nmol/L and 2 nmol/L for risperidone and 9-hydroxyrisperidone, respectively.
Serum concentration thresholds, based on consensus guidelines [7], were used as an informative measure to assess whether exposure to antipsychotic was adequate. These were clozapine 1070 nmol/L (350 µg/L) and risperidone active moiety 50 nmol/L (20 µg/L, risperidone plus 9-hydroxyrisperidone).
Genotyping and imputation
As previously described in detail [8], DNA extracted from the whole blood was genotyped using the Human Omni Express-24 v.1.1 (Illumina Inc., San Diego, CA, USA), at deCODE Genetics (Reykjavik, Iceland), following standard Illumina protocols. Quality control and phasing of chromosome-wide haplotypes were performed with PLINK v1.93 [9, 10] and Eagle2 [11, 12], respectively. The first release of the haplotype reference consortium reference set was used for imputation of missing variants with Minimac3 [13]. Following imputation, exclusion criteria were variants with (1) minor allele frequency < 1% or (2) departure from Hardy–Weinberg equilibrium (P value < 1 × 10−6), or individuals with (3) high rates of missing genotypes (> 5%), that exhibit relatedness (pairwise Identity-By-Descent ^ π > 0.2 according to PLINK v1.9). 1286 individuals with ~ 5.6 million common variants were included for further statistical analysis.
Statistical analysis
The statistical analyses were performed using R statistics [14] for demographic characteristics of the study sample and follow-up analysis of the lead variant identified. Distribution of sex and age were compared between TRS and non-TRS patients using Pearson’s Chi-squared test and Welch’s two sample t-test, respectively. If risperidone patients were prescribed long-acting injectable formulations, total daily dosing was estimated by dividing the prescribed doses by dosing intervals. Estimated means of serum concentrations (nmol/L) and daily dose (mg/day) were calculated using linear mixed models with unique patient identifier as random effects.
The GWAS of TRS versus non-TRS was conducted using logistic regression analyses implemented in PLINK v1.9 [9, 10], controlling for participant age, sex, the first 10 genetic principal components and genotyping batch. Standard GWAS threshold (5 × 10−8) was used to define statistical significance of the GWAS analysis. LDproxy module provided by the LDlink platform [15] was used to perform linkage equilibrium analysis of the lead variant.
For the expression quantitative trait loci (eQTL) analysis, genotype and post-mortem frontal brain cortex gene expression data were obtained from the CommonMind Consortium [16]. Only data from schizophrenia patients with European ethnicity (n = 214) were included in the study. Genotype QC and imputation were performed by the Consortium. The expression data was filtered and normalized using the DESeq2 R package [17]. The R package MatrixEQTL [18] was used for the cis-eQTL analysis, adjusting for age, gender and post-mortem interval. Variant-gene associations within 1 Mb of the identified lead variant were considered. P < 0.05 defined statistical significance of patient/treatment characteristics and gene expression analyses.
For the GWAS sample, a schizophrenia polygenic risk score (PRS) was calculated based on the latest schizophrenia GWAS performed by the PGC [19] using the meta-analysis of European samples comprising 53,386 cases and 77,258 controls. The PRS was calculated using the PRS continuous shrinkage (PRS-cs) approach [20], adjusting for linkage disequilibrium (LD) based on the LD structure of the European sample of the 1000 Genomes Phase III [21] with default options and a shrinkage parameter of phi = 1 [20]. To facilitate the interpretability of the results, the PRS was standardized (mean = 0, SD = 1) before statistical analysis. To investigate if the PRS for schizophrenia as well as smoking, age, and sex is associated with TRS, a logistic regression analysis including TRS (yes/no) as the dependent variable was performed. The model included the PRS, age, sex, smoking (yes/no), as well as genotyping batch and the first 5 principal components for genetic ancestry for adjustment.
Results
Sample characteristics are presented in Table 1. The study population (n = 1286) included n = 478 TRS patients and n = 808 non-TRS patients. There was a higher proportion of males among the TRS group compared to the non-TRS group (59% vs 53%, p = 0.009), but no difference in age distribution (Table 1). Mean daily dose estimates were 335 and 3.3 mg/day for patients who were treated with clozapine (TRS) and risperidone (non-TRS), respectively (Table 1). Furthermore, estimated serum concentration measurements for patients in the TRS and non-TRS group were 1201 nmol/L (393 ng/ml) for clozapine and 64 nmol/L for risperidone (active moiety including 9-hydroxyrisperidone), respectively (Table 1). Among the TRS group, median number of clozapine measurements during the study period was 14 (IQR: 5, 36) spanning 4.3 years (IQR: 0.7, 11.8) of TDM, whereas TDM history of the non-TRS group showed median of 3 (IQR: 1.8, 7) risperidone measurements over 0.8 years (IQR: 0.04, 4.8; Table 1).
Table 1.
Demographic and treatment characteristics of the study population
| Variables | TRS | non-TRS | p |
|---|---|---|---|
| Number of patients, n | 478 | 808 | – |
| Male, n (%) | 284 (59) | 430 (53) | 0.031 |
| Age, years; mean (SD) | 48.7 (15.0) | 48.7 (16.9) | 0.99 |
| Serum concentration, nmol/L; mean (95%CI) | 1201 (1162,1240) | 64 (30, 98) | – |
| Dose, mg/day; mean (95%CI) | 335 (320, 350) | 3.3 (3.2, 3.4) | – |
| History of TDM measurements, n; median (IQR) | 14 (5, 36) | 3 (1.8, 7) | – |
| TDM duration, years; median (IQR) | 4.3 (0.7, 11.8) | 0.8 (0.04, 4.8) | – |
IQR Interquartile range, SD Standard deviation, TDM Therapeutic drug monitoring, TRS treatment resistant schizophrenia. TRS and non-TRS groups comprised of patients who were treated with clozapine and risperidone, respectively
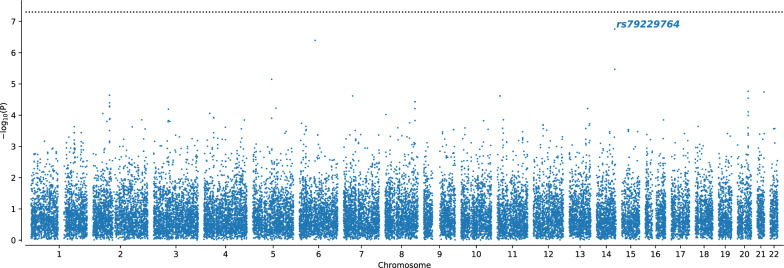
The GWAS on TRS did not show any associations reaching genome-wide significance (p < 5 × 10−8), however identified a single nucleotide polymorphism (SNP; rs79229764 C > T) on chromosome 14 with a p-value just above the genome-wide significance threshold (p = 1.8 × 10−7; Fig. 1). We observed a 2.1-fold higher proportion of the risk allele carriers among TRS patients compared to non-TRS patients with allele frequencies of 3.7% and 1.7%, respectively; OR = 4.89, 95% CI = [2.6, 8.5]). Frequency of the minor allele variant is 4.2% among European populations (https://www.ensembl.org/). LD structure from the 1000 Genomes phase 3-genome browser showed a low LD region (Supplementary Fig. 1) with variants that are in low to moderate LD with the rs79229764 haplotype among the European populations (R2 < 0.54; Supplementary Table 1).
Fig. 1.
Manhattan plot showing the associations with risk of TRS. The − log10 transformed p-values for each SNP are shown on the y-axis and chromosomal positions are indicated on the x-axis. The dashed black line represents the standard GWAS p-value threshold of 5 × 10−8. The most significant SNP is highlighted by text
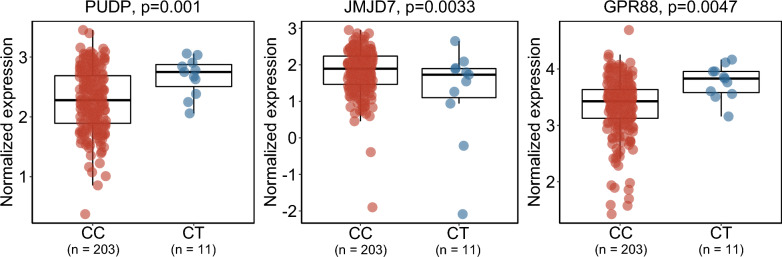
The rs79229764 variant is located within the exon of the long intergenic non-coding RNA 523 (LINC00523) gene. NHGRI-EBI GWAS Catalog did not reveal any previous associations with the rs79229764 variant (https://www.ebi.ac.uk/gwas/). Expression quantitative trait loci (eQTL) analysis of rs79229764 using the CommonMind Consortium post-mortem brain expression data [16] from n = 214 (CC: n = 203, CT: n = 11) schizophrenia patients revealed gene associations with nominal significance. The top 3 most significant risk allele-gene relationships included increased expression of PUDP (p = 0.001) and GPR88 (p = 0.0047), and decreased expression of JMJD7 (p = 0.003; Table 2 and Fig. 2).
Table 2.
The 3 most significant brain tissue gene expression associations of the lead variant rs79229764 C > T from CommonMind Consortium
| Chromosome | Position | Gene name | Beta | p (unadjusted)* | Gene type |
|---|---|---|---|---|---|
| X | 6,667,865–7,148,158 | PUDP | 0.441 | 0.001 | Protein coding |
| 15 | 41,828,092–41,837,581 | JMJD7 | -0.592 | 0.003 | Protein coding |
| 1 | 100,538,139–100,542,021 | GPR88 | 0.435 | 0.005 | Protein coding |
Betas are included from the eQTL analysis which was adjusted for age, gender and post-mortem interval. Only data from schizophrenia patients with European ethnicity were included in the analysis. * p > 0.05 after multiple comparisons corrections
Fig. 2.
Impact of rs79229764 C > T on brain tissue gene expression from CommonMind Consortium data. The 3 most significant gene expression associations were illustrated. Normalized expression results are shown on the y-axis, while rs79229764 C > T genotypes are indicated on the x-axis. Gene names and unadjusted p-values from association analyses are shown at the top part of the plots
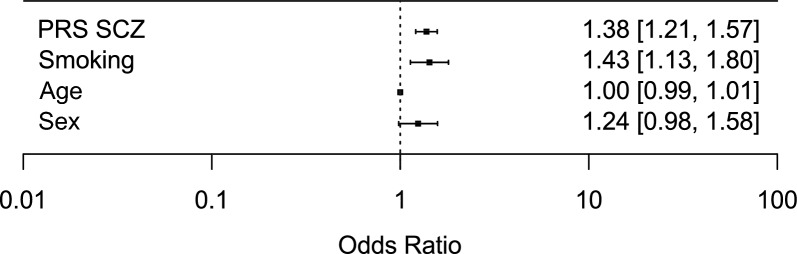
The schizophrenia PRS showed a significant, positive association with TRS, i.e., higher PRS was associated with being a TRS case (OR = 1.4, p = 2.1 × 10−6; Fig. 3 and Supplementary Table 2). In addition, we observed that smoking was significantly associated with TRS (OR = 1.4, p = 0.003; Fig. 3 and Supplementary Table 2). No association was observed between TRS and sex or age (Fig. 3 and Supplementary Table 2).
Fig. 3.
Forest plot showing the association between treatment resistant schizophrenia and schizophrenia polygenic risk score (PRS SCZ), smoking, age, and sex. Effects are reported as odds ratios (95% confidence interval)
Discussion
In the current study, we conducted a genome-wide investigation of TRS based on treatment history of clozapine (TRS) and risperidone (without clozapine history; non-TRS) at therapeutic serum concentrations in a well-phenotyped study cohort (n = 1286). We found that genetic liability to schizophrenia, as measured by schizophrenia PRS was significantly associated with TRS. We also identified a suggestive variant in the LINC00523 gene (rs79229764 C > T) with strong enrichment in clozapine vs. risperidone patients (OR = 4.89) and a significance level close to the genome-wide threshold (p = 1.8 × 10−7). Follow-up eQTL analysis in post-mortem brain samples from schizophrenia donors (n = 214) suggested transcriptional effects of rs79229764 on different genes.
Although previous studies investigating the association between schizophrenia PRS and TRS had conflicting results [22–25], our finding is in line with a recent systematic review and meta-analysis that showed significant associations between schizophrenia PRS and TRS [26]. We have previously observed that higher schizophrenia PRS is associated with antipsychotic prescription pattern [27, 28]. Together with the findings from the current study, these results suggest that schizophrenia PRS may have potential utility to aid in therapeutic decision making on antipsychotic treatment. Furthermore, this shows the ability of the study to use a combinatorial approach investigating pharmacogenomics of TRS at both overall and detailed levels.
Interestingly, tobacco smoking was also significantly associated with TRS. Approximately 60% of patients with schizophrenia are smokers [29, 30], and the “self-medication” hypothesis states that nicotine consumption via tobacco smoking reduces the negative and cognitive symptoms in patients with schizophrenia [31]. Supporting this hypothesis, among patients with TRS, smokers show worse cognitive performances and more severe negative symptoms compared to nonsmokers [32] which may be indicative of self-medication behaviour to alleviate these symptoms. While smoking is known to induce metabolism and reduce serum levels of clozapine, the present findings suggest that smoking of nicotine-containing products may be indicative of a higher disease burden, corresponding non-response to antipsychotics and TRS.
The rs79229764 variant, identified as the closest to genome-wide significant association with TRS in our sample, is located in the exon of the LINC00523 gene on the chromosome 14 open reading frame 70, C14orf70), which encodes an intergenic lncRNA (LINC00523). LINC00523 is highly expressed in basal ganglia (particularly in nucleus accumbens), testis, and adrenal gland (https://www.gtexportal.org/). Functional effect of the rs79229764 variant on LINC00523 expression is currently unknown, however LINC00523 has been previously shown to be downregulated in patients with type 2 diabetes mellitus (T2DM) [33]. It can be speculated that changes in the expression may be of clinical importance since T2DM, as well as tobacco smoking, are among the major risk factors of cardiovascular disease (CVD), which is reported to be the cause of death for 25% of patients with schizophrenia [34]. The contribution of unhealthy lifestyle [35] and metabolic adverse effects related to antipsychotic drugs (especially clozapine) [36] among patients with TRS are well-known, and current evidence also suggest that there may be a shared genetic liability between schizophrenia and CVD [37].
Investigation of the transcriptional effects of rs79229764 variant in post-mortem brain data of schizophrenia patients suggested upregulation of GRP88 and PUDP, and downregulation of JMJD7. Of particular interest may be the GPR88 gene, which encodes an orphan G-protein coupled receptor of the class A rhodopsin family (GPR88) that has previously been suggested to be a candidate susceptibility gene for sporadic cases of schizophrenia and bipolar disorder [38, 39]. GPR88 is robustly expressed in the GABAergic medium spiny neurons (MSN) of the striatum [40–42] and central extended amygdala [43]. MSNs of the striatum are involved in dopaminergic and glutamatergic signalling pathways [44], which have been indicated in the pathophysiology of schizophrenia and treatment-resistance [45]. Furthermore, GPR88 expression has been previously shown to be altered after administration of mood stabilizers [46, 47], antidepressants [48–50], and drugs related to treatment of addiction [43] which highlights its relevance for the treatment of psychiatric disorders. Future studies should therefore elucidate the potential impact of the rs79229764 polymorphism and/or GPR88 expression for the risk of TRS.
PUPD (or HDHD1) encodes a phosphatase which might be involved in RNA degradation [51]. Previous studies suggest involvement in cancer formation [52] and intellectual disability [53]. Associations with intellectual disability is in line with the latest evidence from the larger GWAS on TRS which showed polygenic correlations between TRS and poor cognitive performance and intelligence [4] and might indicate role of neurodevelopmental processes in formation of TRS [54].
One limitation of the current study is the relatively small sample size for a GWAS on complex traits such as TRS. To prevent concentration-dependent serious adverse effects and ensure adequate clinical exposure, TDM of clozapine is strongly recommended and performed more often compared to other antipsychotics. In the current study, this suggests that the clozapine-taking TRS group is more likely to represent general TRS population. On the other hand, since TDM of risperidone is usually performed on physician’s request, the risperidone taking non-TRS patients who are referred to TDM and pharmacogenetic investigation usually suffer from more serious psychiatric conditions and require closer follow-up. Therefore, non-TRS patients with gene variants associated with risk of treatment failure are likely to be overrepresented compared to general risperidone-taking patient population. Although not possible to quantify the magnitude of this effect, the high odds ratio of being carrier of the suggestive variant among clozapine-treated patients in our analysis may be underestimated. Another limitation is that we did not have access to clinical information of patients, which precluded a clinical assessment of treatment response status. However, the study utilizes longitudinal TDM data to include well-defined phenotypes of TRS and non-TRS patients where appropriate drug use was quantitatively evidenced by serum concentrations within the therapeutic ranges for the respective drugs [7]. With access to smoking status on the TDM requisition forms we were also able to identify and adjust for smoking as a predictor of TRS. Thus, the methodological approach in the current study facilitates use of robust phenotypes by accounting for smoking and eliminating pseudoresistance, an important confounding factor in TRS research, which may be driven by poor adherence or other factors [55]. Therefore, the strengths of the study, at least partially, may outweigh the above limitations.
Conclusions
We conducted a GWAS of TRS that utilized a therapeutic drug monitoring patient sample, where smoking status was known, and therapeutic concentrations of clozapine and risperidone confirmed. Although the study did not reveal any genome-wide significant associations, we identified a variant in the LINC0053 gene that was marginally associated with higher risk of TRS. Transcriptional effects of this variant suggest a role of striatal regulation and possible neurodevelopmental underpinnings in the pathophysiology of TRS. We also show that polygenic liability for schizophrenia was significantly associated with TRS. Our results suggest a candidate locus for future follow-up studies to elucidate the molecular underpinnings of TRS and demonstrate the value of combining single variant and polygenic association analyses to suggest potential utility of genetics in predicting TRS.
Supplementary Information
Author contributions
HÇL drafted the manuscript and performed the research; HÇL, RLS, and KSO retrieved the data; HÇL analysed the TDM data, KSO performed the GWAS, EK performed polygenic risk score analyses, and IAA performed the eQTL analysis; EM designed the study; KSO, EK, RLS, IAA, SD, OAA, and EM assisted in writing the manuscript.
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital). This work was supported by South-Eastern Norway Regional Health Authority (Grant No. 2020019) and the European Union's Horizon 2020 research and innovation program (REALMENT; Grant No. 964874).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The Regional Committee for Medical and Health Research Ethics and the Hospital Investigational Review Board approved the study. The study used anonymized data and residual blood samples from already performed routine analyses, therefore study inclusion did not pose any burden to the patients and written informed patient consent was not required.
Consent for publication
Not applicable.
Competing interests
OAA is consultant to Cortechs.ai and Precision Health AS and received speaker’s honoraria from Lundbeck, Sunovion, Janssen and Otsuka. EM received speaker’s honoraria from Lundbeck and Otsuka. The other authors have no competing interests to declare that are relevant to the content of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kane JM, Agid O, Baldwin ML, Howes O, Lindenmayer J-P, Marder S, et al. Clinical guidance on the identification and management of treatment-resistant schizophrenia. J Clin Psychiatry. 2019. 10.4088/JCP.18com12123. [DOI] [PubMed] [Google Scholar]
- 2.Huhn M, Nikolakopoulou A, Schneider-Thoma J, Krause M, Samara M, Peter N, et al. Comparative efficacy and tolerability of 32 oral antipsychotics for the acute treatment of adults with multi-episode schizophrenia: a systematic review and network meta-analysis. The Lancet. 2019;394:939–51. 10.1016/S0140-6736(19)31135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aringhieri S, Carli M, Kolachalam S, Verdesca V, Cini E, Rossi M, et al. Molecular targets of atypical antipsychotics: from mechanism of action to clinical differences. Pharmacol Ther. 2018;192:20–41. 10.1016/j.pharmthera.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Pardiñas AF, Smart SE, Willcocks IR, Holmans PA, Dennison CA, Lynham AJ, et al. Interaction testing and polygenic risk scoring to estimate the association of common genetic variants with treatment resistance in schizophrenia. JAMA Psychiat. 2022;79:260–9. 10.1001/jamapsychiatry.2021.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillespie AL, Samanaite R, Mill J, Egerton A, MacCabe JH. Is treatment-resistant schizophrenia categorically distinct from treatment-responsive schizophrenia? A systematic review. BMC Psychiatry. 2017;17:12. 10.1186/s12888-016-1177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruderfer DM, Charney AW, Readhead B, Kidd BA, Kähler AK, Kenny PJ, et al. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry. 2016;3:350–7. 10.1016/S2215-0366(15)00553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiemke C, Bergemann N, Clement HW, Conca A, Deckert J, Domschke K, et al. Consensus guidelines for therapeutic drug monitoring in neuropsychopharmacology: update 2017. Pharmacopsychiatry. 2018;51: e1. 10.1055/s-0037-1600991. [DOI] [PubMed] [Google Scholar]
- 8.Smith RL, O’Connell K, Athanasiu L, Djurovic S, Kringen MK, Andreassen OA, et al. Identification of a novel polymorphism associated with reduced clozapine concentration in schizophrenia patients—a genome-wide association study adjusting for smoking habits. Transl Psychiatry. 2020;10:1–10. 10.1038/s41398-020-00888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh P-R, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane KH, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–8. 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loh P-R, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet. 2016;48:811–6. 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: A Language and Environment for Statistical Computing [Internet]. R Foundation for Statistical Computing; 2021 [cited 2024 Feb 12]. Available from: https://www.R-project.org/
- 15.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinforma Oxf Engl. 2015;31:3555–7. 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman GE, Bendl J, Voloudakis G, Montgomery KS, Sloofman L, Wang Y-C, et al. CommonMind consortium provides transcriptomic and epigenomic data for schizophrenia and bipolar disorder. Sci Data. 2019;6:180. 10.1038/s41597-019-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinforma Oxf Engl. 2012;28:1353–8. 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8. 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776. 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legge SE, Dennison CA, Pardiñas AF, Rees E, Lynham AJ, Hopkins L, et al. Clinical indicators of treatment-resistant psychosis. Br J Psychiatry. 2020;216:259–66. 10.1192/bjp.2019.120. [DOI] [PubMed] [Google Scholar]
- 23.Werner MCF, Wirgenes KV, Haram M, Bettella F, Lunding SH, Rødevand L, et al. Indicated association between polygenic risk score and treatment-resistance in a naturalistic sample of patients with schizophrenia spectrum disorders. Schizophr Res. 2020;218:55–62. 10.1016/j.schres.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Frank J, Lang M, Witt SH, Strohmaier J, Rujescu D, Cichon S, et al. Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol Psychiatry. 2015;20:150–1. 10.1038/mp.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowalec K, Lu Y, Sariaslan A, Song J, Ploner A, Dalman C, et al. Increased schizophrenia family history burden and reduced premorbid IQ in treatment-resistant schizophrenia: a Swedish National Register and Genomic Study. Mol Psychiatry. 2021;26:4487–95. 10.1038/s41380-019-0575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Facal F, Costas J. Polygenic risk scores for schizophrenia and treatment resistance: new data, systematic review and meta-analysis. Schizophr Res. 2023;252:189–97. 10.1016/j.schres.2023.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Kappel DB, Legge SE, Hubbard L, Willcocks IR, O’Connell KS, Smith RL, et al. Genomic stratification of clozapine prescription patterns using schizophrenia polygenic scores. Biol Psychiatry. 2023;93:149–56. 10.1016/j.biopsych.2022.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch E, Kämpe A, Alver M, Sigurðarson S, Einarsson G, Partanen J, et al. Polygenic liability for antipsychotic dosage and polypharmacy - a real-world registry and biobank study. Neuropsychopharmacology. 2024;49:1113–9. 10.1038/s41386-023-01792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Leon J, Diaz FJ. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophr Res. 2005;76:135–57. 10.1016/j.schres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Lenk HÇ, Løvsletten Smith R, O’Connell KS, Jukić MM, Kringen MK, Andreassen OA, et al. Impact of NFIB and CYP1A variants on clozapine serum concentration—a retrospective naturalistic cohort study on 526 patients with known smoking habits. Clin Transl Sci. 2023;16:62–72. 10.1111/cts.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding JB, Hu K. Cigarette smoking and schizophrenia: etiology, clinical, pharmacological, and treatment implications. Schizophr Res Treat. 2021;2021:7698030. 10.1155/2021/7698030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iasevoli F, Balletta R, Gilardi V, Giordano S, de Bartolomeis A. Tobacco smoking in treatment-resistant schizophrenia patients is associated with impaired cognitive functioning, more severe negative symptoms, and poorer social adjustment. Neuropsychiatr Dis Treat. 2013;9:1113–20. 10.2147/NDT.S47571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansoori Z, Ghaedi H, Sadatamini M, Vahabpour R, Rahimipour A, Shanaki M, et al. Downregulation of long non-coding RNAs LINC00523 and LINC00994 in type 2 diabetes in an Iranian cohort. Mol Biol Rep. 2018;45:1227–33. 10.1007/s11033-018-4276-7. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen RE, Uggerby AS, Jensen SOW, McGrath JJ. Increasing mortality gap for patients diagnosed with schizophrenia over the last three decades—a Danish nationwide study from 1980 to 2010. Schizophr Res. 2013;146:22–7. 10.1016/j.schres.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen RE, Banner J, Jensen SE. Cardiovascular disease in patients with severe mental illness. Nat Rev Cardiol. 2021;18:136–45. 10.1038/s41569-020-00463-7. [DOI] [PubMed] [Google Scholar]
- 36.Pillinger T, McCutcheon RA, Vano L, Mizuno Y, Arumuham A, Hindley G, et al. Comparative effects of 18 antipsychotics on metabolic function in patients with schizophrenia, predictors of metabolic dysregulation, and association with psychopathology: a systematic review and network meta-analysis. Lancet Psychiatry. 2020;7:64–77. 10.1016/S2215-0366(19)30416-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rødevand L, Rahman Z, Hindley GFL, Smeland OB, Frei O, Tekin TF, et al. Characterizing the shared genetic underpinnings of schizophrenia and cardiovascular disease risk factors. Am J Psychiatry. 2023;180:815–26. 10.1176/appi.ajp.20220660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Zompo M, Severino G, Ardau R, Chillotti C, Piccardi M, Dib C, et al. Genome-scan for bipolar disorder with sib-pair families in the Sardinian population: a new susceptibility locus on chromosome 1p22–p21? Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1200–8. 10.1002/ajmg.b.31092. [DOI] [PubMed] [Google Scholar]
- 39.Del Zompo M, Deleuze J-F, Chillotti C, Cousin E, Niehaus D, Ebstein RP, et al. Association study in three different populations between the GPR88 gene and major psychoses. Mol Genet Genomic Med. 2014;2:152–9. 10.1002/mgg3.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghate A, Befort K, Becker JAJ, Filliol D, Bole-Feysot C, Demebele D, et al. Identification of novel striatal genes by expression profiling in adult mouse brain. Neuroscience. 2007;146:1182–92. 10.1016/j.neuroscience.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima K, Miyamoto Y, Tsukahara F, Hirai M, Sakaki Y, Ito T. A novel G-protein-coupled receptor gene expressed in striatum. Genomics. 2000;69:314–21. 10.1006/geno.2000.6340. [DOI] [PubMed] [Google Scholar]
- 42.Massart R, Guilloux J-P, Mignon V, Sokoloff P, Diaz J. Striatal GPR88 expression is confined to the whole projection neuron population and is regulated by dopaminergic and glutamatergic afferents. Eur J Neurosci. 2009;30:397–414. 10.1111/j.1460-9568.2009.06842.x. [DOI] [PubMed] [Google Scholar]
- 43.Befort K, Filliol D, Ghate A, Darcq E, Matifas A, Muller J, et al. Mu-opioid receptor activation induces transcriptional plasticity in the central extended amygdala. Eur J Neurosci. 2008;27:2973–84. 10.1111/j.1460-9568.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- 44.Lobo MK. Chapter 1 - Molecular Profiling of Striatonigral and Striatopallidal Medium Spiny Neurons: Past, Present, and Future. Int Rev Neurobiol [Internet]. Academic Press; 2009 [cited 2023 Apr 17]. p. 1–35. Available from: https://www.sciencedirect.com/science/article/pii/S007477420989001610.1016/S0074-7742(09)89001-6 [DOI] [PubMed]
- 45.Potkin SG, Kane JM, Correll CU, Lindenmayer J-P, Agid O, Marder SR, et al. The neurobiology of treatment-resistant schizophrenia: paths to antipsychotic resistance and a roadmap for future research. Npj Schizophr. 2020;6:1–10. 10.1038/s41537-019-0090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brandish PE, Su M, Holder DJ, Hodor P, Szumiloski J, Kleinhanz RR, et al. Regulation of gene expression by lithium and depletion of inositol in slices of adult rat cortex. Neuron. 2005;45:861–72. 10.1016/j.neuron.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, et al. Candidate genes, pathways and mechanisms for bipolar (manic–depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–29. 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- 48.Boehm C, Newrzella D, Herberger S, Schramm N, Eisenhardt G, Schenk V, et al. Effects of antidepressant treatment on gene expression profile in mouse brain: cell type-specific transcription profiling using laser microdissection and microarray analysis. J Neurochem. 2006;97:44–9. 10.1111/j.1471-4159.2006.03750.x. [DOI] [PubMed] [Google Scholar]
- 49.Conti B, Maier R, Barr AM, Morale MC, Lu X, Sanna PP, et al. Region-specific transcriptional changes following the three antidepressant treatments electro convulsive therapy, sleep deprivation and fluoxetine. Mol Psychiatry. 2007;12:167–89. 10.1038/sj.mp.4001897. [DOI] [PubMed] [Google Scholar]
- 50.Logue SF, Grauer SM, Paulsen J, Graf R, Taylor N, Sung MA, et al. The orphan GPCR, GPR88, modulates function of the striatal dopamine system: a possible therapeutic target for psychiatric disorders? Mol Cell Neurosci. 2009;42:438–47. 10.1016/j.mcn.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Preumont A, Rzem R, Vertommen D, Van Schaftingen E. HDHD1, which is often deleted in X-linked ichthyosis, encodes a pseudouridine-5’-phosphatase. Biochem J. 2010;431:237–44. 10.1042/BJ20100174. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Zhang W, Ding D, Hu Y, Guo G, Wang J, et al. Bioinformatics analysis combined with experiments predicts PUDP as a potential prognostic biomarker for hepatocellular carcinoma through its interaction with tumor microenvironment. Front Oncol. 2022;12: 830174. 10.3389/fonc.2022.830174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gholipour F, Yoshiura K-I, Hosseinpourfeizi M, Elmi N, Teimourian S, Safaralizadeh R. Whole exome sequencing reveals pathogenic variants in KL and PUDP genes as the cause of intellectual disability in an Iranian family. Gene Rep. 2021;24: 101299. 10.1016/j.genrep.2021.101299. [Google Scholar]
- 54.Owen MJ, Legge SE, Rees E, Walters JTR, O’Donovan MC. Genomic findings in schizophrenia and their implications. Mol Psychiatry. 2023. 10.1038/s41380-023-02293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howes OD, Thase ME, Pillinger T. Treatment resistance in psychiatry: state of the art and new directions. Mol Psychiatry. 2022;27:58–72. 10.1038/s41380-021-01200-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.